Abstract
Mammalian CD38 and its Aplysia homolog, ADP-ribosyl cyclase (cyclase), are two prominent enzymes that catalyze the synthesis and hydrolysis of cyclic ADP-ribose (cADPR), a Ca2+ messenger molecule responsible for regulating a wide range of cellular functions. Although both use NAD as a substrate, the cyclase produces cADPR, whereas CD38 produces mainly ADP-ribose (ADPR). To elucidate the catalytic differences and the mechanism of cyclizing NAD, the crystal structure of a stable complex of the cyclase with an NAD analog, ribosyl-2′F-2′deoxynicotinamide adenine dinucleotide (ribo-2′-F-NAD), was determined. The results show that the analog was a substrate of the cyclase and that during the reaction, the nicotinamide group was released and a stable intermediate was formed. The terminal ribosyl unit at one end of the intermediate formed a close linkage with the catalytic residue (Glu-179), whereas the adenine ring at the other end stacked closely with Phe-174, suggesting that the latter residue is likely to be responsible for folding the linear substrate so that the two ends can be cyclized. Mutating Phe-174 indeed reduced cADPR production but enhanced ADPR production, converting the cyclase to be more CD38-like. Changing the equivalent residue in CD38, Thr-221 to Phe, correspondingly enhanced cADPR production, and the double mutation, Thr-221 to Phe and Glu-146 to Ala, effectively converted CD38 to a cyclase. This study provides the first detailed evidence of the cyclization process and demonstrates the feasibility of engineering the reactivity of the enzymes by mutation, setting the stage for the development of tools to manipulate cADPR metabolism in vivo.
Cyclic ADP-ribose is a novel cyclic nucleotide with Ca2+-mobilizing activity targeting the endoplasmic reticulum. Its activity was first described in sea urchin eggs (1, 2), and cADPR3 has since been established as a second messenger molecule responsible for regulating a wide range of physiological functions, from fission in the dinoflagellate (3) to social behavior in mice (Ref. 4 and reviewed in Refs. 5 and 6). The Aplysia ADP-ribosyl cyclase (cyclase) was the first protein identified that uses NAD, a linear substrate, and ligates its two ends to produce cADPR, with the release of the terminal moiety, nicotinamide (7). The cyclase is a soluble protein of 30 kDa and is present in large amounts in Aplysia ovotestis (7). It is also present in the neurons of the Aplysia buccal ganglion, where it is responsible for the synthesis of endogenous cADPR and the regulation of the evoked synaptic transmission (8). Recently, it is shown that depolarization of Aplysia neurons induces the translocation of the cyclase from the cytosol into the nucleus, providing a mechanism for fine tuning of nuclear Ca2+ signals in neurons (9).
CD38 is the major mammalian homolog of the cyclase and is responsible for regulating a wide range of physiological functions. Deletion of the CD38 gene in mice produces multiple defects, including impairment of insulin secretion (10), neutrophil chemotaxis (11), and oxytocin release (4). Catalytically, CD38 is quite different from the cyclase. Although both use NAD as substrate, CD38 produces only a small amount of cADPR, whereas the major product is ADP-ribose (ADPR) instead (12–15) (Fig. 1a). It can also use cADPR as a substrate and hydrolyze it to ADPR (12–15). Ablation of the CD38 gene in mice, nevertheless, results in a large reduction in endogenous cADPR in many tissues (10, 11). CD38 is thus responsible for both the synthesis and the hydrolysis of cADPR in mammalian cells.
FIGURE 1.
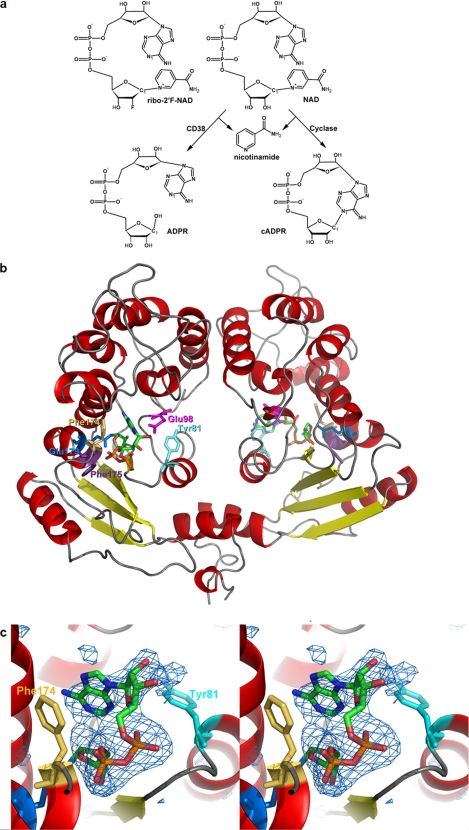
Crystal structure of the complex of cyclase with ribo-2′F-NAD. a, chemical structure of the substrate ribo-2′F-NAD and the reactions catalyzed by CD38 and the cylase. b, crystal structure of the cyclase dimer with the intermediates at each of the active sites of the monomer. The color scheme for the secondary structures is: red, α-helix; yellow, β-sheet; gray, coil. The color scheme for the residues is: cyan, Tyr-81; beige, Phe-174; blue, Glu-179; magenta, Glu-98; purple, Phe-175. The intermediates are colored by their elements: green, carbon; red, oxygen; orange, phosphorus; blue, nitrogen; light green, fluorine. c, stereo view of the folded conformation with electron density from an omit Fo − Fc map contoured at 2.7 σ and shown as blue wire mesh. Other color schemes are the same as in b. The average B-factor is 76 Å2 for the folded intermediate.
