Abstract
Tumors of the exocrine pancreas have a poor prognosis. Several proteins are overexpressed in this cancer type, including the MET tyrosine kinase receptor and the transcription factor PAX6. In this report, we find that PAX6(5a), an alternately spliced variant form of PAX6, is expressed in pancreatic carcinoma cell lines at higher levels than the canonical PAX6 protein. Both protein forms of PAX6 bind directly to an enhancer element in the MET promoter and activate the expression of the MET gene. In addition, inhibition of PAX6 transcripts leads to a decline in cell growth and survival, differentiation, and a concurrent reduction of MET protein expression. These data support a model for a neoplastic pathway, where expression of a transcription factor from development activates the MET receptor, a protein that has been directly linked to protumorigenic processes of resisting apoptosis, tumor growth, invasion, and metastasis.
Pancreatic cancer is an aggressive and deadly disease, with an average median survival of less than a year (1). Several genetic pathways have been identified as being active in the progression of this tumor, including signaling through MET (MET tyrosine kinase receptor protein). The MET gene encodes a tyrosine kinase receptor for the ligand hepatocyte growth factor/scatter factor. The MET gene produces a partially glycosylated 170-kDa precursor protein. This precursor is glycosylated further and cleaved into a 50-kDa α chain and a 140-kDa β chain to create a mature receptor (2). The MET receptor is essential for normal development and plays a role in cell migration, growth, survival, differentiation, angiogenesis, and tube formation/branching morphogenesis (reviewed in Ref. 3). MET has also been implicated in cancer progression and is directly involved in metastasis, resistance to apoptosis, and tumor growth.
MET is expressed in the developing pancreatic bud of the embryo and marks candidate stem/progenitor cells in the embryonic and adult pancreas (4–6). MET expression is expressed at very low levels in normal adult differentiated pancreatic cells (7). MET is overexpressed in pancreatic cancer cells and has been linked to the aggressiveness of this tumor in terms of growth, invasion, and metastasis (7–10).
Although mutations in the MET locus have been identified, overexpression of MET occurs mainly due to aberrant transcriptional regulation (3). The MET gene is regulated by several transcription factors that can either activate or repress expression. Activators include HIF1 (hypoxia-induced factor 1) in response to oxygen deficiency (11), ETS1 (12), Sp1 (13), AP1 (14), Smads downstream of transforming growth factor-β signaling (13), and the p53 protein (15) as well as the basic helix-loop-helix protein MITF and a related family member TFE3 (16, 17). Most repressors of MET function act by inhibiting Sp1-mediated MET induction, including interferon-α (18), androgen receptor (19), and oxidative stress (20). MET expression is also inhibited by Notch signaling through HES1 (21) and the HDAC scaffold protein Daxx (22).
The transcription factor PAX3 can also activate MET expression during the embryonic development of muscle cells (23). PAX3 belongs to the PAX gene family, and most of what is known about these related proteins is their role during development. Only recently has the expression of PAX proteins in adult stem cells and in disease been discovered. We found the PAX3-related protein PAX6 expressed in pancreatic cancer frequently, with expression in 32 of 46 (69.6%) of primary tumors and 9 of 10 established cell lines (24). In our studies, we find that PAX6 is linked to inhibiting differentiation and growth arrest. Inducing differentiation in pancreatic cancer cells triggers a down-regulation of PAX6 expression, whereas a direct inhibition of the PAX6 transcript produces the same differentiated phenotype. Although our data suggest that PAX6 is active in the cancer phenotype, the specific molecular pathways through which PAX6 acts are unknown.
Here, we find transcripts for both the canonical and the alternately spliced 5a transcript of the PAX6 gene in pancreatic carcinoma cell lines. The canonical PAX6 protein contains a central homeodomain and a N-terminal paired DNA binding domain composed of two subdomains, the PAI and RED moieties. Due to an alternate splice insertion of 14 amino acids into the PAI subdomain, the PAX6(5a) variant has DNA binding specificity different from that of the canonical PAX6 protein. Both proteins can bind to DNA through the homeodomain and the paired domain, but whereas PAX6 can bind to sites through both the PAI and RED subdomains, PAX6(5a) relies mostly on the RED domain to bind to paired consensus sites (25–28). The majority of what is known thus far about PAX6(5a) function is its role during development, most of which is focused in eye development. Both transcripts are essential for normal eye development, where loss or overexpression of either of the transcripts leads to specific eye defects (25, 27, 29–31). A role for the PAX6(5a) protein in the pancreas or pancreatic cancer has not been described.
We find that PAX6 and PAX6(5a) directly activate the tyrosine kinase receptor MET through an enhancer element in the 5′-proximal MET promoter. Although both proteins activate a reporter construct containing the MET promoter and both transcripts are expressed in two representative pancreatic carcinoma cell lines, the PAX6(5a) protein is the dominant protein expressed in these cells. Inhibition of PAX6 expression induces a parallel reduction of MET expression in pancreatic carcinoma cells, and this is coupled with a decline in cell growth and survival. We present here in this report a cancer pathway, where aberrant overexpression of PAX6/PAX6(5a) leads to MET expression, a receptor directly involved in the cancer phenotype of metastasis, resistance to apoptosis, and tumor growth.
EXPERIMENTAL PROCEDURES
Immunohistochemistry and Immunofluorescence
PAX6 protein expression was detected in histologically and clinically confirmed pancreatic adenocarcinoma specimens using PAX6 monoclonal mouse antibody (1:500 dilution; University of Iowa Hybridoma Bank, Iowa City, IA). Following primary antibody incubation, slides were incubated with biotinylated secondary antibody, treated with streptavidin immunoperoxidase complex (Vectastain Elite ABC kit, Vector Laboratories, Burlingame, CA), and stained with diaminobenzidine chromagen. For MET receptor detection, slides were incubated with MET receptor polyclonal rabbit primary antibody (1:100 dilution; catalog number sc-10; Santa Cruz Biotechnology, Santa Cruz, CA). The secondary antibody was fluorescein-conjugated goat anti-rabbit (1:200 dilution; catalog number 31635; Pierce). For scoring and analysis of PAX6 and MET expression, slides were purely graded as “positive” or “negative” for expression and scored blinded for sample type.
Statistical Analysis
Differences in sample sets were determined by Student's t test or χ2 analysis using statistical software (GraphPad Prism, version 5.0; GraphPad Software Inc., La Jolla, CA). A p value of less than 0.05 was considered statistically significant.
Western Analysis
For Western analysis detection of PAX6, MET, and vinculin, cell lysates (40 μg/well) were loaded on acrylamide gels, transferred to polyvinylidene difluoride membranes, probed with PAX6 (1:5000 dilution; antibody 5790; Abcam Inc., Cambridge, MA) or MET (1:200 dilution; catalog number sc-10; Santa Cruz Biotechnology) rabbit polyclonal antibodies, and Western blotted (Western Breeze Chemiluminescent kit; Invitrogen). The membranes were stripped using Restore Western blot stripping buffer (Pierce), checked for complete removal of antibodies, and reanalyzed for vinculin (1:5,000 mouse monoclonal antibody clone hVIN-1; Sigma) or β-actin (1:400 mouse monoclonal antibody E7, University of Iowa Hybridoma Bank) expression as a loading control.
