Abstract
Cyclic ADP-ribose (cADPR) is a universal calcium messenger molecule that regulates many physiological processes. The production and degradation of cADPR are catalyzed by a family of related enzymes, including the ADP-ribosyl cyclase from Aplysia california (ADPRAC) and CD38 from human. Although ADPRC and CD38 share a common evolutionary ancestor, their enzymatic functions toward NAD and cADPR homeostasis have evolved divergently. Thus, ADPRC can only generate cADPR from NAD (cyclase), whereas CD38, in contrast, has multiple activities, i.e. in cADPR production and degradation, as well as NAD hydrolysis (NADase). In this study, we determined a number of ADPRC and CD38 structures bound with various nucleotides. From these complexes, we elucidated the structural features required for the cyclization (cyclase) reaction of ADPRC and the NADase reaction of CD38. Using the structural approach in combination with site-directed mutagenesis, we identified Phe-174 in ADPRC as a critical residue in directing the folding of the substrate during the cyclization reaction. Thus, a point mutation of Phe-174 to glycine can turn ADPRC from a cyclase toward an NADase. The equivalent residue in CD38, Thr-221, is shown to disfavor the cyclizing folding of the substrate, resulting in NADase being the dominant activity. The comprehensive structural comparison of CD38 and APDRC presented in this study thus provides insights into the structural determinants for the functional evolution from a cyclase to a hydrolase.
Cyclic ADP-ribose (cADPR)3 is a calcium messenger ubiquitous in mammals as well as in invertebrates and plants and is responsible for regulating many physiological processes ranging from the simple function of calcium channel operation to the complex higher level organization of hormone secretion and autism (reviewed in Lee (1), Schuber and Lund (2), and Malavasi et al. (3)). The enzymatic production of cADPR from the substrate nicotinamide adenine dinucleotide (NAD) requires first the removal of the nicotinamide moiety followed by a cyclization reaction in which both ends of the remaining nucleotide are annealed (Fig. 1A). ADP-ribosyl cyclase (ADPRC) from Aplysia california was the first enzyme discovered to possess this function (cyclase) (4). Based on sequence homology (5), two human antigens, CD38 and CD157, were identified to also have the cyclase activity (6–8). However, different from ADPRC, which produces only cADPR from NAD, CD38/CD157 has evolved more like an NADase, producing mainly ADP-ribose (ADPR) from NAD, with cADPR being a minor product. The acquisition of the NADase and the cADPR hydrolysis activities of CD38 make it an important signaling enzyme in regulating NAD and cADPR homeostasis (9–11). Genetic analysis, as well as the conservation of sequence and disulfide bonds among these enzymes, establish that they all evolved from a common ancestor (12). Little is known of why this conserved family of enzymes has evolved divergently in their catalytic metabolism of NAD and cADPR.
FIGURE 1.
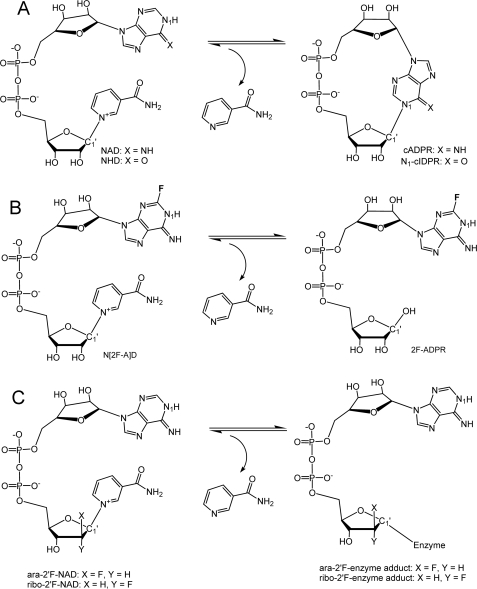
Schemes of cADPR formation and mechanistic analogs for substrate and product. A, the cyclization reaction producing cADPR from NAD is catalyzed by both ADPRC and CD38. The structural difference between cADPR and N1-cIDPR lies at the 6-position of purine ring (6-NH for cADPR; 6-O for N1-cIDPR). B, an analog of the substrate NAD, N(2F-A)D, is enzymatically converted to 2F-ADPR by ADPRC instead of cyclized to c(2F-A)DPR. The formation of cADPR from NAD requires the intramolecular attack of the reaction intermediate by the adenine N1 atom. The addition of a fluorine atom on the adjacent C2 atom of adenine prevents the cyclization from occurring. C, ara-2′F-NAD and ribo-2′F-NAD are analogs of NAD that inhibit the cyclization reaction by producing covalent adducts during the catalysis by CD38. Both analogs differ only in the orientation of their fluorine atoms at the 2′-position of the adenine ribose.
