Abstract
DAX-1 (dosage-sensitive sex reversal adrenal hypoplasia congenital critical region on X chromosome, gene 1) is an atypical member of the nuclear receptor family and acts as a corepressor of a number of nuclear receptors. HNF4α (hepatocyte nuclear factor 4α) is a liver-enriched transcription factor that controls the expression of a variety of genes involved in cholesterol, fatty acid, and glucose metabolism. Here we show that DAX-1 inhibits transcriptional activity of HNF4α and modulates hepatic gluconeogenic gene expression. Hepatic DAX-1 expression is increased by insulin and SIK1 (salt-inducible kinase 1), whereas it is decreased in high fat diet-fed and diabetic mice. Coimmunoprecipitation assay from mouse liver samples depicts that endogenous DAX-1 interacts with HNF4α in vivo. In vivo chromatin immunoprecipitation assay affirms that the recruitment of DAX-1 on the phosphoenolpyruvate carboxykinase (PEPCK) gene promoter is inversely correlated with the recruitment of PGC-1α and HNF4α under fasting and refeeding, showing that DAX-1 could compete with the coactivator PGC-1α for binding to HNF4α. Adenovirus-mediated expression of DAX-1 decreased both HNF4α- and forskolin-mediated gluconeogenic gene expressions. In addition, knockdown of DAX-1 partially reverses the insulin-mediated inhibition of gluconeogenic gene expression in primary hepatocytes. Finally, DAX-1 inhibits PEPCK and glucose-6-phosphatase gene expression and significantly lowers fasting blood glucose level in high fat diet-fed mice, suggesting that DAX-1 can modulate hepatic gluconeogenesis in vivo. Overall, this study demonstrates that DAX-1 acts as a corepressor of HNF4α to negatively regulate hepatic gluconeogenic gene expression in liver.
The nuclear receptor superfamily comprises a diverse group of transcription factors including conventional receptors with known ligands and orphan nuclear receptors without ligands (1, 2). Among nuclear receptors, DAX-1 belongs to a group of atypical nuclear receptors that do not possess classical DNA binding domains and is closely related to the same family member SHP (small heterodimer partner) (3–5). DAX-1 was identified through a screening for genes linked to adrenal hypoplasia critical, a disease that affects the normal development of the adrenal cortex and is often associated with hypogonadism (6). DAX-1 has a putative ligand binding domain (LBD)4 in the C terminus, although no ligand has been identified yet (3), whereas N-terminal region contains a unique repeat domain that is involved in single-stranded DNA and RNA binding and protein-protein interactions (7, 8). This N-terminal region comprises three LXXLL motif-like sequences that are necessary for the interaction with estrogen receptors (ERs) (9). It is well established that DAX-1 functions as a coregulatory protein rather than a transcriptional factor, since it inhibits the transcriptional activity of other nuclear receptors, such as steroidogenic factor 1 (10), ER (9), androgen receptor (11), progesterone receptor (12), liver receptor homolog-1 (13), peroxisome proliferator-activated receptor γ (PPARγ) (14), estrogen receptor-related receptor γ (ERRγ) (15), and Nur77 (nerve growth factor-inducible gene B) (16). It has been reported that activation of cyclic AMP signaling by ACTH, follicle-stimulating hormone, or luteinizing hormone leads to the down-regulation of DAX-1 gene expression (16–18).
HNF4α (hepatocyte nuclear factor 4α) is classified as an orphan nuclear receptor with no identifiable ligands (19). It is a strong constitutive transcriptional activator and can bind as a homodimer to a direct repeat element (AGGTCA) with a 1- or 2-nucleotide spacer (20). It is expressed at high levels in liver and to a lesser degree in kidney, small intestine, colon, and pancreatic β-cells (21). The absence of HNF4α in the adult liver results in marked metabolic dysregulation and increased mortality (22). Mutations in the human HNF4α gene lead to the maturity onset diabetes of the young subtype I, which is characterized by autosomal dominant inheritance and impaired glucose-stimulated insulin secretion from pancreatic β-cells (23–25). HNF4α recruits several coactivators that are able to modulate chromatin, including SRC-1, GRIP-1, PGC-1α (peroxisome proliferator-activated receptor γ coactivator-1α), and CREB-binding protein, all of which may promote histone-3 acetylation of target gene promoters, leading to the formation of transcription-permissive chromatin remodeling events (26–30). HNF4α controls the expression of genes involved in the transport and metabolism of nutrients, including cholesterol, fatty acids, and glucose (22–25); hepatocyte polarization (26); and liver development and differentiation (27). The elevated cellular activity of PGC-1α is responsible for the HNF4α-dependent activation of hepatic target genes, such as Pepck (phosphoenolpyruvate carboxykinase) and G6pase (glucose-6-phosphatase), that leads to the increased gluconeogenesis (28).
Glucose homeostasis is regulated primarily by the opposing actions of insulin and glucagon, hormones that are secreted by pancreatic islets from β-cells and α-cells, respectively (31–33). Glucagon stimulates an increase in cAMP concentration and activates cAMP-dependent protein kinase, whereas insulin suppresses hepatic gluconeogenesis through the activation of Akt (34). In addition, AMP-activated protein kinase has been shown to inhibit gluconeogenesis and lipogenesis by promoting both fatty acid oxidation and lipolysis. Activation of AMP-activated protein kinase by 5-aminoimidazole-4-carboxamide-1-β-d-ribofuranoside and metformin has been shown to inhibit the expression of two key hepatic gluconeogenic genes, Pepck and G6pase (35). Transcription factors, such as FOXO1 and HNF4α, are required for transcriptional activation of gluconeogenic genes, such as Pepck and G6pase, in an insulin-sensitive manner. CREB-binding protein, CRTC2 (CREB-regulated transcription coactivator 2)/TORC2, and PGC-1α are well known coactivators of fasting-induced glucose production. CRTC2/TORC2 can be inactivated during fasting by SIK1 (salt-inducible kinase 1) through an autoregulatory negative feedback loop that attenuates gluconeogenesis by exporting CRTC2/TORC2 from the nucleus (36, 37).
In this study, we have observed that DAX-1 inhibits the transcriptional activity of HNF4α to control hepatic gluconeogenic gene expression. DAX-1 expression is increased under refeeding conditions in mouse liver, which was recapitulated with either treatment of insulin or expression of SIK1 in primary hepatocytes. We demonstrated a physical interaction and colocalization of these two proteins in vivo. DAX-1 competes with the coactivator PGC-1α for binding to HNF4α. Overexpression of DAX-1 decreased HNF4α-mediated expression of G6Pase and PEPCK and reduced glucose production in hepatocytes. Hepatic DAX-1 expression was decreased in high fat diet-induced or genetic (db/db) insulin resistant mice, suggesting that the reduced expression of this protein might be correlated with hyperglycemia that is associated with this condition. Indeed, overexpression of DAX-1 led to the decreased hepatic PEPCK and G6Pase expression and the reduced blood glucose levels in the high fat diet-fed mice. Collectively, these findings suggest that DAX-1 acts a repressor of HNF4α to negatively regulate the hepatic gluconeogenesis in mammals.
EXPERIMENTAL PROCEDURES
Plasmids
The reporter plasmids, 8×HNF4RE-Luc (27), human G6Pase-Luc (G6Pase −1227/+57), and mouse Pepck-Luc were described previously (27, 38, 39). Rat hnf4α WT, 1–128, 1–370, 1–360, 1–339, 128–360, and 128–370 were subcloned into pcDNA3-HA (Invitrogen) and pLeXA (BD Biosciences, Clontech) at EcoRI and XhoI sites. GFP-hDAX-1, FLAG-hDAX-1, pGEX-4T (GST)-hnf4α, and pcDNA3-HA-Pgc-1α were described previously (14, 16, 30). DAX-1 was subcloned into pEBG (GST) vector using BamHI and NotI. The deletion constructs of mouse Dax-1 promoters (15) and Sik1 (36) were described earlier. G6pase, hnf4α, and DAX-1 probes for Northern blot analysis were prepared as described previously (16, 21, 35).
Cell Culture and Transient Transfection Assay
HepG2, 293T, HeLa, and H4IIE cells were maintained in Dulbecco's modified Eagle's medium, and AML12 cells were maintained in Dulbecco's modified Eagle's medium/F-12 (Invitrogen), supplemented with 10% fetal bovine serum (Cambrex Bioscience Walkersville, Inc., Walkersville, MD) and antibiotics (Invitrogen). Cells were split into 24-well plates at densities of 2–8 × 104 cells/well the day before transfection. Transient transfections were performed using the SuperFect transfection reagent (Qiagen, Valencia, CA) according to the manufacturer's instructions. Cells were cotransfected with the indicated reporter plasmids together with expression vectors encoding various transcription factors. Total DNA used in each transfection was adjusted to 1 μg/well by adding an appropriate amount of empty vector, and cytomegalovirus (CMV)-β-galactosidase plasmids were cotransfected as an internal control. Cells were harvested ∼40–48 h after the transfection for luciferase and β-galactosidase assays. The luciferase activity was normalized with β-galactosidase activity.
