Abstract
Loss of bone mass with advancing age in mice is because of decreased osteoblast number and is associated with increased oxidative stress and decreased canonical Wnt signaling. However, the underlying mechanisms are poorly understood. We report an age-related increase in the lipid oxidation product 4-hydroxynonenal (4-HNE) as well as increased expression of lipoxygenase and peroxisome proliferator-activated receptor-γ (PPARγ) in the murine skeleton. These changes together with decreased Wnt signaling are reproduced in 4-month-old mice bearing a high expressing allele of the lipoxygenase Alox15. The addition of 4-HNE to cultured osteoblastic cells increases oxidative stress, which in turn diverts β-catenin from T-cell-specific transcription factors to Forkhead box O (FoxO) transcription factors, thereby attenuating the suppressive effect of β-catenin on PPARγ gene expression. Oxidized lipids, acting as ligands of PPARγ, promote binding of PPARγ2 to β-catenin and reduce the levels of the latter, and they attenuate Wnt3a-stimulated proliferation and osteoblast differentiation. Furthermore, oxidized lipids and 4-HNE stimulate apoptosis of osteoblastic cells. In view of the role of oxidized lipids in atherogenesis, the adverse effects of lipoxygenase-mediated lipid oxidation on the differentiation and survival of osteoblasts may provide a mechanistic explanation for the link between atherosclerosis and osteoporosis.
Age-related bone loss is primarily because of an insufficient number of osteoblasts (1, 2) attributed to exhaustion of multipotential mesenchymal stem cell progenitors (3, 4) and the diversion of these progenitors toward the adipocyte lineage (5–11). Increased osteoblast apoptosis may also be involved as we recently demonstrated that loss of bone mass and strength in C57BL/6 (B6)3 mice with advancing age is associated with an increase in the prevalence of apoptotic osteoblasts and a corresponding decrease in osteoblast number and bone formation rate (12). Moreover, these changes are accompanied by increased oxidative stress and diminished canonical Wnt signaling, a critical regulator of bone formation.
Wnts are secreted proteins that bind to a Frizzled receptor and a low density lipoprotein receptor-related protein 5 (LRP5) or LRP6 co-receptor, resulting in a rise in the level of β-catenin by preventing its degradation by the proteasome. Binding of β-catenin to members of TCF family converts them from repressors to activators of a variety of genes, including those involved in the differentiation and survival of osteoblasts (13–16). Importantly, however, β-catenin is also an essential partner of the FoxO family of transcription factors, which defend against oxidative stress by stimulating the transcription of oxidant scavenging enzymes such as manganese superoxide dismutase and catalase (17). In fact, we have elucidated that oxidative stress compromises β-catenin/TCF-mediated transcription and osteoblastogenesis because of competition between FoxO and TCF for a limited pool of β-catenin (18). Such competition for β-catenin may explain how the increased oxidative stress that occurs with advancing age leads to a deficit in osteoblast number.
Down-regulation of Wnt signaling is permissive for diversion of mesenchymal stem cells to the adipocyte instead of the osteoblast lineage. However, adipogenesis also requires activation of the nuclear hormone receptor PPARγ (19). Lipoxygenases oxidize polyunsaturated fatty acids (PUFAs) to form products that bind to and activate PPARγ, including 12-hydroxyeiscosatetraenoic acid (12-HETE), 15-HETE, 9-hydroxyoctadecadienoic acid (9-HODE), and 13-HODE (20–22). Both PPARγ and lipoxygenases influence skeletal homeostasis as evidenced by the increased bone mineral density in mice lacking Alox15 (23) or which are haplo-insufficient for PPARγ (24). Conversely, mice bearing a high expressing allele of Alox15 have low bone mineral density (23).
Lipoxygenases could increase oxidative stress in the skeleton because hydroxy radicals are generated during the conversion of PUFAs to hydroxy-PUFAs by these enzymes (25, 26). Moreover, hydroperoxy-PUFAs generated either enzymatically by lipoxygenases or by the actions of reactive oxygen species (ROS) can non-enzymatically decompose into α,β-unsaturated aldehydes, of which 4-hydroxynonenal (4-HNE) is a prototype (27). Such α,β-unsaturated aldehydes indirectly increase ROS by reacting with glutathione, thereby depleting cells of this critical component of the coupled glutathione reductase/glutathione peroxidase antioxidant system (28, 29).
Based on the above and evidence that lipid oxidation increases with age (30), we hypothesized that increased lipoxygenase expression increases oxidative stress and reduces Wnt signaling, thereby decreasing the number of osteoblasts. We show that the expression of the lipoxygenases Alox12, Alox12e, and Alox15 increases in the bone of B6 mice with advancing age. These changes are associated with increased levels of lipid oxidation and increased expression of PPARγ. These same changes along with increased oxidative stress and decreased Wnt signaling are reproduced in 4-month-old mice bearing a high expressing allele of Alox15. In addition, we present in vitro evidence for an oxidized PUFA-induced ROS/FoxO/PPARγ/β-catenin cascade. This provides a mechanistic explanation for how increased lipid oxidation can compromise the canonical Wnt signaling required for the differentiation and survival of osteoblasts.
EXPERIMENTAL PROCEDURES
Chemicals, Reagents, and Plasmids
N-Acetyl cysteine (NAC), LiCl, and H2O2 were purchased from Sigma-Aldrich. 4-HNE, 9-HODE, 12-HETE, 13-HODE, and 15-HETE were from Cayman Chemical Co. (Ann Arbor, MI). To prepare 500 μm stock solutions of oxidized PUFAs, ethanol solvent was evaporated with a nitrogen stream, 33 mg/ml fatty acid-free BSA in PBS was added, and the compounds were dissolved by overnight shaking. Rosiglitazone was obtained from Tularik, Inc. (South San Francisco, CA). Wnt3a recombinant protein was from R&D Systems (Minneapolis, MN). pcDNA was from Invitrogen. PPARγ2 and PPRE-luciferase (PPRE-luc) expression plasmids were provided by B. Spiegelman, Harvard Medical School, Boston (Addgene plasmid 8895 and 1015, respectively). A reporter plasmid containing three TCF binding sites upstream of a minimal c-Fos promoter driving the firefly luciferase gene (TCF-luc) (31) and a construct expressing constitutively active β-catenin (S33Y) (32) were provided by B. Vogelstein, John Hopkins University Medical Institutions, Baltimore, MD. A reporter plasmid containing six copies of daf-16 family protein binding element (FoxO-luc) in the pGL3-basic firefly luciferase vector with a minimal TATA box (33) was provided by B. Burgering, University Medical Center, Utrecht, Netherlands.
Animal Experimentation
Animal use protocols were approved by the Institutional Animal Care and Use Committees of the University of Arkansas for Medical Sciences and the Central Arkansas Veterans Healthcare System. B6 mice were purchased from Harlan Inc. from a cohort maintained with support from the National Institute of Aging. Measurements of 4-HNE and certain transcripts were made using bones from 4-, 8-, 16-, and 25-month-old female B6 mice that were obtained during the conduct of a previously published experiment (12). 4-HNE determinations were also made on bones from 2 additional experiments, one comprising 6- and 24-month-old female B6 mice and the other comprising 4- and 19-month-old B6 mice. DBA/2 (D2) mice were from The Jackson Laboratory (Bar Harbor, ME). D2.B6-Alox15 congenic mice bearing an 82-megabase region of chromosome 11 introgressed from B6 were progeny of breeders originally provided by Robert Klein (University of Oregon, Portland, OR) (23). Animals were fed ad libitum with pellets manufactured by Research Diets, Inc. (New Brunswick, NJ). Histomorphometric determination of adipocyte number was done as previously described (5).
Immunoprecipitation and Western Blot Analysis
C2C12 cell lysates were immunoprecipitated with anti-IgG, or anti-PPARγ2, and protein A/G Plus-agarose (Santa Cruz Biotechnology, Santa Cruz, CA). Immunoprecipitates were analyzed by SDS-PAGE followed by Western blotting using mouse monoclonal anti-β-catenin (BD Biosciences), mouse monoclonal anti-PPARγ2, or normal mouse IgG (Santa Cruz Biotechnology). The phosphorylation status of p66shc was determined by immunoblotting of lysates from cultured cells or fifth lumbar vertebral lysates as previously described (12) using a mouse monoclonal antibody recognizing Ser-36-phosphorylated p66shc (Calbiochem). Protein levels of p66shc and β-actin were determined using a rabbit polyclonal antibody recognizing p66shc (BD Biosciences) and a mouse monoclonal antibody recognizing β-actin (Sigma). Quantification of the intensity of the bands in the autoradiograms was performed using a VersaDocTM imaging system (Bio-Rad).