In fact, both CD38 and the cyclase are multifunctional enzymes that can also use NADP as a substrate and, in the presence of nicotinic acid, produce nicotinic acid adenine dinucleotide phosphate (NAADP) via a base-exchange reaction (16). NAADP was first shown to have Ca2+-mobilizing activity in sea urchin eggs (17) and has since been established as another Ca2+ messenger molecule targeting yet another intracellular Ca2+ store, the lysosome, in a variety of cell types (18–20).
To elucidate the mechanism of cyclizing a long linear substrate such as NAD to a compact cyclic product, cADPR, here we present the crystal structure of a stable complex of the cyclase with a substrate analog of NAD. The structure clearly identified critical residues for the cyclization process, which were verified by site-directed mutagenesis. The results demonstrate that catalysis by CD38 or the cyclase is controlled by one or two critical residues and that mutating them can interconvert the reactivities of the two enzymes. This study sets the stage for engineering enzymes with specific activity toward cADPR for expression in cells, which should be valuable tools for manipulating the function and metabolism of this novel Ca2+ messenger.
EXPERIMENTAL PROCEDURES
Protein Production and Mutagenesis
A yeast expression system, including the pPICZαA expression vector and Pichia pastoris yeast (Invitrogen), was used to prepare the wild-type cyclase and CD38 as reported previously (21, 22). Mutants of the cyclase and CD38 were prepared using a QuikChange II XL site-directed mutagenesis kit from Stratagene. The mutations were confirmed by DNA sequencing. The mutated plasmids were linearized by treatment with SacI and electroporated into yeast. Following growth in glucose-enriched medium, the yeast were treated with methanol for 1–3 days to induce protein expression. The yeast media containing the specific proteins were recovered and stored frozen until processed. All proteins were purified by a two-step chromatographic procedure including phenyl-Sepharose and SP-Sepharose cation exchange columns (Sigma-Aldrich). Protein concentrations were determined by Bradford assay (23).
Crystallization and Complex Formation
All crystals were obtained by the hanging drop vapor diffusion technique. Crystallization was achieved by mixing 1 μl of protein sample with 1 μl of precipitant (0.1 m imidazole, pH 7.5, 12–24% polyethylene glycol 4000). Depending on polyethylene glycol 4000 concentration, wild-type cyclase crystals can be P1, P21, or P61. NAD was purchased from Sigma. Ribo-2′F-NAD was synthesized accordingly to a published procedure (24). To obtain the cyclase complex, preformed crystals were soaked with the ribo-2′F-NAD at 4 °C and were immediately flash-frozen with liquid nitrogen.
Crystallographic Data Collection and Structural Refinement
X-ray diffraction data were collected at 100 K, at the A1 station of the Cornell High Energy Synchrotron Source (CHESS). All data sets were processed by using the program HKL2000 suite(25). The crystallographic statistics are listed in Table 1. The protein/nucleotide structure was determined by difference Fourier calculation with the starting phases derived from the wild-type cyclase (Protein Data Bank (PDB) ID: 1LBE). The models for ribo-2′F-ADPR were built manually in O (26) based on the σA weighted Fo − Fc difference electron density maps. Structure refinements for these complexes were performed using the program REFMAC with manually modified stereochemical restraints generated from the program PRODRG (27). TLS group refinements were introduced to model data anisotropy. Solvents were added automatically by Arp/warp (28) and manually inspected and modified in the program O. The refinement results and model statistics are listed in Table 1.
TABLE 1.
Crystallographic data and refinement statistics for WT ADPRC/ribo-2′F-ADPR
Values in parentheses are from the highest resolution shell.
| Data collection | |
| Cell dimensions a, b, c (Å), α,β,γ (°) | 56.7, 56.7, 360, 90, 90, 120 |
| Space group | P61 |
| Resolution (Å) | 50.0-3.0 (3.11-3.0) |
| Unique reflections | 12475 |
| Multiplicity | 6.6 (6.7) |
| I/σ | 11.84 (2.7) |
| Rmergea (%) | 13.3 (48.8) |
| Completeness (%) | 95.3 (100.0) |
| Refinement | |
| Resolution (Å) | 50.0-3.0 |
| R-factor (%) | 22.1 |
| Rfreeb factor (%) | 26.8 |
| Protein atoms | 4024 |
| Ligands | 2 |
| Mean B-factor for protein atoms (Å2) | 45.5 |
| B-factor for ligands (Å2) | 76 (folded conformation); 70 (extended conformation) |
| r.m.s.c deviations | |
| Bond lengths (Å) | 0.02 |
| Bond angles (°) | 1.99 |
aRmerge = Σ|I − 〈I〉|/ΣI, where I is the integrated intensity of a given reflection.
b Rfree = Σ Fobs| − |Fcalc
Fobs| − |Fcalc /Σ| Fobs|. Rfree was calculated using 5% of data excluded from refinement.