Confocal Microscopy
Confocal microscopy was performed as described previously (40). Briefly, HeLa cells were grown on uncoated glass coverslips and transfected with pEGFP-DAX-1 and pCDNA3/HA-hnf4α by the Lipofectamine method (Invitrogen). At 24 h after transfection, cells were fixed with 3.7% formaldehyde for 40 min, mounted on glass slides, and observed with a laser-scanning confocal microscope (Olympus Corp., Lake Success, NY). For detection of pCDNA3/HA-Hnf4α, cells were permeabilized with 2 ml of PBS containing 0.1% Triton X-100 and 0.1 m glycine at room temperature, incubated for 15 min, washed three times with 1× PBS, and blocked with 3% (w/v) bovine serum albumin in PBS for 10 min at room temperature. Cells were directly incubated with primary anti-HA antibody (40) for 1 h at room temperature, washed three times with 1× PBS, and then mounted on a slide and observed under the microscope.
Preparation of Recombinant Adenovirus
For the ectopic expression of transgene, the adenoviral vector systems were used as described previously (36). Briefly, the cDNA encoding HA-hnf4α was cloned into pAdTrack shuttle vector. The HA-rat hnf4α cDNA was cloned by insertion of a KpnI/XhoI-digested fragment into the KpnI/XhoI site of pAdTrack-CMV vector. Recombination of AdTrack-CMV-HA-hnf4α with adenoviral gene carrier vector was performed by transformation into pretransformed AdEasy-BJ21 competent cells. Recombinant adenoviruses expressing GFP only or SIK1 or nonspecific RNA interference control (US) were described earlier (36). Adenoviruses for mouse Dax-1 and Shp were described previously (16, 40). Oligonucleotides for short hairpin RNA of mouse Dax-1 (ctgtaccgctgctgcttctgcggagaa) were synthesized by IDT (Coralville, IA). Adenovirus for mouse shDax-1 was generated as described previously (36).
Isolation and Culture of Primary Hepatocytes
Primary hepatocytes were prepared from 200–300-g Sprague-Dawley rats by the collagenase perfusion method as described (37). After attachment, cells were infected with adenoviruses for 16 h. Subsequently, cells were maintained in serum-free Medium 199 (Mediatech) overnight and treated with 100 nm dexamethasone and 10 nm forskolin for 2 h with or without 100 nm insulin for 16 h.
Animal Experiments
Male, 7-week-old C57BL/6 mice or db/db diabetic mice were purchased from Charles River Laboratories. In some experiments, mice were fed a high fat diet from the start of the weaning period for 8 weeks before being injected with adenoviruses. Recombinant Adenovirus (0.5 × 109 plaque-forming units) was delivered by tail vein injection into mice. To measure fasting blood glucose levels, animals were fasted for 16 h with free access to water. Blood glucose was monitored from tail vein blood using an automatic glucose monitor (One Touch; Lifescan). Insulin was measured by a mouse insulin enzyme-linked immunosorbent assay kit (U-Type; Shibayagi Corp.). All procedures were approved by the Sungkyunkwan University School of Medicine Institutional Animal Care And Use Committee.
Northern Blotting
HepG2 cells were maintained in the indicated media. Cells were infected with adenovirus DAX-1 at a multiplicity of infection of 20 or 40. Cells were further serum-starved for 24 h and were treated with Fsk for 12 h alone or followed by 1 h of insulin treatment. The total RNA was isolated from each sample and used for Northern blot analysis as described previously (16). The probe labeling of each of the cDNAs for hnf4α, G6pase, and DAX-1 with [α-32P]dCTP was performed using a random primer DNA-labeling system (Amersham Biosciences). The expression of all transcripts was normalized to GAPDH levels. All Northern blot analyses were performed identically unless otherwise stated. The data are representative of a minimum of three independent experiments.
siRNA Experiments
The siRNAs for hDAX-1 (si-hDAX-1-I and si-hDAX-1-II) were chemically synthesized (Invitrogen), deprotected, annealed, and transfected according to the manufacturer's instructions. HepG2 cells were transfected with siRNA using Oligofectamine reagent (Qiagen). Forty-eight hours after transfection, total RNA was isolated for RT-PCR for hDAX-1 (30 cycles) and for β-actin (25 cycles) as a control. The sequences of siRNA are as follows: si-hDAX-1-I, 5′-UCACUGAGUAUUUGCUGAGUUCCC-3′; si-hDAX-1-II, 5′-GGGAACUCAGCAAAUACU CAGUGAA-3′.
Quantitative RT-PCR
Total RNAs were extracted from either tissue samples or rat primary hepatocytes under various conditions with TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The mRNAs for dAX-1, hnf4α, G6pase, and Pepck were analyzed by RT-PCR, as described previously (36). The primers used for PCR of human/rat G6Pase, mouse DAX-1, rat PEPCK, and rat β-actin are as follows: human G6Pase, forward (5′-GTGAATTACCAAGACTCCCAG-3′) and reverse (5′-GCCCATGGCATGGCCAGAGGG-3′); rat g6pase, forward (5′-GTGGGTCCTGGACACTGACT-3′) and reverse (5′-AAAGTGAGCCGCAAGGTAGA-3′); human/mouse DAX-1, forward (5′-AGGGCAGCATCCTCTACAAC-3′) and reverse (5′-TGGTCTTCACCACAAAAGCA-3′); rat pepck, forward (5′-CTGGTTCCGGAAAGACAAAA) and reverse (5′-GCTCGGAGCTCCCTCTCTAT); rat β-actin, forward (5′-CGTGAAAAGATGACCCAGATCATGTT) and reverse (5′-GCTCATTGCCGATAGTGATGACCTG-3′). The sizes of PCR products of mouse/rat DAX-1, mouse/rat G6Pase, mouse CYP7A1 (cholesterol 7 α-hydroxylase), rat/mouse PEPCK, and mouse/rat β-actin were 370, 305/274, 350, 297, and 350 bp, respectively.
In Vivo Chromatin Immunoprecipitation (ChIP) Assay and Real Time PCR
The ChIP assay was performed as described previously (36). Briefly, 12 h after fasting or refeeding, mouse livers (40 mg/sample) were homogenized and centrifuged at 2000 rpm for 2 min at 4 °C. Then pellets were resuspended in nuclear lysis buffer (5 mm HEPES, pH 8, 85 mm KCl, 0.5% Nonidet P-40) and incubated at 4 °C for 15 min with rotation, and it was centrifuged at 2000 rpm for 2 min at 4 °C. Cells were fixed with 1% formaldehyde and further processed using the Upstate ChIP assay kit as described previously (36). Soluble chromatin was immunoprecipitated with anti-DAX-1 (H-300; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), HNF4α (H-171; Santa Cruz Biotechnology), and PGC-1α (H-300; Santa Cruz Biotechnology) IgG antibodies. After purification, DNA samples were quantified by quantitative real time PCR using two pairs of primers encompassing proximal (−500/−270 bp) or distal (−1299/−1010 bp) region of the mouse Pepck promoter. The primers used for PCR are as follows: proximal, forward (5′- GATGTAGACTCTGTCCTG-3′) and reverse (5′-GATTGCTCTGCTATGAGT-3′); distal, forward (5′-CACTTGGCCAGGCATGGTGG-3′) and reverse (5′-TTTGAAACAGGGTCTCACTCT-3′).
GST Pull-down Assay
GST pull-down assay was performed according to the method described previously (39). Briefly, HNF4α and PPARγ were labeled with [35S]methionine using a TNT-coupled reticulocyte lysate system (Promega Corp., Madison, WI) according to the manufacturer's instructions. GST alone and GST-fused DAX-1 (GST-DAX-1) proteins were prepared as described previously (14). GST fusion proteins were prebound with glutathione-Sepharose beads (Amersham Biosciences) and then incubated with in vitro-translated [35S]methionine-labeled HNF4α and PPARγ proteins in binding. The beads were washed three times with the binding buffer, analyzed by SDS-PAGE, and visualized by a phosphor image analyzer (BAS-1500; Fuji, Japan).
In Vivo Interaction and Co-immunoprecipitation Assays
293T Cells grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum were plated in 6-well flat-bottomed microplates at a concentration of 2 × 105 cells/well the day before transfection as described previously (40). Briefly, 1 μg of each plasmid was transfected into 293T cells with a calcium phosphate precipitation method. Forty-eight hours after transfection, cells were solubilized, and the cleared lysates were mixed with 15 μl of glutathione-Sepharose beads and rotated for 2 h at 4 °C. The bound proteins were eluted by boiling in SDS sample buffer, subjected to SDS-PAGE, and then transferred to polyvinylidene difluoride membranes (Millipore Corp., Bedford, MA). The membranes were probed with anti-HA or anti-GST antibody and then developed using the ECL kit. For co-immunoprecipitation assays, 750 μg of total protein extract from fasted (12 h) or refed (12 h) mouse livers were immunoprecipitated using HNF4α (H-171; Santa Cruz Biotechnology) or DAX-1 (H-300; Santa Cruz Biotechnology) antibody and then blotted with DAX-1 or HNF4α antibody and anti-rabbit IgG (Trueblot; eBioscience) Signals were detected with ECL-plus (Amersham Biosciences).