Cell Culture
OB-6 cells, an osteoblastic cell line derived from the murine bone marrow (34), were cultured in α-minimum Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), penicillin (100 units/ml), streptomycin (100 μg/ml), and glutamine (292 μg/ml). C2C12 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, antibiotics as above, and 1% sodium pyruvate. OB-6γ2 cells expressing murine PPARγ2 under the control of doxycycline were made as follows. Murine PPARγ2 cDNA was excised from a vector provided by Dr. J. M. Gimble (35) and cloned into the BamHI site of the pRev-TRE eukaryotic expression vector. OB-6 cells were transduced with tetracycline-controlled transcriptional activator (tTA) vector (BD Biosciences/Clontech) using 4 μg/ml Polybrene. After selection with 500 μg/ml G418, cells were transduced with the pRT-PPARγ2 vector and selected using 120 μg/ml hygromycin to obtain stably transduced cells. The OB-6γ2 monoclonal cell line was obtained by limiting dilution of the stably transduced cells. These cells express PPARγ2 only in the absence of doxycycline (supplemental Fig. S1A). The addition of rosiglitazone (RGL) stimulates their differentiation into adipocytes but inhibits their differentiation into osteoblasts (supplemental Fig. S1, B–D).
To quantify adipogenesis, cells were cultured to 70% confluence, and the media were supplemented with the ligand to be tested or with 3.3% BSA in PBS as the vehicle control. Medium containing ligand was changed every 2 days for 6 days. Cells were fixed with 10% formalin in PBS, rinsed, and stained for 30 min with 0.15% Oil Red O (Sigma) in a 55:45 mix of isopropanol and water. Cells were counterstained with 0.5% methyl green (Fisher) in 0.1 m sodium acetate, pH 4. Photomicrographs of Oil Red O-stained cells were obtained after culture of cells as described above on Lab-Tek glass slides (Nalge Nunc International, Naperville, IL). Oil Red O staining was quantified after extraction of the dye with 1 ml of isopropanol, and absorbance determination was at 500 nm.
Osteoblast differentiation was analyzed using freshly isolated murine bone marrow cells cultured in 12-well tissue culture plates at 5 × 106 cells per well in α-minimum Eagle's medium containing 10% fetal bovine serum for 10 days. One half of the medium was replaced every 5 days. Fetal bovine serum was then reduced to 2%, and 25 ng/ml Wnt3a was added in the presence or absence of PPARγ ligands. Two days later 10 mm β-glycerophosphate was added to the medium, and the cultures were maintained for an additional 2 weeks. The mineralized matrix was stained with 40 mm alizarin red, pH 4.2. Alizarin red was quantified after extraction with 10 mm sodium phosphate, 10% cetylpyridinium chloride, pH 7, and absorbance determination was at 562 nm against a known alizarin red standard.
Quantitative Reverse Transcription-PCR
Total RNA was extracted from vertebrae, calvaria, or cultured cells using Ultraspec RNA (Biotecx Laboratories, Houston, TX) and reverse-transcribed using the High-capacity cDNA Archive kit (Applied Biosystems, Foster City, CA). Primers and probes for the different genes were manufactured by the TaqMan® Gene Expression Assays service (Applied Biosystems). Unless otherwise indicated, all samples were normalized to ribosomal protein S2.
Transient Transfections
Luciferase reporter constructs and other plasmids were introduced into cells by transient transfection using Lipofectamine Plus (Invitrogen). C2C12 or OB-6 cells were plated in 48-well plates and transfected 16 h later with a total of 0.4 μg of DNA. Luciferase activity assays were performed as previously described (16). Reporter luciferase activity was normalized with Renilla-luc values for transfection efficiency.
Other Assays
4-HNE adducts in bone extracts were determined using an adaptation of a previously described enzyme-linked immunosorbent assay (36). Dissected bones were stored at −80 °C and then pulverized with Wollenberger tongs and homogenized in 20 mm potassium phosphate, pH 7.0, 1.4 mm 2-mercapthoethanol containing a protease inhibitor mixture (Roche Diagnostics). After centrifugation and determination of soluble protein, extracts were incubated with a 1:4000 dilution of goat polyclonal anti-4-HNE antiserum (Novus, Littleton, CO) for 15 min at room temperature and then added to 96-well plates coated with 5 μg of 4-HNE-derivatized casein. After rinsing, a 1:4000 dilution of rabbit anti-goat horseradish peroxidase conjugate was added for 15 min. Color development was done with 3,3′,5,5′-tetramethylbenzidine (Promega, Madison, WI). The amount of 4-HNE was determined using a standard curve prepared with 4-HNE-derivatized casein and is reported as μg of 4-HNE-casein equivalents.
Intracellular ROS were quantified in freshly isolated marrow cells using dichlorodihydrofluorescein diacetate dye as previously described (37). Bromodeoxyuridine incorporation was quantified with a kit from Roche Diagnostics. Apoptosis in cultured cells was determined by measuring caspase 3 activity by cleavage of the fluorogenic substrate Ac-DEVD-aminofluoromethylcoumarin (Biomol, Plymouth Meeting, PA) as previously described (38).
Statistical Analysis
ANOVA or Student's t test was used to detect age-related changes and the effects of various in vivo and in vitro treatments after establishing that the data were normally distributed and exhibited equivalency of variances. The Bonferroni method was used to perform appropriate pairwise comparisons of treatment groups. When the requirements for performing ANOVA were not met, Kruskal-Wallis ANOVA on Ranks test was used followed by Dunn's method to perform pairwise comparisons. Experiments were repeated at least once. All data are reported as the mean ± S.D.
RESULTS
Lipid Oxidation and the Expression of Lipoxygenases and PPARγ Increase with Advancing Age in the Murine Skeleton
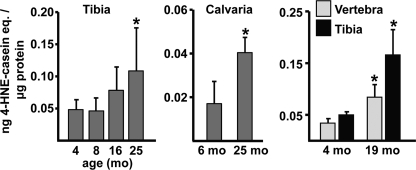
4-HNE reacts with lysine, histidine, and cysteine residues of proteins to form a covalently linked cyclic hemiacetal adduct that can be measured by enzyme-linked immunosorbent assay (39). Using this assay, we found in three separate experiments that bone extracts from tibiae, lumbar vertebrae, or calvaria from old (19 or 25 month) female B6 mice contained significantly higher levels of 4-HNE as compared with extracts from young (4 or 6 month) mice (Fig. 1).
FIGURE 1.
Age-related increase in lipid oxidation in bone of B6 mice. 4-HNE adducts in bone extracts from B6 female mice from three separate experiments were measured by enzyme-linked immunosorbent assay (n = 5–8/group). *, p < 0.05 versus 4 or 6 month.
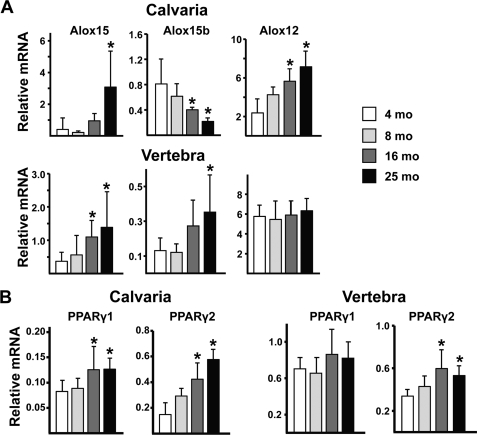
We next examined whether the expression of the lipoxygenases that generate oxidized PUFAs also increase with age. We determined that Alox12, Alox15, and Alox15b are the most abundant lipoxygenases in 4-month-old vertebral and calvarial bone (Fig. 2A and supplemental Fig. S2), whereas Alox12b and Alox12e are 1–2 orders of magnitude less abundant (not shown). The expression of Alox12 and Alox15 progressively increased in calvaria by ∼5-fold between 4 and 25 months of age. Alox15b declined by a similar magnitude, but the overall effect was increased lipoxygenase expression. Alox15 also increased with aging in vertebral bone, but unlike the situation in calvaria, vertebral Alox12 expression did not change, and Alox15b increased. Similarly, PPARγ2, the predominant functional isoform of PPARγ (40), increased by 2–3-fold in both calvaria and vertebrae (Fig. 2B and supplemental Fig. S2), whereas PPARγ1 increased with age by 2-fold in calvaria but did not increase in vertebrae.