Reverse Transcription-PCR Analysis
RNA was isolated using TRIzol reagent (Invitrogen). RNAs were treated with deoxyribonuclease to remove any contaminating DNA. cDNA was synthesized using reverse transcriptase (Invitrogen). To confirm the absence of contaminating genomic DNA, samples were prepared without reverse transcriptase. Primers for PAX6 are located in the paired box, TTC AGA GCC CCA TAT TCG AG and TGC TAG TCT TTC TCG GGC AA, amplifying 787 bp (base pairs) for the canonical PAX6 transcript and 829 bp (42 inserted bp) for PAX6(5a). PCR products were sequenced to confirm correct amplification. Control primers for glyceraldehyde-3-phosphate dehydrogenase are TCA AGG TCG AGT CAA CGG ATT TGG T and CAT GTG GGC ATG AGG TCC ACC AC, amplifying 983 bp, as described (Clontech), and control primers for β2-microglobulin are GGG TTT CAT CCA TCC GAC AT and GAT GCT GCT TAC ATG TCT CGA, amplifying 224 bp, as described (32).
Measurement of DNA Content by Flow Cytometry
Cells were trypsinized, washed in phosphate-buffered saline, fixed in ice-cold 70% ethanol, and stored at −20 °C until analysis of DNA content by flow cytometry. After centrifugation, cells were suspended in 0.2 m phosphate citrate buffer (pH 7.8) and incubated at 37 °C for 30 min. Cells were washed with phosphate-buffered saline and resuspended in staining solution (0.1% Triton X-100, 200 μg/ml RNase A, 20 μg/ml propidium iodide in phosphate-buffered saline) for 30 min. Cells were filtered through a mesh and analyzed on a FACSCalibur flow cytometer.
Luciferase Assays, Cell Culture, and Vectors
Pancreatic carcinoma (PANC-1, MIAPACA-2, and SW979) and control (mouse fibroblast 3T3 and human kidney 293T) cell lines were obtained from the American Type Culture Collection (Manassas, VA). The MET promoter luciferase reporter construct, containing a fragment of the MET promoter with 297 bp 5′ to the transcriptional start to 22 bp of the 5′-UTR (33), was amplified from normal human genomic DNA and subcloned into pGL2-basic vector (Promega) to create the reporter plasmid WT2 hMET pm. The MET1 consensus site (23) for paired protein binding was mutated by site-directed mutagenesis, replacing the sequence GTCCCGC with ACTAGTC, thereby destroying the MET1 site and creating an SpeI restriction site (hMET pm Δ). The human PAX6 expression constructs were kindly provided by Dr. Ales Cvekl (Albert Einstein College of Medicine, New York). 293T and PANC-1 cells were transfected with either empty pGL2-basic, WT hMET pm, or hMET pm Δ reporter construct and an internal control β-galactosidase-expressing construct pCMV (Clontech) in the presence or absence of PAX6/PAX6(5a)-expressing constructs. The total amount of DNA transfected was kept constant by the addition of pBluescript plasmid (Stratagene, La Jolla, CA). Cells were transfected according to the manufacturer's protocols (Effectene reagent; Qiagen, Valencia, CA). After 48 h post-transfection, luciferase and β-galactosidase assays (Promega) were performed. Luciferase activity was normalized to β-galactosidase activity levels as an internal control. All experiments were performed minimally in triplicate.
Electrophoretic Mobility Shift Assay (EMSA)
Whole cell extracts were obtained from 3T3 cells transfected with PAX6 and PAX6(5a) constructs prepared in 20 mm HEPES (pH 7.5), 400 mm KCl, 0.5 mm EDTA, 0.1 mm EGTA, 1 mm dithiothreitol, and 20% glycerol. EMSA was performed in the presence of 40% glycerol, 1 mm MgCl2, 0.3 mm EDTA, 0.5 mm dithiothreitol, 50 mm NaCl, 10 mm Tris/HCl pH 7.5, 50 μg/ml poly(dI-dC)·poly(dI-dC) (Amersham Biosciences). Primers to create probes for EMSA are shown in Fig. 5D. Probes were annealed together and end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) and purified on Microspin G-50 columns (Amersham Biosciences). Competition assays were performed in the presence of a 100-fold excess of unlabeled probe to the labeled probe. Samples were electrophoresed on 5% native gels, dried, and exposed to Biomax MS film (Eastman Kodak Co.).
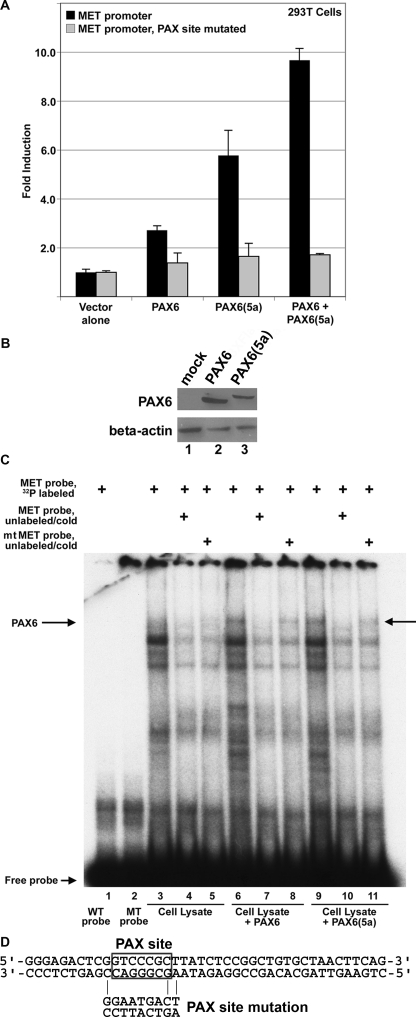
FIGURE 5.
PAX6 and PAX6(5a) activate the proximal MET promoter. A, transfection of PAX6-non-expressing 293T cells with MET promoter reporter constructs, wild type (black bars) or the paired site mutated (gray bars), with expression constructs for PAX6 and/or PAX6(5a). Total DNA transfected is equal for each sample, and the set contained both PAX6 and PAX6(5a) expression construct DNA at a 1:1 ratio. Fold induction is calculated as luciferase activity (measured in arbitrary light units) of each sample (with PAX6 and/or PAX6(5a) added) divided by luciferase levels of the reporter constructs alone. PAX6 proteins activate the MET promoter 2.7 ± 0.2-fold (canonical PAX6), 5.8 ± 0.9-fold (PAX6(5a)), or 9.7 ± 0.3-fold (both). Each bar represents n = 6. B, Western analysis for PAX6 expression in transfected 293T cells. These cells are utilized for the luciferase experiments shown in A. Cells transfected with empty vector (lane 1), a triple-FLAG-tagged PAX6 (lane 2), and a triple-FLAG-tagged PAX6(5a) (lane 3) are tested for expression of PAX6 and β-actin control. C, PAX6 and PAX6(5a) bind to the MET enhancer as shown by an EMSA. A slow migrating band (arrow labeled PAX6) is present when a radioactive probe containing the MET enhancer is combined with PAX6 protein (lane 6) or PAX6(5a) protein (lane 9). This band is absent when WT (lane 1) or mutant (MT) probe is used alone or when mock-transfected lysate is utilized (lanes 3–5). Competition with unlabeled wild type probe (lanes 4, 7, and 10) competes away PAX6/PAX6(5a) binding (lanes 7 and 10). Competition with unlabeled mutant probe (lanes 5, 8, and 11) is not as efficient in competing away protein binding to the radiolabeled probe (lanes 8 and 11), suggesting binding specificity. D, the sequence of the wild type probe used in the EMSA, with the PAX site boxed. The mutant probe is the identical sequence, except with the PAX site mutated, as shown.