In Vivo Immunofluorescent Staining
Paraffin sections were used for immunofluorescent staining. After deparaffinizing in xylene and rehydratation, the sections were treated with sodium citrate buffer (pH 6.5) and microwaved for 15 min. After cooling for 30 min at room temperature, the sections were incubated with 3% bovine serum albumin for 1 h. The tissue sections were incubated overnight at 4 °C with rabbit polyclonal anti-DAX-1 (H-300; Santa Cruz Biotechnology) and anti-HNF4α antibody (1:100; H-171; Santa Cruz Biotechnology). Alexa Fluor 488- and 546-nm goat monoclonal anti-rabbit antibody (1:300; Invitrogen) was then applied for 1 h at 37 °C in the dark. After each step, coverslips were washed three times for 5 min in PBS. The coverslips were then mounted by ProLong Gold Antifade reagent (Invitrogen) and analyzed under a dual fluorescence microscope (LSM 510 META; Carl Zeiss, Jena, Germany). Control coverslips were processed the same way without primary antibodies.
Western Blot Analysis
Western blot analysis was performed as described previously (40). Briefly, 293T cells were transfected with the indicated expression vectors or siRNA oligonucleotides. Forty-eight hours after transfection, cell lysates were prepared and separated on 12% SDS-polyacrylamide gel. About 80% confluences of H4IIE and rat primary hepatocytes were treated with insulin, cells were harvested at different time points, and proteins were transferred to nitrocellulose membrane (Amersham Biosciences). The membranes were probed with an anti-HA, FLAG (40), DAX-1 (16), or β-actin (40) antibody and developed after secondary antibody incubation using the ECL kit (Amersham Biosciences) according to the manufacturer's instructions.
Yeast Two-hybrid Assay
Yeast two-hybrid interaction assays were performed as described previously (16). Briefly, the LexA-DNA binding domain-fused DAX-1 or SHP constructs and B42-Ad-fused Nur77 or HNF4α constructs were co-transformed into yeast strain EGY48 cells. The transformants were selected on plates (Ura−, His−, and Trp−) with appropriate selection markers and assayed for β-galactosidase activity. The liquid β-galactosidase assay was carried out as described previously (16).
Statistical Analyses
Results are shown as mean ± S.D. The comparison of different groups was carried out using two-tailed unpaired Student's t test, and differences of p < 0.05 were considered statistically significant and reported in the figure legends (6–8).
RESULTS
DAX-1 Represses the Transcriptional Activity of HNF4α
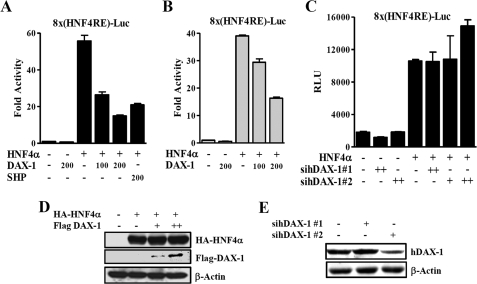
Since DAX-1 is structurally and functionally similar to its family member SHP (1), we have attempted to test DAX-1 interacts with several known SHP-interacting proteins. By a yeast two-hybrid system, we identified HNF4α as a prominent interacting protein with DAX-1 (data not shown). To determine whether DAX-1 affects the transcriptional activity of HNF4α, we performed transient transfection assay using a reporter gene containing multiple HNF4α binding site (8×(HNF4α)RE-Luc) and expression vectors for HA-hnf4α and FLAG-DAX-1 in HepG2 and 293T cells. DAX-1 dose-dependently decreased the transcriptional activity of HNF4α in both cell lines (Fig. 1, A and B), indicating that the repressive effect of DAX-1 is not cell type-specific. SHP was used as a positive control for repressing HNF4α. Western blot analysis showed that increase in FLAG-DAX-1 protein did not affect the expression level of HA-HNF4α, indicating that this repressive effect was not due the reduction in HNF4α protein level (Fig. 1D). To investigate the effect of endogenous DAX-1 on HNF4α transactivity, we knocked down the endogenous DAX-1 using small interfering RNAs. We transfected 8×HNF4αRE-reporter construct along with or without small interfering RNAs of DAX-1 (siRNA-hDAX-1 1 and 2) in HepG2 cells. The basal transactivity of HNF4α was increased about 30% by sihDAX-1 2 (Fig. 1C). Our Western blot analysis in HepG2 cells showed that siRNA 2 significantly reduced the endogenous expression of DAX-1, whereas siRNA 1 did not alter the expression of endogenous DAX-1 (Fig. 1E). Overall, these observations suggest that DAX-1 acts as a novel nuclear corepressor of HNF4α.
FIGURE 1.
DAX-1 represses the transcriptional activity of HNF4α. A and B, HepG2 (A) and 293T (B) cells were transfected with pcDNA3-HA-hnf4α (300 ng), HA-DAX-1 (50, 100, and 200 ng), and 8×(HNF4α)-Luc (200 ng). Effect of DAX-1 alone with basal reporter activity was also shown. C, HepG2 cells were transfected with sihDAX-1 1 and 2, and 24 h later HA-HNF4α (200 ng) and 8×(HNF4α)-Luc (200 ng). After 24 h, the cells were harvested, and we performed luciferase and β-galactosidase assays. D, 293T cells were transfected with HA-HNF4α (10 μg) and FLAG-hDAX-1 (5 and 10 μg). E, HepG2 cells were transfected with siRNA-hDAX-1 1 and 2. After a 48-h transfection, cells (D and E) were harvested for Western blot analysis with the indicated antibodies.
DAX-1 Interacts with HNF4α in Vitro and in Vivo
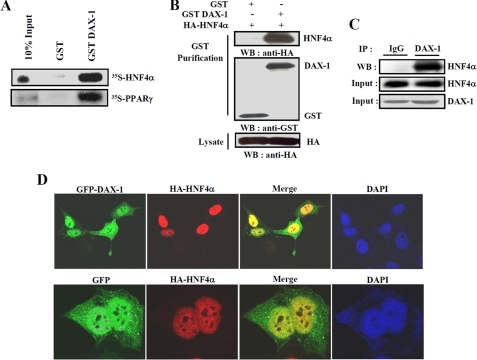
To confirm the interaction between DAX-1 and HNF4α, we performed in vitro and in vivo GST pull-down assays. Our in vitro GST pull-down assay showed that 35S-labeled Hnf4α was bound to bacterially expressed GST-DAX-1 (Fig. 2A). Similarly, labeled PPARγ also interacted with GST-DAX-1, as reported previously (14). Purity controls for in vitro translated and GST-purified proteins were shown (supplemental Fig. 1). Overall, these results suggest that DAX-1 interacts with HNF4α in vitro. To confirm this interaction in vivo, we performed in vivo GST pull-down assays. We transfected HA-hnf4α with mammalian expression vectors GST (pEBG) or GST-DAX-1 (pEBG-hDAX-1) within 293T cells. After GST purification, HA-Hnf4α was detected in the coprecipitates only when coexpressed with GST-DAX-1 but not with the negative control GST alone (Fig. 2B). Similar results were obtained when we performed the assay using pEBG only or pEBG-Hnf4α together with HA-DAX-1 (data not shown). To further examine the endogenous interaction of these two proteins, we performed the coimmunoprecipitation assay using mouse liver samples under normal as well as fasting and refeeding conditions. Endogenous HNF4α and DAX-1 proteins were found to be co-precipitated with DAX-1 and HNF4α, respectively, under normal condition (Fig. 2C) as well as fasting and refeeding conditions (data not shown). These results confirm the endogenous interaction between DAX-1 and HNF4α in mouse liver, although no significant changes in interaction between these two proteins were observed under fasting and refeeding conditions. In order to investigate the subcellular locacalization of DAX-1 and HNF4α, confocal microscopic analysis was performed after transfecting GFP-DAX-1 and HA-hnf4α in HeLa cells. As expected, GFP-DAX-1 was scattered predominantly in the nucleus and weakly in the cytoplasm, consistent with the previous report (14), whereas HA-Hnf4α was distributed only in the nucleus, as expected (38). Moreover, our merge image indicated that these two proteins were found to be colocalized in the nucleus (Fig. 2D). Taken together, these results demonstrate that DAX-1 physically interacts with HNF4α in vivo, and they are colocalized in the nucleus.
FIGURE 2.