FIGURE 2.
Age-related increase in lipoxygenase expression in bone of B6 mice. Expression levels of lipoxygenases (A) and PPARγ1 and PPARγ2 (B) by quantitative PCR in calvaria and vertebrae (L5) of B6 female mice from the experiment are shown in the left panel of Fig. 1 (n = 5–9/group). Data shown are normalized to expression of ribosomal protein S2. Essentially identical results were obtained when calvaria transcripts were normalized to glyceraldehyde-3-phosphate dehydrogenase (supplemental Fig. S2). *, p < 0.05 versus 4 month.
Increased Lipid Oxidation, Oxidative Stress, and PPARγ Expression and Decreased Canonical Wnt Signaling in the Bones of DBA/2 (D2) Mice with Elevated Lipoxygenase Expression
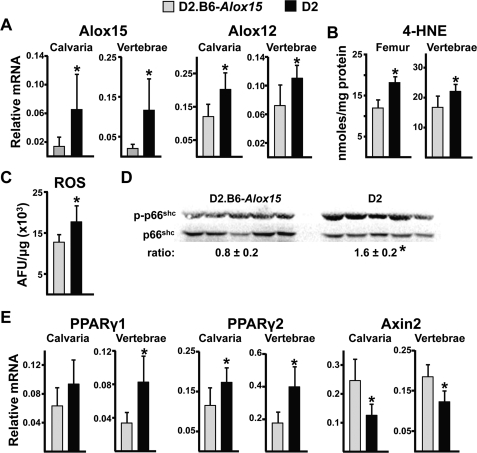
D2 mice, which bear a high expressing allele of Alox15 (23), were used to investigate whether increased lipoxygenase expression causes increased oxidative stress. Congenic D2.B6-Alox15 mice that express the “normal” B6 allele of Alox15 were used as controls. Consistent with earlier findings (23), Alox15 transcripts were elevated 10–20-fold in calvaria and vertebrae of D2 mice compared with D2.B6-Alox15 mice; however, Alox12 was also increased by ∼50% (Fig. 3A). Expression of Alox12b, Alox12e, and Alox15b were indistinguishable in the two strains (not shown). As expected, the increased Alox15 expression in D2 mice was associated with increased levels of 4-HNE (Fig. 3B), indicative of increased lipid oxidation. More important, these changes were accompanied by an increased level of ROS in femoral bone marrow (Fig. 3C) and vertebral levels of phosphorylated p66Shc (Fig. 3D), a reliable and robust index of oxidative stress (41).
FIGURE 3.
Increased Alox15 expression raises the level of oxidative stress, increases PPARγ expression, and decreases Axin2 expression in bone. Shown are Alox transcripts (A), 4-HNE adducts (B), ROS in the bone marrow (C), and phosphorylated p66shc (D) levels in vertebral extracts (each lane represents one animal) and PPARγ1, PPARγ2, and Axin2 transcripts from 4 month-old D2.B6-Alox15 or D2 mice (n = 6/group) (E). Combined data from male and female mice are presented because there was no effect of gender on the indices examined. *, p < 0.05 versus D2.B6-Alox15.
Similar to the situation in aged B6 mice, the increased oxidative stress and lipoxygenase expression seen in D2 mice was associated with increased expression of PPARγ1 and PPARγ2 in vertebral and calvarial bone, as compared with D2.B6-Alox15 controls (Fig. 3E). Moreover, expression of the classical β-catenin/TCF target gene Axin2 was decreased in D2 mice.
4-HNE-induced Oxidative Stress Activates FoxO-dependent Transcription and Decreases β-Catenin/TCF Activity
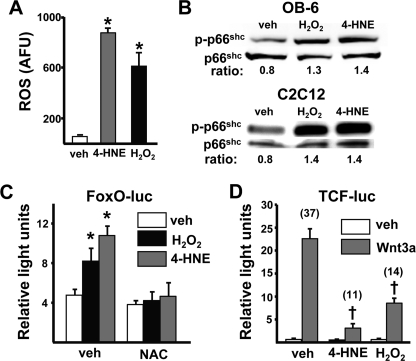
Based on the above findings, we proceeded to investigate in vitro whether the lipid oxidation product 4-HNE increases ROS in osteoblastic cells and, if so, the consequences for Wnt signaling. The addition of 20 μm 4-HNE increased the level of ROS in cultured osteoblastic OB-6 cells to approximately the same extent as 100 μm H2O2 (Fig. 4A). The increase in ROS was associated with an increase in phosphorylated p66Shc (Fig. 4B) in osteoblastic OB-6 cells as well as C2C12 cells, a model of uncommitted mesenchymal cells capable of differentiating toward the osteoblast lineage. The addition of 4-HNE to C2C12 cells also activated FoxO-mediated transcription as measured by a FoxO-luciferase reporter construct (Fig. 4C). The magnitude of the 4-HNE effect was similar to that of H2O2 and was prevented by the anti-oxidant NAC, indicating that the 4-HNE-induced oxidative stress caused activation of FoxO-mediated transcription. Moreover, 4-HNE attenuated Wnt3a-induced activation of β-catenin/TCF-mediated transcription as measured by TCF luciferase activity (Fig. 4D), as we had previously seen with H2O2-induced oxidative stress (18).
FIGURE 4.
4-HNE-induced oxidative stress stimulates FoxO-mediated transcription and decreases β-catenin/TCF-mediated transcription in C2C12 and osteoblastic OB-6 cells. A, ROS levels in OB-6 cells incubated with vehicle (PBS), 4-HNE (20 μm), or H2O2 (100 μm) for 15 min. AFU, arbitrary fluorescence units. B, phosphorylated p66shc levels by Western blot analysis in OB-6 or C2C12 cells incubated with vehicle, H2O2 (100 μm), or 4-HNE (20 μm) for 1 h. The ratio of phosphorylated to total protein is depicted numerically in the bottom of each lane. C, luciferase activity in C2C12 cells transfected with the FoxO-luc reporter construct after pretreatment with vehicle (PBS) or NAC (10 mm) for 1 h then with vehicle, H2O2 (50 μm), or 4-HNE (20 μm) for 24 h. D, luciferase activity in C2C12 cells transfected with a TCF-luc reporter construct after pretreatment with vehicle, 4-HNE, or H2O2, as in B, then with vehicle or Wnt3a (50 ng/ml) for 24 h. Bars represent the mean ± S.D. of triplicate determinations. Numbers in parentheses represent -fold change versus respective vehicle control. *, p < 0.05 versus vehicle; †, p < 0.05 versus Wnt3a alone.
ROS Attenuate Wnt-activated Suppression of PPARγ Expression
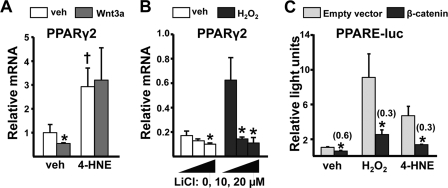
In view of evidence that Wnt signaling suppresses expression of PPARγ (42), we next examined whether the ROS/FoxO-induced decrease in the availability of β-catenin leads to increased PPARγ expression. As expected, the addition of Wnt3a to C2C12 cell cultures suppressed PPARγ2 expression by about 50% (Fig. 5A). Moreover, 4-HNE not only increased the basal levels of PPARγ2 but also prevented the negative effect of Wnt3a on PPARγ2 expression. Conversely, pretreatment with LiCl, which constitutively stabilizes β-catenin, decreased expression of PPARγ2 and blocked the stimulatory effect of H2O2 on PPARγ2 expression (Fig. 5B). Finally, and consistent with the opposing effects of oxidative stress and β-catenin on PPARγ2 expression, both H2O2 and 4-HNE increased the activity of a PPARγ reporter construct, and this effect was sharply attenuated by cotransfection with β-catenin (Fig. 5C).
FIGURE 5.
