Abstract
The function of the mitochondrial phospholipid cardiolipin (CL) is thought to depend on its acyl chain composition. The present study aims at a better understanding of the way the CL species profile is established in Saccharomyces cerevisiae by using depletion of the acyl-CoA-binding protein Acb1p as a tool to modulate the cellular acyl chain content. Despite the presence of an intact CL remodeling system, acyl chains shorter than 16 carbon atoms (C16) were found to accumulate in CL in cells lacking Acb1p. Further experiments revealed that Taz1p, a key CL remodeling enzyme, was not responsible for the shortening of CL in the absence of Acb1p. This left de novo CL synthesis as the only possible source of acyl chains shorter than C16 in CL. Experiments in which the substrate specificity of the yeast cardiolipin synthase Crd1p and the acyl chain composition of individual short CL species were investigated, indicated that both CL precursors (i.e. phosphatidylglycerol and CDP-diacylglycerol) contribute to comparable extents to the shorter acyl chains in CL in acb1 mutants. Based on the findings, we conclude that the fatty acid composition of mature CL in yeast is governed by the substrate specificity of the CL-specific lipase Cld1p and the fatty acid composition of the Taz1p substrates.
Cardiolipin (CL)5 is a unique anionic glycerophospholipid with dimeric structure containing four acyl chains, which is almost exclusively localized to the mitochondrial inner membrane in eukaryotic cells (1, 2). CL has been shown to co-isolate with, and to be required for optimal activity of a number of enzymes in the respiratory chain (3–5), and it has been implicated in the stability and assembly of protein (super)complexes (6–8). In the presence of divalent cations and dependent on the acyl chain composition, CL has a propensity for membrane negative curvature, a property that may be important in, e.g. membrane fusion and fission (9, 10). In addition, CL is thought to serve as a proton trap in oxidative phosphorylation (11). In recent years, CL has also been implicated in apoptosis (12, 13).
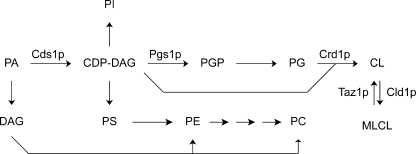
CL is synthesized in the inner mitochondrial membrane by condensation of PG and CDP-DAG, catalyzed by the cardiolipin synthase Crd1p (see Fig. 1; reviewed in Ref. 4). Compared with the other phospholipid classes, CL is enriched in unsaturated acyl chains, and the molecular species of CL possess a high degree of molecular symmetry (14). The CL-specific acyl chain pattern originates from substrate preferences during biosynthesis and subsequent remodeling by acyl chain exchange (15). The finding of an aberrant CL species profile in patients suffering from Barth syndrome, which results from mutations in the tafazzin gene (16), revealed the importance of CL remodeling, and set the stage for the identification of tafazzin as the acyltransferase involved (17, 18). The Drosophila homologue of tafazzin was shown to be a CoA-independent phospholipid transacylase with substrate preference for CL and PC (19).
FIGURE 1.
The cardiolipin biosynthetic pathway in the context of phospholipid biosynthesis in yeast. The enzymes of the CL biosynthetic pathway identified at the gene level are indicated: Cds1p, CDP-DAG synthase; Pgs1p, phosphatidylglycerolphosphate synthase; Crd1p, CL synthase; Taz1p, Tafazzin; Cld1p, CL-specific deacylase.
The biosynthesis and remodeling of CL have been extensively studied in the yeast Saccharomyces cerevisiae. After synthesis by Crd1p, CL is subject to deacylation and reacylation, which involves the yeast homologue of tafazzin encoded by the TAZ1 gene. The yeast taz1 mutant has defects similar to those found in Barth syndrome, including reduced CL content, an aberrant CL species profile, and an accumulation of monolyso-CL (20). The bioenergetic coupling of isolated mitochondria from a taz1 mutant is compromised (21), which may be accounted for by the impaired assembly of the III2IV2 supercomplex (22). Recently, the CL-specific phospholipase Cld1p was identified, which functions upstream of Taz1p (23).
Because the acyl chain composition of CL is important for its function, we investigated how the molecular species profile of CL is attained by using depletion of the 10-kDa cytosolic acyl-CoA-binding protein Acb1p as a tool to modify the cellular acyl chain content. Deletion of the ACB1 gene increases the cellular levels of C14 and C16 fatty acids at the expense of C18, without having adverse effects on cell growth or on the rate of glycerophospholipid synthesis (24–26). The changes in fatty acid composition are reflected to varying extents in the molecular species profile of phospholipids in Acb1p-depleted cells as determined by electrospray ionization-mass spectrometry (ESI-MS) (27, 28). We first determined by mass spectrometry that in the absence of Acb1p acyl chains shorter than C16 accumulate in CL as in the other phospholipid classes despite the Cld1p-Taz1p remodeling system. Using appropriate mutants and analysis by mass spectrometry, we investigated two possible origins of the shorter acyl chains in CL: (i) remodeling by Taz1p and (ii) de novo synthesis of CL from PG and CDP-DAG.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, Media, and Culture Conditions
The yeast strains listed in Table 1 were maintained on YPD agar plates (1% yeast extract, 2% bactopeptone, and 2% glucose). Strains harboring the pYPGK18 or the p416 plasmids (Table 1) were obtained according to the “rapid transformation protocol” (29), and maintained on agar plates containing synthetic glucose medium (SD) lacking leucine or uracil, respectively. Synthetic medium contained per liter: 6.7 g of yeast nitrogen base without amino acids (Difco), 20 mg of adenine, 20 mg of arginine, 20 mg of histidine, 60 mg of leucine, 230 mg of lysine, 20 mg of methionine, 300 mg of threonine, 20 mg of tryptophan, 40 mg of uracil, and either 30 g of glucose (SD) or 22 ml of 90% (v/v) lactic acid and 1 g of glucose (synthetic lactate medium (SL), adjusted to pH 5.5 using KOH). Strains were cultured aerobically in SL medium at 30 °C to ensure optimal mitochondrial development. Growth was monitored by measuring the OD at 600 nm, using a Unicam Helios Epsilon spectrophotometer. Cells were harvested at late-log phase (A600 between 0.8 and 1.0) by centrifugation (3 min, 3000 × g), washed with water, and freeze-dried.
TABLE 1.
Yeast strains and plasmids used in this study
| Strain/plasmid | Genotype/characteristics | Source/Ref. |
|---|---|---|
| BY4742 (wild type) | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Euroscarf |
| BY4741 acb1 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 acb1 ::kanMX4 | Euroscarf |
| BY4742 crd1 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 crd1 ::kanMX4 | Euroscarf |
| BY4741 taz1 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 taz1 ::kanMX4 | Euroscarf |
| BY4742 crd1acb1 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 crd1 ::kanMX4 acb1 ::HIS5 (S. pombe) | This study |
| BY4741 taz1acb1 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 taz1 ::kanMX4 acb1 ::HIS5 (S. pombe) | This study |
| p416CYC | Low copy (CEN/ARS) shuttle vector containing the CYC1 promoter and URA3 | N. Faergeman/59 |
| p416CYC-ACB1 | P416CYC derivative vector containing ACB1 gene | N. Faergeman |
| pYPGK18 | Multicopy (2μ) shuttle vector containing the PGK1 promoter and LEU2 | 37 |
| pYPGK18-TAZ1 | pYPGK18 derivative vector containing TAZ1 gene | 37 |
To analyze growth phenotypes of yeast deletion strains, cells grown in YPD to mid-log phase (A600 between 0.6 and 0.8) were harvested by centrifugation, washed twice with sterile water, and resuspended in sterile water to an A600 value of 1, followed by serial dilution to A600 values of 10−1, 10−2, 10−3, 10−4, and 10−5. From each dilution, 6 μl was spotted onto agar plates, containing 1% (w/v) yeast extract, 2% (w/v) peptone, and either 2% (w/v) glucose (YPD) or 3% (v/v) glycerol (YPG). The plates were incubated at 30 or 37 °C for 2–6 days.