Interaction between DAX-1 and HNF4α both in vitro and in vivo. A, in vitro GST pull-down assay. Hnf4α and PPARγ proteins were labeled with [35S]methionine by in vitro translation and incubated with glutathione-Sepharose beads containing bacterially expressed GST alone and GST-DAX-1 fusion proteins (top). B, in vivo interaction between DAX-1 and HNF4α. 293T cells were cotransfected with expression vectors for HA-hnf4α together with pEBG-DAX-1 (GSTDAX-1) or GST alone (pEBG) as a control. The complex formation (top, GST puri.) and the amount of HA-hnf4α used for the in vivo binding assay (bottom, Lysate) were determined by anti-HA antibody immunoblot (WB). The same blot was stripped and reprobed with an anti-GST antibody (middle) to confirm the expression levels of the GST fusion protein (GST-hDAX-1) and the GST control (GST). C, co-immunoprecipitation assays with liver extracts (n = 4) demonstrate the functional association between the HNF4α and DAX-1. Protein extracts from livers were immunoprecipitated using DAX-1 antibody and Western blotted with HNF4α (top) antibody. Expression of HNF4α (middle) and DAX-1 (bottom) from 10% of lysate were analyzed by Western blotting with specific antibodies. D, subcellular localization of DAX-1 and HNF4α. HeLa cells were transiently transfected with pEGFP-DAX-1 or pEGFP with pCDNA3/HA-hnf4α. The yellow stain in the merged image depicts colocalization of DAX-1 and HNF4α. Data shown are representative cells from one of three independent experiments. DAPI, 4′,6-diamidino-2-phenylindole.
Interaction Domain Mapping of DAX-1 and HNF4α
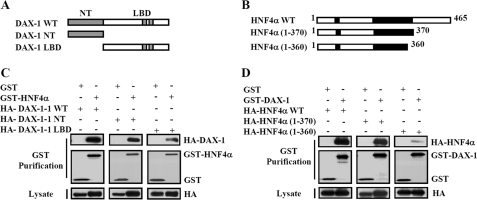
Next, we tried to examine the interaction domain of DAX-1 with HNF4α. We thus performed yeast two-hybrid assays using LexA-fused wild-type (LexA hDAX-1 WT) or deletion constructs of DAX-1(LexA hDAX-1 NT and LBD) and B42 fused wild-type HNF4α (B42 hnf4α WT). Interestingly, all three constructs of DAX-1 interacted with wild-type Hnf4α, indicating that the entire protein is required for the interaction with Hnf4α. To map the domain within HNF4α required for the interaction with DAX-1, we performed yeast two-hybrid and in vivo GST pull-down assays. We used B42-fused wild-type (B42-hnf4α WT) or deletion constructs of hnf4α (B42-HNF4α 1–128, 1–370, 1–360, 1–339, 128–360, and 128–370) and LexA DAX-1 WT for the yeast two-hybrid interaction assays. We have observed strong interaction between LexA-DAX-1 and B42-Hnf4α 1–370 constructs, which was similar to that of the interaction with B42-Hnf4α WT. However, the interaction of LexA-DAX-1 with B42- Hnf4α 1–360 was reduced to 50%, indicating that the motif containing 10 amino acids (amino acids 360–370) of HNF4α is essential for the interaction. For positive controls, Nu77 (14) and SHP (45) were used with the deletion constructs of DAX-1 and HNF4α, respectively (supplemental Fig. S2).
Next, we confirmed the result of domain mapping in vivo by cotransfecting the mammalian expression vector encoding either GST (pEBG) alone or GST-Hnf4α (pEBG-hDAX-1) along with HA-DAX-1 WT or deletion constructs (HA-NT and HA-LBD) in 293T cells. Similar to the result of in vitro mapping, all three HA-DAX-1 proteins (HA-DAX-1 WT, NT, and LBD) were detected in the coprecipitates with GST-Hnf4α but not with GST alone (Fig. 3C). Similarly, we cotransfected the mammalian expression vectors encoding GST empty or GST-DAX-1 with wild-type or deletion constructs of Hnf4α (HA-Hnf4α WT, 1–370, and 1–360) with pEBG-DAX-1 WT. In agreement with the yeast two-hybrid result, HNF4α 1–360 weakly interacted with GST-DAX-1 (Fig. 3D). This result is consistent with the previous report showing that the AF-2 domain of HNF4α was essential for the interaction with coactivators and its transactivation potential (41). Collectively, these in vivo results also indicate that the entire structure of DAX-1 contributes to the interaction with the AF-2 region of HNF4α (residues 360–370), which is essential for its transcriptional activity and coactivator binding.
FIGURE 3.
Mapping of interaction domain between DAX-1 and HNF4α. A and B, schematic representation of DAX-1 and HNF4α deletion constructs. C, in vivo GST pull-down assay was performed using HA-DAX-1 WT and deletion constructs with pEBG-HNF4α. 293T cells were cotransfected with the mammalian expression vector encoding either GST (pEBG) alone or GST-hnf4α (pEBG-hDAX-1) along with HA-DAX-1 WT and deletion constructs (HA-NT and HA-LBD). Lysates were immunoprecipitated with GST beads, and we performed Western blotting using anti-HA or anti-GST antibodies. D, in vivo GST pull-down assay was performed using HA-hnf4α WT, 1–370, and 1–360 with pEBG and pEBG-DAX-1. 293T cells were cotransfected with mammalian expression vectors encoding GST alone or GST-DAX-1 with wild-type and some deletion constructs of HNF4α (HA-HNF4α WT, 1–370, and 1–360) with pEBG-DAX-1 WT. Then cell lysates were immunoprecipitated with GST beads, and we performed Western blotting using anti-HA or anti-GST antibodies.
DAX-1 Inhibits the Transactivity of HNF4α by Competing with the Coactivator PGC-1α
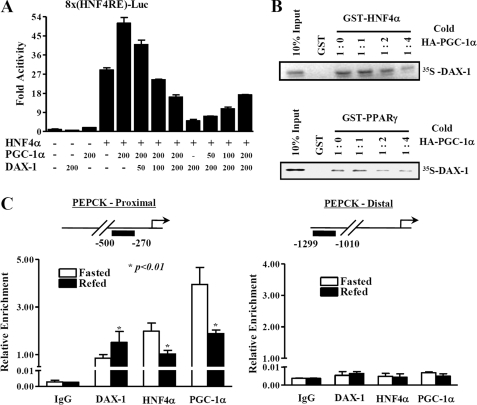
Since DAX-1 interacts with the AF-2 domain of HNF4α which is known to interact with coactivators, such as PGC-1α, we thus investigated whether DAX-1 competes with coactivator PGC-1α for binding to HNF4α. We cotransfected the mammalian expression vectors encoding hnf4α, Pgc-1α, and DAX-1 in 293T cells with different combinations. PGC-1α coactivated the HNF4α transactivity as expected, whereas DAX-1 strongly repressed the transactivity of HNF4α. DAX-1 repressed the coactivation of PGC-1α in a dose-dependent manner, whereas increasing amount of PGC-1α dose-dependently released the DAX-1-mediated repression on HNF4α transactivity (Fig. 4A). Transfection of DAX-1 alone substantially inhibited the basal reporter activity, whereas Pgc-1α increased the basal activity. Similar results were observed from an in vitro GST pull-down competition assay. Increasing amounts of unlabeled full-length PGC-1α competed with 35S-labeled DAX-1 for the binding with GST- Hnf4α (Fig. 4B, top) or with the positive control GST-PPARγ (Fig. 4B, bottom). It is well known that HNF4α can bind to the promoters and regulate the expression of target genes, such as Pepck, G6Pase, Cyp7a1 (42), and Pdk4 (pyruvate dehydrogenase kinase 4) (43). Therefore, we next investigated whether DAX-1 forms a complex with HNF4α on the promoter of its target gene. We performed in vivo ChIP assay from the liver of mice with 12 h of fasting and refeeding. We found that the recruitment of DAX-1 on the HNF4α binding region of the Pepck promoter was significantly increased under refeeding compared with fasting conditions. However, the recruitment of Hnf4α and Pgc-1α on the proximal promoter was high under fasting, and it was significantly decreased under the refeeding state (Fig. 4C, left). No recruitment of any of these proteins was observed in the nonspecific distal region of the Pepck promoter (Fig. 4C, right). Overall, these results suggest the existence of competition mechanism between DAX-1 and PGC-1α for repressing HNF4α transactivation.
FIGURE 4.
DAX-1 competes with PGC-1α for HN4α transactivation. A, 293T cells were transfected using 200 ng of 8×(HNF4α)-Luc with the indicated amount of hnf4α, Pgc-1α, and DAX-1. Cells were harvested 40 h after transfection, and lysates were utilized for luciferase and β-galactosidase assay. The results shown are means of the β-galactosidase value from three independent experiments. Effects of DAX-1 and PGC1α alone on the basal reporter activity were also shown. B, in vitro GST competition assay was performed using in vitro translated labeled DAX-1 and unlabeled PGC-1α with bacterially expressed GST-Hnf4α (top) or with control GST-PPARγ (bottom). The ratios of labeled DAX-1 and unlabeled PGC-1α are indicated. C, quantitative analysis of the relative amounts of DAX-1, PGC-1α, and HNF4α on Pepck promoter regions using ChIP real time PCR. Recruitment of Dax-1 on the Pepck promoter is inversely correlated with the recruitment of PGC-1α and HNF4α under fasting and refeeding conditions. ChIP assays were performed using fasted (n = 4) and refed (n = 4) mouse liver samples. Liver extracts from fasted (12 h) and refed (12 h) mouse livers were chromatin-immunoprecipitated using DAX-1, HNF4α, and PGC-1α antibodies, and purified DNA samples were quantified using real time PCR using primers encompassing the proximal (lower left) and nonspecific distal (lower right) regions of the Pepck promoter (*, p < 0.01; n = 4). The upper panels show the schematic representation of the proximal (upper left) and distal (upper right) regions of the Pepck promoter and the primers used for the in vivo ChIP assay. Promoter occupancies of the indicated proteins were quantified compared with input. Data are expressed as relative enrichment of indicated proteins and Pepck promoter regions relative to that in input chromatin.