4-HNE or H2O2 increase PPARγ2 expression in C2C12 cells. PPARγ2 gene expression by quantitative PCR in C2C12 cells pretreated with vehicle (PBS), 4-HNE (20 μm), or H2O2 (100 μm) for 1 h followed by vehicle or Wnt3a (50 ng/ml) for 5 h (A) or in cells pretreated with vehicle or H2O2 as above followed by vehicle or indicated concentration of LiCl for 5 h (B). C, luciferase activity in C2C12 cells transfected with a PPARE-luc reporter construct and co-transfected with either an empty vector (pcDNA) or a constitutively active β-catenin mutant. Cells were then treated with vehicle, 4-HNE, or H2O2 as in A for 24 h. Bars represent the mean ± S.D. of triplicate determinations. Numbers in parentheses represent -fold change versus respective vehicle control. *, p < 0.05 versus respective vehicle control (A and B) or empty vector control (C). †, p < 0.05 versus cells given neither 4-HNE nor Wnt3a.
Activation of PPARγ by Oxidized PUFA Ligands Promotes Binding to β-Catenin and Suppression of β-Catenin/TCF-mediated Transcription
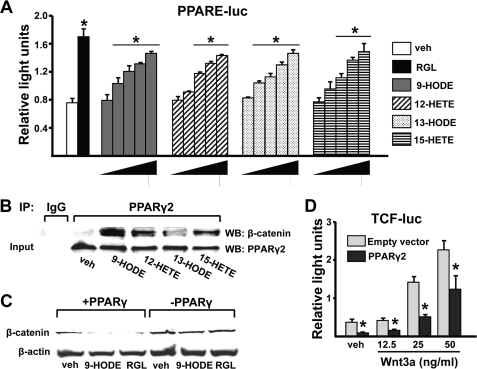
We then explored potential mechanisms by which oxidized PUFA ligands of PPARγ themselves, as opposed to their downstream 4-HNE product, could attenuate pro-osteogenic canonical Wnt signaling. The oxidized PUFAs 9-HODE, 13-HODE, 12-HETE, or 15-HETE activated PPARγ in a dose-dependent fashion, as determined by a PPARE luciferase reporter assay in C2C12 cells (Fig. 6A). These ligands also promoted the association of PPARγ with β-catenin in C2C12 cells as determined by coimmunoprecipitation with an anti-PPARγ2 antibody (Fig. 6B), consistent with previous observations using the synthetic PPARγ ligand RGL (43). In OB6-γ2 cells expressing murine PPARγ2 under the control of a Tet-OFF-regulated tetracycline response element (supplemental Fig. S1), 9-HODE as well as RGL caused a PPARγ2-dependent reduction in β-catenin protein levels (Fig. 6C). Finally, β-catenin/TCF-mediated transcription in OB-6 cells was decreased by cotransfection of PPARγ2 in the absence of exogenous ligand (Fig. 6D), suggesting that sequestration of β-catenin by a high level of PPARγ2 is sufficient to suppress Wnt signaling.
FIGURE 6.
Peroxidized PUFAs promote PPARγ-mediated transcription and binding of PPARγ to β-catenin and suppress β-catenin/TCF-mediated transcription. A, luciferase activity in C2C12 cells transfected with a PPARE-luc reporter construct and treated with vehicle (3.3% BSA in PBS), RGL (5 μm), or the indicated peroxidized PUFAs (3, 10, 30, 60, 100 μm) for 24 h. Bars represent the mean ± S.D. of triplicate determinations. *, p < 0.05 versus vehicle by ANOVA. B, C2C12 cells were incubated with vehicle or the indicated peroxidized PUFAs (10 μm) for 1 h. Cell lysates were immunoprecipitated (IP) with protein A-Sepharose in combination with the non-immune IgG or an anti-PPARγ2 antibody. Immunoprecipitates were analyzed by Western blotting (WB) using anti-β-catenin or anti-PPARγ2 antibodies. C, β-catenin protein levels by Western blotting of OB-6γ2 cell extracts. Cells were cultured as described under “Experimental Procedures” to induce or repress PPARγ2 gene expression and then incubated with vehicle, 9-HODE (10 μm), or RGL (5 μm) for 4 h. D, luciferase activity in C2C12 cells transfected with a TCF-luc reporter construct and co-transfected with either an empty vector (pcDNA) or a PPARγ2 expression construct followed by treatment with vehicle (PBS) or Wnt3a for 24 h. Bars represent the mean ± S.D. of triplicate determinations. *, p < 0.05 versus respective empty vector control.
Oxidized PUFA Ligands Suppress Wnt3a-induced Activation of β-Catenin/TCF-mediated Transcription as Well as the Proliferation and Differentiation of Osteoblast Progenitors
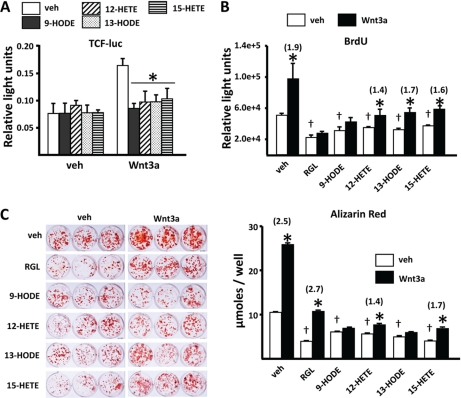
In view of the strong suppressive effects of oxidized lipid ligands of PPARγ on β-catenin, we examined whether these agents attenuate the stimulatory effect of Wnts on osteoblast differentiation. The addition of 9-HODE, 12-HETE, 13-HODE, or 15-HETE to C2C12 cells suppressed β-catenin/TCF-mediated transcription stimulated by 10 ng/ml Wnt3a (Fig. 7A). However, only 9-HODE was effective at 25 ng/ml Wnt3a (not shown), suggesting that the higher levels of β-catenin obtained with higher levels of Wnt3a can overcome the negative effects of the available activated PPARγ. Oxidized PUFAs as well as RGL also attenuated the proliferation of C2C12 cells (Fig. 7B), and both RGL and 9-HODE prevented Wnt3a-induced proliferation. The oxidized PUFA ligands also attenuated the differentiation of osteoblasts in primary cultures of murine marrow cells as measured by mineral deposition (Fig. 7C), and they prevented the stimulatory effect of Wnt3a on osteoblastogenesis.
FIGURE 7.
Peroxidized PUFAs prevent Wnt3a-induced transcription, proliferation, and osteoblastogenesis. A, luciferase activity in C2C12 cells transfected with a TCF-luc reporter construct. Cells were pretreated with vehicle (3.3% BSA in PBS) or the indicated peroxidized PUFAs (100 μm) for 1 h, then with Wnt3a (10 ng/ml) for 24 h. B, proliferation of C2C12 cells, determined by bromodeoxyuridine (BrdU) incorporation. Cells were pretreated with vehicle, RGL (5 nm), or the indicated peroxidized PUFAs (10 μm) for 1 h followed by Wnt3a (25 ng/ml) for 3 days. C, mineralized matrix visualized and quantified by alizarin red staining. Cultures of femoral bone marrow cells were established as described under “Experimental Procedures,” then incubated with RGL (1 nm) or the indicated peroxidized PUFAs (100 μm) without or with Wnt3a (25 ng/ml) for 18 days. Mineralized matrix was visualized by staining with alizarin red (left panel) and quantified after extraction (right panel). Bars represent the mean ± S.D. of triplicate determinations. Numbers in parentheses represent -fold change versus respective vehicle control. *, p < 0.05 versus respective vehicle control. †, p < 0.05 versus cells given neither PPARγ ligands nor Wnt3a.