Chromatin Immunoprecipitation (ChIP) Assay
Plated PANC-1 cells were fixed in 1% formaldehyde and quenched in 0.125 m glycine and then processed according to the manufacturer's protocol (Upstate Biotechnology, Inc., Temecula, CA). For immunoprecipitation of PAX6-DNA complexes, 1 μl of mouse monoclonal antibody (University of Iowa Hybridoma Bank) was added per experimental reaction. A nonspecific antibody, normal IgG (Sigma), was used as a negative control against nonspecific DNA precipitation by an antibody. Nested PCR was performed with primers to the MET enhancer, first with TCC GCC TCT AAC AAT GAA CTC C and AAG GTG AAA CTT TCT AGG TGG and then a second PCR reaction with primers TGC CCA AAT CTC TCT AAA CCC and AAG TTT TCT CGC CCT GGC TGC G. All ChIP samples were tested for false positive PCR amplification using primers that amplify sequence located within the fourth large coding exon of the β-tubulin gene to control against genomic DNA contamination. The nested primer set for this control was AAA GGC CAC TAC ACA GAG GG and TAC CAA CTG ATG GAC GGA GAG G for the first PCR and then TTG ATT CTG TCC TGG ATG TGG and TCA GAC ACT TTG GGT GAA GGC for the second PCR round. For the first PCR, 10% of the ChIP sample was utilized as template. For the nested second reaction, 5% of the first PCR was used as template.
Inhibition of PAX6/PAX6(5a) Expression with RNA Interference
PANC-1 cells were transfected with siRNA specific for both PAX6 and PAX6(5a) transcripts or control siRNA (100 pmol; Dharmacon, Lafayette, CO) using Lipofectamine 2000 (Invitrogen), following the manufacturer's protocols. The transfection efficiency for these experiments was greater than 80%, as calculated by the number of fluorescing cells successfully transfected with a green fluorescent protein-labeled control siRNA. Cell lysates were collected at 48 h post-siRNA transfection.
RESULTS
PAX6 and MET Are Expressed in Primary Pancreatic Adenocarcinoma Tissues and Cell Lines
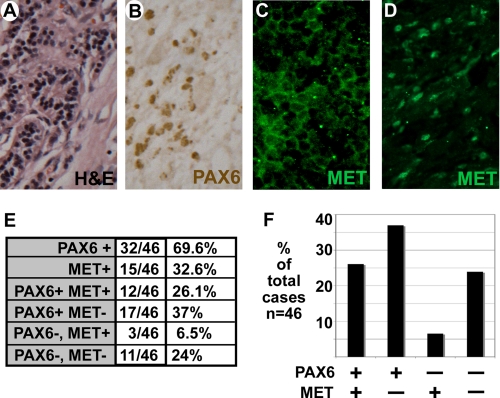
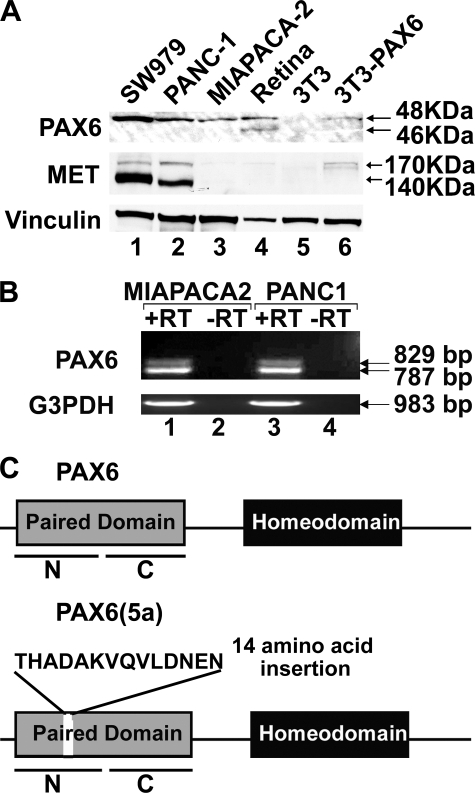
We have previously found PAX6 expressed in pancreatic carcinoma cell lines and primary tissue (24). Here, we compared the presence of PAX6 expression to MET expression in a sample set of pancreatic adenocarcinoma tissue (Fig. 1). We find that 32 of 46 of the samples express PAX6 (69.6%) and 15 of 46 express MET (32.6%) (Fig. 1E). Most of the MET-expressing tissues demonstrated cytoplasmic and/or membranous staining of the receptor (Fig. 1C), but eight of the samples had at least some degree of nuclear staining as well (Fig. 1D). 12 of 15 samples that express MET also expressed PAX6. MET expression coupled with an absence of PAX6 was an infrequent finding in comparison with the other group combinations (Fig. 1F), and this is a statistically significant finding (p < 0.05). The pancreatic carcinoma cell lines SW979, PANC-1, and MIAPACA-2 also expressed PAX6 (Fig. 2A, lanes 1–3), whereas the mouse fibroblast cell line 3T3 did not (Fig. 2A, lane 5). Both the cell lines SW979 and PANC-1 expressed abundant amounts of MET, whereas MIAPACA-2 and 3T3 cells did not express detectable levels. The predominant form of MET is the mature form (140-kDa β chain) as well as some expression of the 170-kDa precursor protein. The lack of MET expression in MIAPACA-2 cells has been previously observed (34). 3T3 cells are utilized in these experiments as a negative control, since the cells do not express both MET and PAX6.
FIGURE 1.
PAX6 and MET are expressed in primary pancreatic adenocarcinoma tissues. A–D, representative histology of pancreatic adenocarcinoma tumor tissue. A, hematoxylin and eosin (H&E) tissue stain. B, immunohistochemical stain for nuclear PAX6 expression (brown). C and D, immunofluorescence stain for MET receptor expression (green). MET expression is cytoplasmic (C) and/or nuclear (D). E and F, summary of immunohistochemical staining of PAX6 and MET receptor in 46 unrelated tumor samples in a table (E) and a graph (F). The graph represents the last four sample sets of the chart (PAX6 and MET receptor co-expression analysis).