DAX-1 Decreases HNF4α-mediated Target Gene Expression, and Its Expression Is Regulated under Different Nutritional Conditions
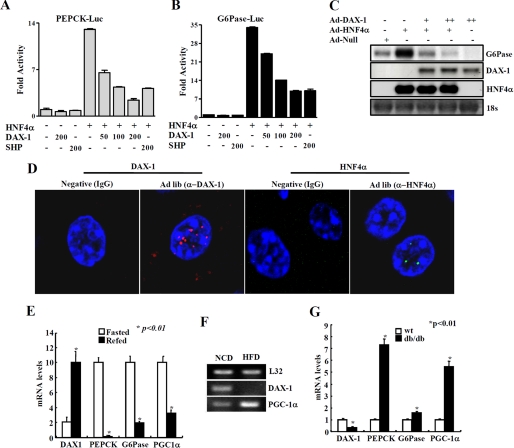
To further investigate whether DAX-1 represses the natural target gene promoters of HNF4α, transient transfection assays were performed using reporters containing Pepck and G6Pase gene promoters along with expression vectors for HA-HNF4α and FLAG-DAX-1 in HepG2 cells. As expected, HNF4α significantly increased both Pepck and G6Pase luciferase reporter activities. The expression of DAX-1 decreased the HNF4α-mediated transactivation of both Pepck and G6Pase promoters in a dose-dependent manner (Fig. 5, A and B), indicating that DAX-1 inhibits HNF4α-mediated transactivation of Pepck and G6Pase promoter activities. In addition, transfection of DAX-1 considerably inhibited the basal transcriptional activities of Pepck and G6Pase reporters. In order to examine whether DAX-1 can also repress the HNF4α-mediated expression of endogenous gluconeogenic genes, we prepared a recombinant adenovirus for HNF4α (Ad-HNF4α). As expected, infection of Ad-HNF4α significantly increased the expression of G6Pase in HepG2 cells. Infection of adenovirus for DAX-1 dose-dependently decreased Ad-HNF4α-mediated G6Pase gene expression (Fig. 5C). Overall, these results indicate that DAX-1 reduces HNF4α-mediated transcription of gluconeogenic genes.
FIGURE 5.
DAX-1 inhibits HNF4α-mediated target genes and regulation of DAX-1 gene expression under different nutritional conditions. DAX-1 inhibits HNF4α target gene promoter activity and gene expression. A and B, HepG2 cells were transfected with Pepck-Luc (A) and G6Pase-Luc (B) independently with hnf4α, DAX-1, and SHP, and cell lysates were utilized for luciferase and β-galactosidase assays. Effects of DAX-1 and PGC-1α alone on the basal reporter activity were also shown. The results shown are the mean of the β-galactosidase value from three independent experiments. C, Northern blot analysis using adenovirus null, HNF4α, and DAX-1. Adenovirus for HNF4α was infected alone or with different combinations of adenovirus for DAX-1 in HepG2 cells. Then total RNA was isolated and utilized for Northern blot analysis. The same blot was hybridized with labeled G6Pase, DAX-1, and hnf4α probes. D, paraffin sections of normal (ad libitum; Ad lib) mouse liver sample were used for immunofluorescent staining. The hepatic DAX-1 and HNF4α proteins were detected with anti-DAX-1 and HNF4α antibodies and visualized with red fluorescence for DAX-1 and green fluorescence for HNF4α. Pictures are shown at ×400 magnification with a confocal microscope. DAX-1 expression under different nutritional conditions. E, quantitative PCR analysis of hepatic mRNA levels for DAX-1, PGC-1α, PEPCK, and G6Pase from fasted (24 h) or refed (24 h) mice (*, p < 0.01; n = 3). F, RT-PCR analysis of hepatic DAX-1 and PGC-1α from mice under normal chow diet or high fat diet for 8 weeks (n = 3 each). G, quantitative PCR analysis of hepatic mRNA levels for DAX-1, PGC-1α, PEPCK, and G6Pase from WT or db/db mice (*, p < 0.01; n = 3). F and G, data in E, G, and F are represented as mean ± S.D.
To investigate the physiological relevance of DAX-1 in the liver, we examined the hepatic DAX-1 expression using RT-PCR and Western blot analysis in mouse tissue samples. We found that DAX-1 is moderately expressed in liver when compared with testis (data not shown). Our co-immunoprecipitation result also evidenced that DAX-1 is expressed in liver. We also performed immunofluorescent staining in a normal mouse liver sample. We have confirmed that DAX-1 is expressed in liver. As a positive control, we showed the staining of HNF4α in the same liver samples (Fig. 5D). Our in vivo ChIP assay data revealed the increased recruitment of Dax-1 in the HNF4α binding region of the Pepck promoter under refeeding conditions, suggesting that the expression pattern of Dax-1 may be inversely correlated with that of gluconeogenic genes under different nutritional conditions. Indeed, our quantitative PCR analysis showed that the expression of Dax-1 was significantly higher under refeeding conditions compared with fasting conditions (Fig. 5E). Furthermore, hepatic expression of DAX-1 was decreased in high fat fed mice (Fig. 5F) or db/db mice (Fig. 5G) when compared with normal mice, showing a stark contrast with the expression pattern of gluconeogenic genes. Overall, these results suggest a potential role of the DAX-1 gene in the regulation of the gluconeogenic pathway in liver.
The DAX-1 Gene Is Induced by Insulin and SIK1
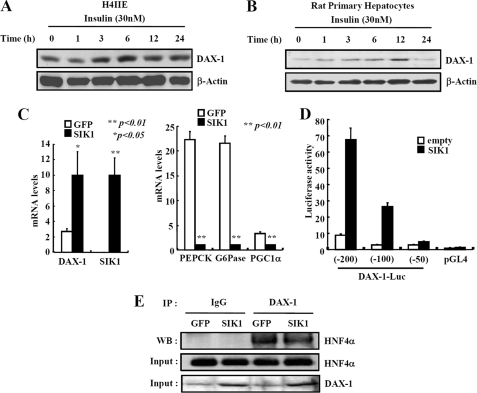
Since DAX-1 expression is inversely correlated with gluconeogenic gene expression under fasting and refeeding conditions, we intended to find out whether DAX-1 can be induced by any negative regulators of gluconeogenesis. Interestingly, the treatment of insulin moderately increased the expression of DAX-1 in H4IIE (Fig. 6A) and rat primary hepatocytes (Fig. 6B). It has been reported that SIK1 can inhibit the hepatic gluconeogenesis via phosphorylation-dependent modulation of CRTC2 (also known as TORC2) (36). Therefore, we speculated that SIK1 may be involved in the regulation of DAX-1 expression. We infected adenovirus for Sik1 in rat primary hepatocytes and analyzed the expression of Dax-1 and gluconeogenic genes. Our result indicated that SIK1 induced Dax-1 gene expression and decreased Pepck, G6pase, and Pgc-1α gene expression in rat primary hepatocytes (Fig. 6C). Moreover, a reporter assay revealed that the DAX-1 promoter activity was also increased about 7-fold by the expression of SIK1 (Fig. 6D). Further dissection of the DAX-1 promoter suggested that the region between −100 and −50 bp from the transcriptional start site is responsible for SIK1-mediated activation. We are currently investigating the identity of a potential transcription factor that mediates SIK-dependent activation of DAX-1 transcription. To further examine whether induction of Dax-1 gene expression by Ad-Sik1 can increase the interaction ability of DAX-1 with HNF4α, we delivered SIK1 in mouse liver via adenovirus tail vein injection. Although Sik1 increases the endogenous DAX-1 protein levels in the input panel, there is no significant change in interaction between DAX-1 and HNF4α (Fig. 6E). Collectively, these results indicated that DAX-1 can be induced by insulin and SIK signaling.
FIGURE 6.
