Abstract
VILIP-1 (gene name VSNL1), a member of the neuronal Ca2+ sensor protein family, acts as a tumor suppressor gene by inhibiting cell proliferation, adhesion, and invasiveness. VILIP-1 expression is down-regulated in several types of human cancer. In human non-small cell lung cancer, we found that down-regulation was due to epigenetic changes. Consequently, in this study we analyzed the VSNL1 promoter and its regulation. Serial truncation of the proximal 2-kb VSNL1 promoter (VP-1998) from its 5′ terminus disclosed that the last 3′ terminal 100-bp promoter fragment maintained similar promoter activity as compared with VP-1998 and therefore was referred to as VSNL1 minimal promoter. When the 5′ terminal 50 bp were deleted from the minimal promoter, the activity was dramatically decreased, suggesting that the deleted 50 bp contained a potential cis-acting element crucial for promoter activity. Deletion and site-directed mutagenesis combined with in silico transcription factor binding analysis of VSNL1 promoter identified nuclear respiratory factor (NRF)-1/α-PAL as a major player in regulating VSNL1 minimal promoter activity. The function of NRF-1 was further confirmed using dominant-negative NRF-1 overexpression and NRF-1 small interfering RNA knockdown. Electrophoretic mobility shift assay and chromatin immunoprecipitation provided evidence for direct NRF-1 binding to the VSNL1 promoter. Methylation of the NRF-1-binding site was found to be able to regulate VSNL1 promoter activity. Our results further indicated that NRF-1 could be a regulatory factor for gene expression of the other visinin-like subfamily members including HPCAL4, HPCAL1, HPCA, and NCALD.
VILIP-13 (visinin-like protein-1, gene name VSNL1) is a member of the visinin-like subfamily of neuronal Ca2+ sensor (NCS) proteins, which include neurocalcin-δ (NCALD), hippocalcin (HPCA), VILIP-1, VILIP-2 (HPCAL4), and VILIP-3 (HPCAL1) (1–3). In the nervous system, VILIP-1 was consistently found to be expressed in hippocampal neurons, cerebellar granular cells, interneurons, cortical pyramidal cells, as well as in other neurons (4, 5). The deregulation of VILIP-1 expression implicates it in certain pathological processes of the nervous system such as Alzheimer disease and schizophrenia (1).
Although VILIP-1 was originally identified in the brain, its expression can be detected in the peripheral tissues and organs such as heart, lung, liver, and testis (6), suggesting a potential role in maintaining normal tissue homeostasis in both nervous tissue and non-neural tissues. Recently, VILIP-1 was found to be expressed in insulin-expressing pancreatic β-cells and glucagon-expressing α-cells (7). Basal epidermal keratinocytes of normal murine skin express VILIP-1, whereas its expression is markedly decreased in aggressive and invasive squamous cell carcinomas (8). Enforced expression of VILIP-1 led to inhibition of cell adhesion and migration by down-regulating fibronectin receptors, suggesting a tumor suppressor function for VILIP-1 (9). Similarly, VILIP-1 expression is present in normal squamous epithelium of esophagus and bronchial mucociliary epithelium. Conversely VILIP-1 was down-regulated in squamous cell carcinomas from these two sites (10, 11). Despite the recent reports that highlight the role of VILIP-1 in physiological and pathological processes, the molecular mechanisms controlling its expression remain unknown.
Recently, we reported the cloning of a 2-kb VSNL1 promoter from the 5′-untranslated region of the gene (11). The VSNL1 2-kb promoter contains two CpG islands that are the targets of methylation modification, constituting one of the epigenetic mechanisms contributing to the silencing of VSNL1 gene expression in lung and other cancer cells. Increased acetylation of histones 3 and 4 around the VSNL1 promoter by the histone deacetylase inhibitor, trichostatin A, released the inhibition of gene expression, thus leading to the reactivation of VSNL1 expression (11). To further understand VSNL1 gene regulation and identify the transcriptional elements of the human VSNL1 promoter, we characterized the 2-kb VSNL1 promoter by deletion, mutation, and in vitro DNA binding and chromatin immunoprecipitation assays. Using dominant-negative construct transfection and siRNA knockdown techniques, we identified Nuclear respiratory factor 1 (NRF-1) as a major trans-acting element regulating VSNL1 promoter. In addition, we found that NRF-1 could be the regulatory factor for the promoters of all the other visinin-like subfamily genes.
EXPERIMENTAL PROCEDURES
Cell Culture
Two non-small cell lung cancer cell lines, NCI-H522 and NCI-H520, which express different levels of VILIP-1, and normal human bronchial epithelial cells were cultured as previously described (11).
Deletion and Mutation Constructs of VSNL1 Promoter
pGL4.10[luc2] vector and pGL4.73 luciferase reporter vectors (Promega, Madison, WI) were used for VSNL1 promoter reporter assays. VP-1998 (VP2kb) was constructed as described (11). All of the deletion constructs of VSNL1 promoter were derived from VP-1998 by PCR using the same reverse primer: 5′-GAAGATCTGCAGATTGGGAATCCCATAG. The forward primers were as follows: for VP-354, 5′-GGGGTACCAGGTTCGGTAACCACTGGG; for VP-174, 5′-GGGGTACCTGCGCCATCGCCAGGCG; for VP-100, 5′-GGGGTACCAAGAGAGGAAAGGGGAGGG; for VP-90 5′-GGGGTACCAGGGGAGGGGGTGCCTG; for vp-80, 5′-GGGGTACCGTGCCTGGAGAGGCGGAG; for VP-70, 5′-GGGGTACCAGGCGGAGGCTCGCGCG; for VP-60, 5′-GGGGTACCTCGCGCGCCTGCGCATC; and for VP-50, 5′-GGGGTACCGCGCATCCAGCTCCAGGG. For VP-100 and VP-90 containing Sp1 mutations and VP-60 containing NRF-1 mutation, the same reverse primer as above was used. The forward primers were: for VP-100Sp1m, 5′-GGGGTACCAATTTAGGAAAGGGGAGGGGGTG; for VP-90Sp1m, 5′-GGGGTACCAGTTTAGGGGGTGCCTGGAGAGG; and for VP-60NRFm, 5′-GGGGTACCTCGCGCGGCATTCCATCCAGCTCC. The PCR products were digested with KpnI and BglII and then inserted into pGL4.10 basic vector. VP-174NRFm, in which the NRF-1-binding site was mutated, was made by using QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) with the following primers: forward, 5′-GGCGGAGGCTCGCGCGGCATTCCATCCAGCTCCAGGG; and reverse, 5′-CCCTGGAGCTGGATGGAATGCCGCGCGAGCCTCCGCC. The underlined bases were mutated within NRF-1 sites. All of the constructs had been verified by DNA sequencing.
Construction of Dominant-negative (DN) NRF-1 Expression Vector
The expression plasmid of DN NRF-1 was constructed according to Chang and Huang (12). The DN NRF-1 consisted of the first N-terminal 304 residues of NRF-1, which contained the DNA-binding and nuclear localization domains and lacked the transactivation domain. DN NRF-1 cDNA was amplified from RNA extracted from normal human bronchial epithelial cells by using the SuperScript One-Step reverse transcription-PCR system (Invitrogen). The following primers were used: 5′-TTAAGCTTGCGCAGCCGCTCTGAGAAC and 5′-GACTCGAGTCACTGTGATGGTACAAGATGAGC. The underlined regions indicate the restriction enzyme sites. The cDNA fragments were digested with HindIII and XhoI and inserted into pcDNA3.1B+Myc vector (Invitrogen) to make pCDNA3.1 Myc-DN-NRF-1. The transfection of this construct resulted in the expression of Myc-tagged DN NRF-1 (therefore named Myc-DN-NRF-1).