Deletion of ACB1
The ACB1 gene was deleted via homologous recombination with a PCR construct containing the Schizosaccharomyces pombe his5+ open reading frame flanked by coliphage loxP sites (kindly provided by Dr. J. C. Holthuis, Utrecht University) that were created using primers 5′-GACTAAAACTCTAAAATTAGTTAAACTAGTGTTTTCAGCAAAATGAGGAGGGCTTTTGTAGAAAG-3′ and 5′-CTAGGCCAAAACTCCTTACATGGAGCTAGTATACCCCTTTTTTACAACACTCCCTTCGTGCTTG-3′, with the underlined sequences corresponding to nucleotides −42 to −1 upstream and to nucleotides 1 to 41 downstream (reverse complementary) of the ACB1 gene, respectively. Yeast cells were transformed with the PCR product of 734 bp according to the “high efficiency transformation protocol” (29), and transformants were selected on SD plates lacking histidine. Correct integration of the PCR fragment was verified by colony PCR.
Phospholipid Extraction and TLC Analysis
Total lipid extracts were prepared of freeze-dried cells as described (15). To 10 mg of cells (dry weight), 3 ml of chloroform/methanol (2:1, v/v) was added, and the suspension was sonicated for 20 min in a Branson B1200 bath sonicator (Bransonic Ultrasonics, Danbury, CT) containing ice water. Subsequently, 1 ml of water was added, and, if appropriate, 0.4 nmol of tetramyristoyl-CL or 0.064 nmol of dimyristoyl-PG (both from Avanti Polar Lipids, Alabaster, AL) dissolved in 50 μl of chloroform were added as the internal standard. The mixture was shaken vigorously for 1 min, and incubated on ice for 15 min. The lower organic layer was collected after centrifugation for 10 min at 1000 × g. Residual lipids in the upper layer were extracted with 3 ml of chloroform/methanol (2:1, v/v), and the combined organic layers were evaporated under a stream of nitrogen. This procedure was applied to different amounts of cells, with volumes adjusted proportionally. Total lipid extracts were separated by thin layer chromatography (TLC) as described (30). The phospholipid containing spots identified by running the appropriate standards were scraped off and their quantities determined as described (31).
Analysis of Phospholipid Molecular Species by Mass Spectrometry
Total lipid extracts obtained from 10 mg of cells (dry weight) corresponding to 150 to 200 nmol of phospholipid phosphorous were dissolved in 150 μl of chloroform/methanol/water (50:45:5, v/v/v) containing 0.01% (w/v) NH4OH, and 5 μl of this solution was analyzed by HPLC-MS as described (15). Briefly, phospholipids were separated on a silica HPLC column using a linear gradient between chloroform/methanol (97:3, v/v) and methanol/water (85:15, v/v). The HPLC eluent was introduced into a TSQ Quantum AM mass spectrometer (Thermo Electron Corporation). Mass spectra of CL, PI, and PG were recorded in the negative ion mode with the following settings: source collision-induced dissociation, 10 V; spray voltage, 3.0 kV; and capillary temperature, 300 °C. PC, PE, and PS were measured in the positive ion mode, using a parent ion scan for m/z 184.1, and neutral loss scans for m/z 141.1 and 185.1, respectively. Other settings were: source collision-induced dissociation, 10 V; spray voltage, 3.6 kV; pressure of collision gas (argon), 0.5 mTorr; collision energy, 40 V for the parent ion scan and 25 V for the neutral loss scans; and capillary temperature, 300 °C. Mass spectra of each lipid were acquired during the corresponding retention time in the HPLC elution profile. The acyl chain composition of short CL species was determined by daughter scan analysis recorded in the negative ion mode with a collision energy of 50 V, Q1 peak width of 0.3, and Q3 peak width of 0.7 (FWHM).
Molecular Species Composition of Newly Synthesized PG
To label newly synthesized PG with deuterium-labeled glycerol (1,1,2,3,3-d5-glycerol; Sigma) the crd1 and crd1acb1 strain were cultured on SL medium. When the A600 of the cultures had reached ∼0.8, d5-glycerol was added to the medium to a concentration of 1.2% (w/v). After 10, 30, 60, and 180 min cells were harvested, washed with water by centrifugation (3 min at 3000 × g), and the cell pellets were freeze-dried. Lipids were extracted from 20 mg of freeze-dried cells as described above.
To determine the molecular species composition of d5-labeled PG, precursor scans of m/z 232 were recorded on a 4000 QTRAP mass spectrometer (Applied Biosystems/MDS Sciex, Concord, ON, Canada). Lipids were sprayed from chloroform/methanol/water (5:10:4, v/v/v) at a flow rate of 5 μl/min. Ionization was performed at 300 °C at an ion spray voltage of −4200 V. The declustering potential was set to −120 V and the collision energy to −55 V. Other parameters were optimized for a maximum signal-to-noise ratio. For analysis of unlabeled PG molecular species, precursor scans of m/z 227 (32) were recorded with identical settings as above. The identity of the molecular species was confirmed by recording product spectra, operating the second mass filter in linear ion-trap mode with dynamic fill time. In these experiments, a collision energy of 50 V with a spread of 15 V was used, which enabled the clear identification of the [M − H]− molecular ion, the corresponding lyso-lipid fragment ions, the subsequent loss of the head group from this fragment ion, the fatty acyl-derived carboxylate ions, and the glycerophosphate ion (data not shown).
Fatty Acid Analysis
Aliquots of total lipid extracts corresponding to 1 μmol of lipid phosphorous obtained from about 50 mg of freeze-dried cells were each dissolved in 2 ml of CH3OH/H2SO4 (40:1, v/v), and transesterified by heating at 70 °C for 2 h. After cooling to room temperature, 2 ml of water was added and the fatty acid methylesters were extracted three times with 2 ml of hexane. The composition of the fatty acid methylester mixture was determined on a Chrompack CP-9001 gas chromatograph equipped with a capillary column CP-WAX58 CB, in a 20-min run using the following temperature profile. After 2 min at 100 °C, the column was heated to 200 °C at a rate of 10 °C per min. Fatty acid methylesters were identified and signal intensities were calibrated using an equimolar mixture of the methylesters of C8:0, C10:0, C12:0, C12:1, C14:0, C14:1, C16:0, C16:1, C18:0, and C18:1 (Nu-ChekPrep, Elysian, MN).
In Vitro Crd1p Activity Assay
Crd1p activity of sucrose gradient-purified mitochondria isolated from wild type strain BY4742 (33) was determined based on published methods (15, 34) with the 50 μl of standard reaction mixture containing 0.1 m BisTris propane/HCl buffer (pH 9), 20 mm MgCl2, 24 μm CDP-dioleoyl-[U-14C]glycerol (42 dpm/pmol), 150 μm dioleoyl-PG, and 1 to 8 μg of mitochondrial protein. PG substrate specificity of Crd1p was tested in reaction mixtures containing increasing concentrations of the PG species indicated (diC16:0 PG and diC18:1 PG from Sigma; diC12:0 PG, diC14:0 PG, and C14:1/C17:0 PG from Avanti Polar Lipids, Alabaster, AL), whereas the CDP-DAG substrate selectivity of the enzyme was determined by the degree of inhibition of the labeling of CL caused by increasing concentrations of different unlabeled CDP-DAG species (Sigma) in the standard reaction mixture. Reactions were stopped after incubation for 1 h at 37 °C, and reaction products were extracted and analyzed as described before (15).