Insulin and SIK1 increases the DAX-1 gene expression. A and B, insulin increases DAX-1 gene expression. H4IIE (A) and rat primary hepatocytes (B) were cultured for 24 h under serum starvation. The cells were then treated with insulin (100 nm) for various time periods. DAX-1 gene expression was analyzed using Western blotting and normalized with β-actin. SIK1 induces DAX-1 gene expression. C, quantitative PCR analysis shows the effects of Sik1 on mRNA levels of dax-1, pepck, g6pase, and pgc-1α in rat primary hepatocytes. Cells were infected with Ad-GFP or Ad-Sik1 for 48 h and then exposed to forskolin or DMSO for 2 h (*, p < 0.05; **, p < 0.01; n = 3). D, HepG2 cells were cotransfected with 300 ng of various pGL4-mouse DAX1-luc constructs with 100 ng of pcDNA3-FLAG-SIK-1 or 100 ng of pcDNA3 empty vector for 48 h (*, p < 0.05; n = 3). Effects of SIK1 expression on mouse DAX-1 luciferase reporter activity in HepG2 cells were shown. E, co-immunoprecipitation assays with liver extracts from adenovirus for GFP and SIK1. Protein extracts from adenovirus for GFP- or SIK1-infected mouse livers were immunoprecipitated (IP) using DAX-1 antibody and blotted with HNF4α antibody. Expression of HNF4α and DAX-1 from 10% lysate were analyzed by Western blotting with specific antibodies. Data in C and D are represented as mean ± S.D.
DAX-1 Decreases the Expression of Key Gluconeogenic Genes and Blood Glucose Levels
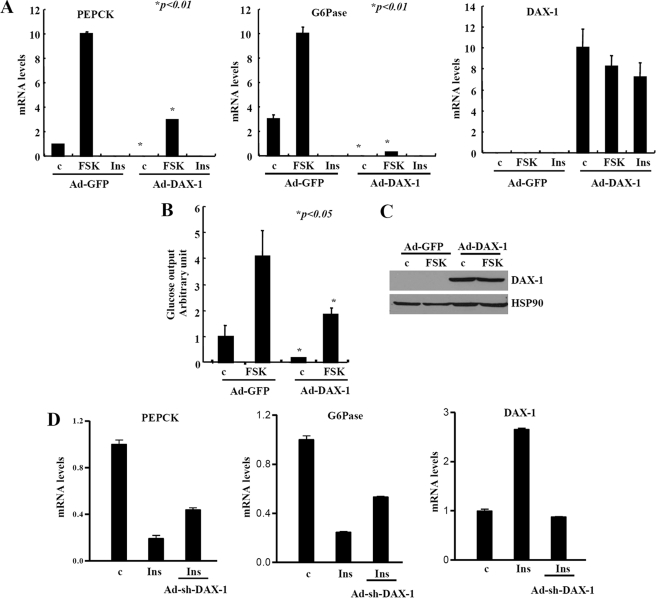
Having seen the inhibitory effects of DAX-1 on gluconeogenic gene expression in hepatocytes, we next attempted to investigate whether DAX-1 expression would inhibit hepatic glucose production. Ad-Dax-1 infection significantly inhibits both basal and cAMP-induced expression of gluconeogenic genes, such as Pepck and G6pase, in rat primary hepatocytes (Fig. 7A). Furthermore, glucose production in rat primary hepatocytes was significantly decreased by Ad-Dax-1 when compared with that of Ad-GFP control (Fig. 7B), suggesting that DAX-1-mediated repression of gluconeogenic gene expression led to the decreased hepatic glucose output.
FIGURE 7.
DAX-1 reduces hepatic gluconeogenesis. A, quantitative PCR analysis shows the effects of Ad-Dax-1 infection on hepatic expression of gluconeogenic genes. Primary hepatocytes were infected with adenoviruses for GFP or DAX-1 for 48 h and then exposed to 10 μm FSK for 2 h or 100 nm insulin for 12 h (*, p < 0.01; n = 3). Total RNA was isolated, and quantitative PCR was performed using specific primers for Pepck, g6pase, and dax-1 genes. B, adenovirus for Dax-1 decreases hepatic glucose output in rat primary hepatocytes. Cells were infected with adenoviruses for GFP or DAX-1 for 48 h and then exposed to 10 μm FSK plus 100 nm dexamethasone for 8 h (*, p < 0.05; n = 3), and we performed a glucose output assay (left). C, Western blot analysis shows the expression of Ad-DAX1 (FLAG-mDAX1). D, insulin-mediated effect on gluconeogenic genes were partially reversed after knockdown of DAX-1. Primary hepatocytes were treated with 100 nm insulin for 12 h with prior infection of either Ad-US or Ad-sh-Dax-1 in a 60-mm dish. Total RNA was isolated, and RT-PCR was performed. The graphs represent the quantitation of RT-PCR products. Data in A and B are represented as mean ± S.D.
In order to investigate whether endogenous DAX-1 has any effect on hepatic gluconeogenesis, we have constructed adenovirus for Dax-1 short hairpin RNA (Ad-sh-DAX-1). As expected, we observed that insulin strongly decreased the expression of pepck and g6pase genes in primary hepatocytes. However, knockdown of dax-1 by Ad-sh-Dax-1 significantly reversed the insulin-mediated inhibition of pepck and g6pase gene expression. As a control for dax-1 knockdown, we also showed that insulin-mediated induction of dax-1 was totally abolished by Ad-sh-DAX-1 (Fig. 7D). Overall, these results indicated that knockdown of DAX-1 partially reverses the insulin-mediated down-regulation of gluconeogenic gene expression.
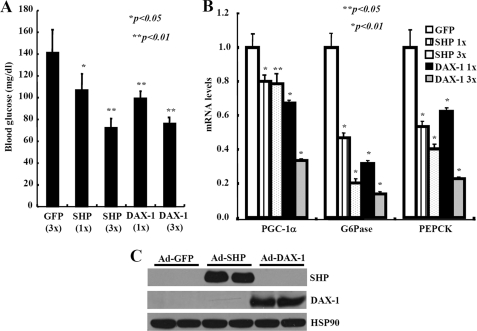
To further assess the functional consequences of DAX-1-mediated repression of gluconeogenic gene expression, we performed adenoviral delivery of Dax-1 in high fat diet-fed mice. Adenoviruses for GFP, Dax-1, or Shp were injected via the tail vein of mice that were previously fed a high fat diet for 8 weeks. Ad-Dax-1 or Ad-Shp infection significantly reduced the fasting (16 h) blood glucose in a dose-dependent manner. The maximum reduction of blood glucose reached 50% of the basal level (Fig. 8A). Concomitantly, there was a significant reduction in mRNA levels of hepatic gluconeogenic genes, such as Pgc-1α, Pepck, and G6pase, in DAX-1 mice compared with control GFP mice (Fig. 8B). Our Western blot analysis indicated that both adenovirus Shp and Dax-1 were successfully delivered in mouse liver (Fig. 8C). Taken together, these results indicated that activation of DAX-1 pathway could reduce plasma glucose level in vivo by repressing the expression of hepatic gluconeogenic genes.
FIGURE 8.
Hepatic expression of DAX-1 lowers the blood glucose levels in mice. Adenovirus for DAX-1 lowers the blood glucose levels in high fat-fed mice. A, 16-h fasting glucose levels from high fat-fed mice injected with either Ad-GFP (n = 4), Ad-Shp (1×, n = 3; 3×, n = 3), or Ad-Dax-1 (1×, n = 3; 3×, n = 3) (*, p < 0.05; **, p < 0.01). B, quantitative PCR analysis showing the effect of Ad-GFP, Ad-Shp, or Ad-Dax-1 infection on hepatic expression of Pepck, G6pase, and Pgc-1α genes in high fat-fed mice fasted for 16 h. (*, p < 0.05; **, p < 0.01; n = 3). C, Western blot analysis showing the expression levels of SHP or DAX-1 from 3× samples. Data in A and B are represented as mean ± S.D.
DISCUSSION
In this study, DAX-1 was found to interact with Hnf4α and inhibit its transcriptional activity by competing with PGC-1α for binding to the AF-2 domain of HNF4α. Expression of DAX-1 in liver is inhibited during fasting or insulin-resistant conditions. Moreover, we have found that Sik1 and insulin, the two signaling pathways that inhibit hepatic gluconeogenesis, can increase the expression of Dax-1. Indeed, Dax-1 can form a complex with Hnf4α on the promoter of its target genes and repress both HNF4α-mediated and forskolin-induced expression of PEPCK and G6Pase genes. We have also shown that DAX-1 overexpression could inhibit endogenous gluconeogenic gene expression and blood glucose levels in animal models of diet-induced insulin resistance, accentuating the significance of our finding.
DAX-1 has been shown to repress the transcriptional activity of various nuclear receptors, including androgen receptor, GR, liver receptor homolog-1, PPARγ, and Nur77 (14). It is well known that transcription factors, such as FOXO1, GR, and HNF4α, are required for transcriptional activation of the gluconeogenic program (44). Since DAX-1 has already been shown to repress the GR transactivity (14) in an LXXLL-dependent manner, we further focused on the potential role of DAX-1 on HNF4α transactivity. Here, we found that DAX-1 represses the transcriptional activity of HNF4α, which is not cell type-specific (Fig. 1). This same phenomenon was also observed in the case of SHP for repressing HNF4α (45).