Transfection and Dual-Luciferase Assay
For all experiments, the cells were transfected by Lipofectamine 2000 (Invitrogen) using the manufacturer's protocol. For reporter gene assay, DNA mixture containing 0.8 μg of VSNL1 promoter constructs and 8 ng of pGL4.73, a transfection efficiency control, was diluted in 50 μl of Opti-Mem I medium (Invitrogen) and mixed with 2 μl of Lipofectamine 2000 diluted in 50 μl of Opti-Mem I medium. 100 μl of DNA-Lipofectamine 2000 complexes were added to each well of 24-well plates after 20 min of incubation at room temperature, and the transfected cells were left in the incubator for 24 h before the lysis step with 150 μl of passive lysis buffer. Reporter gene activity was measured in 96-well plates according to the protocol from Dual-Luciferase reporter 1000 assay system kit (Promega) by using an Envision plate reader (PerkinElmer Life Sciences). For assessing the effect of DN NRF-1 on VSNL1 promoter activity, different amounts of pCDNA3.1 Myc-DN-NRF-1 or vector were co-transfected with 0.8 μg of pGL4.10 VP-174 and 8 ng of pGL4.73, and luciferase activity was measured in 1 or 2 days.
For siRNA transfections, NRF-1 siRNA (SC-38105) and scrambled control siRNA (SC-37007) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Different amounts of NRF-1 siRNA were tested for knocking down NRF-1 protein, and 50 pmol was finally used in the reporter gene assay with 0.8 μg of VP-174 and 8 ng of pGL4.73 in 24-well plate. Luciferase activity was measured in 3 days. For immunoblot analysis of VILIP-1 protein after NRF-1 knockdown, scrambled (D-001810-10-05) and NRF-1 (L-017924-00-0005) siRNAs from Thermo SCIENTIFIC were also used. All of the data were obtained from at least three individual experiments.
Electrophoretic Mobility Shift Assays (EMSA) and Chromatin Immunoprecipitation (ChIP)
Nuclear proteins were extracted from cells using NE-PER nuclear and cytoplasmic extraction reagents (Pierce) according to the protocol provided by the company. The following NRF-1 oligonucleotides were used: forward, 5′-TCGCGCGCCTGCGCATCCAG, and reverse, 5′-GCTGGATGCGCAGGCGCGCG; for methylated NRF-1 M1: forward, 5′ TCGCMetGCMetGCCTGCMetGCATCCAG, and reverse, 5′-GCTGGATGCMetGCAGGCMetGCMetGCG; and for methylated NRF-1 M2: forward, 5′-TCGCGCMetGCCTGCMetGCATCCAG, and reverse, 5′-GCTGGATGCMetGCAGGCMetGCGCG. CMet represents the methylated cytosine. The equal volumes of the forward and reverse oligonucleotides with the same molar concentration were mixed and boiled for 10 min and were annealed at room temperature overnight. The probe labeling and EMSA were performed using a gel shift assay system (Promega). Briefly, the annealed double-stranded oligonucleotides were labeled using T4 polynucleotide kinase before purification with Microspin G-25 columns (GE Healthcare, Buckinghamshire, UK). The DNA binding was conducted at 4 °C for 30 min in a mixture containing 5 μg of nuclear extract, 1× gel shift binding buffer, and 1 μl of 32P-labeled probe. In the competition experiment, 60× unlabeled double-stranded oligonucleotides were used as competitors and incubated with nuclear extract and binding buffer for 30 min before the addition of 32P-labeled probe. The reactions were further incubated at 4 °C for 30 min. In supershift assays, the antibodies were also incubated with the reaction mixture at 4 °C for 30 min before the addition of the 32P-labeled probe. Anti-NRF-1 antibodies were kindly provided by Dr. Claude Piantadosi (13) and Dr. Kimitoshi Kohno (14). The control sc2025 mouse IgG was from Santa Cruz Biotechnology. Finally the reaction mixtures were analyzed on a 6% DNA retardation gel (Invitrogen) at 10 v/cm on ice for 1.5 h. The gel was dried and analyzed by autoradiography.
ChIP was performed as before (11) using a chromatin immunoprecipitation assay kit (Upstate Biotechnology, Lake Placid, NY) following the manufacturer's protocol. Briefly, the proteins were cross-linked to DNA by incubating 1 × 106 cells with 1% formaldehyde for 2.5 or 10 min at 37 °C. The cell pellets were resuspended in 200 μl of SDS lysis buffer followed by DNA sonication for a total of 16 times (each time for 20 s at 30% of maximal power) by using the Ultrasonic Processor (Cole Parmer, Vernon Hills, IL). Immunocomplexes were captured from the rest of the lysates with 10 μg of antibody. After the cross-linking was reversed by heating the sample at 65 °C for 4 h, DNA was then extracted with phenol/chloroform and precipitated with ethanol. PCR was performed by using 1% of immunoprecipitated material and the same primers as used before (11). The following PCR program was used: 95 °C for 5 min followed by 35–40 cycles of 95 °C for 35 s, 54 °C for 45 s, 72 °C for 40 s, and finally 72 °C for 10 min. Rabbit anti-NRF-1 serum was kindly provided by D. Reines (15). Antibodies from Santa Cruz Biotechnology include: normal rabbit IgG (sc-2027), MBD4 (sc-10753), HDAC1 (sc-7872 X), HDAC2 (sc-7899 X), and HDAC3 (sc-11417 X). Antibodies against four MBP family proteins were from Abcam, including MBD1 (ab3753), MBD2a (ab3754), MBD3 (ab3755), and MeCP2 (ab3752). Anti-human α1-antitrypsin rabbit serum (A0409) was from Sigma.
Methylation of VSNL1 Promoter
The promoter-specific methylation of VSNL1 was performed according to a published protocol (11). In this case we used HhaI DNA methyltransferase (New England Biolabs, Ipswich, MA) in the methylation-modifying treatment. VP-100 was used because it contains the NRF-1 site, which is the only target site within VP-100 for HhaI DNA methyltransferase. The HhaI methylation of the whole plasmids pGL4.10 and pGL4.10VP-100 was done as follows: 20 μg of plasmids was treated with HhaI DNA methyltransferase in the presence or absence of S-adenosylmethionine and gel-purified by using a QIAquick gel extraction kit (Qiagen). The complete methylation of VP-100 or plasmids was confirmed by digestion with HhaI restriction enzymes (New England Biolabs). The modified plasmids were co-transfected with pGL4.73 as described above.
Western Blot Analysis
Immunoblotting analysis of protein was performed as described (11). The expression of DN NRF-1 was detected by using 25 μg of nuclear proteins and anti-Myc antibody (R950-25; Invitrogen). NRF-1 anti-serum (15) was kindly provided by Dr. D. Reines (Emory University, Atlanta, GA) and used to detect NRF-1 protein.