FIGURE 2.
Expression of PAX6 and PAX6(5a) in pancreatic carcinoma cell lines. A, Western analysis of cell lysates. Lysate samples include pancreatic cancer cell lines SW979 (lane 1), PANC-1 (lane 2), and MIAPACA-2 (lane 3), mouse retina (lane 4), PAX6-non-expressing mouse fibroblast cell line 3T3 (lane 5), and 3T3 cells transfected with 3×FLAG-PAX6 (lane 6). Samples are tested for expression of Vinculin (loading control), MET receptor, and PAX6. Expected sizes for PAX6 proteins are 46 kDa (canonical PAX6) and 48 kDa (PAX6(5a) and 3×FLAG-PAX6). Retinal tissues express both forms of PAX6 and are a size control for both isoforms (45). B, reverse transcription-PCR analysis for the expression of PAX6 and PAX6(5a) transcripts in MIAPACA-2 (lanes 1 and 2) and PANC-1 (lanes 3 and 4). The reverse transcription reactions occurs in the presence (lanes 1 and 3) or absence (lanes 2 and 4) of reverse transcriptase. The resulting cDNAs are amplified for primers recognizing glyceraldehyde-3-phosphate dehydrogenase (G3PDH; positive control) or the paired domain of PAX6. The amplicon for canonical PAX6 is 787 bp, and the amplicon for PAX6(5a) is 829 bp. C, schematic of the PAX6 and PAX6(5a) proteins. Proteins are identical (with paired and homeodomain DNA binding domains) except that the PAX6(5a) protein contains a 14-amino acid insertion in the PAI N-terminal subdomain (N) of the paired domain. The paired domain also contains a C-terminal RED subdomain (C).
Pancreatic Cancer Cell Lines Predominantly Express the 5a Variant of PAX6
Using primers in the paired box of the PAX6 transcript, reverse transcription-PCR analysis of MIAPACA-2 and PANC-1 cells revealed two major PCR products, the expected 787 bp band for the canonical PAX6 transcript and a slower migrating band at 829 bp (Fig. 2B). Both PCR products were sequenced, confirming the amplicons as the canonical PAX6 transcript and a major splice variant of PAX6, PAX6(5a). This variant protein is identical to the canonical version, with the exception of a 42-bp insertion in the paired box, adding 14 amino acids to the PAI subdomain of the Paired DNA binding domain (Fig. 2C). Since the pancreatic cancer cell lines predominantly show only one band in Western analysis, the cancer cell lysates were run in parallel with protein derived from mouse retina as a size control. Mouse and human retina have significant expression of both isoforms (30). Although the retina protein shows two distinct bands on a Western analysis for PAX6 expression at 46 kDa for PAX6 and 48 kDa for PAX6(5a) (Fig. 2A, lane 4), the three pancreatic carcinoma cell lines predominantly show only the 48 kDa band. The PAX6 protein expressed in the cancer cells also matches the size for the 48-kDa 3×FLAG-PAX6 protein produced in transfected in 3T3 cells (Fig. 2A, lane 6).
Inhibition of PAX6 Expression Leads to Apoptosis and a Change in Cellular Morphology
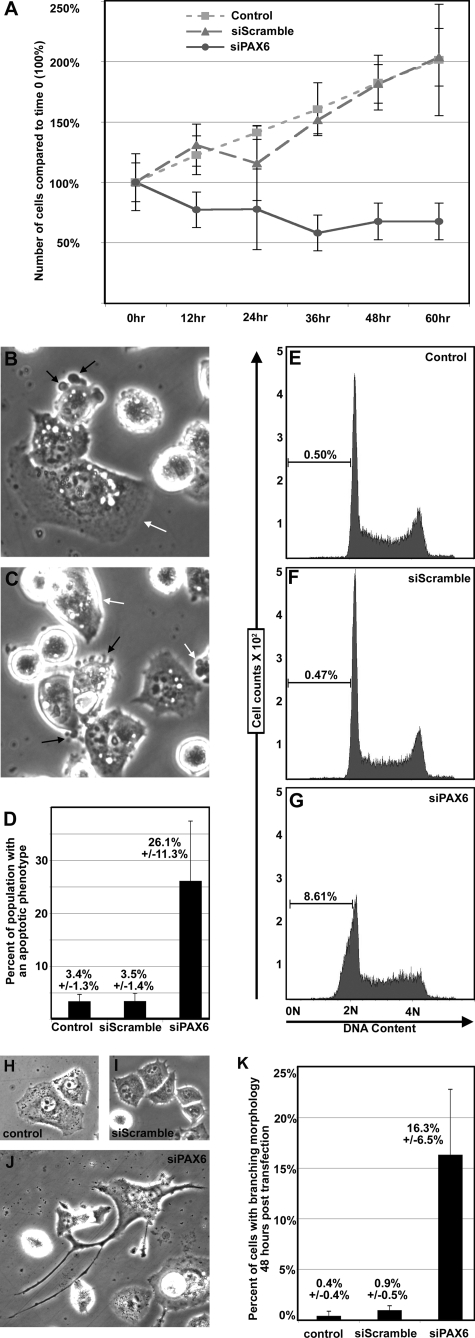
PAX6 is expressed in a significant number of pancreatic tumors. To test whether PAX6 plays a role in the growth and survival of pancreatic cancer cells, the expression of PAX6 was specifically inhibited through RNA interference in PANC-1 cells. Expression of PAX6 in this cell line can be inhibited 50–70% with gene-specific siRNA but not with random siRNA (siScramble) (Fig. 7). Reduction of PAX6 expression is due to specific targeting of the PAX6 transcript and does not affect non-related genes, such as β2-microglobulin (supplemental Fig. S1). Over a time course, parental and siScramble-transfected PANC-1 cells show healthy growth and a population doubling at around 60 h (Fig. 3A). However, PANC-1 cells transfected with siPAX6 demonstrate a loss of cells over the same time course. Morphologically, 26.1 ± 11.3% of the cells appear to undergo apoptosis, round up, and detach from the plate (Fig. 3, B–D). The untransfected and siScramble-transfected PANC-1 cells have significantly fewer cells with this phenotype, 3.4 ± 1.3 and 3.5 ± 1.4%, respectively. A hallmark feature of apoptosis is DNA fractionation, and this can be detected by examining DNA content by flow cytometry. The DNA content of cells in G0/G1 will be 2 n, whereas cells in G2 will be 4 n, and cells undergoing apoptosis will be less than 2 n. Although control cells have less than 1% of the cell population with a DNA content of less than 2 n, 8.61% of the siPAX6-transfected cells fall into this category (Fig. 3, E–G). Evidence of late stage apoptosis occurs at 9–12 h post-siPAX6 transfection. At 24 h post-transfection, there is evidence of large branching cells in 16.3 ± 6.5% of the remaining cells (Fig. 3J), but these cells are rare in the control cells (Fig. 3, H–K). It is possible that the percentage of these branching cells in the siPAX6-transfected populations would be higher but are diluted due to untransfected cells in the plate.