/Σ| Fobs|. Rfree was calculated using 5% of data excluded from refinement.
c r.m.s., root mean square.
Enzyme Activity Assays
The cyclase or CD38 mutants (0.5–5 μg/ml) were incubated with NAD at concentrations ranging from 50 to 900 μm in 25 mm Tris-HCl, pH 8. The reaction times ranged from 1 to 10 min, and the incubations were stopped with 0.2% SDS and analyzed by HPLC using a column of AG MP-1 resin (10 × 120 mm). The nucleotides were eluted with a gradient of trifluoroacetic acid as described previously (21). Vmax and Km values were determined by nonlinear regression analysis and are shown as average ± S.E., n = 4 determinations. All assays were performed at room temperature (24 °C).
The GDP-ribosyl cyclase activities of the expressed proteins (1–5 μg/ml) were determined with 50–400 μm nicotinamide guanine dinucleotide (NGD) (Sigma) in 50 mm Tris-HCl, pH 8, in a fluorescence plate reader (excitation, 300 nm, emission, 410 nm). The assay depends on the catalytic conversion by cyclase or CD38 of the nonfluorescent NGD substrate to the fluorescent cyclic GDP-ribose product. The kinetic constants were determined by nonlinear regression and represent average values ± S.E., n = 4.
Incubations with cADPR as a Substrate
cADPR was prepared by incubating 1 mm NAD with cyclase (0.5 μg/ml) for 16 h in 20 mm Tris-HCl, pH 7, and purified by HPLC. Wild-type CD38, T221F/E146A, and T221F (all at 5 μg/ml) were incubated with 1 mm cADPR in 50 mm Tris-HCl, pH 8, for 10, 120, or 120 min, respectively. The reactions were stopped by adding 150 mm HCl. The cADPR remaining was analyzed by converting it to NADH as described previously (29, 30). A 10-μl aliquot of an acid-stopped sample was added to 200 μl of a reagent consisting of 50 mm Tris-HCl, pH 8, 10 mm nicotinamide, 1% ethanol, wild-type cyclase (0.2 μg/ml), and alcohol dehydrogenase (10 μg/ml). NADH production was monitored in a fluorescence plate reader (excitation, 355 nm, emission, 460 nm). The rate of cADPR hydrolysis was calculated from the decrease in fluorescence of samples incubated with enzyme as compared with control incubations without enzyme.
RESULTS AND DISCUSSION
It is intuitively clear that to cyclize a linear substrate to a cyclic product, the active site of the enzyme must bind and fold the substrate into a ring so that the two ends can be linked. We have previously solved the crystal structures of the cyclase and its complex with co-substrates of the base-exchange reaction, such as nicotinamide (21, 31, 32). To visualize the cyclization process, however, we had to choose an appropriate substrate that can form a stable complex with the cyclase without being consumed and released as product. One possibility is ara-2′F-NMN, which has been shown to form a covalent intermediate with CD38 and inhibit its enzyme activity (33, 34). However, ara-2′F-NMN cannot form a cyclic product because it is missing the adenine group. The cyclization site in cADPR is between the N-1 of adenine and the anomeric carbon of the terminal ribose (Fig. 1a). We thus, instead, chose an analog of NAD, ribo-2′-F-NAD (Fig. 1a), for this purpose. We have previously shown that ribo-2′F-NAD and its isomer ara-2′F-NAD both form covalent intermediates with CD38 (35). Both analogs were tested with the cyclase, and only ribo-2′F-NAD, but not ara-2′F-NAD, was found to form a stable complex after infusing the analog into preformed cyclase crystals. The crystallographic data and refinement statistics of the complex are shown in Table 1.
As we have shown before, the cyclase is a homodimer, and the intermediate is at the active site near the middle of each monomer (21, 32) (Fig. 1b). We have reported a number of different crystal forms of the cyclase, including PDB ID 1LBE, which is in P212121; 1ROS, which is in P21, and 1R15, which is in P1. In all these crystal forms, the basic assembly is always a homodimer. In the present study, the crystal form is P61, which is a new form. There is no obvious conformational difference among these forms. In the P61 form, the symmetry manipulation of the asymmetric unit shows no crystallographic packing on the side of the active site for both molecules of the dimer. Both active sites face directly to solvent, allowing access of the substrate. The fact that a reaction adduct (ribo-2′F-ADP-ribose) can be obtained by soaking the preformed crystals with the substrate suggests that there is no obvious adverse influence of crystal packing on the catalytic activity of the active sites.
The alignment of the two chains of the dimer in the present structure gives a root mean square of 0.122 Å for all CA atoms. Therefore, the differences between the two molecules are small. Fig. 1c shows the simulated annealing omit Fo − Fc difference electron density of the intermediate contoured at 2.7 σ. The nicotinamide group of the substrate was released, and the ribose unit formed close association with the catalytic residue, Glu-179, of the cyclase (22). The close proximity suggests a covalent linkage, as we have previously shown for CD38 (35), although the resolution of the structure of the complex (3.0 Å) does not permit a definitive determination of the nature of the linkage in cyclase.