Construction of the Promoter Fragments of NCALD, HPCA, HPCAL4, and HPCAL1
The fragments containing the potential NRF-1-binding site were amplified by PCR using the following primers. For NCALD, the forward primer is 5′-GGGGTACCACCATCTGGAATGGGAGTTG; and the reverse primer is 5′-GAAGATCTAGTATGCTGTGGCACAAACG. For HPCA, the forward primer is 5′-GGGGTACCTGAGGGCGTCCCCTTCTC; and the reverse primer is 5′-GAAGATCTGACTGCGCAGGGAAGGCG. For HPCAL4, the forward primer is 5′-GGGGTACCGCAGAGATGTGGACTGCTG; and the reverse primer is 5′-GAAGATCTCCGGTGGGTTTGTTGC. For HPCAL2, the forward primer is 5′-GGGGTACCCTGGCCCCTCCCGGGC; and the reverse primer is 5′- GAAGATCTGCAAAGAGCCGGATCGCAG. The fragments were subsequently subcloned to pGL4.10[luc2] vector.
Bioinformatics and Statistics
Transcription factor binding sites were searched by using the following programs: TESS (Current Protocols in Bioinformatics), AliBaBa2.1 (Biostatics and Bioinformatics Facility, Fox Chase Cancer Center), and MatInspector (Genomatix). For comparing the relative activity of different reporter constructs, unpaired Student's t test for paired comparisons was performed, and a p value <0.05 was considered as significant.
RESULTS
Identification of a Core Promoter Essential for Transcription of VSNL1
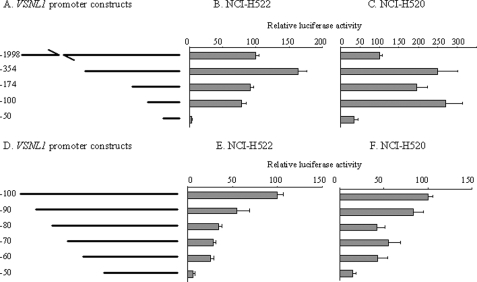
In our previous study, we had cloned a 2-kb fragment of VSNL1 promoter (VP-1998) from normal human bronchial epithelial cells and showed that its regulation by epigenetic mechanisms included promoter hypermethylation and histone acetylation (11). To define the boundaries of the minimal promoter and to identify cis elements that govern the transcriptional activity of VSNL1, we prepared a series of truncation constructs (Fig. 1A) and tested them in a transient luciferase reporter system. As shown in Fig. 1 (B and C), VP-354 was observed to have stronger transcriptional activity than VP-1998 in two non-small cell lung cancer cell lines (NCI-H522 and NCI-H520), indicating that putative negative cis-acting regulatory elements are located between the −1998 and −354. A 5′-deletion of 180 bp (VP-174) reduced promoter activity to the level comparable with that of VP-1998 in NCI-H522 cells. However, in the other non-small cell lung cancer cell line (NCI-H520), the reduction of activity was not as remarkable as in NCI-H522 cells. Further deletion of 74 bp from VP-174 caused a 14% decrease in activity when using NCI-H522 cells. In contrast, a 37% increase in activity was noted with NCI-H520 cells. VP-50 containing only the last 50 bp displayed the lowest luciferase activity in both cell lines. Its activity was 4 and 35% of VP-1998 in NCI-H522 and NCI-H520 cells, respectively, strongly indicating that the region from VP-100 to VP-50 is critical to VSNL1 promoter activity.
FIGURE 1.
Identification of the VSNL1 core promoter. A, schematic representation of VP-1998 deletion constructs. pGL4.10VP-354, -174, -100, and -50 were constructed as described under “Experimental Procedures.” B and C, VP-1998 deletion constructs were co-transfected with pGL4.73 into NCI-H522 and NCI-H520 cell lines, respectively. Luciferase activity was expressed as the relative activity relative to VP-1998. Each value represents the average of triplicate wells from at least three independent transfections in this and the following experiments. D, schematic representation of VP-100 deletion constructs. E and F, VP-100 deletion constructs were transfected into NCI-H522 and NCI-H520, respectively, and luciferase activity was calculated as in B and C.
Identification of cis-Acting Elements in the VSNL1 Core Promoter
To further examine which part within the truncated 50 bp could sustain transcriptional activity, we designed 10-bp stepwise deletion constructs (Fig. 1D) and tested them in the same assay system. The stepwise 10-bp deletion caused a gradual loss of promoter activity, and VP-60 still maintained 26 and 43% of VP-100 activity in NCI-H522 and NCI-H520, respectively (Fig. 1, E and F). Nevertheless, removal of 10 bp from VP-60 dramatically decreased the activity to 6 and 14% of VP-100 in NCI-H522 and NCI-H520 cells, respectively.
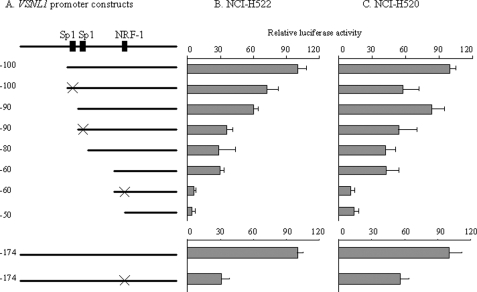
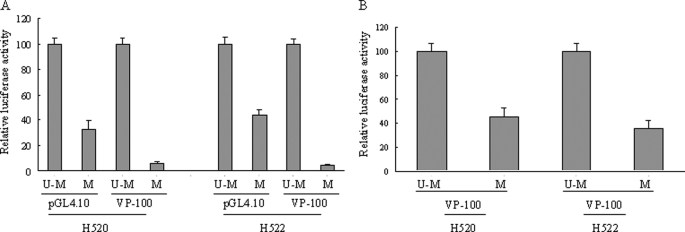
We searched the sequence of VP-100 for homology to known regulatory elements by using the TESS, AliBaBa2.1, and MatInspector bioinformatics programs. There were a number of putative binding sites for transcriptional factors in this region (Fig. 2), among which Sp1 and NRF-1 were frequently identified. Combining this in silico analysis with the result from the 10-bp stepwise deletion experiment, it could be concluded that the first Sp1 site was located between VP-100 and VP-90 and the second Sp1 site was located between VP-90 and VP-80 and that a NRF-1 site spanning between VP-60 and VP-40 could have regulatory effect on VSNL1 promoter activity. To confirm these findings, we introduced point mutations to the sites and tested the mutant constructs in both cell lines (Fig. 3A). Comparable with the 10-bp stepwise deletion results, mutation of two Sp1 sites demonstrated that these sites played a positive regulatory role in VSNL1 transcription activity (Fig. 3, B and C). The putative NRF-1 site in the VSNL1 promoter is an 11-base tandem repeat sequence, GCGCCTGCGCA. When the core CCTGCG was replaced by GCATTC, an 80 and 74% drop in promoter activity was observed as compared with intact VP-60 in NCI-H522 and NCI-H520 cells, respectively. The effect of mutation of the NRF-1-binding site was further verified in a longer VSNL1 promoter setting (VP-174). This mutation caused a 69% decrease of VP-174 activity in NCI-H522 cells and a 44% decrease in NCI-H520 cells. These results suggested that the NRF-1-binding sequence is a very important regulatory element in the VSNL1 promoter.
FIGURE 2.
Alignment of VP-100 sequences from human, mouse, and rat. The transcription factor binding sites and Exon 1 of VSNL1 gene are underlined and labeled.
FIGURE 3.
Deletion and mutation analyses of NRF-1 and Sp1 sites within the VSNL1 gene promoter. A, schematic representation of the VSNL1 promoter and mutant constructs. The filled boxes represent Sp1- and NRF-1-binding sites, respectively. The × symbols on VP-100 and VP-90 indicate where the Sp1 sites were mutated in VP-100 and VP-90. The × symbols on VP-60 and VP-174 indicate where the NRF-1 sites were mutated in VP-60 and VP-174, respectively. B and C, VSNL1 wild type and mutant plasmids were transfected into NCI-H522 and NCI-H520 cells. The luciferase activity was normalized to VP-100 (upper panel) and VP-174 (lower panel). Note that the mutation of NRF-1 site resulted in a pronounced decrease in promoter activity (∼80%) relative to VP-60.