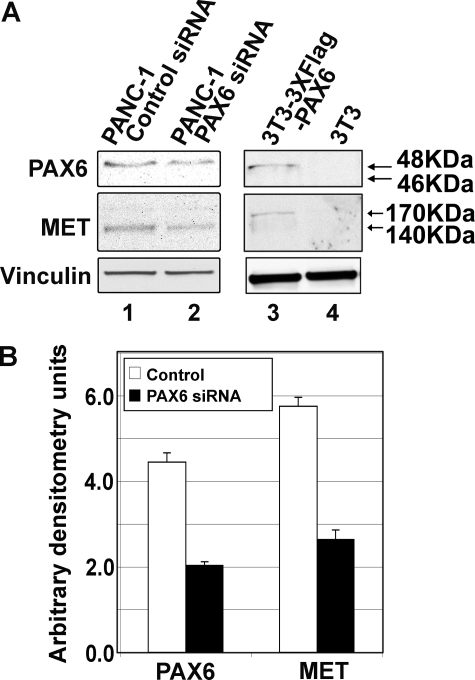
FIGURE 7.
Inhibiting PAX6 expression in PANC-1 pancreatic carcinoma cells leads to a reduction of MET receptor protein levels. A, Western analysis of cell lysates. Samples include PANC-1 cells transfected with control siRNA (random siRNA siScramble, negative control; lane 1) or siRNA specific to PAX6/PAX6(5a) (lane 2) and control lysates of 3T3 mouse fibroblast cells with transfected 3×FLAG-PAX6 (positive control for PAX6; lane 3) and untransfected 3T3 (negative control for PAX6; lane 4), Expression for vinculin (loading control), MET receptor, and PAX6/PAX6(5a) is measured. B, densitometry of lanes 1 and 2 in Western analysis shown in A. Samples are normalized against vinculin expression. The band intensity for PAX6/PAX6(5a) and MET receptor is measured for the control (siScramble-transfected; white bars) and PAX6/PAX6(5a)-targeted (black bars) samples. In comparison with mock-transfected cells, PAX6/PAX6(5a) expression is 46.7% (from 4.5 arbitrary densitometry units to 2.1) and MET expression is 47.5% (from 5.9 arbitrary densitometry units to 2.8 of controls). The graph represents two independent experiments.
FIGURE 3.
Inhibition of PAX6 expression leads to a reduction of cell numbers, apoptosis, and a change in cellular morphology. A, cell numbers over a 60-h time course. PANC-1 cells that are untransfected (squares) or transfected with a random siRNA (siScramble; triangles) or with PAX6 gene-specific siRNA (siPAX6; circles) are counted at 12-h intervals to 60 h (x axis). Cells are counted at the time of transfection, and this number is set at 100% of starting levels. Cell numbers are presented as percentage of 0 h levels (y axis). B and C, morphology of siPAX6-transfected cells 9 h post-transfection. A number of the cells display blebbing (black arrows), rounding up, and a loss of adhesion to the culture plate. There are also cells that maintain the parental phenotype (white arrows) (magnification, ×400). D, quantification of cells shown in B and C with an apoptotic morphology. In six random fields, total cells and cells with overt signs of apoptosis (blebbing, rounding up) are counted and graphed. The average percentage of the population with an apoptotic phenotype for untransfected, siScramble-transfected, and siPAX6-transfected PANC-1 cells is 3.4 ± 1.3%, 3.5 ± 1.4%, and 26.1 ± 11.3%, respectively. E–G, measurement of DNA content by flow cytometry. DNA from PANC-1 cells that are untreated or are 9 h post-transfected with siScramble or siPAX6 is stained with propidium iodide. DNA from cells in G0/G1 is measured as 2 n, and G2 DNA is measured as 4 n. DNA fragmented during apoptosis is seen as a population less than 2 n. This population is 0.5, 0.47, and 8.61% of the total cell counts for the untransfected, siScramble-transfected, and siPAX6-transfected PANC-1 cells, respectively. H, phenotype of the untransfected cells at the 24 h time point. I, phenotype of the siScramble-transfected cells at the 24 h time point. J, phenotype of the siPAX6-transfected cells at the 24 h time point. Although the majority of cells in the control groups have an epithelioid phenotype, the siPAX6-transfected cells have several branchlike outgrowths (H, I, and J; magnification, ×200). K, quantification of cells shown in H–J with a branching morphology. In six random fields, total cells and cells with overt signs of branching are counted and graphed. The average percentage of the population with a branching phenotype for untransfected, siScramble-transfected, and siPAX6-transfected PANC-1 cells is 0.4 ± 0.4, 0.9 ± 0.5, and 16.3 ± 6.5%, respectively.
PAX6 and PAX6(5a) Activate the Proximal MET Promoter
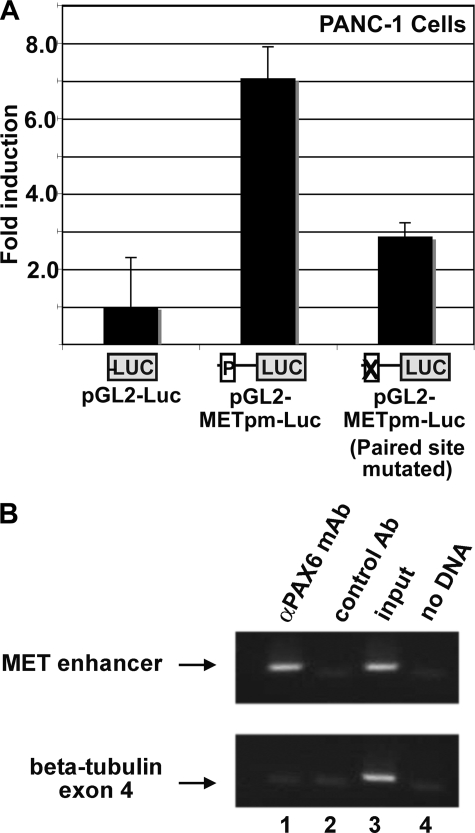
The PAX6-related protein, PAX3, can activate MET expression during muscle development (23). The PAX3 response element, called MET1, is located in the MET gene promoter region within the 500 bp 5′-proximal to the transcriptional start (Fig. 4). This is a response element for the paired DNA binding domain although not an ideal consensus for the PAX6 protein, as determined by binding site selection assays (26). To determine if PAX6 and PAX6(5a) activate a reporter construct containing the MET1 enhancer, the plasmid WT hMET pm was transfected into 293T cells with empty pCDNA3 vector, a PAX6 expression construct, and/or a vector expressing PAX6(5a). The WT hMET pm construct contains a fragment of the 5′-proximal MET promoter driving expression of the reporter gene luciferase. The advantage of using the 293T line as a cell system here is that these cells do not express endogenous PAX6. Both versions of the PAX6 protein moderately activated reporter expression from the MET1 enhancer alone while synergistically activating expression together (Fig. 5, A and B, black bars). To determine if this activation is dependent on the presence of the MET1 enhancer, the luciferase assay is repeated with a mutant reporter construct, hMET pm Δ, where the MET1 site is replaced with a SpeI restriction site. Both PAX6 and PAX6(5a) were unable to drive expression of the luciferase reporter from this construct (Fig. 5A, gray bars).