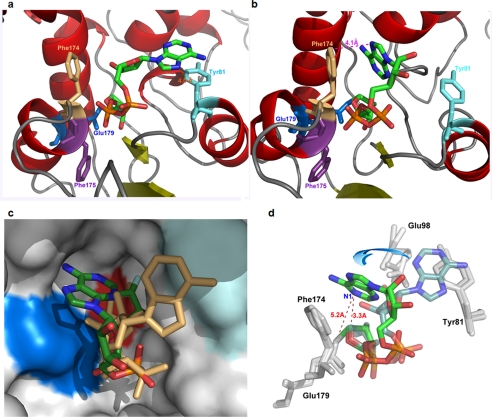
At the other end of the substrate, the adenine ring was found to have two different binding sites at each of the two monomers of the cyclase dimer. One site was close to Tyr-81 (Fig. 2a). Binding to this site, the substrate was in an extended configuration, and the two ends were too far apart for cyclization. The other site was close to Phe-174, where the adenine ring was only 4.1 Å from and in hydrophobic stacking with the aromatic ring of the residue, as shown in Fig. 2b. Binding to this site, the N1 of the adenine ring, the cyclization site, was within 3.3 Å from the C1′ of the terminal ribose and 5.2 Å from the glutamate oxygen of the catalytic residue (Glu-179), well positioned for a nucleophilic attack on the intermediate that would result in cyclization (Fig. 2d). That the two monomers in a crystallographic asymmetric unit can interact with the substrate differently has previously been shown for CD38 using NGD as substrate (36) and provides a convincing way to observe distinct reaction states in a self-consistent manner.
FIGURE 2.
Two conformations of the intermediate from ribo-2′F-NAD. a, the extended conformation. The orange dashed line shows that the distance between the adenine and Tyr-81 is 4.0 Å. Phe-175 is colored in purple. Other color schemes are as in Fig. 1b. b, the folded conformation. The magenta dashed line shows that the distance between the adenine and Phe-174 is 4.1 Å. c, surface view of the active site pocket with the two conformations of intermediate superimposed. The folded conformation is colored by its elements as in Fig 1a. The extended conformation is colored beige. Surface color is as follows: deep blue, Phe-174; cyan, Tyr-81; red, Glu-179; white, all other residues. d, rotational conversion between the two conformations. The folded conformation is brightly colored by its elements as in Fig. 1a. The distance between N1 of the adenine is 5.2 Å from Glu-179 in this conformation. The extended conformation is lightly colored by its elements.
Fig. 2c shows the surface view of the active site, with the two forms of the substrate superimposed. The surface of the two binding sites, Tyr-81 and Phe-174, are colored light and dark blue, respectively. Tyr-81 is at the rim of the active site pocket, and the adenine that binds to it is extended out of the active site. In contrast, Phe-174 is located deeper into the pocket, and the interacting adenine is close to the bottom of the active site, where the catalytic residue, Glu-179 (colored red), is located. The two conformations of the substrate are interconvertible by a series of rotations, as indicated in Fig. 2d. These rotations involve only single bonds, and the movement of the adenine ring does not produce any clashes with the residues of the active site, as shown in the supplemental movie. That the two conformations can be mapped rotationally provides a strong support for the self-consistency of the structural determination.
The structural results suggest the following reaction scenario. The binding of the adenine ring to the Tyr-81 site may well be the first step. The nicotinamide end of the substrate then enters the active site pocket and interacts with the catalytic residue, Glu-179, at the bottom of the pocket, resulting in the hydrolysis of the glycosidic bond and release of the nicotinamide. Through thermodynamic single-bond rotations, the adenine ring folds back and enters the active site. The observed hydrophobic stacking with Phe-174 stabilizes this folded conformation and increases the chance of nucleophilic linkage between the ribosyl end of the intermediate and N1 of the adenine, finally resulting in cyclization and production of cADPR.
This scenario was tested by site-directed mutagenesis. We first mutated the Tyr-81 to a charged residue, lysine. This resulted in a more than 12-fold decrease in the cyclization activity (Vmax = 49,000 ± 4,000 nmol/mg/min) as compared with the wild type (Vmax = 600,000 ± 43,000 nmol/mg/min). Changing the residue to an uncharged residue, alanine, resulted in less reduction in activity (Vmax = 170,000 ± 16,000 nmol/mg/min). The results are consistent with Tyr-81 being a binding site for the substrate.
Another corollary of the scenario described above is that if Phe-174 is mutated to reduce the hydrophobic interaction, the binding of the adenine to the site should decrease. The consequence would be a reduction in the cyclization activity, whereas the hydrolysis activity should be correspondingly enhanced because of the greater chance of the alternate attack by water on the intermediate.