RESULTS
Effect of Deleting the ACB1 Gene on the Molecular Species Profiles of the Major Membrane Phospholipids
Deletion of the ACB1 gene (24, 25) or depletion of Acb1p (26–28) is known to affect the fatty acid composition of yeast cell; the average length of the fatty acids decreases, whereas the degree of unsaturation increases. Using mass spectrometry, we investigated how these changes translate to the molecular species compositions of the separate membrane phospholipid classes.
The molecular species profiles of PC, PE, PI, and PS in wild type and acb1 mutant cells as determined by ESI-(MS/)MS can be found in supplemental Fig. S1. As yeast cells have a limited repertoire of fatty acids with saturated and monounsaturated C16 and C18 acyl chains as main components (25), the majority of the PC, PE, PI, and PS species in wild type cells belonged to the C32 and C34 clusters (cf. Ref. 35). Comparison of the acb1 strain to the wild type revealed that the relative abundance of the C32 clusters was increased at the expense of the C34 clusters, in agreement with the average length of the acyl chains of all phospholipid classes tested being shorter in the mutant (supplemental Fig. S1), as reported earlier (27, 28). Moreover, the levels of species with 30 or less carbon atoms in their acyl chains increased to extents depending on the phospholipid class, which is consistent with the rise in the abundance of fatty acids shorter than 16 carbon atoms (24, 25). For example, in PS an increased content of the C30 cluster was observed compared with wild type, whereas in the species profile of PC significant amounts of C28 and even C26 species were also found in acb1 cells. The deletion of the ACB1 gene did not significantly influence the degree of saturation of PC and PE. In contrast, in acb1 cells PI contained more unsaturated acyl chains, as observed previously (27, 28), whereas for PS the content of unsaturated species was decreased.
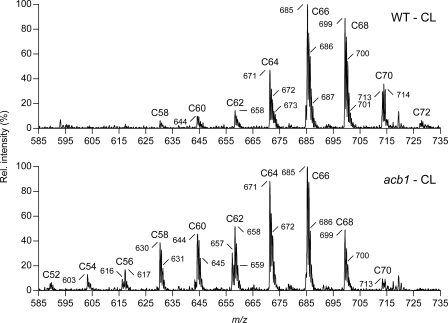
The molecular species profiles of CL in wild type and acb1 cells are shown in Fig. 2. To interpret the mass spectra of CL it should be realized that this phospholipid is measured as a doubly charged anion, implying that the measured m/z values correspond to half the molecular mass. In wild type cells, the CL species profile is dominated by clusters in the range of C64 to C70, as found previously (20) and consistent with the abundance of C16 and C18 acyl chains in yeast. In wild type cells, the most abundant clusters are dominated by the peak with the lowest m/z value, i.e. the one that corresponds to the tetra-unsaturated species. This peak is followed in decreasing order of intensity by the peaks representing tri- (m/z increased by 1) and diunsaturated species (m/z increased by 2).
FIGURE 2.
The effect of deleting the ACB1 gene on the species profile of CL in yeast. Total lipid extracts were prepared from wild type and acb1 cells cultured on SL medium to late log-phase. Phospholipid classes were separated by HPLC, and subjected to ESI-MS in the negative ion mode to determine the species composition of CL. The major doubly charged species of CL are indicated by their m/z values. Clusters of CL species are indicated by the total number of C-atoms in their acyl chains. In both panels, the intensity of the highest peak was set at 100%. See supplemental Table S3 for the molecular species assignment per cluster.
In the absence of Acb1p, the CL species profile was profoundly changed as illustrated by the increase in the summed relative abundance of the signal intensity of the clusters smaller than C64 from ∼10 to 40% (based on the highest peak in each cluster). The clusters smaller than C64 contain acyl chains shorter than C16, demonstrating a shortening of the acyl chains in CL. The increase in shorter CL species was at the expense of the C18-containing C68–C72 clusters, with the C70 and C72 species being virtually absent in acb1. Examination of the relative peak intensities within clusters indicated that the shorter CLs were on average more saturated than the longer species (Fig. 2, lower panel).
Yeast lacking the ACB1 gene has been reported to undergo a so far uncharacterized adaptation (26). To check whether the lipid species profiles shown in supplemental Figs. S1 and S2 were affected by this adaptation, acb1 and wild type cells were transformed with plasmid p416CYC-ACB1 (Table 1), carrying the wild type ACB1 gene, or with the empty plasmid p416CYC as control. The phospholipid species profiles of the acb1 cells were restored to wild type upon introducing the ACB1 gene, whereas the species profiles of the acb1 cells transformed with the empty vector and the wild type cells were not affected (data not shown). These results render it unlikely that the species profiles were affected by the adaptation, if any, of the acb1 mutant in the BY4742 background. Moreover, the lipid profiles recorded for the acb1 strain in this study were comparable with those found for the non-adapted yeast strain with a conditional ACB1 knock-out (28).
The profound effect of ACB1 deletion on the species profile of CL was remarkable, because the acyl chain composition of CL is thought to be important for CL function (14), and because yeast cells possess a CL remodeling system to replace inappropriate acyl chains. In the following, two possible origins of the acyl chains shorter than C16 in CL were investigated, remodeling of CL by Taz1p and de novo synthesis of CL, involving the precursors PG and CDP-DAG.
Are the Acyl Chains Shorter Than C16 Introduced in CL in acb1 via Remodeling by Taz1p?
After synthesis, CL is remodeled to obtain the correct acyl chain configuration, and the transacylase Taz1p is involved in this process (19, 21, 36). To examine whether Taz1p is required for the introduction of acyl chains <C16 in CL in an acb1 mutant, the ACB1 gene was deleted in a taz1 strain, yielding the taz1acb1 strain.
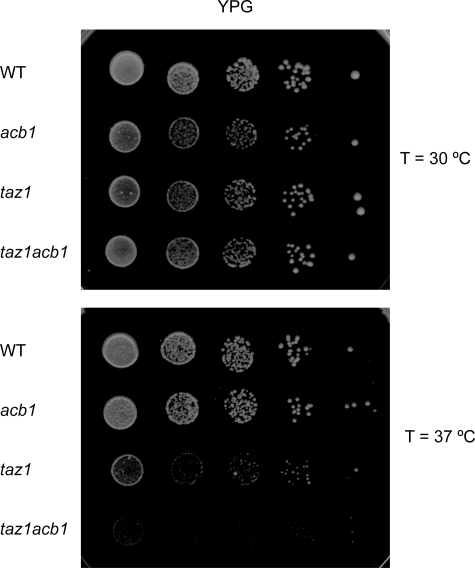
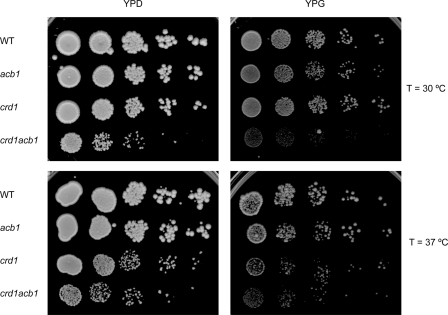
Growth phenotypes on YPD at 30 and 37 °C were indistinguishable between wild type, acb1, taz1, and acb1taz1 cells (data not shown). However, on a non-fermentable carbon source the taz1 cells showed a temperature-dependent growth defect (cf. Refs. 22 and 37) that was exacerbated in the taz1acb1 double mutant (Fig. 3).
FIGURE 3.
Growth of wild type, acb1, taz1, and taz1acb1 strains on YPG plates after preculture on YPD. The plates were incubated at 30 and 37 °C as indicated for 6 days. Results from a typical experiment are shown.