FIGURE 4.
Sequence of the proximal human MET promoter. This promoter segment, which includes 297 bases directly 5′-upstream of the transcriptional start site and 22 bases directly 3′, corresponds with the DNA segment contained in the MET promoter luciferase reporter construct utilized in Figs. 5 and 6. The PAX/paired site is highlighted with a black box (−252 to −244). The transcriptional start site is marked with an arrow, and previously described Sp1 and Smad-binding element (SBE) sites are labeled (20).
The ability of PAX6 and PAX6(5a) to directly bind to this enhancer sequence was tested using an EMSA (Fig. 5C) with probes containing endogenous and mutated sequences (shown in Fig. 5D). Lysates from cells transfected with empty vector or expression vectors driving expression of either PAX6 or PAX6(5a) were utilized for these experiments (Figs. 2A, 5B, and 7). PAX6 and PAX6(5a) were able to bind to the PAX site (Fig. 5C, lanes 6 and 9) and could be competed off the site with unlabeled wild type probe (lanes 7 and 10) but not cold mutant probe (lanes 8 and 11). Hence, both full-length isoforms of PAX6 bind to the MET enhancer in a sequence-specific fashion.
Empty pGL2-Luc vector, WT hMET pm, and hMET pm Δ were also transfected into PANC-1 cells. Here, the PAX6 gene was endogenously expressed, and both PAX6 and PAX6(5a) were present (Fig. 2B, lane 3), but the dominant protein form expressed was PAX6(5a) (Fig. 2A, lane 2). In PANC-1 cells, transfection of WT hMET pm reporter resulted in significant luciferase activity (7.1 ± 0.8-fold over pGL2-Luc vector alone). Mutating the MET1 in this reporter construct decreased the levels of luciferase activity produced after transfection to 2.9 ± 0.4-fold (Fig. 6A).
FIGURE 6.
PAX6/PAX6(5a) activate MET receptor expression in PANC-1 pancreatic carcinoma cells. A, transfection of PANC-1 pancreatic carcinoma cells with empty pGL2-Luc vector or pGL2-Luc containing either wild type or paired site-mutated MET promoter sequence. The average light units for the pGL2-Luc samples are set as the standard, or “1-fold” light unit. The fold luciferase activity in samples with the pGL2-METpm-Luc is 7.1 ± 0.8-fold and 2.9 ± 0.4-fold when the paired site is mutated. Each bar represents n = 6. B, ChIP analysis of PANC-1 pancreatic carcinoma cells. The top gel panel utilizes primers for the MET promoter enhancer, and the bottom gel panel uses primers located in exon 4 of the β-tubulin gene (negative control). Lane 1, precipitation with mouse anti-PAX6 antibody (Ab). Lane 2, precipitation with normal mouse IgG. Lane 3, cell lysate input, pre-ChIP. Lane 4, no sample, water blank PCR negative control.
Additional support for the premise that PAX6 can activate MET expression is the presence of some expression of MET protein in the mouse 3T3 cell line after the addition of PAX6. This cell line does not normally express either PAX6 or MET (Fig. 2A, lane 5, and Fig. 7A, lane 4). A triple-flagged PAX6-expressing construct was transfected into these cells, and this led to both the expression of PAX6 and MET protein (Fig. 2A, lane 6, and Fig. 7A, lane 3). However, unlike the PANC-1 and SW979 cells that express predominantly the processed 140-kDa β-MET fragment and the 170-kDa glycosylated precleaved precursor, the transfected 3T3 cells only produce the precursor protein.
PAX6 Drives MET Reporter Expression in PANC-1 Pancreatic Carcinoma Cells
An antibody that recognizes both forms of PAX6 (canonical and 5a variant) was able to precipitate a complex containing the MET enhancer in a ChIP analysis using cross-linked PANC-1 cell lysate (Fig. 6B, lane 1). To test against a PCR-amplified false positive reading, a nonspecific antibody (normal mouse IgG) and primers against a genomic region without PAX6 binding sites (exon 4 of the β-tubulin gene) were also tested in this assay.
To test if MET expression has at least some dependence on the expression of PAX6, the expression of PAX6 in PANC-1 cells was specifically inhibited through RNA interference. PAX6 gene-specific siRNA reduced PAX6 protein levels in PANC-1 cells (Fig. 7A, lane 2) compared with PANC-1 cells transfected with siRNA randomers (Fig. 7A, lane 1). The bands were measured using densitometry software (NIH ImageJ), showing a decrease of PAX6 protein levels to 46.7% of the control levels (Fig. 7B). This reduction in PAX6 protein levels was mirrored with a decrease in MET protein levels (Fig. 7A, lane 2) measured to 47.5% compared with control cells (Fig. 7B). This suggests that PAX6 binds the MET1 enhancer and plays a role in MET expression in this cell line.
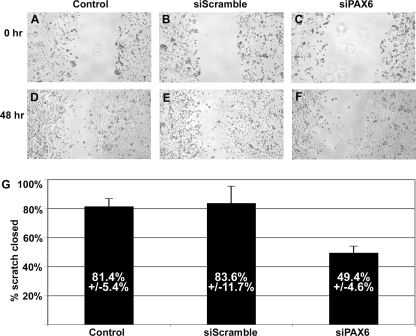
Since MET has been implicated in cell migration, a scratch assay was performed with untransfected and siScramble- and siPAX6-transfected PANC-1 cells. Unlike the growth assays that use low confluence cells (10–20%), the scratch assay utilizes semiconfluent monolayers of cells. A scratch (or wound) in the monolayer was made with a fine point pipette 10 h after transfection. Cells in all three groups were photographed just after scratch (0 h; Fig. 8, A–C) and 48 h later (Fig. 8, D–F). The scratch width was measured at both time points, and the percentage of the scratch area closed was calculated as (width at t0 − width at t48)/width at t0 × 100. Untransfected and siScramble-transfected PANC-1 cells closed the scratch 81.4 ± 5.4 and 83.6 ± 11.7%, respectively (Fig. 8G). PANC-1 cells transfected with siPAX6 closed the monolayer wound 49.4 ± 4.6%. Although this cell population was less efficient at migrating into the scratch area, an inhibition of growth rather than migration cannot be ruled out, although PANC-1 cells in monolayer do have some cell to cell contact inhibition when grown in a monolayer.
FIGURE 8.
Scratch assay measuring cell migration. A–F, photographs of cells for scratch assay. Semiconfluent monolayers of untransfected (Control), random siRNA-transfected (siScramble), and PAX6 gene-specific siRNA-transfected (siPAX6) PANC-1 cells are scratched and are photographed just after scratch (0 h; A–C) and 48 h later (D–F). G, graph of change in scratch width over 48 h. For each group, the scratch width was measured at both time points, and the percentage of the scratch area closed was calculated as (width at t0 − width at t48)/width at t0 × 100. The percentage of scratch closed is 81.4 ± 5.4% for control, 83.6 ± 11.7% for siScramble, and 49.4 ± 4.6% for siPAX6 groups. Each bar represents n = 8.