NRF-1 Is a Transcription Factor Regulating VSNL1 Promoter Activity
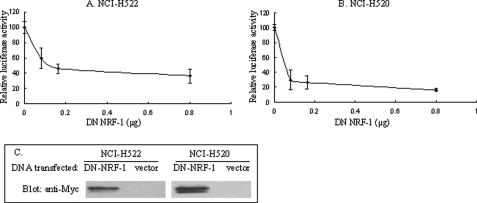
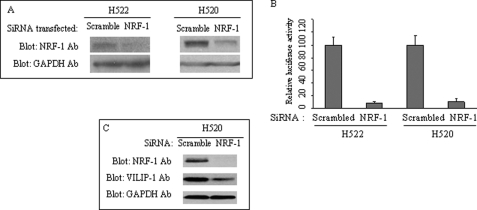
The requirement of NRF-1 for effective VSNL1 promoter activity was verified by co-transfection of the Myc-tagged DN NRF-1 expression vector with VP-174 reporter construct. Overexpression of Myc-DN-NRF-1, containing only the N-terminal DNA-binding and nuclear localization domains, should compete with the binding of endogenous NRF-1 to the NRF-1-binding site on the VSNL1 promoter and therefore interrupt the activating function of endogenous NRF-1 because it does not contain the C-terminal transactivation domain. As shown in Fig. 4(A and B), the VP-174 activity was inhibited upon the expression of Myc-DN-NRF-1, exhibiting ∼64 and 84% inhibition (0.8 μg) in NCI-H522 and NCI-H520 cells, respectively. The expression of Myc-DN-NRF-1 was confirmed by immunoblotting analysis with anti-Myc antibody (Fig. 4C). The second evidence for NRF-1 regulation of the VSNL1 promoter was obtained by NRF-1 siRNA treatment. Transfection of 50 pmol of NRF-1 siRNA resulted in a strong inhibition of NRF-1 protein synthesis as compared with that of scrambled siRNA control (Fig. 5A). When the same amount of NRF-1 siRNA was co-transfected with VP-174, an approximately 90% reduction in promoter activity in both cell lines was observed (Fig. 5B). In addition, the essential requirement of NRF-1 for VILIP-1 transcription was corroborated by the observation that NRF-1 silencing led to a significant down-regulation of VILIP-1 protein (Fig. 5C).
FIGURE 4.
Functional studies showed the critical role of NRF-1 in the regulation of the VSNL1 promoter. A and B, DN NRF-1 inhibited the VSNL1 gene promoter. Different amounts of pCDNA3.1 Myc-DN-NRF-1 were co-transfected into NCI-H522 (A) and NCI-H520 (B) cells together with pGL4.10 VP-174 reporter, luciferase activity was normalized to vector (pCDNA3.1 Myc) transfection. C, detection of overexpressed Myc-DN-NRF-1 in nuclear extracts by Western blot analysis. 0.8 μg of pCDNA3.1 Myc-DN-NRF-1 or vector was transfected, and the nuclear extracts were obtained from the transfected cells. 25 μg of nuclear proteins were loaded into each well, and Myc-DN-NRF-1 was detected with anti-Myc antibody.
FIGURE 5.
NRF-1 is required for VILIP-1 expression. A, knockdown of NRF-1 expression. NRF-1 siRNA (50 pmol) or the same amount of scrambled control siRNA was transfected, and the cells were lysed in 3 days. NRF-1 was detected with anti-NRF-1 antibody, and equal loading of total protein was demonstrated by Western blot analysis of glyceraldehyde-3-phosphate dehydrogenase. B, VSNL1 promoter activity was suppressed by down-regulation of NRF-1. NRF-1 siRNA (50 pmol) or the same amount of scrambled control siRNA was co-transfected with pGL4.10 VP-174 reporter. Luciferase activity was assayed in 3 days and normalized to scrambled control for both cell lines. C, suppression of VILIP-1 expression by NRF-1 silencing. Either scrambled or NRF-1 siRNA was transfected into NCI-H520 cells, and the expression of NRF-1, VILIP-1, and glyceraldehyde-3-phosphate dehydrogenase was analyzed 3 days post-transfection.
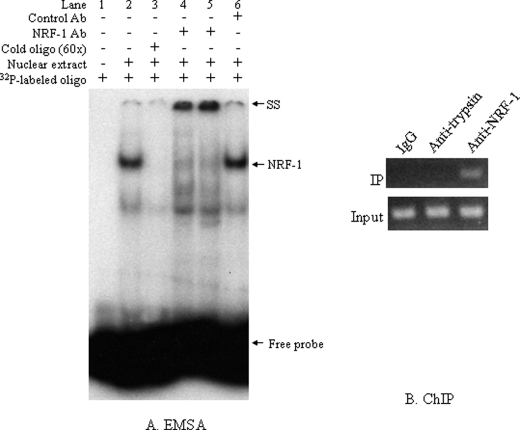
To determine whether the NRF-1 sequence is effectively recognized by cellular NRF-1 protein, we performed EMSA with the nuclear proteins extracted from NCI-H520 cells. Two major bands were observed (Fig. 6A, lane 2) that were abolished by cold (unlabeled) NRF-1 oligonucleotides (lane 3), indicating that these DNA-protein complexes contain proteins able to recognize and bind to the NRF-1 response element. The upper band represented the major NRF-1-containing DNA-protein complex because it was specifically supershifted by using two different NRF-1 antibodies from Drs. Claude Piantadosi and Kimitoshi Kohno, respectively (lanes 4 and 5), whereas the control antibody was unable to supershift it (lane 6).
FIGURE 6.
In vitro and in vivo binding of NRF-1. A, EMSA analysis demonstrated binding of NRF-1 to its binding site. 5 μg of nuclear proteins extracted from NCI-H520 cells were incubated with 32P-labeled NRF-1 oligonucleotides. Mixtures containing no nuclear extracts were used as negative controls. 60× unlabeled oligonucleotides were used as competitors and incubated with nuclear extract and binding buffer for 30 min before the addition of the 32P-labeled probe. For supershift, NRF-1 antibody from either Dr. Piantadosi's lab (lane 4) or Dr. Kohno's lab (lane 5) or control mouse IgG was incubated with the reaction mixture before the addition of the 32P-labeled probe. SS, supershift; NRF-1, NRF-1-specific band. B, ChIP assay was performed by immunoprecipitating (IP) the DNA-protein complexes with rabbit anti-NRF-1 antibody. Normal rabbit IgG and anti-human α1-antitrypsin rabbit serum were used as negative controls (upper panel). The lower panel corresponds to the input control for PCR amplification of the materials from 1% of the total sample before precipitation.
To determine the in vivo occupancy of the VSNL1 promoter by NRF-1, we performed a ChIP assay. As negative controls, a normal rabbit IgG and a nonrelated anti-α1-trypsin antiserum were included for immunoprecipitation using the same batch of cell lysates as used in NRF-1 antibody immunoprecipitation. The VSNL1 promoter was co-immunoprecipitated only with NRF-1 antibody (Fig. 6B), indicating the specific binding of VSNL1 promoter by NRF-1 in vivo.