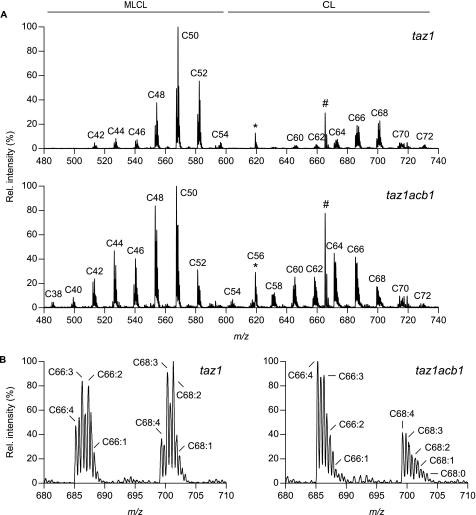
The CL species profile of the taz1acb1 mutant was compared with that of the taz1 strain (Fig. 4A). Almost all CL molecules in the taz1 mutant were found in clusters C64 and up, as in wild type cells (cf. Fig. 2). However, the degree of unsaturation was lower in taz1 cells, as evidenced by a shift of the signal intensity from the m/z value corresponding to the tetraunsaturated species toward that of the tri- and diunsaturated species in each of the clusters (Fig. 4B), in agreement with previous reports (20, 38). Another major difference was the high content of monolyso-CL (MLCL) in taz1, as had been observed before (20, 37). In the absence of Taz1p, deletion of the ACB1 gene resulted in a similar CL profile as found in acb1 (Fig. 2) with increased levels of clusters containing short CL species. Moreover, the MLCL profile in taz1acb1 resembles the profile of CL in (taz1)acb1 cells with one C16 acyl chain subtracted (Figs. 2, lower panel, and 4A, lower panel), implying that MLCL contains relatively more chains <C16 than CL. To illustrate this, if one C16 chain is removed from CL molecules that on average contain n acyl chains <C16 and hence 4-n normal acyl chains, then the produced MLCL will on average still have n acyl chains <C16, but only 3-n normal acyl chains. The relative content of acyl chains <C16 in the total pool of CL and MLCL is therefore higher in the absence of Taz1p than in its presence. The results shown in Fig. 4 therefore indicate that Taz1p is not responsible for the presence of acyl chains <C16 in CL in acb1, but they do not exclude that Taz1p is able to attach shorter acyl chains to MLCL.
FIGURE 4.
The effect of deleting the ACB1 gene on the species profile of CL in a taz1 strain. A, total lipid extracts of the taz1 and taz1acb1 strains were obtained from late-log phase cells cultured on SL medium, and the CL species profiles were determined by HPLC-MS. Peaks at m/z 619.5 and 665.5 indicated by the asterisk (*) and the number sign (#) represent the internal standards TMCL and DMPG, respectively. The horizontal lines mark the m/z ranges of the MLCL and CL clusters that are indicated by the number of C-atoms in their acyl chains. For each panel, the intensity of the highest peak was set at 100%. B, enlargements of the C66 and C68 clusters of CL in taz1 and taz1acb1. The doubly charged species of CL are indicated. The intensity of the highest peak in each panel was set at 100%. Results from a typical experiment are shown. See supplemental Table S3 for the molecular species assignment per cluster.
Interestingly, comparison of the MLCL and CL spectra in taz1 and taz1acb1 cells revealed that deletion of the ACB1 gene in the taz1 background resulted in a partial restoration of the taz1 phenotype: within the major clusters, the balance shifted from the more saturated species toward the tetraunsaturated species (Fig. 4B) as also found for wild type CL (Fig. 2, upper panel). However, with regard to the CL and MLCL contents, no significant differences were observed between taz1acb1 and taz1 strains by TLC analysis (data not shown).
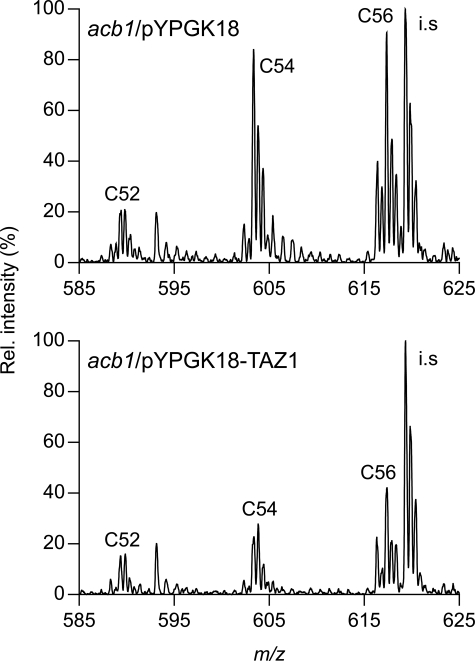
The notion that Taz1p is not required for the presence of acyl chains <C16 in CL in the acb1 mutant was corroborated by an experiment in which acb1 cells were transformed with the pYPGK18 plasmid carrying the TAZ1 gene under control of the strong PGK1 promoter. If Taz1p were responsible for the acyl chains <C16 in the short CL molecules, overexpression of this protein would be expected to increase the abundance of the short CL species. However, the opposite effect was observed. Whereas transformation with the empty plasmid did not affect the levels of the C52–C56 clusters in the CL species profile of the acb1 strain (data not shown), overexpression of Taz1p from pYPGK18 decreased the levels of C54 and C56 CL by at least 50% relative to the internal standard (Fig. 5). Episomal expression of Taz1p in acb1 cells did not affect the species profiles of the other major membrane phospholipids, nor did it affect growth (data not shown). We conclude that Taz1p is not required for the introduction of acyl chains <C16 into CL species in acb1 cells, and may to some extent contribute to their replacement.
FIGURE 5.
The influence of episomal expression of Taz1p under control of the constitutive PGK1 promoter on the content of the short CL species (up to C56) in acb1 cells. The CL species profiles of acb1 cells transformed with plasmid pYPGK18-TAZ1 and with the empty vector as control, were determined by HPLC-MS. Only the parts of the mass spectra corresponding to the m/z 585–625 range are shown. Clusters of CL species are indicated by the total number of C-atoms in their acyl chains. The intensity of the internal standard TMCL (i.s.) was set at 100%. Results from a typical experiment are shown. See supplemental Table S3 for the molecular species assignment per cluster.
Do Acyl Chains Shorter Than C16 in CL in acb1 Originate from PG and/or CDP-DAG via de Novo Synthesis?
The other route via which the shorter acyl chains could end up in CL is the de novo synthesis of CL. Crd1p synthesizes CL by transferring the activated phosphatidyl moiety from CDP-DAG to PG (Fig. 1). The low cellular content of these two CL precursors due to high turnover rates interferes with the determination of the species profiles that are representative for the pools of substrates available to Crd1p (39–41). The existence of two separate intracellular pools of CDP-DAG further complicates the analysis of this intermediate in CL synthesis (42–44). For PG, the problem of low abundance can be circumvented by using a crd1 strain, in which CL synthesis is blocked, resulting in increased PG levels (41, 45). To examine whether PG exhibits a similar accumulation of relatively short acyl chains in the absence of CL synthesis as CL in the acb1 strain, a crd1acb1 double deletion strain was constructed. Growth phenotypes of the crd1acb1 strain, the congenic single deletion mutants, and the wild type strain are shown in Fig. 6.
FIGURE 6.
Growth of wild type, acb1, crd1, and crd1acb1 on YPD and YPG plates after preculture on YPD. The plates were incubated at 30 or 37 °C for 2–5 days. Results from a typical experiment are shown.
On YPD and YPG at 30 °C, growth of the crd1acb1 mutant was somewhat impaired compared with that of the acb1 and crd1 single mutants and the parental wild type. At 37 °C, the poorer growth phenotype of the crd1 strain compared with acb1 and wild type (cf. Refs. 46 and 47) was slightly exacerbated in the crd1acb1 mutant.