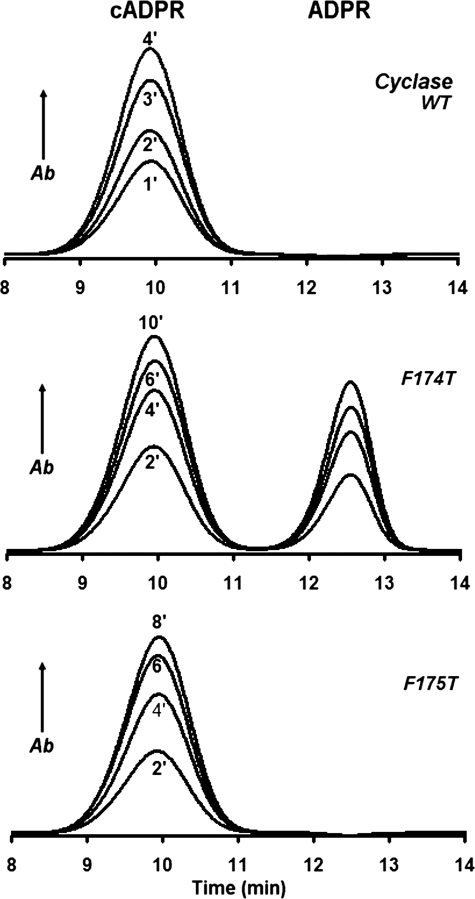
This change in activity was found to be the case, as shown in Fig. 3. Wild-type cyclase was incubated with 0.1 mm NAD for various time periods, and the products were analyzed by HPLC. The only product observed was cADPR (Fig. 3, top panel). Changing Phe-174 to Thr greatly increased the hydrolysis activity, and about 40% of the product was now ADP-ribose (Fig. 3, middle panel). The effect was specific for Phe-174 because changing the adjacent Phe-175 to Thr produced no such enhancement of hydrolysis, and the product remained only cADPR (Fig. 3, bottom panel). Consistent with these results, as shown in Fig. 2a, Phe-175 (colored purple) is directed away from and does not interact closely with the intermediate.
FIGURE 3.
HPLC analyses of the enzymatic products from NAD produced by the cyclase and its mutants. WT cyclase (upper panel, 0.1 μg/ml), F174T (middle panel, 1 μg/ml), or F175T (lower panel, 1 μg/ml) was incubated with 100 μm NAD for the indicated times and analyzed by HPLC. The part of the HPLC chromatogram that includes only the cADPR and ADPR peaks is shown for clarity. Of the three proteins, only the F174T mutant produces ADPR. Ab, absorbance.
Table 2 shows the enzyme kinetic data of the various cyclase mutants. The wild-type cyclase could only cyclize NAD to produce cADPR and did not hydrolyze NAD to ADPR. Changing Phe-174 to Ala, Gly, Ser, or Thr bestowed the cyclase with the new NAD hydrolysis activity (NAD → ADPR) to varying degree, the most active being the Ala mutant. In contrast and as a specificity control, mutation of the adjacent Phe-175 did not confer any NADase activity. The ratio of cADPR to ADPR produced from NAD was lowest for the Phe-174 to Thr (F174T) mutant, reaching a value of 1.3 and indicating that 43% of the product was ADPR (compare Fig. 3). The various mutations also dramatically inhibited the cyclase activity (NAD → cADPR) as compared with the wild-type cyclase (Table 2). The inhibitory effect of the mutations was specific and was apparent only when NAD was used as a substrate. When NGD was used as a substrate, the cyclase converted it into cyclic GDP-ribose (NGD → cGDPR), and this reaction was only minimally affected by the mutations on Phe-174, as shown in Table 3. Consistently, we have previously shown that the cyclization site in cGDPR is at the N-7 of the guanine ring in contrast to the N-1 of the adenine ring in cADPR (37, 38). The folding of the guanine is thus likely to involve a residue different from Phe-174.
TABLE 2.
Effects of mutating Phe-174 or Phe-175 on the enzymatic activities of the cyclase with NAD as substrate
| Cyclase | NAD→ADPR |
cADPR/ADPR | NAD→cADPR |
||
|---|---|---|---|---|---|
| Vmax | Km | Vmax | Km | ||
| nmol/mg/min | μm | nmol/mg/min | μm | ||
| WT | 0 | ∞ | 600,000 ± 43,000 | 200 ± 40 | |
| F174A | 5,900 ± 400 | 600 ± 90 | 6.6 | 39,000 ± 1,900 | 400 ± 50 |
| F174G | 1,800 ± 800 | 300 ± 120 | 2.2 | 4,000 ± 700 | 200 ± 80 |
| F174S | 3,800 ± 300 | 300 ± 60 | 2.9 | 11,000 ± 700 | 100 ± 20 |
| F174T | 5,000 ± 800 | 600 ± 180 | 1.3 | 7,000 ± 900 | 300 ± 80 |
| F175A | 0 | 22,000 ± 1,600 | 20 ± 8 | ||
| F175T | 0 | 21,000 ± 700 | 70 ± 6 | ||
| F175G | 0 | 58,000 ± 5,000 | 100 ± 20 | ||
TABLE 3.