Lipid Oxidation Stimulates Apoptosis of Osteoblastic Cells via Both PPARγ-dependent and -independent Mechanisms
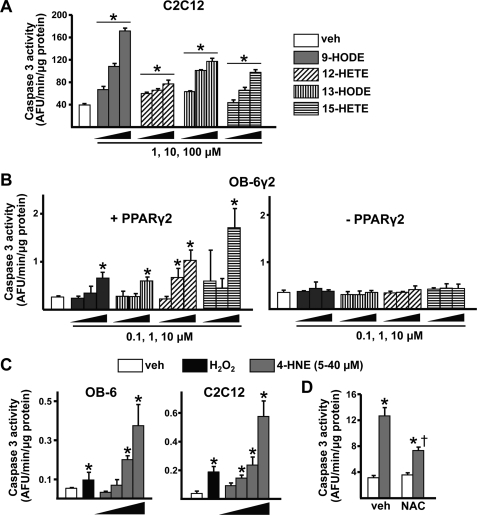
To determine whether the increased prevalence of osteoblast apoptosis we reported previously in aging B6 mice (12) could be because of increased lipid oxidation, we measured the effects of oxidized PUFAs and 4-HNE on osteoblast apoptosis in vitro. Each of the oxidized PUFA ligands induced apoptosis of C2C12 cells in a dose-dependent fashion, as measured by caspase 3 activity (Fig. 8A). Moreover, the proapoptotic effect of oxidized PUFAs was only seen in OB-6γ2 cells when PPARγ2 was expressed (Fig. 8B). We also determined that 4-HNE dose-dependently induced the apoptosis of osteoblastic in OB-6 cells and C2C12 cells (Fig. 8C). As expected, the toxic effect of 4-HNE did not depend on the presence of PPARγ2 in OB6-γ2 cells (not shown). Finally, pretreatment of C2C12 cells with NAC prevented 4-HNE-induced cell death, indicating that the proapoptotic effect of this agent is mediated by oxidative stress (Fig. 8D).
FIGURE 8.
Oxidized PUFAs and 4-HNE promote apoptosis of osteoblastic cells. A and B, caspase-3 activity in cells incubated with vehicle (3.3% BSA in PBS) or the indicated concentrations of peroxidized PUFAs for 6 h. A, C2C12 cells. B, OB-6γ2 cells were cultured as described under “Experimental Procedures” to induce or repress PPARγ2 gene expression. C and D, caspase-3 activity in OB-6 or C2C12 cells incubated with H2O2 (50 μm) or 5, 10, 20. or 40 μm 4-HNE for 6 h (C) or C2C12 cells pretreated for 1 h with NAC (10 mm) followed by 4-HNE (20 μm) for 6 h (D). Bars represent the mean ± S.D. of triplicate determinations. *, p < 0.05 versus vehicle control. †, p < 0.05 versus cells treated with 4-HNE alone.
Adipogenesis Is Not an Inevitable Accompaniment of Increased Lipid Oxidation
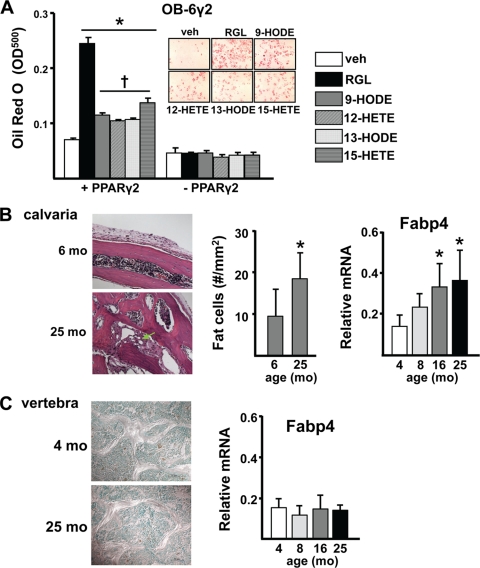
Consistent with their PPARγ-activating property, each of the oxidized PUFAs also stimulated the development of adipocytes in OB6-γ2 cell cultures in a PPARγ2-dependent fashion, albeit the pro-adipogenic effect of RGL was greater than that of the oxidized PUFAs (Fig. 9A). In view of the age-related increase in lipid oxidation in the murine skeleton, we examined whether adipocytes were increased in the bone marrow of aged B6 mice. Calvaria of 25-month-old B6 mice contained 2-fold more adipocytes than 6-month-old mice and a comparable increase in expression of the adipocyte marker adipocyte fatty acid-binding protein 4 (Fabp4) (Fig. 9B). In contrast, there were few if any adipocytes in vertebral bone sections obtained from either 4- or 25-month-old mice (Fig. 9C). Consistent with this, expression of Fabp4 remained low in vertebral bone during aging.
FIGURE 9.
Adipogenesis is not an inevitable accompaniment of increased lipid oxidation. A, OB-6γ2 cells were cultured to induce or repress PPARγ2 gene expression and then treated with vehicle (PBS), RGL (1 nm), or the indicated oxidized PUFAs (100 μm) for 8 days. Lipid was visualized with Oil Red O (inset, PPARγ2-expressing cells) and quantified as described under “Experimental Procedures.” *, p < 0.05 versus vehicle control. †, p < 0.05 versus RGL by ANOVA. B, adipocytes (arrow, left panel) in calvaria bone of B6 mice were quantified by histomorphometry (middle panel) of hematoxylin- and eosin-stained decalcified sections. Quantification of the adipocyte marker Fabp4 was done by quantitative PCR after normalization to glyceraldehyde-3-phosphate dehydrogenase in a separate experiment (right panel). C, adipocytes in toluidine blue-stained nondecalcified sections (left panel) and Fabp4 expression (right panel) number was determined in vertebral bone as in B. Bars represent the means ± S.D.; *, p < 0.05 versus 4 or 6 months.
DISCUSSION
The results described in this report show that lipoxygenase expression in the skeleton of B6 mice increases with age and that this increase is associated with an increase in lipid oxidation and PPARγ expression. These changes coincide with a rise in oxidative stress, a reduction of canonical Wnt signaling, and a decrease in osteoblast number, which we previously reported in these mice (12). The evidence for increased lipid oxidation, oxidative stress, and PPARγ expression as well as decreased canonical Wnt signaling were reproduced in 4-month-old D2 mice expressing high levels of Alox15. These mice exhibit decreased bone mass compared with their congenic controls (23). Collectively, the findings of this report establish that increased lipoxygenase expression causes oxidative stress in the skeleton.
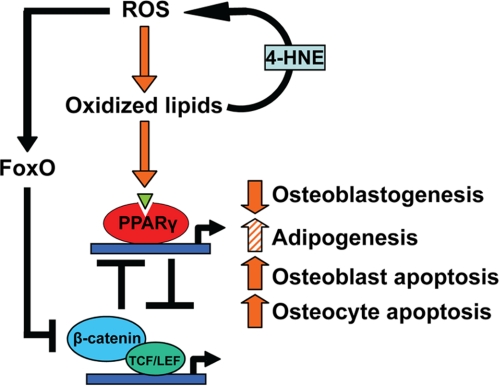
Using cultured cell models, we demonstrate the existence of an oxidized PUFA-activated ROS/FoxO/PPARγ/β-catenin cascade, which explains how a rise in oxidized lipids causes increased oxidative stress, increased PPARγ expression and reduced canonical Wnt signaling in osteoblasts and osteoblast progenitors. As depicted in the model shown in Fig. 10, lipid oxidation initiates this cascade by generating 4-HNE, which increases ROS and thereby activates FoxO. This results in diversion of β-catenin from pro-osteogenic TCF-mediated transcription to anti-oxidant FoxO-mediated transcription, as we had previously described for H2O2-induced oxidative stress (18). The decrease of β-catenin not only attenuates canonical Wnt signaling but also unleashes the expression of PPARγ, which is normally suppressed by β-catenin (42, 44, 45). The increase in PPARγ levels serves as an additional β-catenin sink by sequestering it and activating its proteasomal degradation (43). The increased lipoxygenase and PPARγ expression is probably auto-amplified as evidenced by the increased expression of PPARγ, Alox12, and Alox15 in vascular smooth muscle cells treated with oxidized PUFAs or RGL (46). Despite earlier reports that FoxOs suppress expression of PPARγ in preadipocytes (47, 48), the findings reported here make it clear that the pathways leading to increased PPARγ expression predominate in bone cells. Taken together with evidence for the essential role of canonical Wnt signaling in the regulation of osteoblast number (13–16) and the negative role of Alox15 and PPARγ in this process (23, 24), our findings strongly suggest that activation of this cascade accounts, at least part, for the osteoblast deficit and the loss of bone with age. In studies reported elsewhere, we have elucidated that FoxOs play a critical role in skeletal homeostasis as evidenced by an increase in oxidative stress and osteoblast apoptosis in bone of mice lacking FoxO1, FoxO3, and FoxO4, the three principal members of the FoxO family.4 Therefore, the decline of β-catenin below the critical threshold required for FoxO-dependent transcription of anti-oxidant enzymes could further amplify lipoxygenase-induced oxidative stress.