To check whether the crd1 strain provides a representative background to study the effect of ACB1 deletion on the sorting of acyl chains, the fatty acid compositions of total lipid extracts were compared between wild type and crd1 cells, and between acb1 and crd1acb1 cells. No significant differences were observed (see supplemental Table S1), validating the crd1acb1 mutant strain as a model for investigating the sorting of acyl chains.
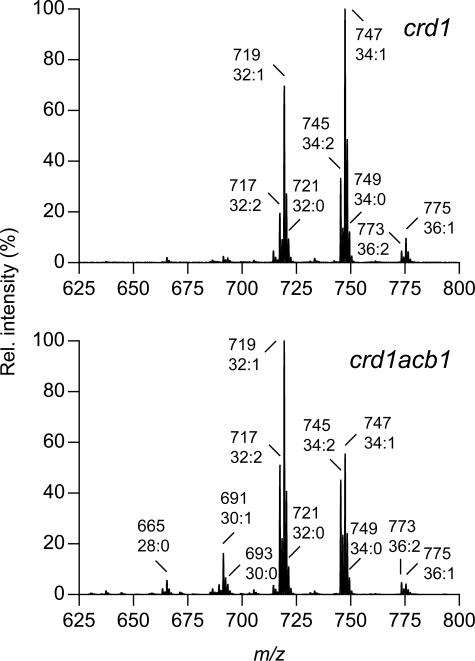
The species profile of PG in the crd1acb1 double mutant was determined by mass spectrometry and compared with that in the crd1 strain (Fig. 7). In the latter strain, the PG profile was dominated by C32 and C34 clusters with a minor contribution of C36. In the crd1acb1 mutant, the PG molecules were on average shorter with C32 as the most abundant cluster. In addition, about 10% of the PG signal originated from the C28 and C30 clusters, which were virtually absent in crd1. These results are in line with the earlier observations for the other phospholipid classes in the acb1 mutant (see Figs. 2 and supplemental S1). With respect to the degree of saturation, most PG species in the crd1 cells were monounsaturated; deletion of the ACB1 gene resulted in a slight increase in the relative levels of diunsaturated species.
FIGURE 7.
The effect of deleting the ACB1 gene on the species profile of PG in the crd1 strain. Total lipid extracts were obtained from late-log phase crd1 and crd1acb1 cells, and the species composition of PG was analyzed by HPLC-MS, as detailed under “Experimental Procedures.” The major [M − H]− species of PG are indicated. The intensity of the highest peak was set at 100%. Representative results are shown. See supplemental Table S2 for the molecular species assignment per cluster.
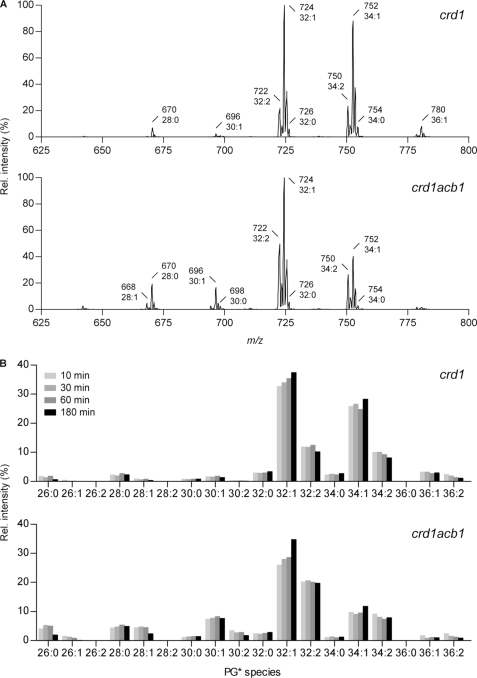
Considering that the shortest PG in acb1 cells contained at least 28 carbon atoms in its combined acyl chains, PG is unlikely to be the sole source of acyl chains shorter than C16 in CL in view of the presence of the C52–C58 clusters in the acb1 CL profile (Fig. 2). CDP-DAG must also contribute acyl chains <C16 to CL, unless the PG profile of the crd1acb1 mutant is not representative for the pool of PG available as substrate for Crd1p in the acb1 mutant due to species selective turnover (48) or remodeling of PG. To test the latter possibility, the species profile of newly synthesized PG was monitored over time by stable isotope labeling in conjunction with tandem mass spectrometry (49). Logarithmically growing crd1 and crd1acb1 cells were pulsed for different periods of time with d5-glycerol to label PG. The incorporation of d5-glycerol in the backbone or head group of PG increases the m/z values of the PG species by 5, thus enabling the selective detection of newly synthesized PG by scanning for precursors of m/z 232, corresponding to the d5-glycerophosphate glycerol minus H2O fragment ion (32). The parent ion scans of newly synthesized PG obtained after 3 h of labeling of crd1 and crd1acb1 cells (Fig. 8A) are consistent with the ESI-MS spectra of unlabeled PG (Fig. 7) when taking into account that the chances of ion fragmentation decrease with increasing m/z value, resulting in a more sensitive detection of parent ions as the m/z value decreases. Unfortunately, the species profiles of unlabeled PG could not be obtained by parent ion scanning (m/z 227) because of interference by C14 acyl chains that yield a fragment ion with the corresponding m/z value (data not shown), rendering a direct comparison impossible.
FIGURE 8.
The effect of deleting ACB1 on the species profile of newly synthesized PG in crd1. To selectively detect newly synthesized PG, crd1 and crd1acb1 cells cultured to mid-log phase in SL medium were labeled with d5-glycerol for the times indicated. The species compositions of the labeled PG (denoted by PG*) were analyzed in total lipid extracts by MS/MS as detailed under “Experimental Procedures.” A, parent ion scans at m/z 232 of d5-labeled PG in total lipid extracts of crd1 and crd1acb1 cells after 3 h of incubation with d5-glycerol. The major [M − H]− species of PG* are indicated. The intensity of the highest peak is set at 100%. B, the time-dependent evolution of the species profiles of labeled PG in crd1 and crd1acb1 strains during labeling with d5-glycerol. The panels show the relative signal intensities of the molecular species indicated, with the total PG signal intensity set at 100%. Representative results are shown. See supplemental Table S2 for the molecular species assignment per cluster.
Fig. 8B shows the time course of the relative intensities of d5-labeled PG species after different times of labeling with d5-glycerol. In the crd1 strain, the species profile of d5-labeled PG shows only minor changes over time, arguing against extensive species selective turnover or acyl chain remodeling of PG. The same holds true for the crd1acb1 strain in which, with the possible exceptions of C26:0, C28:1, and C32:1, no large changes in the d5-PG species distribution occur. Although newly synthesized PG apparently contained slightly shorter species than steady state PG (C26 versus C28, respectively), the results corroborate the above conclusion that CDP-DAG also serves as a source of acyl chains shorter than C16 in CL in acb1.
Because CDP-DAG is a precursor of PG, it might seem a trivial notion that acyl chains <C16 end up in CL via both PG and CDP-DAG, but it should be kept in mind that Crd1p might prefer long species of one substrate and short species of the other (cf. Ref. 15), and/or that Pgs1p and Crd1p (Fig. 1) might have access to different CDP-DAG pools. To gain more insight into the relative contributions of CDP-DAG and PG to the acyl chains <C16 in CL in acb1 cells, the substrate specificity of Crd1p was examined in vitro in isolated mitochondria from the wild type strain. First, to assess whether substrate specificity of Crd1p could have led to a preferred use of short PG over short CDP-DAG or vice versa, CL synthesis by isolated wild type mitochondria in the presence of various species of both CL precursors was evaluated.