Effects of mutating Phe-174 on the cyclization of NGD
| Cyclase | NGD→cGDPR |
|
|---|---|---|
| Vmax | Km | |
| nmol/mg/min | μm | |
| WT | 44,000 ± 2,000 | 11 ± 2 |
| F174A | 73,000 ± 5,000 | 400 ± 40 |
| F174G | 31,000 ± 4,000 | 600 ± 100 |
| F174S | 50,000 ± 3,000 | 400 ± 40 |
| F174T | 23,000 ± 400 | 400 ± 90 |
The overall rates of NAD utilization of the Phe-174 mutants were less than the wild-type cyclase but are comparable with that of the wild-type CD38 (Table 4). For example, the Vmax values of the F174A mutant were 39,000 nmol/mg/min for the cyclization reaction and 5,900 nmol/mg/min for the hydrolysis reaction (Table 2). The total rate of the two reactions was about 55% of the Vmax value of the wild-type CD38. This is consistent with the fact that mutations at Phe-174 are converting the cyclase toward an enzyme that is catalytically more CD38-like.
TABLE 4.
Illustration of mutational conversion of CD38 to a cyclase-like enzyme
ND; not determined due to low activity.
| CD38 | NAD→cADPR |
NAD→ADPR |
||
|---|---|---|---|---|
| Vmax | Km | Vmax | Km | |
| nmol/mg/min | μm | nmol/mg/min | μm | |
| WT | ∼0 | ND | 81,000 ± 3,000 | 11 ± 3 |
| T221F/E146A | 2,000 ± 300 | 4 ± 3 | 30 ± 2 | ND |
| T221F | 70 ± 4 | ND | 3,000 ± 200 | 30 ± 6 |
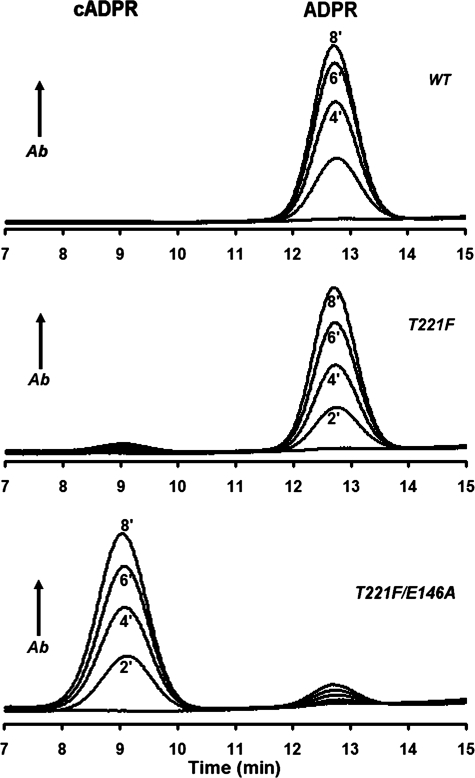
In CD38, Thr-221 is the residue that corresponds to Phe-174 of cyclase. As shown in Fig. 3, changing Phe-174 to Thr converted the cyclase to more CD38-like, producing ADPR from NAD. It was thus of interest to know whether changing Thr-221 to Phe converts CD38 to more cyclase-like, thereby enhancing the cyclization activity of CD38. The HPLC chromatograph in Fig. 4 shows that the product of incubating wild-type CD38 with NAD was mainly ADPR. Cyclic ADP-ribose was also produced, but the amounts were generally not detectable by HPLC (top panel). Changing Thr-221 to Phe did modestly enhance the cyclization reaction, and the mutant enzyme produced significant amounts of cADPR, which are definitely detectable by HPLC (middle panel). Essentially complete conversion of CD38 to a cyclase-like enzyme could be achieved, however, by a double mutation, Thr-221 to Phe and Glu-146 to Ala. As shown in of Fig. 4, in the bottom panel, in this case over 94% of the catalytic product from NAD was cADPR. We have previously shown that Glu-146 is important in controlling the access of water to the active site in CD38 (39). Changing it to a less hydrophilic Ala reduces water access, which together with the enhancement of the cyclization attack by the adenine, because of the Thr-221 mutation, enables the essentially complete conversion of CD38 to a cyclase-like enzyme. These results on mutational interconversion indicate that the basic catalytic mechanisms of CD38 and the cyclase are indeed very similar. The residue in the cyclase equivalent to Glu-146 is Glu-98, and its location is shown in Figs. 1b and 2d.
FIGURE 4.
HPLC analyses of the enzymatic products from NAD produced by CD38 and its mutants. WT CD38 (upper panel, 0.2 μg/ml), T221F (middle panel, 5 μg/ml), or T221F/E146A (lower panel, 2 μg/ml) was incubated with 100 μm NAD for the indicated times and analyzed by HPLC. There was no detectable cADPR production by WT CD38 under these conditions, the T221F mutant produced some detectable cADPR, and the T221F/E146A double mutant produced predominantly cADPR. Ab, absorbance.