In summary, PAX6 is expressed in pancreatic carcinoma cell lines and primary tumor tissue and actively plays a role in cancer cell survival. One possible mechanism for this is through the direct activation of the MET gene.
DISCUSSION
A major splice form of PAX6 is the PAX6(5a) variant. The two proteins exhibits different DNA-binding properties due to a 14-amino acid insertion into the Paired domain that reduces binding through the N-terminal region and favors C-terminal binding (Fig. 2C) (27). Ratios between these two PAX6 protein forms are tightly regulated during brain and eye development, and the maintenance of levels is critical for normal eye development (28, 35, 36). In addition, PAX6 protein can heterodimerize with the PAX6(5a) isoform and synergistically activate downstream genes, and data here suggest that this may be true for MET as well (Fig. 5A) (37). Loss of total Pax6 expression or just the canonical Pax6 leads to significant defects in murine development, including the absence of one or both eyes, central nervous system defects, and pancreatic abnormalities (25, 31). Mice with a specific deletion for PAX6(5a) have a milder phenotype. This mouse has iris hypoplasia and defects in the cornea, lens, and retina as well as cataract formation (31). The overall development of the eye in the PAX6(5a) mutants is relatively normal, suggesting that this protein variant does not play a significant role in eye specification during development but may be involved with maintenance of eye tissues later in embryogenesis and in the adult. The findings from murine models are supported by the phenotype of four patients with a missense mutation in the seventh codon of the 5a exon (GTC to GAC, Val to Asp) (38). In these individuals, eye development is normal overall, but there is incidence of Peters and Axenfeldt anomaly, congenital cataract, and hypoplasia of the fovea. The function of PAX6(5a) in tissue maintenance is not known, but it may play a role in iris contraction, as suggested by Saunders and co-workers (31). The presence of an alternatively spliced isoform is mainly restricted to vertebrates and is highly conserved. A distinctive feature of the vertebrate eye is the ability to regulate the levels of incoming light through contractile pupils, and the PAX(5a) transcript may be a late evolutionary acquirement to accomplish this ability. A role for PAX6(5a) in pancreatic development and tissue maintenance has not been examined.
A presence of PAX6 expression in tumors is a recent finding. PAX6 is expressed in a subset of glioblastomas, and this expression is correlated with longer patient survival (39). In this tumor type, mutations in the PAX6 gene are absent, but expression is correlated with a tumor suppressor function by slowing overall growth and repressing expression of the proinvasiveness gene matrix matalloproteinase-2 (40–42). In other tissue types, PAX6 is correlated with an oncogenic phenotype. PAX6 is expressed in retinoblastoma and in tumors of the pancreas and the intestine (24, 43, 44). Very little is known about the expression of the PAX6(5a) variant in tumors. The only study that tested for the presence of this version of PAX6 in cancer cells involved gliomas, where PAX6 has tumor-suppressive qualities (42). In these tumor cells, PAX6(5a) is absent.
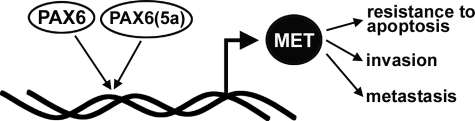
Here, we find that PAX6 is expressed in cell lines and primary tumors. PAX6 plays an active role in the tumor phenotype and that down-regulation of the gene leads to apoptosis and differentiation. We also demonstrate that PAX6 directly up-regulates the MET receptor gene. We propose a model where PAX6 promotes the expression of MET, and this receptor promotes the tumor phenotype (Fig. 9). MET promotes protumorigenic cellular functions, such as resistance to apoptosis, invasion, and the ability to metastasize. Not all consequences of PAX6 inhibition in pancreatic cells can be explained solely by MET expression, however, such as the induction of neural-like differentiation, as reported previously (24). This suggests that PAX6 (and/or the PAX6(5a) variant) regulates other genes in this cell type that affect cellular differentiation.
FIGURE 9.
A model for PAX6 promotion of a cancer phenotype in pancreatic carcinoma cells. Here we present a pathway for PAX6 protumorigenic function in cancer cells. PAX6 and/or its alternative splice variant directly activate the MET tyrosine kinase receptor gene through an upstream enhancer. The MET receptor promotes the tumor phenotype by promoting survival/resisting apoptosis and an ability for the cells to be locally invasive as well as to metastasize.
Supplementary Material
Acknowledgments
We are grateful for the generosity of Ales Cvekl (Albert Einstein College of Medicine, New York), Bernhard Ortel and YuYing He (University of Chicago), and Bruce Ruggeri (Cephalon, Inc., West Chester, PA) for reagents and/or scientific input.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA130202, R01CA100750, R01CA125541, and R21CA140003. This work was also supported by the Schweppe Foundation and the University of Chicago Cancer Center Pilot Program, Grant P30 CA014599.
This paper is dedicated to the memory of Gary Capps.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- WT
- wild type
- ChIP
- chromatin immunoprecipitation
- EMSA
- electrophoretic mobility shift assay
- siRNA
- small interfering RNA
- hMET
- human MET
- pm
- promoter.