ADPRC, however, is not solely a cyclase because it can also catalyze the hydrolysis of NMN into ribose-5-phosphate and nicotinamide (13, 14). The catalytic outcome of this novel enzyme is thus determined not by the enzyme alone but also by the specific interactions between the active site and a particular substrate. Consistently, using an NAD analog, N(2F-A)D, as substrate, Zhang et al. (15) showed that the hydrolase activity of ADPRC can be dominantly revealed, whereas its cyclase activity is suppressed beyond detection (Fig. 1B). Likewise, human CD38 can be converted to a ADPRC-like enzyme by mutation of a single residue, Glu-146, at the active site (16). In this study, we determined the structural determinants critical for the catalytic characteristics of ADPRC and CD38 by comparing the crystal structures of the complexes of ADPRC and CD38 bound with various catalytically revealing substrates and products (Fig. 1, A–C). The results identify residues Phe-174 in the cyclase and Thr-221 in CD38 as the main determinants for the cyclase and hydrolysis activities of the enzymes. All together, these structures provide insights into the structural requirements for functional evolution from a cyclase to a hydrolase.
EXPERIMENTAL PROCEDURES
Crystallization and Complex Formation
Production of recombinant CD38 and Aplysia ADPRC were as described previously (17, 18). All crystals were obtained by the hanging drop vapor diffusion technique. Crystallization of ADPRC was done by mixing 1 μl of protein sample with 1 μl of precipitants (0.1 m imidazole, pH 7.5, 12–24% polyethylene glycol 4000). Dependent on polyethylene glycol 4000 concentration, wild-type ADPRC crystals can be crystallized in space group P1, P21, or P61, whereas E179GADPRC mutant crystals are always in space group P21. NAD was purchased from Sigma-Aldrich. N(2F-A)D, ara-2′F-NAD, and ribo-2′F-NAD were synthesized according to established procedures (15, 19). N1-cIDPR was obtained by chemoenzymatic routes as illustrated (20).
The E179GADPRC·NAD complex was obtained by soaking crystals with NAD (10 mm) in mother liquor containing 25% glycerol for about 2 min. The WTADPRC·2F-ADPRI· Nic complex was obtained by soaking WTADPRC crystals with a solution of N(2F-A)D (33 mm) for 3 min, whereas the E179GADPRC·N1-cIDPR complex was obtained by soaking mutant ADPRC crystals in 12 mm N1-cIDPR solution for 8 min. All soaking experiments were carried out at 4 °C for optimal results. After soaking, crystals were immediately flash-frozen with liquid nitrogen.
Wild-type CD38 was crystallized as described previously (21). The WTCD38·ara-2′F-ADPR and WTCD38·ribo-2′F-ADPR complexes were obtained by soaking crystals in a solution containing either 5.2 mm ara-2′F-NAD or 3 mm ribo-2′F-NAD and 100 mm MES, pH 6.0, 15% polyethylene glycol 4000, and 30% glycerol. For these two NAD analogs, covalent adducts are formed after nicotinamide cleavage, resulting in inhibition of the enzyme activity. The soaking is thus insensitive to time (1 min or several days) and temperature (4 or 24 °C).