Regulation of VSNL1 Promoter by Methylation of the NRF-1-binding Site
Methylation is one of the major epigenetic mechanisms governing VSNL1 gene expression (11). Methylation of all the CpG islands in VP-1998 with M.SssI treatment nearly abolished VSNL1 promoter activity (decreased to 11%) (11). Within VP-100, the NRF-1-binding site is the only sequence containing two GCGC sequences. To further study the epigenetic regulation of VSNL1 expression and the regulation of VSNL1 promoter activity by methylation of the NRF-1-binding site, we took advantage of HhaI DNA methyltransferase that methylates the cytosine residue in the GCGC sequence. Therefore, the methylating effect observed can be attributable exclusively to methylation of the NRF-1 site. Whole plasmids (pGL4.10 and pGL4.10VP-100) were modified by HhaI, and the complete modification was confirmed by digestion with restriction enzyme HhaI (data not shown). Methylation of the GCGC sequence of pGL4.10VP-100 almost completely inhibited VP-100 activity (leading to 94 and 95% decrease in NCI-H520 and NCI-H520 cells, respectively) (Fig. 7A). Nevertheless, methylation of the vector itself also suppressed its basal activity to 30 and 44% of the unmethylated counterparts because the vector backbone harbors GCGC sequences. To further confirm the methylation effect, a promoter-specific methylation approach was utilized, and the ligated plasmids were introduced into cells. A decrease of 55% in NCI-H520 cells and 65% in NCI-H520 cells was observed because of methylation of the NRF-1-binding site within VP-100 (Fig. 7B).
FIGURE 7.
VSNL1 promoter activity is regulated by methylation of the NRF-1-binding site. A, the effect of whole plasmid methylation: pGL4.10 VP-100 or vector was methylated (M) or unmethylated (U-M) as described under “Experimental Procedures.” The resulting plasmids were introduced into NCI-H520 and NCI-H522 cells, and the relative luciferase activity of methylated plasmids was normalized with respect to the unmethylated ones. B, the effect of promoter-specific methylation. VP-100 fragments excised from pGL4.10 VP-100 were subjected to HhaI treatment under methylating or unmethylating conditions and ligated back to pGL4.10 vector. The resulting plasmids were introduced into NCI-H520 and NCI-H522 cells, and the relative luciferase activity of the vector with methylated VP-100 (M) was normalized to that of the vector with unmethylated VP-100 (U-M).
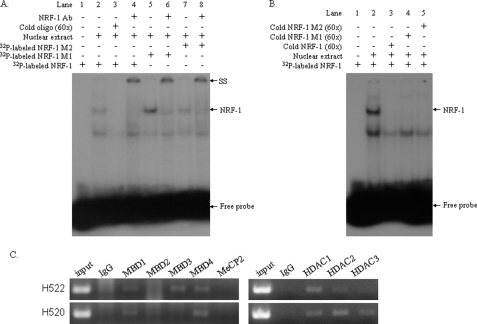
The mechanisms underlining the regulation of transcription by DNA methylation remain unclear. We first investigated whether the methylation of NRF-1-binding sites could block its binding by performing an EMSA assay. Two oligonucleotides in which two (M2) or three (M1) CpGs within the NRF-1 sites were methylated were used. When the EMSA was performed with the unmethylated NRF-1 probe, we observed two major bands (Fig. 8A, lane 2). The upper band was supershifted by NRF-1 antibody (lane 4), as in Fig. 6A. However, methylation of two or three CpGs of NRF-1-binding sites failed to abolish the formation of these two bands (lanes 5 and 7), and similarly, the upper bands represented the NRF-1-containing complex because they were supershifted by the NRF-1 antibody (lanes 6 and 8). Furthermore, the formation of the NRF-1 containing complex (Fig. 8B, lane 2) was not observed when either the unlabeled NRF-1 oligonucleotides (lane 3) or the unlabeled methylated counterparts (lanes 4 and 5) were used, indicating that the methylation of the CpG sites within the NRF-1-binding site has no effect on the binding of NRF-1 to the VSNL1 promoter. Methylation of the CpG sites within gene promoters can recruit MBP and histone deacetylases, thus leading to the alteration of chromatin structure (16, 17). We next examined the association of all five members of MBP family and class I HDACs with VSNL1 promoter by using ChIP assay. As shown in Fig. 8C, MBD1 and MBD4 were able to bind VSNL1 promoter in NCI-H522 and NCI-H520 cells, whereas MBD3 binding was detected predominantly in NCI-H522 cells. Interaction of MBD2 and MeCP2 with VSNL1 promoter was not observed in any cell line. Binding of HDAC1, HDAC2, and HDAC3 to VSNL1 promoter was found in NCI-H520 cells, nevertheless, among these three HDACs, it appears that the association of HDAC1 was the most intense in NCI-H522 cells, which do not express endogenous VILIP-1 (Fig. 8C).
FIGURE 8.
The mechanisms of the regulation of VSNL1 transcription. A, EMSA analysis of the effect of CpG methylation on binding of NRF-1 to its binding site. 5 μg of nuclear proteins extracted from NCI-H520 cells were incubated with 32P-labeled NRF-1, NRF-1 M1, or NRF-1 M2 oligonucleotides. 60× unlabeled oligonucleotides were used as competitors and incubated with nuclear extract and binding buffer for 30 min before the addition of the 32P-labeled probe. For supershift, NRF-1 antibody was incubated with the reaction mixture before the addition of the 32P-labeled probe. B, competing of NRF-1 binding by NRF-1 M1 or NRF-1 M2 oligonucleotides. Nuclear proteins extracted from NCI-H520 cells were incubated with 32P-labeled NRF-1 oligonucleotides. 60× unlabeled NRF-1, NRF-1 M1, or NRF-1 M2 oligonucleotides were used as competitors. C, the interaction of MBP and HDAC proteins with VSNL1 promoter was analyzed by ChIP assay.