Our co-immunoprecipitation results indicated that Dax-1 interacted with Hnf4α in the liver. However, interaction of Dax-1 with Hnf4α did not significantly change under fasting or refeeding conditions (data not shown). This might be due to changes in HNF4α protein level, since it has been reported that insulin can decrease the expression of HNF4α (46). We have also found that DAX-1 interacts with the AF-2 domain of HNF4α, which is consistent with the interaction of SHP with AF-2 domain of HNF4α (45). However, our recent report indicated that DAX-1 binds to the DNA binding domain/hinge region of PPARγ (14). On the other hand, the entire DAX-1 protein is involved in the interaction with Hnf4α (Fig. 3, C and D) and PPARγ (14), whereas SHP is known to utilize its interaction domain for the interaction with Hnf4α (45).
The coactivator PGC-1α is transcriptionally induced by cAMP-response element-binding protein and acts as an important coactivator of HNF4α, GR, and FOXO for inducing the expression of PEPCK and G6pase genes (44). Our current study demonstrated that DAX-1 represses the transcriptional activity of HNF4α by competing with the coactivator PGC-1α (Fig. 4). It is also consistent with our previous study, where DAX-1 competes with PGC-1α for repressing the PPARγ transcriptional activity (14). Similar competitions were also observed in the cases of SHP and SMRT, which interact with the same HNF4α surface recognized by transcriptional coactivators and competes with them for binding in vivo (47). Although DAX-1, SMRT, and SHP have similar repressive mechanisms for repressing HNF4α, they are different by competing with coactivator PGC-1α, GRIP1, and SRC-3, respectively (14, 44, 47). In addition, recruitment of Dax-1 on the Hnf4α binding region of the PEPCK promoter under fasting was increased significantly under refeeding, and it is inversely correlated with the recruitment of Hnf4α and Pgc-1α. The recruitment and ability of DAX-1 to repress the transactivity of HNF4α by competing with PGC-1α would constitute the mechanism underlying the suppression of gluconeogenic genes by DAX-1.
Our expression analysis has shown that DAX-1 was moderately expressed in liver when compared with testis (data not shown). The expression of DAX-1 in the liver was further evidenced by in vivo co-immunoprecipitation and ChIP assays from mouse liver samples. Hepatic DAX-1 expression was further confirmed using immunofluorescent staining of DAX-1 in mouse liver samples. These observations are reminiscent of the previous reports showing that expression of DAX-1 has been observed in the livers of fish (48), chicken (49), and frog (50). The expression of Dax-1 was decreased during fasting and increased upon refeeding, which indicated that either the cAMP or insulin pathway may play a role for regulating the expression of Dax-1. It is consistent with the previous reports that activation of cAMP pathway by ACTH, follicle-stimulating hormone, or luteinizing hormone (16–18) leads to the down-regulation of Dax-1 gene expression. These observations prompted us to check whether DAX-1 can be induced by some negative regulator of gluconeogenesis. Insulin is the most important hormone that inhibits gluconeogenesis and acts predominantly by suppressing the expression of key gluconeogenic enzymes PEPCK and G6Pase (51). Indeed, we were able to show that the treatment of insulin increased the expression of DAX-1 protein. Previously, we have reported that metformin-induced SHP inhibits hepatic glucose production by directly regulating the expression of gluconeogenic genes, such as PEPCK and G6Pase, in response to increased AMP-activated protein kinase activity (35). AMP-activated protein kinase or its related kinases, SIKs were shown to inhibit hepatic gluconeogenesis by serine 171 phosphorylation and the cytoplasmic retention of CRTC2 (36). Interestingly, we have found that overexpression of Sik1 induced the expression of Dax-1 in primary hepatocytes (Fig. 6C), whereas AMP-activated protein kinase expression did not show any significant changes in DAX-1 expression (data not shown).
Dax-1 promoter analysis revealed that SIK kinases appear to regulate DAX-1 expression at the transcriptional level, and the promoter sequences from −100 to −50 bp from the transcriptional start site are essential for SIK-mediated activation (Fig. 6D). We are currently screening for the potential SIK-mediated transcriptional factor that enhances the DAX-1 gene expression in liver. It is tempting to speculate that insulin and SIK pathways might converge on the same factor to regulate the expression of DAX-1 in liver, based on the recent report suggesting the Akt-mediated activation of SIK2 activity (52). Although we observed significant increase of DAX-1 gene expression after hepatic delivery of SIK1 in mice, we could not find any significant increase in DAX-1 interaction with HNF4α. This might be due to the significant difference in the abundance of DAX-1 and HNF4α expression even after the induction by SIK1 or the presence of more DAX-1 binding partners in liver other than HNF4α. Although there is no significant change in interaction between DAX-1 and HNF4α, the increase in Dax-1 expression under specific stimuli, such as SIK1, insulin, or refeeding is enough for its recruitment on gluconeogenic enzyme promoters (Fig. 4C) and for its inhibition of gluconeogenic gene expression (Figs. 7 and 8). Moreover, knockdown of dax-1 partially reverses the insulin-mediated inhibition of gluconeogenic gene expression (Fig. 7C), indicating that DAX-1 may also mediate the insulin effect along with previously known mediators.
This study demonstrated that DAX-1 decreased HNF4α-mediated expression of PEPCK and G6Pase, which is consistent with the previous report that SHP can inhibit the expression of these gluconeogenic genes via the repression of HNF4α (21). Moreover, cAMP-stimulated expression of gluconeogenic genes and blood glucose level were also decreased by DAX-1, which is also similar to our previous study, where overexpression of SHP reduces blood glucose level as well as the expression of PEPCK and G6Pase (34). Collectively, our results suggest that DAX-1 can decrease both HNF4α- and cAMP-stimulated expression of gluconeogenic genes. A recent study has demonstrated that DAX1 and SHP can form homodimers as well as heterodimers (53), suggesting the possibility of novel functions independent of their coregulatory roles. Hence, combinatorial expression or knock-out of these two orphan nuclear receptors in liver will be necessary to fully understand the molecular mechanism underlying the repression of gluconeogenic genes. The repression of gluconeogenic genes under control and fasting conditions by DAX-1 suggests the existence of more DAX-1 target proteins other than HNF4α and PGC-1α. Moreover, DAX-1 may also inhibit the expression of target genes involved in cholesterol and bile acid metabolism other than gluconeogenesis. Therefore, the search for a potential inducer of DAX-1 under feeding conditions will be useful to better understand the role of DAX-1 in liver metabolism. Overall, this study expands the function of DAX-1 in liver other than gonads and adrenal gland.
In summary, our study indicated that DAX-1 acts as a novel corepressor of HNF4α by competing with coactivator PGC-1α to regulate hepatic gluconeogenic gene expression. Overall, this study suggests that DAX-1 inhibits HNF4α- and cAMP-mediated expression of PEPCK and G6Pase genes to control blood glucose levels in mammals.
Supplementary Material
Acknowledgments
We thank Gwang Sik Kim, Don-Kyu Kim, and Sun Myung Park for technical assistance and Dr. Young-Chul Lee for helpful discussions.
This study was supported by the Korea Science and Engineering Foundation through the National Research Laboratory Program funded by Ministry of Science and Technology Grant M10500000047-06J0000-04710 and Korea Research Foundation Grant 2006-005-J03003.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- LBD
- ligand binding domain
- ACTH
- adrenocorticotropic hormone
- ER
- estrogen receptor
- GR
- glucocorticoid receptor
- PPARγ
- peroxisome proliferator-activated receptor γ
- ERRγ
- estrogen receptor-related receptor γ
- CREB
- cAMP-response element-binding protein
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- CMV
- cytomegalovirus
- NT
- N terminus
- PBS
- phosphate-buffered saline
- RT
- reverse transcription
- siRNA
- small interfering RNA
- SMRT
- silencing mediator of retinoic acid and thyroid hormone receptor
- ChIP
- chromatin immunoprecipitation
- WT
- wild type
- Ad
- adenovirus
- HA
- hemagglutinin.