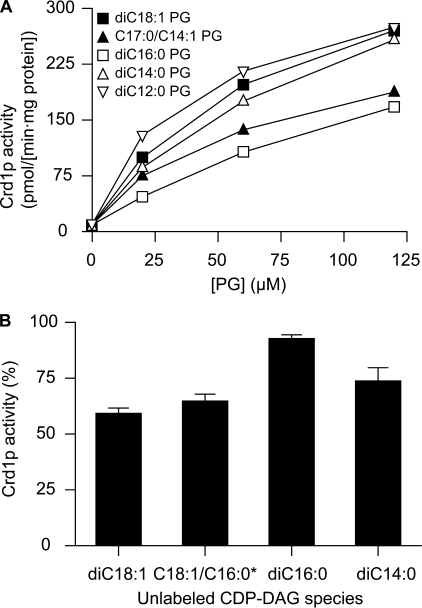
The PG species specificity of Crd1p was analyzed by determining the CL synthesis rates from labeled CDP-dioleoylglycerol and increasing concentrations of various PG species. As depicted in Fig. 9A, Crd1p displayed the highest activity with the shortest and the most unsaturated PG species tested, and the lowest activity with the dipalmitoyl species. The observed PG species specificity of Crd1p was less distinct than those of CL synthases in more complex eukaryotes (15, 50, 51). CDP-DAG competition experiments revealed that Crd1p also preferentially used the CDP-DAG species with shorter or unsaturated acyl chains (Fig. 9B). Again, the selectivity of the yeast enzyme was only weak in comparison to the selectivity of CL synthases from mammalian cells (15) and higher plants (51). Hence, the results suggest that in yeast mitochondria de novo synthesized CL species can to a large extent be determined by the substrate species composition available to the Crd1p. In addition, based on the observed similar species specificity of Crd1p for both its substrates, it is unlikely that Crd1p preferentially sources acyl chains shorter than C16 from either PG or CDP-DAG.
FIGURE 9.
Substrate species specificity of Crd1p. A, formation rates of CL by Crd1p as a function of the concentrations of diC18:1 (■), C17:0/C14:1 (▴), diC16:0 (□), diC14:0 (▵), and diC12:0 (▿) PG species under otherwise standard assay conditions. B, incorporation of 24 μm CDP-dioleoyl[U-14C]glycerol into CL remaining in the presence of 24 μm of the indicated unlabeled CDP-DAG species under otherwise standard assay conditions. The amount of labeled CL produced in the absence of unlabeled CDP-DAG is set at 100%. The error bars represent the S.D. (n = 3). Similar results were obtained when the assays were conducted with 2–3-fold higher concentrations of unlabeled CDP-DAG species or 5-fold lower concentrations of labeled CDP-dioleoylglycerol (data not shown). *18:1/16:0 CDP-DAG is a mixture of CDP-DAG species prepared from egg lecithin, which predominantly consists of the 18:1/16:0 species.
To shed more light on the relative contributions of acyl chains <C16 by PG and CDP-DAG, the dominant CL species in the C52–C56 clusters were subjected to MS/MS analysis. Based on the acyl chains that were identified, the most likely acyl chain compositions were determined for each species (Table 2), taking into account the mass and degree of saturation. Surprisingly, all CL molecules were found to contain two regular (C16 or C18) and two short (C10 and/or C12) chains. No C14-containing molecules were found. Because it is highly improbable that the two shorter chains both originated from a single precursor, the shortest PG and CDP-DAG species used for CL synthesis are apparently C26 and C28, consistent with the results obtained for PG (Fig. 8). The data further imply that short PG and CDP-DAG species predominantly have chains with unequal lengths, for instance, C10 and C16 instead of C12 and C14. This remarkable finding was confirmed by daughter scan analysis of C28:0 PG from crd1acb1, which revealed C12:0 and C16:0 as most abundant acyl chains (data not shown). We conclude that PG and CDP-DAG are to similar extents responsible for the presence of acyl chains shorter than C16 in CL in the acb1 strain.
TABLE 2.
Molecular structures of short CL species in acb1 cells as resolved by MS/MS analysis of the peak of highest intensity in the C52–C56 clusters
| CL species | Parent m/z | Most abundant acyl chainsa | Possible acyl chain compositionsb |
|---|---|---|---|
| C52:2 | 589.37 | C10:0 and C16:1 | (C10:0)2(C16:1)2 |
| C54:2 | 603.38 | C10:0, C12:0 and C16:1 | (C10:0)(C12:0)(C16:1)2 |
| C56:2 | 617.40 | C10:0, C12:0, C16:1 and C18:1 | (C10:0)(C12:0)(C16:1)(C18:1) and (C12:0)2(C16:1)2 |
a Based on peak heights.
bAcyl chain compositions were determined, based on the acyl chains that were identified and the mass and number of double bonds of each species.
DISCUSSION
The present study revealed that deletion of the ACB1 gene leads to an altered molecular species profile of CL, as was observed previously for other phospholipid classes (27, 28). Because the acyl chain composition is thought to be important for the functioning of CL (14, 52, 53), we expected beforehand that the CL profile would be relatively resistant to changes, also in view of the available CL remodeling system. Therefore, the mechanism by which acyl chains shorter than C16 accumulate in CL in the absence of Acb1p was elucidated using MS analysis of lipid extracts from various yeast mutant strains.
The results showed that CL remodeling by Taz1p was not required for the accumulation of acyl chains <C16 in CL. Analysis of PG in crd1acb1 cells (steady state and newly synthesized) demonstrated PG species containing acyl chains <C16, but not to the extent that PG could solely account for the shortest CL molecules in acb1 cells. Therefore, a substantial portion of the acyl chains <C16 in CL had to stem from the other CL precursor, i.e. CDP-DAG. The conclusion that both CL precursors contribute to comparable extents to acyl chains <C16 in CL in acb1 cells was supported by the species selectivity of Crd1p in vitro, and the distinct acyl chain compositions found for short CL species in acb1 cells. We will discuss the implications of our findings for CL remodeling in yeast.
Why did the CL remodeling enzymes not restore the wild type species profile of CL in the absence of Acb1p? It could be argued that CL remodeling depends on the supply of acyl-CoA by Acb1p. However, because Acb1p does not localize to mitochondria (54), and because the only CL remodeling acyltransferase identified so far, Taz1p, is thought to be a transacylase that does not require acyl-CoA (19), we consider the direct involvement of Acb1p in CL remodeling unlikely. An indirect effect of Acb1p on CL remodeling, e.g. resulting from a possible requirement for Acb1p of the re-acylation of acyl chain donors of CL remodeling, cannot be excluded. CL remodeling could have been hampered by a shortage of appropriate acyl chains. Phospholipids most likely serve as acyl chain donors in Taz1p-mediated remodeling (19). In acb1 the vast majority of phospholipids belongs to the C32 and C34 mono- and diunsaturated species (supplemental Fig. S1), rendering a shortage of C16 and C18 chains highly improbable. Even if other compounds than phospholipids serve as donor in a transacylation, no impact on CL remodeling is expected based on the fatty acid composition in acb1 total lipid extracts (supplemental Table S1). In view of the absence of even the slightest accumulation of MLCL in acb1 cells, and because overexpression of Taz1p had only a minor effect on the overall CL profile in acb1 cells, it is unlikely that the remodeling capacity of Taz1p is insufficient.