FIGURE 10.
Suppression of β-catenin by an oxidized lipid-activated ROS/FoxO/PPARγ/β-catenin cascade, leading to decreased bone formation. Oxidized PUFAs generated by ROS and lipoxygenases increase the oxidative burden of the skeleton via the generation of 4-HNE. Oxidative stress activates the FoxO family of transcription factors, which in turn attenuate β-catenin/TCF-mediated transcription, leading to derepression of PPARγ transcription. Oxidized PUFAs activate PPARγ and promote association with β-catenin, resulting in a further decrease in β-catenin. Via this cascade, lipid oxidation contributes to the decline in osteoblast number and bone formation that occurs with aging by attenuating the canonical Wnt signaling required for the differentiation and survival of osteoblasts. However, PPARγ-mediated diversion of progenitors from the osteoblast to the adipocyte lineage may or may not occur depending on the skeletal site. LEF, lymphoid enhancer-binding factor.
The findings of this report indicate that oxidized PUFAs attenuate the stimulatory effects of Wnt3a on the proliferation of C2C12 cells, the differentiation of marrow-derived osteoblast progenitors, and the development of adipocytes from osteoblastic OB-6γ2 cells. This evidence is consistent with previous reports that activation of PPARγ with synthetic thiazolidinedione ligands in vitro (5, 34, 49) and in rodents (5, 50, 51) cause reciprocal changes in osteoblast and adipocyte differentiation. Moreover, patients receiving thiazolidinediones for the management of type II diabetes exhibit decreased bone mass and increased incidence of fractures (52–54). Taken together, our results support the view that the combined effects of increased lipid oxidation, PPARγ expression, and reduced Wnt signaling that occur with advancing age favor adipogenesis at the expense of osteoblastogenesis. Thus, the mechanisms described herein may account for the increased number of adipocytes in the marrow of humans and rodents with involutional osteoporosis (6–11).
Despite the age-related increase in lipid oxidation, PPARγ expression, oxidative stress, diminished Wnt signaling, and reduced osteoblast number in vertebrae of aged B6 mice, marrow adipocytes were not increased at this site, as determined by both histologic and molecular measurements. Thus, diversion of mesenchymal stem cells toward adipocytes may not be an obligatory part of the anti-osteogenic cascade activated by lipid oxidation. Indeed, an age-related increase in adipocytes via PPARγ activation would depend on the presence of multipotential mesenchymal stem cell progenitors and/or monopotential adipocyte progenitors in bone. The relative paucity of such progenitors in vertebral bone is supported by our earlier findings that adipocytes are 10-fold less abundant in vertebral marrow of 6-month-old Swiss-Webster mice, as compared with the femoral marrow (5). Moreover, even though administration of RGL increases adipocytes at both sites, the number in vertebral bone remains 5-fold less than in femoral bone.
The lipoxygenase-induced ROS/FoxO/PPARγ/β-catenin cascade may contribute to the increased osteoblast and osteocyte apoptosis seen in bone of aging B6 mice (12). Thus, oxidized PUFAs stimulate apoptosis of osteoblastic OB-6 cells via a PPARγ-dependent mechanism, consistent with previous evidence for a negative impact of RGL on osteoblast survival in vitro (55) and in vivo (51). We found that the proapoptotic effect of 4-HNE is mediated by increased oxidative stress; however, other mechanisms, for example activation of p53 (56), could also be involved. NAC (12), as well as Wnt signaling (15, 16) promote osteoblast survival in vivo. In other work we have found that a rise in endogenous glucocorticoids also contributes to the age-related increase in osteoblast apoptosis in B6 mice (57). Hence, the cause of diminished osteoblast lifespan in bone of aging animals is most likely multifactorial.
Besides its cytotoxic effects, 4-HNE has signaling properties including interference with extracellular signal-regulated kinase 1/2 phosphorylation (58) and activation of PPARβ/δ (59), which is expressed in bone (60). Such actions of 4-HNE may contribute to the deleterious effects of age on skeletal homeostasis.
Numerous clinical and epidemiologic studies (61–66) as well as studies in mice (67) point to a link between osteoporosis and cardiovascular disease. In addition, a vast literature supports the contention that lipoxygenase-dependent formation of oxidized lipids bound to low density lipoprotein plays an essential role in the development of atherosclerosis (30, 68–72). Although lipid oxidation has been noted in other tissues with advancing age (30), our findings are the first to document an age-related increase in lipoxygenase expression and lipid oxidation in the murine skeleton. That lipid oxidation has negative effects on osteoblasts was previously suggested by in vitro evidence that oxidized low density lipoprotein stimulates apoptosis of osteoblastic cells (73, 74) and that oxidized palmitoyl-arachidonyl-phosphatidylcholine inhibits BMP-2-induced osteoblast differentiation (75). The ROS/FoxO/PPARγ/β-catenin cascade elucidated here is most likely responsible for such anti-osteogenic effects because BMP-2-induced osteoblastogenesis depends on Wnt signaling (76). Whether this cascade is also involved in the pathogenesis of atherosclerosis awaits further study. However, vascular calcification has been linked with increased H2O2 levels and decreased expression of catalase, a FoxO target gene (77, 78). Moreover, reduced Wnt signaling contributes to atherogenesis as deletion of LRP5 enhances the formation of atherosclerotic lesions in ApoE-deficient mice (79). Most importantly, early onset coronary artery disease and osteoporosis coexist in individual members of an Iranian family bearing a hypomorphic mutation of LRP6 (80).
In conclusion, the evidence described herein supports the working hypothesis that involutional bone loss is due in part to an age-related rise in lipoxygenase-mediated lipid oxidation leading to increased oxidative stress and a consequent reduction in the canonical Wnt signaling required for the differentiation and survival of osteoblasts. Because lipid oxidation also plays a fundamental pathogenetic role in atherosclerosis, therapies that target lipoxygenases may be effective in the management of both conditions.
Supplementary Material
Acknowledgments
We thank Leslie Climer, Annick DeLoose, Kanan Vyas, Aaron Warren, and Rebecca Wynne for excellent technical assistance, Igor Gubrij for the generation of OB6-γ2 cells, Dr. Piotr Zimniak for advice on the measurement of 4-HNE, and Drs. Robert S. Weinstein and Charles A. O'Brien for helpful advice and suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant P01-AG13918 (to S. C. M.). This work was also supported by the Veterans Affairs merit review grants from the Office of Research and Development of the Department of Veterans Affairs (to R. L. J. and S. C. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
E. Ambrogini, M. Almeida, M. Martin-Millan, J.-H. Paik, R. A. DePinho, L. Han, J. Goellner, R. S. Weinstein, R. L. Jilka, C. A. O'Brien, and S. C. Manolagas, submitted for publication.