REFERENCES
- 1.Giguère V. (1999) Endocr. Rev. 20, 689–725 [DOI] [PubMed] [Google Scholar]
- 2.Aranda A., Pascual A. (2001) Physiol. Rev. 81, 1269–1304 [DOI] [PubMed] [Google Scholar]
- 3.Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer A. K., McCabe E. R. (2004) Mol. Genet. Metab. 83, 60–73 [DOI] [PubMed] [Google Scholar]
- 5.Seol W., Choi H. S., Moore D. D. (1996) Science 272, 1336–1339 [DOI] [PubMed] [Google Scholar]
- 6.Zanaria E., Muscatelli F., Bardoni B., Strom T. M., Guioli S., Guo W., Lalli E., Moser C., Walker A. P., McCabe E. R., Meitinger T., Monaco A. P., Sassone-Corsi P., Camerino G. (1994) Nature 372, 635–641 [DOI] [PubMed] [Google Scholar]
- 7.Zazopoulos E., Lalli E., Stocco D. M., Sassone-Corsi P. (1997) Nature 390, 311–315 [DOI] [PubMed] [Google Scholar]
- 8.Lalli E., Ohe K., Hindelang C., Sassone-Corsi P. (2000) Mol. Cell. Biol. 20, 4910–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H., Thomsen J.S., Johansson L., Gustafsson J. A., Treuter E. (2000) J. Biol. Chem. 275, 39855–39859 [DOI] [PubMed] [Google Scholar]
- 10.Crawford P. A., Dorn C., Sadovsky Y., Milbrandt J. (1998) Mol. Cell. Biol. 18, 2949–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holter E., Kotaja N., Mäkela S., Strauss L., Kietz S., Jänne O. A., Gustafsson J. A., Palvimo J. J., Treuter E. (2002) Mol. Endocrinol. 16, 515–528 [DOI] [PubMed] [Google Scholar]
- 12.Agoulnik I. U., Krause W. C., Bingman W. E., 3rd, Rahman H. T., Amrikachi M., Ayala G. E., Weigel N. L. (2003) J. Biol. Chem. 278, 31136–31148 [DOI] [PubMed] [Google Scholar]
- 13.Suzuki T., Kasahara M., Yoshioka H., Morohashi K., Umesono K. (2003) Mol. Cell. Biol. 23, 238–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim G. S., Lee G.Y., Nedumaran B., Park Y. Y., Kim K. T., Park S. C., Lee Y. C., Kim J. B., Choi H. S. (2008) Biochem. Biophys. Res. Commun. 370, 264–268 [DOI] [PubMed] [Google Scholar]
- 15.Park Y. Y., Ahn S. W., Kim H. J., Kim J. M., Lee I. K., Kang H., Choi H. S. (2005) Nucleic Acids Res. 33, 6756–6768 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Song K. H., Park Y. Y., Park K. C., Hong C. Y., Park J. H., Shong M., Lee K., Choi H. S. (2004) Mol. Endocrinol. 18, 1929–1940 [DOI] [PubMed] [Google Scholar]
- 17.Ragazzon B., Lefrançois-Martinez A. M., Val P., Sahut-Barnola I., Tournaire C., Chambon C., Gachancard-Bouya J. L., Begue R. J., Veyssière G., Martinez A. (2006) Endocrinology 147, 1805–1818 [DOI] [PubMed] [Google Scholar]
- 18.Tamai K. T., Monaco L., Alastalo T. P., Lalli E., Parvinen M., Sassone-Corsi P. (1996) Mol. Endocrinol. 10, 1561–1569 [DOI] [PubMed] [Google Scholar]
- 19.Sladek F. M., Zhong W. M., Lai E., Darnell J. E., Jr. (1990) Genes Dev. 4, 2353–2365 [DOI] [PubMed] [Google Scholar]
- 20.Jiang G., Nepomuceno L., Hopkins K., Sladek F. M. (1995) Mol. Cell. Biol. 15, 5131–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drewes T., Senkel S., Holewa B., Ryffel G. U. (1996) Mol. Cell. Biol. 16, 925–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue Y., Yu A. M., Yim S. H., Ma X., Krausz K. W., Inoue J., Xiang C. C., Brownstein M. J., Eggertsen G., Björkhem I., Gonzalez F. J. (2006) J. Lipid Res. 47, 215–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamagata K., Daitoku H., Shimamoto Y., Matsuzaki H., Hirota K., Ishida J, Fukamizu A. (2004) J. Biol. Chem. 279, 23158–23165 [DOI] [PubMed] [Google Scholar]
- 24.Hattersley A. T. (1998) Diabet. Med. 15, 15–24 [DOI] [PubMed] [Google Scholar]
- 25.Lausen J., Thomas H., Lemm I., Bulman M., Borgschulze M., Lingott A., Hattersley A. T., Ryffel G. U. (2000) Nucleic Acids Res. 28, 430–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J. C., Stafford J. M., Granner D. K. (1998) J. Biol. Chem. 273, 30847–30850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida E., Aratani S., Itou H., Miyagishi M., Takiguchi M., Osumu T., Murakami K., Fukamizu A. (1997) Biochem. Biophys. Res. Commun. 241, 664–669 [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez F. J. (2008) Drug Metab. Pharmacokinet. 23, 2–7 [DOI] [PubMed] [Google Scholar]
- 29.Miao J., Fang S., Bae Y., Kemper J. K. (2006) J. Biol. Chem. 281, 14537–14546 [DOI] [PubMed] [Google Scholar]
- 30.Rada-Iglesias A., Wallerman O., Koch C., Ameur A., Enroth S., Clelland G., Wester K., Wilcox S., Dovey O. M., Ellis P. D., Wraight V. L., James K., Andrews R., Langford C., Dhami P., Carter N., Vetrie D., Pontén F., Komorowski J., Dunham I., Wadelius C. (2005) Hum. Mol. Genet. 14, 3435–3447 [DOI] [PubMed] [Google Scholar]
- 31.Olefsky J. M. (2000) J. Clin. Invest. 106, 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bansal P., Wang Q. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E751–E761 [DOI] [PubMed] [Google Scholar]
- 33.Jiang Y., Cypess A. M., Muse E. D., Wu C. R., Unson C. G., Merrifield R. B., Sakmar T. P. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10102–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whiteman E. L., Cho H., Birnbaum M. J. (2002) Trends Endocrinol. Metab. 13, 444–451 [DOI] [PubMed] [Google Scholar]
- 35.Kim Y. D., Park K. G., Lee Y. S., Park Y. Y., Kim D. K., Nedumaran B., Jang W. G., Cho W. J., Ha J., Lee I. K., Lee C. H., Choi H. S. (2008) Diabetes 57, 306–314 [DOI] [PubMed] [Google Scholar]
- 36.Koo S. H., Flechner L., Qi L., Zhang X., Screaton R. A., Jeffries S., Hedrick S., Xu W., Boussouar F., Brindle P., Takemori H., Montminy M. (2005) Nature 437, 1109–1111 [DOI] [PubMed] [Google Scholar]
- 37.Matsumoto M., Accili D. (2006) Nat. Med. 12, 33–34 [DOI] [PubMed] [Google Scholar]
- 38.Ruse M. D., Jr., Privalsky M. L., Sladek F. M. (2002) Mol. Cell. Biol. 22, 1626–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J. Y., Kim H. J., Kim K. T., Park Y. Y., Seong H. A., Park K. C., Lee I. K., Ha H., Shong M., Park S. C., Choi H. S. (2004) Mol. Endocrinol. 18, 2880–2894 [DOI] [PubMed] [Google Scholar]
- 40.Xie Y. B., Lee O. H., Nedumaran B., Seong H. A., Lee K. M., Ha H., Lee I. K., Yun Y., Choi H. S. (2008) Biochem. J. 416, 463–473 [DOI] [PubMed] [Google Scholar]
- 41.Hadzopoulou-Cladaras M., Kistanova E., Evagelopoulou C., Zeng S., Cladaras C., Ladias J. A. (1997) J. Biol. Chem. 272, 539–550 [DOI] [PubMed] [Google Scholar]
- 42.Li T., Ma H., Chiang J. Y. (2008) J. Lipid Res. 49, 1981–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma K., Zhang Y., Elam M. B., Cook G. A., Park E. A. (2005) J. Biol. Chem. 280, 29525–29532 [DOI] [PubMed] [Google Scholar]
- 44.Puigserver P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., Spiegelman B. M. (2003) Nature 423, 550–555 [DOI] [PubMed] [Google Scholar]
- 45.Lee Y. K., Dell H., Dowhan D. H., Hadzopoulou-Cladaras M., Moore D. D. (2000) Mol. Cell. Biol. 20, 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie X., Liao H., Dang H., Pang W., Guan Y., Wang X., Shyy J. Y., Zhu Y., Sladek F. M. (2009) Mol. Endocrinol. 23, 434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torres-Padilla M. E., Sladek F. M., Weiss M. C. (2002) J. Biol. Chem. 277, 44677–44687 [DOI] [PubMed] [Google Scholar]
- 48.Wang D. S., Kobayashi T., Senthilkumaran B., Sakai F., Sudhakumari C. C., Suzuki T., Yoshikuni M., Matsuda M., Morohashi K., Nagahama Y. (2002) Biochem. Biophys. Res. Commun. 297, 632–640 [DOI] [PubMed] [Google Scholar]
- 49.Smith C. A., Clifford V., Western P. S., Wilcox S. A., Bell K. S., Sinclair A. H. (2000) J. Mol. Endocrinol. 24, 23–32 [DOI] [PubMed] [Google Scholar]
- 50.Sugita J., Takase M., Nakamura M. (2001) Gene 280, 67–74 [DOI] [PubMed] [Google Scholar]
- 51.Hall R. K., Yamasaki T., Kucera T., Waltner-Law M., O'Brien R., Granner D. K. (2000) J. Biol. Chem. 275, 30169–30175 [DOI] [PubMed] [Google Scholar]
- 52.Dentin R., Liu Y., Koo S. H., Hedrick S., Vargas T., Heredia J., Yates J., 3rd, Montminy M. (2007) Nature 449, 366–369 [DOI] [PubMed] [Google Scholar]
- 53.Iyer A. K., Zhang Y. H., McCabe E. R. (2006) Mol. Endocrinol. 20, 2326–2342 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.