Crystallographic Data Collection and Structural Refinement
X-ray diffraction data were collected at 100 K at the Cornell High Energy Synchrotron Source (CHESS) A1 station. All data sets were processed by using the program HKL2000 suite (22). The crystallographic statistics are listed in Table 1. All complex structures were determined by difference Fourier calculation with the starting phases derived from the WTCD38 model (Protein Data Bank (PDB) ID: 1YH3) for CD38 complexes and WTADPRC (PDB ID: 1BLE) for ADPRC complexes. The models for all ligands were built manually in O (23) based on the σA weighted Fo − Fc difference electron density maps. For CD38 complexes, the catalytic residue Glu-226 was modeled as covalently linked to ara-2′F-ADPR and ribo-2′F-ADPR. Structure refinements for these complexes were performed using the program REFMAC5 (24) with manually modified stereochemical restraints generated from the program PRODRG (25). TLS group refinements were introduced to model data anisotropy. Solvents were added automatically by Arp/warp (26) and manually inspected and modified in the program O. The refinement results and model statistics are also listed in Table 1.
TABLE 1.
Crystallographic data and refinement statistics
Values in parentheses are from the highest resolution shell.
| WTADPRC·2F-ADPRI·Nic | E179GADPRC·NAD | E179GADPRC·N1-cIDPR | WTCD38·ara-2′F-ADPR | WTCD38·ribo-2′F-ADPR | |
|---|---|---|---|---|---|
| Data collection | |||||
| Cell dimensions a, b, c (Å)/α, β, γ (°) | 57.1 | 69.5 | 69.6 | 41.6 | 42.0 |
| 57.1 | 58.3 | 58.3 | 53.7 | 54.4 | |
| 364.7/90.0 | 71.5/90.0 | 71.7/90.0 | 65.3/106.3 | 64.3/109.8 | |
| 90.0 | 101.2 | 101.4 | 92.0 | 90.9 | |
| 120.0 | 90.0 | 90.0 | 95.1 | 94.9 | |
| Space group | P61 | P21 | P21 | P1 | P1 |
| Resolution (Å) | 350-2.18 (2.26-2.18) | 30-1.83 (1.90-1.83) | 30-1.75 (1.81-1.75) | 30-1.75 (1.81-1.75) | 50-2.0 (2.07-2.0) |
| Unique reflections | 33,025 | 47,800 | 56,813 | 51,062 | 33,891 |
| Multiplicity | 5.4 (5.3) | 3.4 (2.1) | 7.4 (5.9) | 3.9 (3.3) | 2.7 (2.5) |
| I/σ | 13.3 (2.2) | 19.2 (2.1) | 26.3 (5.0) | 23.8 (5.9) | 9.3 (1.9) |
| Rmergea (%) | 8.8 (42.7) | 5.9 (38.3) | 6.6 (38.5) | 4.8 (19.1) | 9.4 (47.9) |
| Completeness (%) | 93.9 (70.3) | 97.2 (81.4) | 99.8 (97.7) | 96.8 (89.7) | 96.6 (95.6) |
| Refinement | |||||
| Resolution (Å) | 20-2.18 | 20-1.83 | 20-1.75 | 20-1.75 | 20-2.0 |
| R-factor (%) | 19.7 | 17.5 | 16.6 | 19.6 | 21.2 |
| Rfreeb factor (%) | 24.8 | 23.1 | 19.6 | 23.6 | 27.8 |
| Protein atoms | 4024 | 4024 | 4024 | 4100 | 4100 |
| Water molecules | 154 | 474 | 638 | 402 | 212 |
| Ligands | 3 | 1 | 1 | 2 | 2 |
| Mean B (Å2) | 50.0 | 31.1 | 25.5 | 27.5 | 31.8 |
| r.m.s.c deviations | |||||
| Bond lengths (Å) | 0.022 | 0.026 | 0.016 | 0.014 | 0.021 |
| Bond angles (°) | 1.931 | 2.047 | 1.971 | 1.549 | 2.097 |
aRmerge = Σ|I − 〈I〉|/ΣI, where I is the integrated intensity of a given reflection.
b Rfree = Σ Fobs| − |Fcalc
Fobs| − |Fcalc /Σ|Fobs|. Rfree was calculated using 5% of data excluded from refinement.
/Σ|Fobs|. Rfree was calculated using 5% of data excluded from refinement.
c r.m.s., root mean square.