Enzyme kinetic data for the CD38 mutants are listed in Table 4. The double mutation enhanced the cyclase activity by about 30-fold, as compared with the single mutation, whereas the wild type had no detectable cyclase activity by HPLC. Correspondingly, the NADase activity was greatly inhibited by the single mutation and was barely detectable in the double mutant, as compared with the wild type. It has been well established that wild-type CD38 is a multifunctional enzyme that can use not only NAD as substrate but also cADPR, which is hydrolyzed to ADPR (12–15). This cADPR hydrolysis activity was also greatly inhibited by the mutations, as shown in Table 5. The inhibitory effect of the mutations was, however, very specific and was not observed in the cyclization reaction of NGD (Table 5). Instead, it was similarly enhanced, as shown for the cyclization of NAD.
TABLE 5.
Effects of mutating Thr221 on the cADPR hydrolysis and NGD cyclization activities of CD38
| CD38 | cADPR→ADPR Va | NGD→cGDPR |
|
|---|---|---|---|
| Vmax | Km | ||
| nmol/mg/min | nmol/mg/min | μm | |
| WT | 6,000 ± 2,000 | 31,000 ± 400 | 5 ± 1 |
| T221F/E146A | 0 | 100,000 ± 10,000 | 60 ± 10 |
| T221F | 500 ± 20 | 5,000 ± 90 | 10 ± 1 |
aActivities were determined only at 1 mm cADPR, because the activities of the mutants were too low for Vmax/Km determination.
The structural and mutational evidence presented in this study provides the first detailed account of the mechanism of cyclizing NAD to cADPR by the cyclase, the first enzyme identified for the synthesis of cADPR (7). The results also set the stage for engineering the reactivity of CD38 for the specific action on cADPR. Of particular interest is the double mutant listed in Tables 4 and 5 because it has minimal cADPR and NAD hydrolysis activities but can efficiently cyclize NAD to cADPR. Expression of this mutant in cells should lead to increased cellular cADPR levels and should provide a useful tool for assessing the physiological function of cADPR in cells and tissues.
Acknowledgments
The crystallographic data were collected at CHESS, which is supported by the National Science Foundation and NIGMS, National Institutes of Health, under award DMR-0225180. MacCHESS is supported by National Institutes of Health Grant RR01646.
This work was supported, in whole or in part, by National Institutes of Health Grant GM061568 (to H. C. L./Q. H.). This work was also supported by grants from the Hong Kong General Research Fund and the National Science Foundation of China/Research Grant Council of Hong Kong (NSFC/RGC) Joint Research Scheme (to H. C. L.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 3I9O) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and a supplemental movie.
- ADPR
- ADP-ribose
- cADPR
- cyclic ADP-ribose
- NGD
- nicotinamide guanine dinucleotide
- HPLC
- high pressure liquid chromatography
- WT
- wild type
- ribo-2′-F-NAD
- ribosyl-2′F-2′deoxynicotinamide adenine dinucleotide
- ara-2′F-NMN
- arabinosyl-2′-fluoro-2′-deoxynicotinamide mononucleotide
- ribo-2′F-ADP-ribose
- ribosyl-2′-fluoro-2′-deoxy-adenosine diphosphate ribose.
REFERENCES
- 1.Clapper D. L., Walseth T. F., Dargie P. J., Lee H. C. (1987) J. Biol. Chem. 262, 9561–9568 [PubMed] [Google Scholar]
- 2.Lee H. C., Walseth T. F., Bratt G. T., Hayes R. N., Clapper D. L. (1989) J. Biol. Chem. 264, 1608–1615 [PubMed] [Google Scholar]
- 3.Lam C. M., Yeung P. K., Lee H. C., Wong J. T. (2009) Cell Calcium 45, 346–357 [DOI] [PubMed] [Google Scholar]
- 4.Jin D., Liu H. X., Hirai H., Torashima T., Nagai T., Lopatina O., Shnayder N. A., Yamada K., Noda M., Seike T., Fujita K., Takasawa S., Yokoyama S., Koizumi K., Shiraishi Y., Tanaka S., Hashii M., Yoshihara T., Higashida K., Islam M. S., Yamada N., Hayashi K., Noguchi N., Kato I., Okamoto H., Matsushima A., Salmina A., Munesue T., Shimizu N., Mochida S., Asano M., Higashida H. (2007) Nature 446, 41–45 [DOI] [PubMed] [Google Scholar]
- 5.Lee H. C. (2002) Cyclic ADP-ribose and NAADP: Structures, Metabolism and Functions, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 6.Lee H. C. (2004) Curr. Mol. Med. 4, 227–237 [DOI] [PubMed] [Google Scholar]