- B6
- C57BL/6
- LRP
- low density lipoprotein receptor-related protein
- 4-HNE
- 4-hydroxynonenal
- Fabp4
- adipocyte fatty acid-binding protein
- D2
- DBA/2
- FoxO
- Forkhead box O
- HETE
- hydroxyeiscosatetraenoic acid
- HODE
- hydroxyoctadecadienoic acid
- luc
- luciferase
- NAC
- N-acetyl cysteine
- PPARγ
- peroxisome proliferator-activated receptor-γ
- PPARE
- peroxisome proliferator response element
- PUFA
- polyunsaturated fatty acid
- ROS
- reactive oxygen species
- RGL
- rosiglitazone
- TCF
- T-cell-specific transcription factor
- BSA
- bovine serum albumin
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Lips P., Courpron P., Meunier P. J. (1978) Calcif. Tissue Res. 26, 13–17 [DOI] [PubMed] [Google Scholar]
- 2.Parfitt A. M. (1990) in Bone: Volume 1. The Osteoblast and Osteocyte (Hall B. K. ed) pp. 351–429, Telford Press and CRC Press, Boca Raton, FL [Google Scholar]
- 3.Tyner S. D., Venkatachalam S., Choi J., Jones S., Ghebranious N., Igelmann H., Lu X., Soron G., Cooper B., Brayton C., Hee Park S., Thompson T., Karsenty G., Bradley A., Donehower L. A. (2002) Nature 415, 45–53 [DOI] [PubMed] [Google Scholar]
- 4.Jilka R. L. (2002) BoneKey-Osteovision 10.1138/2002024 [DOI] [Google Scholar]
- 5.Ali A. A., Weinstein R. S., Stewart S. A., Parfitt A. M., Manolagas S. C., Jilka R. L. (2005) Endocrinology 146, 1226–1235 [DOI] [PubMed] [Google Scholar]
- 6.Meunier P., Aaron J., Edouard C., Vignon G. (1971) Clin. Orthop. 80, 147–154 [DOI] [PubMed] [Google Scholar]
- 7.Justesen J., Stenderup K., Ebbesen E. N., Mosekilde L., Steiniche T., Kassem M. (2001) Biogerontology 2, 165–171 [DOI] [PubMed] [Google Scholar]
- 8.Verma S., Rajaratnam J. H., Denton J., Hoyland J. A., Byers R. J. (2002) J. Clin. Pathol. 55, 693–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perrien D. S., Akel N. S., Dupont-Versteegden E. E., Skinner R. A., Siegel E. R., Suva L. J., Gaddy D. (2007) Am. J. Physiol. Regul. Integr. Comp. Physiol. 292, R988–R996 [DOI] [PubMed] [Google Scholar]
- 10.Duque G., Rivas D., Li W., Li A., Henderson J. E., Ferland G., Gaudreau P. (2009) Exp. Gerontol. 44, 183–189 [DOI] [PubMed] [Google Scholar]
- 11.Moerman E. J., Teng K., Lipschitz D. A., Lecka-Czernik B. (2004) Aging Cell 3, 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Almeida M., Han L., Martin-Millan M., Plotkin L. I., Stewart S. A., Roberson P. K., Kousteni S., O'Brien C. A., Bellido T., Parfitt A. M., Weinstein R. S., Jilka R. L., Manolagas S. C. (2007) J. Biol. Chem. 282, 27285–27297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodda S. J., McMahon A. P. (2006) Development 133, 3231–3244 [DOI] [PubMed] [Google Scholar]
- 14.Glass D. A., 2nd, Karsenty G. (2006) Curr. Top. Dev. Biol. 73, 43–84 [DOI] [PubMed] [Google Scholar]
- 15.Bodine P. V. (2008) Cell Res. 18, 248–253 [DOI] [PubMed] [Google Scholar]
- 16.Almeida M., Han L., Bellido T., Manolagas S. C., Kousteni S. (2005) J. Biol. Chem. 280, 41342–41351 [DOI] [PubMed] [Google Scholar]
- 17.Kops G. J., Dansen T. B., Polderman P. E., Saarloos I., Wirtz K. W., Coffer P. J., Huang T. T., Bos J. L., Medema R. H., Burgering B. M. (2002) Nature 419, 316–321 [DOI] [PubMed] [Google Scholar]
- 18.Almeida M., Han L., Martin-Millan M., O'Brien C. A., Manolagas S. C. (2007) J. Biol. Chem. 282, 27298–27305 [DOI] [PubMed] [Google Scholar]
- 19.Tontonoz P., Spiegelman B. M. (2008) Annu. Rev. Biochem. 77, 289–312 [DOI] [PubMed] [Google Scholar]
- 20.Funk C. D., Chen X. S., Johnson E. N., Zhao L. (2002) Prostaglandins Other Lipid Mediat. 68–69, 303–312 [DOI] [PubMed] [Google Scholar]
- 21.Nagy L., Tontonoz P., Alvarez J. G., Chen H., Evans R. M. (1998) Cell 93, 229–240 [DOI] [PubMed] [Google Scholar]
- 22.Schild R. L., Schaiff W. T., Carlson M. G., Cronbach E. J., Nelson D. M., Sadovsky Y. (2002) J. Clin. Endocrinol. Metab. 87, 1105–1110 [DOI] [PubMed] [Google Scholar]
- 23.Klein R. F., Allard J., Avnur Z., Nikolcheva T., Rotstein D., Carlos A. S., Shea M., Waters R. V., Belknap J. K., Peltz G., Orwoll E. S. (2004) Science 303, 229–232 [DOI] [PubMed] [Google Scholar]
- 24.Akune T., Ohba S., Kamekura S., Yamaguchi M., Chung U. I., Kubota N., Terauchi Y., Harada Y., Azuma Y., Nakamura K., Kadowaki T., Kawaguchi H. (2004) J. Clin. Invest. 113, 846–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kühn H., Borchert A. (2002) Free Radic. Biol. Med. 33, 154–172 [DOI] [PubMed] [Google Scholar]
- 26.Griesser M., Boeglin W. E., Suzuki T., Schneider C. (June24, 2009) J. Lipid Res. 10.1194/jlr.M900181-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider C., Porter N. A., Brash A. R. (2008) J. Biol. Chem. 283, 15539–15543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchida K., Shiraishi M., Naito Y., Torii Y., Nakamura Y., Osawa T. (1999) J. Biol. Chem. 274, 2234–2242 [DOI] [PubMed] [Google Scholar]
- 29.Lee J. Y., Jung G. Y., Heo H. J., Yun M. R., Park J. Y., Bae S. S., Hong K. W., Lee W. S., Kim C. D. (2006) Toxicol. Lett. 166, 212–221 [DOI] [PubMed] [Google Scholar]
- 30.Spiteller G. (2001) Exp. Gerontol. 36, 1425–1457 [DOI] [PubMed] [Google Scholar]
- 31.He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., da Costa L. T., Morin P. J., Vogelstein B., Kinzler K. W. (1998) Science 281, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 32.Morin P. J., Sparks A. B., Korinek V., Barker N., Clevers H., Vogelstein B., Kinzler K. W. (1997) Science 275, 1787–1790 [DOI] [PubMed] [Google Scholar]
- 33.Furuyama T., Nakazawa T., Nakano I., Mori N. (2000) Biochem. J. 349, 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lecka-Czernik B., Gubrij I., Moerman E. J., Kajkenova O., Lipschitz D. A., Manolagas S. C., Jilka R. L. (1999) J. Cell. Biochem. 74, 357–371 [PubMed] [Google Scholar]
- 35.Robinson C. E., Wu X., Morris D. C., Gimble J. M. (1998) Biochem. Biophys. Res. Commun. 244, 671–677 [DOI] [PubMed] [Google Scholar]
- 36.Satoh K., Yamada S., Koike Y., Igarashi Y., Toyokuni S., Kumano T., Takahata T., Hayakari M., Tsuchida S., Uchida K. (1999) Anal. Biochem. 270, 323–328 [DOI] [PubMed] [Google Scholar]
- 37.Huang X., Frenkel K., Klein C. B., Costa M. (1993) Toxicol. Appl. Pharmacol. 120, 29–36 [DOI] [PubMed] [Google Scholar]
- 38.Kousteni S., Han L., Chen J. R., Almeida M., Plotkin L. I., Bellido T., Manolagas S. C. (2003) J. Clin. Invest. 111, 1651–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siddappa R., Martens A., Doorn J., Leusink A., Olivo C., Licht R., van Rijn L., Gaspar C., Fodde R., Janssen F., van Blitterswijk C., de Boer J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7281–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mueller E., Drori S., Aiyer A., Yie J., Sarraf P., Chen H., Hauser S., Rosen E. D., Ge K., Roeder R. G., Spiegelman B. M. (2002) J. Biol. Chem. 277, 41925–41930 [DOI] [PubMed] [Google Scholar]