The Potential Regulation of the Promoter Activity of Other Visinin-like Subfamily Members by NRF-1
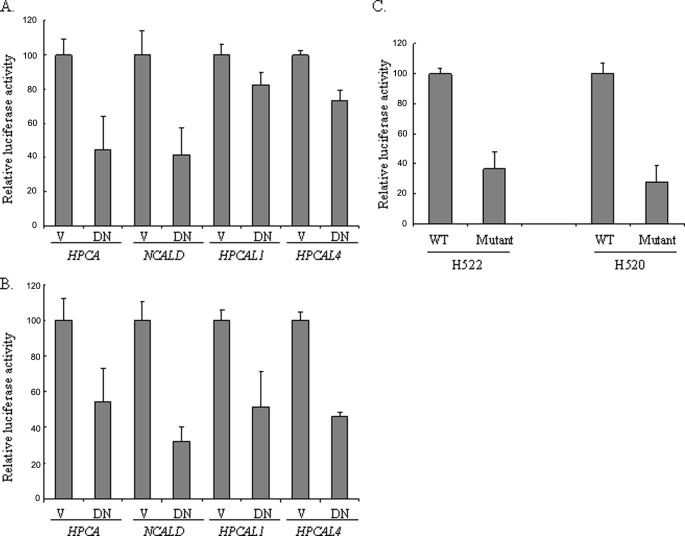
The recent data showing that neurocalcin (18), a calcium sensor in Drosophila neurons, was down-regulated in the ewg (NRF-1 homologue in Drosophila) mutant prompted us to examine whether NRF-1 could be implicated in regulating the expression of each member of the visinin-like subfamily. The promoters of the human visinin-like subfamily were poorly studied, and most of the promoter sequences were unavailable; therefore, we first searched and analyzed ∼2.5 kb upstream and downstream sequence of the first exons of HPCAL4, HPCAL1, and HPCA genes. None of the first exons from these genes is transcribed, and CpG islands were found to exist in all three promoters using the criteria formulated by Gardiner-Garden and Frommer (19), i.e. a 200-bp or greater stretch of DNA with a C/G content of >50% and an observed CpG/expected CpG ratio in excess of 0.6. In silico analysis of these fragments revealed that all of these gene promoters harbor the potential NRF-1-binding sites (Table 1). The analysis of the promoter fragment of NCALD was initially done using a Drosophila genomic DNA sequence (data not shown). A perfect potential ewg binding site was located within the second intron of Drosophila neurocalcin (CG7646). Similarly, we found a NRF-1-binding site with the consensus sequence GCGCATGCGCA within the first intron of the human NCALD gene. After we subcloned the DNA fragments containing the potential NRF-1 site, we studied the effect of DN NRF-1 on the promoter transcriptional activity. Application of DN NRF-1 significantly inhibited the promoter activity of human HPCAL4, HPCAL1, HPCA, and NCALD in both NCI-H522 (Fig. 9A) and NCI-H520 (Fig. 9B) cells, suggesting that NRF-1 could also be the trans-acting element regulating these gene promoters. Among the promoters, we selected NCALD for the study of the effect of a NRF-1 mutation on promoter transcriptional activity because the promoter of NCALD harbors a canonical NRF-1 site, and it showed the most remarkable decrease after DN NRF-1 transfection in our cell systems. A NRF-1 mutant construct in which the core GCA sequence was replaced by TTT demonstrated a decrease of ∼64 and 72% in the promoter activity in NCI-H522 cells and NCI-H520 cells, respectively (Fig. 9C), indicating the direct regulatory effect of NRF-1 on the NCALD promoter.
TABLE 1.
The in silico prediction of the potential NRF-1-binding sites of the gene promoters of the other visinin-like subfamily members compared with VSNL1
The potential binding sequences are in bold type, and the positions of the promoter fragments are numbered relative to the start of the first exon.
| Gene | Position | Sequence |
|---|---|---|
| VSNL1 | −60/−41 | TCGCGCGCCTGCGCATCCAG |
| HPCAL4 | −4/+23 | GGAAGGCGGCTCCGGCGCAGACCTTGG |
| HPCAL1 | +1283/+1310 | AGGCTCCCGCGCAAGCGTGGGGGTCCC |
| NCALD | +43298/+43316 | AAGTGCGCATGCGCAAAGC |
| HPCA | −46/−21 | GCCCCTGCCGCGCGCGCCGCAGCAGC |
FIGURE 9.
A and B, the promoter activity of HPCA, NCALD, HPCAL4, and HPCAL1 was suppressed by the overexpression of DN NRF-1 in NCI-H522 cells (A) and NCI-H520 cells (B). DN NRF-1 transfection and the luciferase assay were performed as described in the legend to Fig. 4. V, vector; DN, DN NRF-1. C, site-directed mutagenesis of the NRF-1-binding site in NCALD promoter inhibited its activity. The promoter constructs containing the wild type (WT) or the mutated (Mutant) NRF-1-binding sites were introduced into the cells, and the luciferase assay was performed.
DISCUSSION
We characterized the VSNL1 promoter by serial promoter deletion experiments and identified a 100-bp fragment that sustained most of the VSNL1 promoter activity. Further deletion of this minimal promoter revealed a cis-acting element at −60 to −50 nucleotides upstream of the first VSNL1 gene exon. Alignment of human, mouse, and rat VSNL1 promoters revealed a high homology of the core promoter among all three species (Fig. 2). By using three transcription factor searching programs, we identified one potential binding site (GCGCCTGCGC) that is a perfect match for the NRF-1-binding site (20). This putative NRF-1-binding site is well conserved in human, mouse, and rat (Fig. 2), suggesting that it plays an essential role in regulating VSNL1 promoter activity. As expected, deletion and mutation of this binding site dramatically decreased the promoter activity in human lung cancer cell lines.
NRF-1 is one of the major transcription factors involved in regulating the expression of nuclear genes essential for mitochondrial biogenesis (21, 22). NRF-1 binding is required for the expression of genes encoding subunits of all the five respiratory complexes (23). Additionally, acting on major regulators of mitochondrial transcription including Tfam and TFB (24, 25), NRF-1 coordinates the respiratory subunit expression with that of the mitochondrial transcriptional machinery. Furthermore, NRF-1 has been implicated indirectly in specific respiratory functions by regulating a number of nuclear genes such as outer membrane transporter TOMM20 (26) and a cytochrome oxidase assembly factor (27). The induction of mitochondrial biogenesis in response to environment signals such as cold exposure and exercise is coordinated by co-factors that interact with NRF-1, including the transcriptional co-activators PGC (peroxisome proliferator-activated receptor γ co-factor)-1α, PGC-1β and PRC (PGC-1-related co-activator) (28–30). These NRF-1 co-activators bind to NRF-1 and trans-activate NRF-1 target genes that are involved in mitochondrial respiration and transcription. The NRF-1-responsive element has been identified to be a GC-rich palindrome with the core sequence YGCGCAYGCGCR (21). Interestingly, the NRF-1 recognition site is one of the seven transcription factor-binding sites that are most frequently found in the proximal promoters of ubiquitous genes (31), indicating a broader spectrum of target genes for NRF-1. A recent study using CHIP-on-chip has revealed a collection of human promoters that are occupied by NRF-1 in vivo (32). Besides the genes that are expected to mediate mitochondrial biogenesis, NRF-1 target genes overlap significantly with those of E2F, suggesting a role in the regulation of DNA replication and cell cycle progression. It is plausible that deregulation of NRF-1 could contribute to cell proliferation during tumorigenesis. Interestingly, it has been noted that certain gene products related to tumor cell growth, differentiation, migration, and invasion are NRF-1-regulated targets. These genes include GalNAc-T3 (14), CAPNS1 (33), E2F6 (34), TOMM34 (35), and VHL (36). Our findings showing that NRF-1 is a key trans-acting factor regulating the transcription of the tumor suppressor VSNL1 further extends this observation. To our knowledge, this is the first characterization of human VSNL1 promoter as well as the first description of NRF-1 as a critical cis-acting element in VSNL1 promoter.
Although targeted disruption of NRF-1 gene in mouse resulted in an embryonic lethal phenotype (37), the data from deletion of NRF-1 homologues in other species implicates NRF-1 in the physiology of the nervous system. Not really finished (nrf), a NRF-1 homologue in zebra fish, is expressed ubiquitously throughout the developing retina and central nervous system and is crucial for the development of the zebra fish outer retina (38). Mutations of the NRF-1 homologue locus in Drosophila, ewg, exhibit severe neural phenotype leading to embryonic death (39). In addition to its prominent function in maintaining the respiratory chain, the extensive EWG protein expression pattern in all neurons of Drosophila suggests its potential role as a transcription factor in the regulation of neuron-specific gene expression. Indeed, recent analysis of ewg) mutant (ewgl1), in which ewg is inactivated, showed that a spectrum of genes other than mitochondrial function-related genes were either up- or down-regulated by EWG in neurons (18). VILIP-1 expression was detected in the hippocampus where VILIP-1 might influence signaling pathways, such as cAMP/cGMP pathway and the activity of neurotransmitter receptors and ion channels, which are related to learning and memory processes (40–42). Interestingly, a number of other neuronal genes, including IAP (12), GluR2 (43), and FMR-1 (15), all containing NRF-1-binding sites in their promoter regions, can also regulate the synaptic plasticity and learning and memory processes, indicating that NRF-1 might participate in these complex activities.