REFERENCES
- 1.Ghaneh P., Costello E., Neoptolemos J. P. (2007) Gut 56, 1134–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giordano S., Ponzetto C., Di Renzo M. F., Cooper C. S., Comoglio P. M. (1989) Nature 339, 155–156 [DOI] [PubMed] [Google Scholar]
- 3.Gentile A., Trusolino L., Comoglio P. M. (2008) Cancer Metastasis Rev. 27, 85–94 [DOI] [PubMed] [Google Scholar]
- 4.Oshima Y., Suzuki A., Kawashimo K., Ishikawa M., Ohkohchi N., Taniguchi H. (2007) Gastroenterology 132, 720–732 [DOI] [PubMed] [Google Scholar]
- 5.Sonnenberg E., Meyer D., Weidner K. M., Birchmeier C. (1993) J. Cell Biol. 123, 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki A., Nakauchi H., Taniguchi H. (2004) Diabetes 53, 2143–2152 [DOI] [PubMed] [Google Scholar]
- 7.Di Renzo M. F., Poulsom R., Olivero M., Comoglio P. M., Lemoine N. R. (1995) Cancer Res. 55, 1129–1138 [PubMed] [Google Scholar]
- 8.Ebert M., Yokoyama M., Friess H., Büchler M. W., Korc M. (1994) Cancer Res. 54, 5775–5778 [PubMed] [Google Scholar]
- 9.Furukawa T., Duguid W. P., Kobari M., Matsuno S., Tsao M. S. (1995) Am. J. Pathol. 147, 889–895 [PMC free article] [PubMed] [Google Scholar]
- 10.Liu N., Furukawa T., Kobari M., Tsao M. S. (1998) Am. J. Pathol. 153, 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pennacchietti S., Michieli P., Galluzzo M., Mazzone M., Giordano S., Comoglio P. M. (2003) Cancer Cell 3, 347–361 [DOI] [PubMed] [Google Scholar]
- 12.Gambarotta G., Boccaccio C., Giordano S., Andŏ M., Stella M. C., Comoglio P. M. (1996) Oncogene 13, 1911–1917 [PubMed] [Google Scholar]
- 13.Zhang X., Yang J., Li Y., Liu Y. (2005) Am. J. Physiol. Renal Physiol. 288, F16–F26 [DOI] [PubMed] [Google Scholar]
- 14.Seol D. W., Chen Q., Zarnegar R. (2000) Oncogene 19, 1132–1137 [DOI] [PubMed] [Google Scholar]
- 15.Seol D. W., Chen Q., Smith M. L., Zarnegar R. (1999) J. Biol. Chem. 274, 3565–3572 [DOI] [PubMed] [Google Scholar]
- 16.Beuret L., Flori E., Denoyelle C., Bille K., Busca R., Picardo M., Bertolotto C., Ballotti R. (2007) J. Biol. Chem. 282, 14140–14147 [DOI] [PubMed] [Google Scholar]
- 17.Tsuda M., Davis I. J., Argani P., Shukla N., McGill G. G., Nagai M., Saito T., Laé M., Fisher D. E., Ladanyi M. (2007) Cancer Res. 67, 919–929 [DOI] [PubMed] [Google Scholar]
- 18.Radaeva S., Jaruga B., Hong F., Kim W. H., Fan S., Cai H., Strom S., Liu Y., El-Assal O., Gao B. (2002) Gastroenterology 122, 1020–1034 [DOI] [PubMed] [Google Scholar]
- 19.Verras M., Lee J., Xue H., Li T. H., Wang Y., Sun Z. (2007) Cancer Res. 67, 967–975 [DOI] [PubMed] [Google Scholar]
- 20.Zhang X., Liu Y. (2003) Am. J. Physiol. Renal Physiol. 284, F1216–F1225 [DOI] [PubMed] [Google Scholar]
- 21.Stella M. C., Trusolino L., Pennacchietti S., Comoglio P. M. (2005) Mol. Cell Biol. 25, 3982–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morozov V. M., Massoll N. A., Vladimirova O. V., Maul G. G., Ishov A. M. (2008) Oncogene 27, 2177–2186 [DOI] [PubMed] [Google Scholar]
- 23.Epstein J. A., Shapiro D. N., Cheng J., Lam P. Y., Maas R. L. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4213–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lang D., Mascarenhas J. B., Powell S. K., Halegoua J., Nelson M., Ruggeri B. A. (2008) Mol. Carcinog. 47, 148–156 [DOI] [PubMed] [Google Scholar]
- 25.Duncan M. K., Kozmik Z., Cveklova K., Piatigorsky J., Cvekl A. (2000) J. Cell Sci. 113, 3173–3185 [DOI] [PubMed] [Google Scholar]
- 26.Epstein J., Cai J., Glaser T., Jepeal L., Maas R. (1994) J. Biol. Chem. 269, 8355–8361 [PubMed] [Google Scholar]
- 27.Epstein J. A., Glaser T., Cai J., Jepeal L., Walton D. S., Maas R. L. (1994) Genes Dev. 8, 2022–2034 [DOI] [PubMed] [Google Scholar]
- 28.Kozmik Z., Czerny T., Busslinger M. (1997) EMBO J. 16, 6793–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azuma N., Tadokoro K., Asaka A., Yamada M., Yamaguchi Y., Handa H., Matsushima S., Watanabe T., Kohsaka S., Kida Y., Shiraishi T., Ogura T., Shimamura K., Nakafuku M. (2005) Hum. Mol. Genet. 14, 735–745 [DOI] [PubMed] [Google Scholar]
- 30.Chauhan B. K., Yang Y., Cveklová K., Cvekl A. (2004) Invest. Ophthalmol. Vis. Sci. 45, 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh S., Mishra R., Arango N. A., Deng J. M., Behringer R. R., Saunders G. F. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6812–6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schag K., Schmidt S. M., Müller M. R., Weinschenk T., Appel S., Weck M. M., Grünebach F., Stevanovic S., Rammensee H. G., Brossart P. (2004) Clin. Cancer Res. 10, 3658–3666 [DOI] [PubMed] [Google Scholar]
- 33.Gambarotta G., Pistoi S., Giordano S., Comoglio P. M., Santoro C. (1994) J. Biol. Chem. 269, 12852–12857 [PubMed] [Google Scholar]
- 34.Qian L. W., Mizumoto K., Inadome N., Nagai E., Sato N., Matsumoto K., Nakamura T., Tanaka M. (2003) Int. J. Cancer 104, 542–549 [DOI] [PubMed] [Google Scholar]
- 35.Jaworski C., Sperbeck S., Graham C., Wistow G. (1997) Biochem. Biophys. Res. Commun. 240, 196–202 [DOI] [PubMed] [Google Scholar]
- 36.Pinson J., Mason J. O., Simpson T. I., Price D. J. (2005) BMC Dev. Biol. 5, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chauhan B. K., Yang Y., Cveklová K., Cvekl A. (2004) Nucleic Acids Res. 32, 1696–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azuma N., Yamaguchi Y., Handa H., Hayakawa M., Kanai A., Yamada M. (1999) Am. J. Hum. Genet. 65, 656–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y. H., Tan F., Hess K. R., Yung W. K. (2003) Clin. Cancer Res. 9, 3369–3375 [PubMed] [Google Scholar]
- 40.Mayes D. A., Hu Y., Teng Y., Siegel E., Wu X., Panda K., Tan F., Yung W. K., Zhou Y. H. (2006) Cancer Res. 66, 9809–9817 [DOI] [PubMed] [Google Scholar]
- 41.Pinto G. R., Clara C. A., Santos M. J., Almeida J. R., Burbano R. R., Rey J. A., Casartelli C. (2007) Genet. Mol. Res. 6, 1019–1025 [PubMed] [Google Scholar]
- 42.Zhou Y. H., Wu X., Tan F., Shi Y. X., Glass T., Liu T. J., Wathen K., Hess K. R., Gumin J., Lang F., Yung W. K. (2005) J. Neurooncol. 71, 223–229 [DOI] [PubMed] [Google Scholar]
- 43.Salem C. E., Markl I. D., Bender C. M., Gonzales F. A., Jones P. A., Liang G. (2000) Int. J. Cancer 87, 179–185 [DOI] [PubMed] [Google Scholar]
- 44.Zhong X., Li Y., Peng F., Huang B., Lin J., Zhang W., Zheng J., Jiang R., Song G., Ge J. (2007) Int. J. Cancer 121, 2125–2131 [DOI] [PubMed] [Google Scholar]
- 45.Stanescu D., Iseli H. P., Schwerdtfeger K., Ittner L. M., Remé C. E., Hafezi F. (2007) Eye 21, 90–93 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.