- 7.Lee H. C., Aarhus R. (1991) Cell Regul. 2, 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mothet J. P., Fossier P., Meunier F. M., Stinnakre J., Tauc L., Baux G. (1998) J. Physiol. 507.2, 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bezin S., Charpentier G., Lee H. C., Baux G., Fossier P., Cancela J. M. (2008) J. Biol. Chem. 283, 27859–27870 [DOI] [PubMed] [Google Scholar]
- 10.Kato I., Yamamoto Y., Fujimura M., Noguchi N., Takasawa S., Okamoto H. (1999) J. Biol. Chem. 274, 1869–1872 [DOI] [PubMed] [Google Scholar]
- 11.Partida-Sánchez S., Cockayne D. A., Monard S., Jacobson E. L., Oppenheimer N., Garvy B., Kusser K., Goodrich S., Howard M., Harmsen A., Randall T. D., Lund F. E. (2001) Nat. Med. 7, 1209–1216 [DOI] [PubMed] [Google Scholar]
- 12.Howard M., Grimaldi J. C., Bazan J. F., Lund F. E., Santos-Argumedo L., Parkhouse R. M., Walseth T. F., Lee H. C. (1993) Science 262, 1056–1059 [DOI] [PubMed] [Google Scholar]
- 13.Kim H., Jacobson E. L., Jacobson M. K. (1993) Science 261, 1330–1333 [DOI] [PubMed] [Google Scholar]
- 14.Lee H. C., Zocchi E., Guida L., Franco L., Benatti U., De Flora A. (1993) Biochem. Biophys. Res. Commun. 191, 639–645 [DOI] [PubMed] [Google Scholar]
- 15.Takasawa S., Tohgo A., Noguchi N., Koguma T., Nata K., Sugimoto T., Yonekura H., Okamoto H. (1993) J. Biol. Chem. 268, 26052–26054 [PubMed] [Google Scholar]
- 16.Aarhus R., Graeff R. M., Dickey D. M., Walseth T. F., Lee H. C. (1995) J. Biol. Chem. 270, 30327–30333 [DOI] [PubMed] [Google Scholar]
- 17.Lee H. C., Aarhus R. (1995) J. Biol. Chem. 270, 2152–2157 [DOI] [PubMed] [Google Scholar]
- 18.Churchill G. C., Okada Y., Thomas J. M., Genazzani A. A., Patel S., Galione A. (2002) Cell 111, 703–708 [DOI] [PubMed] [Google Scholar]
- 19.Galione A. (2006) Biochem. Soc. Trans. 34, 922–926 [DOI] [PubMed] [Google Scholar]
- 20.Guse A. H., Lee H. C. (2008) Sci. Signal. 1, re10. [DOI] [PubMed] [Google Scholar]
- 21.Munshi C., Aarhus R., Graeff R., Walseth T. F., Levitt D., Lee H. C. (2000) J. Biol. Chem. 275, 21566–21571 [DOI] [PubMed] [Google Scholar]
- 22.Munshi C., Thiel D. J., Mathews I. I., Aarhus R., Walseth T. F., Lee H. C. (1999) J. Biol. Chem. 274, 30770–30777 [DOI] [PubMed] [Google Scholar]
- 23.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 24.Sleath P. R., Handlon A. L., Oppenheimer N. J. (1991) J. Org. Chem. 56, 3608–3613 [Google Scholar]
- 25.Otwinowski Z., Minor W. (1997) Macromol. Crystallog., Pt. A. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 27.Schüttelkopf A. W., van Aalten D. M. F. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 28.Morris R. J., Perrakis A., Lamzin V. S. (2003) Macromol. Crystallog., Pt. D. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 29.Graeff R. M., Lee H. C. (2003) Comb. Chem. High Throughput Screen 6, 367–379 [DOI] [PubMed] [Google Scholar]
- 30.Graeff R., Lee H. C. (2002) Biochem. J. 361, 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love M. L., Szebenyi D. M., Kriksunov I. A., Thiel D. J., Munshi C., Graeff R., Lee H. C., Hao Q. (2004) Structure 12, 477–486 [DOI] [PubMed] [Google Scholar]
- 32.Prasad G. S., McRee D. E., Stura E. A., Levitt D. G., Lee H. C., Stout C. D. (1996) Nat. Struct. Biol. 3, 957–964 [DOI] [PubMed] [Google Scholar]
- 33.Sauve A. A., Deng H. T., Angeletti R. H., Schramm V. L. (2000) J. Am Chem. Soc. 122, 7855–7859 [Google Scholar]
- 34.Liu Q., Kriksunov I. A., Jiang H., Graeff R., Lin H., Lee H. C., Hao Q. (2008) Chem Biol 15, 1068–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Q., Graeff R., Kriksunov I. A., Jiang H., Zhang B., Potter B. V. L., Oppenheimer N., Lin H., Lee H. C., Hao Q. (2009) J. Biol. Chem. 284, 27637–27645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Q., Kriksunov I. A., Graeff R., Munshi C., Lee H. C., Hao Q. (2006) J. Biol. Chem. 281, 32861–32869 [DOI] [PubMed] [Google Scholar]
- 37.Graeff R. M., Walseth T. F., Fryxell K., Branton W. D., Lee H. C. (1994) J. Biol. Chem. 269, 30260–30267 [PubMed] [Google Scholar]
- 38.Graeff R. M., Walseth T. F., Hill H. K., Lee H. C. (1996) Biochemistry 35, 379–386 [DOI] [PubMed] [Google Scholar]
- 39.Graeff R., Munshi C., Aarhus R., Johns M., Lee H. C. (2001) J. Biol. Chem. 276, 12169–12173 [DOI] [PubMed] [Google Scholar]