VILIP-1 belongs to a subfamily of NCS proteins including VILIP-1, VILIP-2, VILIP-3, hippocalcin, and neurocalcin-δ (1, 2). The finding that neurocalcin, a calcium sensor in Drosophila neurons, was down-regulated in ewg mutant suggested that VSNL1 might not be the only NRF-1-regulatable member of this subfamily (18). The in silico analysis of the promoters of the other four genes in the same family predicted the presence of NRF-1 cis-acting regulatory elements in most of the promoters of visinin-like subfamily. Blocking the endogenous NRF-1 function with a dominant-negative form led to a significant decrease in the promoter transcriptional activity of HPCA, NCALD, HPCAL4, and HPCAL1, indicating that all of these promoters can be directly or indirectly regulated by NRF-1. Mutation of the NRF-1-binding site within the NCALD promoter suppressed its activity, further substantiating the importance of NRF-1 site as a cis-acting regulatory element in the gene promoters of this NCS subfamily. The potential regulation by NRF-1 could further extend the function of NRF-1 to a spectrum of physiological and pathological processes that are mediated by this NCS subfamily. Interestingly, hippocalcin, another member of the same subfamily highly expressed in the hippocampal pyramidal cells, has also been implicated in hippocampal long term depression (44) and in memory processes because the hippocalcin-deficient mice display defects in spatial and associative memory (45).
VSNL1 gene expression can be regulated epigenetically in human lung cancer (11). Global methylation of CpG in VP-1998 by M.SssI abolished VSNL1 promoter activity, probably through methylation of a group of CG-containing transcription factor binding sites including Sp1, NRF-1 (46), AP2 (47), and NF-κB (48). In this study, we have identified VP-100 as the VSNL1 minimal promoter and NRF-1 site as a key cis-acting regulatory element. The fact that NRF-1 is the only GCGC-containing sequence present within the core promoter allowed us to pinpoint the effect of NRF-1 site methylation on VSNL1 transcription. Using both whole plasmid and promoter-specific methylation approaches, we found that methylation decreased VSNL1 gene expression by 55–65%. Interestingly, the extent of the observed promoter activity suppression is comparable with those found in studies of FMR-1 and hTfam promoters using different methylation methods (49, 50). Comparing the reduced activity induced by NRF-1 site methylation (Fig. 7) with that produced by NRF-1 site mutation (Fig. 3), it appears that both led to similar levels of VSNL1 promoter inhibition, suggesting that methylation of the NRF-1 site in the VSNL1 promoter could block NRF-1 binding. However, failure of the methylated oligonucleotides to block the NRF-1 binding in EMSA (Fig. 8, A and B) excluded the possibility that direct impeding of transcription factor binding by methylation could account for the VSNL1 promoter repression. MBP family proteins can be recruited to hypermethylated promoter and act as important “translators” between DNA methylation and histone-modifier genes, therefore establishing a transcriptionally inactive chromatin environment (16, 51). Except MBD3, all the other MBP proteins, MBD1, MBD2, MBD4, and MeCP2, were able to recognize and bind methylated DNA (52). MBD3 is a bona fide component of a large macromolecular complex, Mi-2·NuRD (nucleosome remodeling and deacetylase), that couples a chromatin-remodeling ATPase and histone deacetylation (53). In addition to MBD3, the Mi-2·NuRD complex consists of other multiple subunits, including at least two Mi-2 proteins, HDAC1 and/or HDAC2, Rbbp4 (p48), Rbbp7 (p46), and MTA proteins, the activity of which implicates it in repressing gene transcription. Our analysis of the association between MBP, class I HDACs, and VSNL1 promoter by using chromatin immunoprecipitation suggests that the repression of VSNL1 promote activity might be mediated through the recruitment of Mi-2·NuRD complex containing MBD3 and HDAC1 and HDAC2 to the VSNL1 promoter.
In summary, we have characterized the human VSNL1 gene promoter and identified the NRF-1-binding site as a major regulatory site for VSNL1 gene transcription. The functional importance of NRF-1 was confirmed by demonstrating VSNL1 promoter repression by DN NRF-1 and NRF-1 siRNA interference and the consequent VILIP-1 protein down-regulation by NRF-1 silencing. EMSA and ChIP assay further demonstrated the direct binding of NRF-1 protein to its binding site. Furthermore, in silico analysis suggested that NRF-1 might be able to regulate the entire visinin-like subfamily of NCS.
Acknowledgments
We appreciate the critical review and comments from Dr. Daniel Reines (Department of Biochemistry, Emory University School of Medicine, Atlanta, Georgia) and Dr. Maureen E. Murphy (Division of Medical Science, Fox Chase Cancer Center) and the help for promoter analysis from Dr. Yan Zhou (Biostatistics and Bioinformatics Facility, Fox Chase Cancer Center). Special thanks to Dr. Daniel Reines, Dr. Kimitoshi Kohno (Department of Molecular Biology, School of Medicine, University of Occupational and Environmental Health Japan), and Dr. Claude Piantadosi (Department of Medicine, Duke University School of Medicine) for providing NRF-1-specific antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grants CA107257 and CA06927. This work was also supported by an appropriation from the Commonwealth of Pennsylvania and by a grant from the Pennsylvania Department of Health.
- VILIP-1
- visinin-like protein-1
- HDAC
- histone deacetylase
- EWG
- erect wing gene
- NCS
- neuronal Ca2+ sensor
- ChIP
- chromatin immunoprecipitation, DN dominant-negative
- NRF-1
- nuclear respiratory factor-1
- EMSA
- electrophoretic mobility shift assay
- VP
- VSNL1 promoter
- siRNA
- small interfering RNA
- MBD
- methyl-CPG-binding domain
- MBP
- methyl-binding proteins.