- 41.Giorgio M., Migliaccio E., Orsini F., Paolucci D., Moroni M., Contursi C., Pelliccia G., Luzi L., Minucci S., Marcaccio M., Pinton P., Rizzuto R., Bernardi P., Paolucci F., Pelicci P. G. (2005) Cell 122, 221–233 [DOI] [PubMed] [Google Scholar]
- 42.Kang S., Bennett C. N., Gerin I., Rapp L. A., Hankenson K. D., Macdougald O. A. (2007) J. Biol. Chem. 282, 14515–14524 [DOI] [PubMed] [Google Scholar]
- 43.Sharma C., Pradeep A., Wong L., Rana A., Rana B. (2004) J. Biol. Chem. 279, 35583–35594 [DOI] [PubMed] [Google Scholar]
- 44.Christodoulides C., Lagathu C., Sethi J. K., Vidal-Puig A. (2009) Trends Endocrinol. Metab. 20, 16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okamura M., Kudo H., Wakabayashi K., Tanaka T., Nonaka A., Uchida A., Tsutsumi S., Sakakibara I., Naito M., Osborne T. F., Hamakubo T., Ito S., Aburatani H., Yanagisawa M., Kodama T., Sakai J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5819–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Limor R., Sharon O., Knoll E., Many A., Weisinger G., Stern N. (2008) Am. J. Hypertens. 21, 219–223 [DOI] [PubMed] [Google Scholar]
- 47.Dowell P., Otto T. C., Adi S., Lane M. D. (2003) J. Biol. Chem. 278, 45485–45491 [DOI] [PubMed] [Google Scholar]
- 48.Armoni M., Harel C., Karni S., Chen H., Bar-Yoseph F., Ver M. R., Quon M. J., Karnieli E. (2006) J. Biol. Chem. 281, 19881–19891 [DOI] [PubMed] [Google Scholar]
- 49.Lecka-Czernik B., Moerman E. J., Grant D. F., Lehmann J. M., Manolagas S. C., Jilka R. L. (2002) Endocrinology 143, 2376–2384 [DOI] [PubMed] [Google Scholar]
- 50.Rzonca S. O., Suva L. J., Gaddy D., Montague D. C., Lecka-Czernik B. (2004) Endocrinology 145, 401–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sorocéanu M. A., Miao D., Bai X. Y., Su H., Goltzman D., Karaplis A. C. (2004) J. Endocrinol. 183, 203–216 [DOI] [PubMed] [Google Scholar]
- 52.Schwartz A. V., Sellmeyer D. E., Vittinghoff E., Palermo L., Lecka-Czernik B., Feingold K. R., Strotmeyer E. S., Resnick H. E., Carbone L., Beamer B. A., Park S. W., Lane N. E., Harris T. B., Cummings S. R. (2006) J. Clin. Endocrinol. Metab. 91, 3349–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meier C., Kraenzlin M. E., Bodmer M., Jick S. S., Jick H., Meier C. R. (2008) Arch. Intern. Med. 168, 820–825 [DOI] [PubMed] [Google Scholar]
- 54.Andersen T. L., Sondergaard T. E., Skorzynska K. E., Dagnaes-Hansen F., Plesner T. L., Hauge E. M., Plesner T., Delaisse J. M. (2009) Am. J. Pathol. 174, 239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim S. H., Yoo C. I., Kim H. T., Park J. Y., Kwon C. H., Kim Y. K. (2006) Toxicol. Appl. Pharmacol. 215, 198–207 [DOI] [PubMed] [Google Scholar]
- 56.Sharma A., Sharma R., Chaudhary P., Vatsyayan R., Pearce V., Jeyabal P. V., Zimniak P., Awasthi S., Awasthi Y. C. (2008) Arch. Biochem. Biophys. 480, 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinstein R. S., Goellner J., Chambers T. M., Hogan E. A., Berryhill S. B., Shelton R., Webb W. W., Wicker C. A., Manolagas S. C. (2008) J. Bone Miner. Res. 23, S25 (abstr) [Google Scholar]
- 58.Sampey B. P., Carbone D. L., Doorn J. A., Drechsel D. A., Petersen D. R. (2007) Mol. Pharmacol. 71, 871–883 [DOI] [PubMed] [Google Scholar]
- 59.Coleman J. D., Prabhu K. S., Thompson J. T., Reddy P. S., Peters J. M., Peterson B. R., Reddy C. C., Vanden Heuvel J. P. (2007) Free Radic. Biol. Med. 42, 1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giaginis C., Tsantili-Kakoulidou A., Theocharis S. (2007) Fundam. Clin. Pharmacol. 21, 231–244 [DOI] [PubMed] [Google Scholar]
- 61.Tankò L. B., Bagger Y. Z., Christiansen C. (2003) Calcif. Tissue Int. 73, 15–20 [DOI] [PubMed] [Google Scholar]
- 62.Farhat G. N., Cauley J. A., Matthews K. A., Newman A. B., Johnston J., Mackey R., Edmundowicz D., Sutton-Tyrrell K. (2006) J. Bone Miner. Res. 21, 1839–1846 [DOI] [PubMed] [Google Scholar]
- 63.Vogt M. T., Wolfson S. K., Kuller L. H. (1992) J. Clin. Epidemiol. 45, 529–542 [DOI] [PubMed] [Google Scholar]
- 64.Hak A. E., Pols H. A., van Hemert A. M., Hofman A., Witteman J. C. (2000) Arterioscler. Thromb. Vasc. Biol. 20, 1926–1931 [DOI] [PubMed] [Google Scholar]
- 65.Kiel D. P., Kauppila L. I., Cupples L. A., Hannan M. T., O'Donnell C. J., Wilson P. W. (2001) Calcif. Tissue Int. 68, 271–276 [DOI] [PubMed] [Google Scholar]
- 66.Schulz E., Arfai K., Liu X., Sayre J., Gilsanz V. (2004) J. Clin. Endocrinol. Metab. 89, 4246–4253 [DOI] [PubMed] [Google Scholar]
- 67.Parhami F., Tintut Y., Beamer W. G., Gharavi N., Goodman W., Demer L. L. (2001) J. Bone Miner. Res. 16, 182–188 [DOI] [PubMed] [Google Scholar]
- 68.Huo Y., Zhao L., Hyman M. C., Shashkin P., Harry B. L., Burcin T., Forlow S. B., Stark M. A., Smith D. F., Clarke S., Srinivasan S., Hedrick C. C., Praticò D., Witztum J. L., Nadler J. L., Funk C. D., Ley K. (2004) Circulation 110, 2024–2031 [DOI] [PubMed] [Google Scholar]
- 69.Cyrus T., Praticò D., Zhao L., Witztum J. L., Rader D. J., Rokach J., FitzGerald G. A., Funk C. D. (2001) Circulation 103, 2277–2282 [DOI] [PubMed] [Google Scholar]
- 70.Navab M., Ananthramaiah G. M., Reddy S. T., Van Lenten B. J., Ansell B. J., Fonarow G. C., Vahabzadeh K., Hama S., Hough G., Kamranpour N., Berliner J. A., Lusis A. J., Fogelman A. M. (2004) J. Lipid Res. 45, 993–1007 [DOI] [PubMed] [Google Scholar]
- 71.Berliner J. A., Watson A. D. (2005) N. Engl. J. Med. 353, 9–11 [DOI] [PubMed] [Google Scholar]
- 72.Steinberg D. (2009) J. Lipid Res. 50, S376–S381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brodeur M. R., Brissette L., Falstrault L., Ouellet P., Moreau R. (2008) Free Radic. Biol. Med. 44, 506–517 [DOI] [PubMed] [Google Scholar]
- 74.Klein B. Y., Rojansky N., Ben-Yehuda A., Abou-Atta I., Abedat S., Friedman G. (2003) J. Cell Biochem. 90, 42–58 [DOI] [PubMed] [Google Scholar]
- 75.Huang M. S., Morony S., Lu J., Zhang Z., Bezouglaia O., Tseng W., Tetradis S., Demer L. L., Tintut Y. (2007) J. Biol. Chem. 282, 21237–21243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rawadi G., Vayssière B., Dunn F., Baron R., Roman-Roman S. (2003) J. Bone Miner. Res. 18, 1842–1853 [DOI] [PubMed] [Google Scholar]
- 77.Towler D. A. (2008) J. Am. Coll. Cardiol. 52, 851–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller J. D., Chu Y., Brooks R. M., Richenbacher W. E., Peña-Silva R., Heistad D. D. (2008) J. Am. Coll. Cardiol. 52, 843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Magoori K., Kang M. J., Ito M. R., Kakuuchi H., Ioka R. X., Kamataki A., Kim D. H., Asaba H., Iwasaki S., Takei Y. A., Sasaki M., Usui S., Okazaki M., Takahashi S., Ono M., Nose M., Sakai J., Fujino T., Yamamoto T. T. (2003) J. Biol. Chem. 278, 11331–11336 [DOI] [PubMed] [Google Scholar]
- 80.Mani A., Radhakrishnan J., Wang H., Mani A., Mani M. A., Nelson-Williams C., Carew K. S., Mane S., Najmabadi H., Wu D., Lifton R. P. (2007) Science 315, 1278–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.