Therefore, we postulated that the acyl chains <C16 in CL may not be recognized as substrate by the CL phospholipases. In support of this idea, a qualitative comparison of the CL and MLCL profiles in taz1acb1 (Fig. 4A) in terms of relative peak heights within the clusters suggested that MLCL was produced by the cleavage of predominantly C16 chains rather than by removal of acyl chains <C16, with e.g. C46 MLCL originating from C62 CL, and C50 MLCL from C66 CL. During the preparation of this article, identification of the yeast CL phospholipase Cld1p was reported (23). Cld1p was shown to have a strong preference for removing C16:0 acyl chains from CL, fully consistent with our hypothesis. Furthermore, circumstantial evidence is provided by the daughter scan of the most intense peak in the C52 CL cluster from acb1 cells, corresponding to C52:2 CL, which revealed that this minor species was built from C10:0 and C16:1 chains (Table 2). Remarkably, within the C26 cluster of PG, C26:0 PG was the most abundant species (data not shown), suggesting that a C16:0 chain originating from PG had been replaced during CL remodeling by a C16:1 chain.
The specificity of Cld1p allows an elegant reduction of the number of different acyl chains in remodeled CL, thereby contributing to the symmetry in CL (14, 52). Selective removal of C16:0 is therefore sufficient to establish a CL pool with mainly C16:1 and C18:1 in wild type cells. The specificity of the CL phospholipase for C16:0 explains why C18-rich clusters C70 and C72 are not dominated by tetraunsaturated species in wild type and acb1 cells (Fig. 2), and also why acyl chains <C16 in acb1 are not adequately removed, resulting in the accumulation of short (and relatively saturated) CL species in acb1 compared with wild type.
Based on our findings and those of Beranek et al. (23), we propose the following mechanism for CL remodeling in yeast: Cld1p is the prime phospholipase acting on CL, and responsible for the recognition and cleavage of C16:0 chains from CL. Subsequently, Taz1p is primarily responsible for the reacylation of the MLCL produced. The transacylase Taz1p uses the acyl chains available from donor phospholipid(s). The combined actions of Cld1p and Taz1p lead to an enrichment in CL of C16:1 and C18:1 (the major acyl chains in yeast next to C16:0 (38)) at the expense of C16:0. Recently, a lack of acyl specificity has been reported in transacylase reactions catalyzed by rat liver, human, and Drosophila tafazzin (55), supporting the proposed mechanism. In conclusion, the fatty acid composition of mature CL in yeast is governed by the specificity of Cld1p and the fatty acid composition of the Taz1p substrates.
The partial restoration of the CL species profile observed in the acb1taz1 double mutant compared with the taz1 mutant can be accounted for by the increased content of unsaturated fatty acids in acb1 cells (24) (see supplemental Table S1). The increased unsaturation is reflected in the de novo synthesized CL rather than in the other phospholipid classes (supplemental Fig. S1), consistent with the preference of Crd1p for unsaturated substrates. Even though the partial restoration of the CL species profile in acb1taz1 seemed to be accompanied by a trend toward restoration of mitochondrial protein (super)complexes,6 it did not suppress the growth defect of taz1 cells cultured on a non-fermentable carbon source at 37 °C (Fig. 3), suggesting that processes other than oxidative phosphorylation limit growth.
Although the pronounced growth defect of crd1acb1 on non-fermentable carbon sources was not explored further, we speculate that the underlying cause for the retarded growth is related to the altered acyl chain composition of PE in this double mutant compared with crd1. The PE content in crd1acb1 was found to be similar to that in crd1 (data not shown). PE has been shown to be essential for the viability of yeast cells lacking CL, probably because PE and CL can both promote hexagonal lipid phases (10, 56), and can substitute for each other in mitochondrial processes where this non-bilayer propensity is required. A functional relationship between PE and CL is further suggested by the concerted regulation of the synthesis of these lipids (57). In crd1acb1 cells, PE is on average shorter than in crd1 (data not shown), which implies a decreased propensity of PE in the double mutant to form non-bilayer structures (58). When CL is present, the impact of the shortening of PE on mitochondrial function might be limited. Because the double mutant crd1acb1 lacks CL, it might be less able to cope with the altered PE species profile, resulting in the severe retardation of growth. An alternative explanation for the growth defect of crd1acb1 would be that the altered acyl chain composition of PG due to Acb1p depletion renders PG less suited to replace CL. However, because this replacement is thought to rely primarily on the net negative charge shared by PG and CL (45), alterations in the acyl chain composition of PG are likely to have less of an impact than the changes in PE.
Supplementary Material
Acknowledgments
We thank Dr. Henry Boumann for performing pilot studies and the mass spectrometry section of the Laboratory Genetic Metabolic Diseases for technical assistance. The p416CYC plasmids were a kind gift of Dr. Nils Færgeman.
This work was supported by an Earth and Life Sciences (ALW) grant with financial aid from the Dutch Organization for Scientific Research (NWO). This work was also supported by Prinses Beatrix Fonds Grant WAR05-0126 and the Barth Syndrome Foundation (to F. M. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1–S3.
R. A. van Gestel, P. J. Rijken, S. Surinova, M. O'Flaherty, A. J. R. Heck, J. A. Killian, A. I. P. M. de Kroon, and M. Slijper, manuscript in preparation.
- CL
- cardiolipin
- CDP-DAG
- cytidinediphosphate diacylglycerol
- DAG
- diacylglycerol
- MLCL
- monolysocardiolipin
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- ESI-MS
- electrospray ionization mass spectrometry
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Daum G. (1985) Biochim. Biophys. Acta 822, 1–42 [DOI] [PubMed] [Google Scholar]
- 2.de Kroon A. I., Dolis D., Mayer A., Lill R., de Kruijff B. (1997) Biochim. Biophys. Acta 1325, 108–116 [DOI] [PubMed] [Google Scholar]
- 3.Shinzawa-Itoh K., Aoyama H., Muramoto K., Terada H., Kurauchi T., Tadehara Y., Yamasaki A., Sugimura T., Kurono S., Tsujimoto K., Mizushima T., Yamashita E., Tsukihara T., Yoshikawa S. (2007) EMBO J. 26, 1713–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlame M., Rua D., Greenberg M. L. (2000) Prog. Lipid Res. 39, 257–288 [DOI] [PubMed] [Google Scholar]
- 5.Palsdottir H., Hunte C. (2004) Biochim. Biophys. Acta 1666, 2–18 [DOI] [PubMed] [Google Scholar]
- 6.Claypool S. M., Oktay Y., Boontheung P., Loo J. A., Koehler C. M. (2008) J. Cell Biol. 182, 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfeiffer K., Gohil V., Stuart R. A., Hunte C., Brandt U., Greenberg M. L., Schägger H. (2003) J. Biol. Chem. 278, 52873–52880 [DOI] [PubMed] [Google Scholar]