REFERENCES
- 1.Braunewell K. H. (2005) Trends Pharmacol. Sci. 26, 345–351 [DOI] [PubMed] [Google Scholar]
- 2.Burgoyne R. D. (2007) Nat. Rev. 8, 182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braunewell K. H., Klein-Szanto A. J. (2009) Cell Tissue Res. 335, 301–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braunewell K. H., Gundelfinger E. D. (1999) Cell Tissue Res. 295, 1–12 [DOI] [PubMed] [Google Scholar]
- 5.Bernstein H. G., Baumann B., Danos P., Diekmann S., Bogerts B., Gundelfinger E. D., Braunewell K. H. (1999) J. Neurocytol. 28, 655–662 [DOI] [PubMed] [Google Scholar]
- 6.Gierke P., Zhao C., Brackmann M., Linke B., Heinemann U., Braunewell K. H. (2004) Biochem. Biophys. Res. Commun. 323, 38–43 [DOI] [PubMed] [Google Scholar]
- 7.Dai F. F., Zhang Y., Kang Y., Wang Q., Gaisano H. Y., Braunewell K. H., Chan C. B., Wheeler M. B. (2006) J. Biol. Chem. 281, 21942–21953 [DOI] [PubMed] [Google Scholar]
- 8.Mahloogi H., González-Guerrico A. M., Lopez De Cicco R., Bassi D. E., Goodrow T., Braunewell K. H., Klein-Szanto A. J. (2003) Cancer Res. 63, 4997–5004 [PubMed] [Google Scholar]
- 9.Gonzalez Guerrico A. M., Jaffer Z. M., Page R. E., Braunewell K. H., Chernoff J., Klein-Szanto A. J. (2005) Oncogene 24, 2307–2316 [DOI] [PubMed] [Google Scholar]
- 10.Wickborn C., Klein-Szanto A. J., Schlag P. M., Braunewell K. H. (2006) Mol. Carcinog. 45, 572–581 [DOI] [PubMed] [Google Scholar]
- 11.Fu J., Fong K., Bellacosa A., Ross E., Apostolou S., Bassi D. E., Jin F., Zhang J., Cairns P., Ibañez de Caceres I., Braunewell K. H., Klein-Szanto A. J. (2008) PLoS ONE 3, e1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang W. T., Huang A. M. (2004) J. Biol. Chem. 279, 14542–14550 [DOI] [PubMed] [Google Scholar]
- 13.Piantadosi C. A., Suliman H. B. (2008) J. Biol. Chem. 283, 10967–10977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izumi H., Ohta R., Nagatani G., Ise T., Nakayama Y., Nomoto M., Kohno K. (2003) Biochem. J. 373, 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith K. T., Coffee B., Reines D. (2004) Hum. Mol. Genet. 13, 1611–1621 [DOI] [PubMed] [Google Scholar]
- 16.Miranda T. B., Jones P. A. (2007) J. Cell. Physiol. 213, 384–390 [DOI] [PubMed] [Google Scholar]
- 17.Ting A. H., McGarvey K. M., Baylin S. B. (2006) Genes Dev. 20, 3215–3231 [DOI] [PubMed] [Google Scholar]
- 18.Haussmann I. U., White K., Soller M. (2008) Genome Biol. 9, R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardiner-Garden M., Frommer M. (1987) J. Mol. Biol. 196, 261–282 [DOI] [PubMed] [Google Scholar]
- 20.Piantadosi C. A., Suliman H. B. (2006) J. Biol. Chem. 281, 324–333 [DOI] [PubMed] [Google Scholar]
- 21.Scarpulla R. C. (2008) Physiol. Rev. 88, 611–638 [DOI] [PubMed] [Google Scholar]
- 22.Scarpulla R. C. (2006) J. Cell. Biochem. 97, 673–683 [DOI] [PubMed] [Google Scholar]
- 23.Kelly D. P., Scarpulla R. C. (2004) Genes Dev. 18, 357–368 [DOI] [PubMed] [Google Scholar]
- 24.Virbasius J. V., Scarpulla R. C. (1994) Proc. Natl. Acad. Sci. U. S. A. 91, 1309–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gleyzer N., Vercauteren K., Scarpulla R. C. (2005) Mol. Cell. Biol. 25, 1354–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blesa J. R., Prieto-Ruiz J. A., Hernández J. M., Hernández-Yago J. (2007) Gene 391, 198–208 [DOI] [PubMed] [Google Scholar]
- 27.Takahashi Y., Kako K., Arai H., Ohishi T., Inada Y., Takehara A., Fukamizu A., Munekata E. (2002) Biochim. Biophys. Acta 1574, 359–364 [DOI] [PubMed] [Google Scholar]
- 28.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. (1999) Cell 98, 115–124 [DOI] [PubMed] [Google Scholar]
- 29.Lin J., Puigserver P., Donovan J., Tarr P., Spiegelman B. M. (2002) J. Biol. Chem. 277, 1645–1648 [DOI] [PubMed] [Google Scholar]
- 30.Andersson U., Scarpulla R. C. (2001) Mol. Cell. Biol. 21, 3738–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.FitzGerald P. C., Shlyakhtenko A., Mir A. A., Vinson C. (2004) Genome Res. 14, 1562–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cam H., Balciunaite E., Blais A., Spektor A., Scarpulla R. C., Young R., Kluger Y., Dynlacht B. D. (2004) Mol. Cell 16, 399–411 [DOI] [PubMed] [Google Scholar]
- 33.Asangani I. A., Rasheed S. A., Leupold J. H., Post S., Allgayer H. (2008) Gene 410, 197–206 [DOI] [PubMed] [Google Scholar]
- 34.Kherrouche Z., De Launoit Y., Monte D. (2004) Biochem. J. 383, 529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blesa J. R., Prieto-Ruiz J. A., Abraham B. A., Harrison B. L., Hegde A. A., Hernández-Yago J. (2008) Biochem. Cell Biol. 86, 46–56 [DOI] [PubMed] [Google Scholar]
- 36.Kuzmin I., Duh F. M., Latif F., Geil L., Zbar B., Lerman M. I. (1995) Oncogene 10, 2185–2194 [PubMed] [Google Scholar]
- 37.Huo L., Scarpulla R. C. (2001) Mol. Cell. Biol. 21, 644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becker T. S., Burgess S. M., Amsterdam A. H., Allende M. L., Hopkins N. (1998) Development 125, 4369–4378 [DOI] [PubMed] [Google Scholar]
- 39.DeSimone S. M., White K. (1993) Mol. Cell. Biol. 13, 3641–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spilker C., Gundelfinger E. D., Braunewell K. H. (2002) Biochim. Biophys. Acta 1600, 118–127 [DOI] [PubMed] [Google Scholar]
- 41.Braunewell K. H., Brackmann M., Manahan-Vaughan D. (2003) Neuropharmacology 44, 707–715 [DOI] [PubMed] [Google Scholar]
- 42.Brackmann M., Zhao C., Kuhl D., Manahan-Vaughan D., Braunewell K. H. (2004) Biochem. Biophys. Res. Commun. 322, 1073–1079 [DOI] [PubMed] [Google Scholar]
- 43.Myers S. J., Peters J., Huang Y., Comer M. B., Barthel F., Dingledine R. (1998) J. Neurosci. 18, 6723–6739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmer C. L., Lim W., Hastie P. G., Toward M., Korolchuk V. I., Burbidge S. A., Banting G., Collingridge G. L., Isaac J. T., Henley J. M. (2005) Neuron 47, 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobayashi M., Masaki T., Hori K., Masuo Y., Miyamoto M., Tsubokawa H., Noguchi H., Nomura M., Takamatsu K. (2005) Neuroscience 133, 471–484 [DOI] [PubMed] [Google Scholar]
- 46.Rozenberg J. M., Shlyakhtenko A., Glass K., Rishi V., Myakishev M. V., FitzGerald P. C., Vinson C. (2008) BMC Genomics 9, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Comb M., Goodman H. M. (1990) Nucleic Acids Res. 18, 3975–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Z., Kone B. C. (2004) J. Biol. Chem. 279, 46954–46961 [DOI] [PubMed] [Google Scholar]
- 49.Kumari D., Usdin K. (2001) J. Biol. Chem. 276, 4357–4364 [DOI] [PubMed] [Google Scholar]
- 50.Choi Y. S., Kim S., Kyu Lee H., Lee K. U., Pak Y. K. (2004) Biochem. Biophys. Res. Commun. 314, 118–122 [DOI] [PubMed] [Google Scholar]
- 51.Esteller M. (2007) Hum. Mol. Genet. 16, R50–R59 [DOI] [PubMed] [Google Scholar]
- 52.Fatemi M., Wade P. A. (2006) J. Cell Sci. 119, 3033–3037 [DOI] [PubMed] [Google Scholar]
- 53.Denslow S. A., Wade P. A. (2007) Oncogene 26, 5433–5438 [DOI] [PubMed] [Google Scholar]