- 8.Zhang M., Mileykovskaya E., Dowhan W. (2005) J. Biol. Chem. 280, 29403–29408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasilenko I., De Kruijff B., Verkleij A. J. (1982) Biochim. Biophys. Acta 684, 282–286 [DOI] [PubMed] [Google Scholar]
- 10.Gohil V. M., Thompson M. N., Greenberg M. L. (2005) J. Biol. Chem. 280, 35410–35416 [DOI] [PubMed] [Google Scholar]
- 11.Haines T. H., Dencher N. A. (2002) FEBS Lett. 528, 35–39 [DOI] [PubMed] [Google Scholar]
- 12.Gonzalvez F., Gottlieb E. (2007) Apoptosis 12, 877–885 [DOI] [PubMed] [Google Scholar]
- 13.Gonzalvez F., Schug Z. T., Houtkooper R. H., MacKenzie E. D., Brooks D. G., Wanders R. J., Petit P. X., Vaz F. M., Gottlieb E. (2008) J. Cell Biol. 183, 681–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlame M., Ren M., Xu Y., Greenberg M. L., Haller I. (2005) Chem. Phys. Lipids 138, 38–49 [DOI] [PubMed] [Google Scholar]
- 15.Houtkooper R. H., Akbari H., van Lenthe H., Kulik W., Wanders R. J., Frentzen M., Vaz F. M. (2006) FEBS Lett. 580, 3059–3064 [DOI] [PubMed] [Google Scholar]
- 16.Vreken P., Valianpour F., Nijtmans L. G., Grivell L. A., Plecko B., Wanders R. J., Barth P. G. (2000) Biochem. Biophys. Res. Commun. 279, 378–382 [DOI] [PubMed] [Google Scholar]
- 17.Hauff K. D., Hatch G. M. (2006) Prog. Lipid Res. 45, 91–101 [DOI] [PubMed] [Google Scholar]
- 18.Schlame M., Ren M. (2006) FEBS Lett. 580, 5450–5455 [DOI] [PubMed] [Google Scholar]
- 19.Xu Y., Malhotra A., Ren M., Schlame M. (2006) J. Biol. Chem. 281, 39217–39224 [DOI] [PubMed] [Google Scholar]
- 20.Gu Z., Valianpour F., Chen S., Vaz F. M., Hakkaart G. A., Wanders R. J., Greenberg M. L. (2004) Mol. Microbiol. 51, 149–158 [DOI] [PubMed] [Google Scholar]
- 21.Ma L., Vaz F. M., Gu Z., Wanders R. J., Greenberg M. L. (2004) J. Biol. Chem. 279, 44394–44399 [DOI] [PubMed] [Google Scholar]
- 22.Brandner K., Mick D. U., Frazier A. E., Taylor R. D., Meisinger C., Rehling P. (2005) Mol. Biol. Cell 16, 5202–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beranek A., Rechberger G., Knauer H., Wolinski H., Kohlwein S. D., Leber R. (2009) J. Biol. Chem. 284, 11572–11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi J. Y., Stukey J., Hwang S. Y., Martin C. E. (1996) J. Biol. Chem. 271, 3581–3589 [DOI] [PubMed] [Google Scholar]
- 25.Schjerling C. K., Hummel R., Hansen J. K., Borsting C., Mikkelsen J. M., Kristiansen K., Knudsen J. (1996) J. Biol. Chem. 271, 22514–22521 [DOI] [PubMed] [Google Scholar]
- 26.Gaigg B., Neergaard T. B., Schneiter R., Hansen J. K., Faergeman N. J., Jensen N. A., Andersen J. R., Friis J., Sandhoff R., Schrøder H. D., Knudsen J. (2001) Mol. Biol. Cell 12, 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faergeman N. J., Feddersen S., Christiansen J. K., Larsen M. K., Schneiter R., Ungermann C., Mutenda K., Roepstorff P., Knudsen J. (2004) Biochem. J. 380, 907–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feddersen S., Neergaard T. B., Knudsen J., Faergeman N. J. (2007) Biochem. J. 407, 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gietz R. D., Woods R. A. (2002) Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 30.Vaden D. L., Gohil V. M., Gu Z., Greenberg M. L. (2005) Anal. Biochem. 338, 162–164 [DOI] [PubMed] [Google Scholar]
- 31.Rouser G., Fkeischer S., Yamamoto A. (1970) Lipids 5, 494–496 [DOI] [PubMed] [Google Scholar]
- 32.Welti R., Wang X., Williams T. D. (2003) Anal. Biochem. 314, 149–152 [DOI] [PubMed] [Google Scholar]
- 33.de Kroon A. I., Koorengevel M. C., Goerdayal S. S., Mulders P. C., Janssen M. J., de Kruijff B. (1999) Mol. Membr. Biol. 16, 205–211 [DOI] [PubMed] [Google Scholar]
- 34.Tamai K. T., Greenberg M. L. (1990) Biochim. Biophys. Acta 1046, 214–222 [DOI] [PubMed] [Google Scholar]
- 35.Schneiter R., Brügger B., Sandhoff R., Zellnig G., Leber A., Lampl M., Athenstaedt K., Hrastnik C., Eder S., Daum G., Paltauf F., Wieland F. T., Kohlwein S. D. (1999) J. Cell Biol. 146, 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y., Kelley R. I., Blanck T. J., Schlame M. (2003) J. Biol. Chem. 278, 51380–51385 [DOI] [PubMed] [Google Scholar]
- 37.Vaz F. M., Houtkooper R. H., Valianpour F., Barth P. G., Wanders R. J. (2003) J. Biol. Chem. 278, 43089–43094 [DOI] [PubMed] [Google Scholar]
- 38.Testet E., Laroche-Traineau J., Noubhani A., Coulon D., Bunoust O., Camougrand N., Manon S., Lessire R., Bessoule J. J. (2005) Biochem. J. 387, 617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klig L. S., Homann M. J., Kohlwein S. D., Kelley M. J., Henry S. A., Carman G. M. (1988) J. Bacteriol. 170, 1878–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loewen C. J., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., Henry S. A., Levine T. P. (2004) Science 304, 1644–1647 [DOI] [PubMed] [Google Scholar]
- 41.Tuller G., Hrastnik C., Achleitner G., Schiefthaler U., Klein F., Daum G. (1998) FEBS Lett. 421, 15–18 [DOI] [PubMed] [Google Scholar]
- 42.Kuchler K., Daum G., Paltauf F. (1986) J. Bacteriol. 165, 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zinser E., Sperka-Gottlieb C. D., Fasch E. V., Kohlwein S. D., Paltauf F., Daum G. (1991) J. Bacteriol. 173, 2026–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen H., Heacock P. N., Clancey C. J., Dowhan W. (1996) J. Biol. Chem. 271, 789–795 [DOI] [PubMed] [Google Scholar]
- 45.Chang S. C., Heacock P. N., Mileykovskaya E., Voelker D. R., Dowhan W. (1998) J. Biol. Chem. 273, 14933–14941 [DOI] [PubMed] [Google Scholar]
- 46.Jiang F., Gu Z., Granger J. M., Greenberg M. L. (1999) Mol. Microbiol. 31, 373–379 [DOI] [PubMed] [Google Scholar]
- 47.Zhong Q., Gohil V. M., Ma L., Greenberg M. L. (2004) J. Biol. Chem. 279, 32294–32300 [DOI] [PubMed] [Google Scholar]
- 48.Simocková M., Holic R., Tahotná D., Patton-Vogt J., Griac P. (2008) J. Biol. Chem. 283, 17107–17115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boumann H. A., Damen M. J., Versluis C., Heck A. J., de Kruijff B., de Kroon A. I. (2003) Biochemistry 42, 3054–3059 [DOI] [PubMed] [Google Scholar]
- 50.Nowicki M., Müller F., Frentzen M. (2005) FEBS Lett. 579, 2161–2165 [DOI] [PubMed] [Google Scholar]
- 51.Frentzen M., Griebau R. (1994) Plant Physiol. 106, 1527–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlame M. (2008) J. Lipid Res. 49, 1607–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen S., He Q., Greenberg M. L. (2008) Mol. Microbiol. 68, 1061–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 55.Malhotra A., Xu Y., Ren M., Schlame M. (2009) Biochim. Biophys. Acta 1791, 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gohil V. M., Greenberg M. L. (2009) J. Cell Biol. 184, 469–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osman C., Haag M., Potting C., Rodenfels J., Dip P. V., Wieland F. T., Brügger B., Westermann B., Langer T. (2009) J. Cell Biol. 184, 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boumann H. A., Gubbens J., Koorengevel M. C., Oh C. S., Martin C. E., Heck A. J., Patton-Vogt J., Henry S. A., de Kruijff B., de Kroon A. I. (2006) Mol. Biol. Cell 17, 1006–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mumberg D., Müller R., Funk M. (1995) Gene 156, 119–122 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.