Site-directed Mutagenesis of ADPRC
The WTADPRC cDNA was cloned into the pPICZaA vector (Invitrogen) and expressed in Pichia pastoris (27). The F174G mutant was prepared from the wild-type plasmid by using a site-directed mutagenesis kit (Stratagene). The forward primer used during the PCR reaction was 5′-GCTTACAGGCCAGACAGTGGCTTTGGCAAGTACGAACT-3′, and the reverse primer was 5′-AGTTCGTACTTGCCAAAGCCACTGTCTGGCCTGTAAGC-3′. The bases in bold represent the changes made to code for glycine. The E179G mutant was prepared as described previously (17). All cyclase proteins and CD38 were purified by cation exchange chromatography.
Enzyme Assays
Hydrolysis of cADPR by ADPRC. The wild-type ADPRC (5 μg/ml) was incubated (10–40 min) at room temperature with 1 mm cADPR buffered by 25 mm Tris-HCl, pH 8. The total volume of the reaction mixture was 0.1 ml, and the reaction was stopped by the addition of SDS (0.15% final concentration). The reaction products were analyzed by HPLC using an AG MP-1 column 10 × 120 mm (Bio-Rad). The elution was performed using a gradient of trifluoroacetic acid at a flow rate of 3 ml/min. The results shown are mean ± S.E. of four determinations.
Cyclization and hydrolysis of NAD by ADPRC. Wild-type (0.1 μg/ml) or F174G (1 μg/ml) ADPRC was incubated with 0.1 mm NAD in 25 mm Tris-HCl, pH 8, for various times, and the products were analyzed by HPLC as described above. The results are the mean ± S.E. of four determinations.
RESULTS AND DISCUSSION
E179GADPRC·NAD Complex
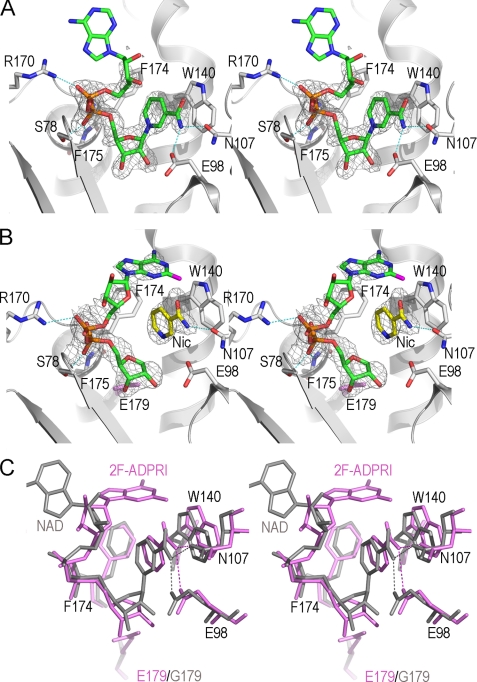
Wild-type ADPRC catalyzes the production of cADPR from NAD. A catalytically inactive mutant E179G (E179GADPRC) (17) was thus used to form a complex with NAD by soaking preformed E179GADPRC crystals with NAD at 4 °C. Due to the greatly reduced catalytic activity in the E179GADPRC mutant (17), NAD accumulated in the active site without being hydrolyzed. The E179GADPRC·NAD complex structure was determined at 1.83 Å resolution, and the active site structure is shown in Fig. 2A. The nicotinamide moiety of NAD has H-bonding with both Glu-98 and Asn-107, and it stacks with the hydrophobic indole ring of Trp-140 in a parallel manner, thereby positioning the N-glycosidic bond into the active site for catalytic cleavage. The diphosphate group of NAD forms H-bonds with residues Arg-170, Ser-78, Phe-174, and Phe-175. These four residues, together with the Glu-98-Asn-107-Trp-140 triplet, seemingly compose a docking site for proper recognition and positioning of NAD into the active site for catalysis. Electron densities for nicotinamide, the nicotinamide ribose, and diphosphate are well defined, whereas those of the adenine terminus, however, cannot be seen because of its out-of-pocket location. It is intuitively clear that in order for the cyclization of NAD to occur, the adenine ring must fold back toward and anneal with the nicotinamide ribose. The complex shown in Fig. 2A, with the adenine ring positioned out of the active site pocket in a disordered manner, is thus not poised for the cyclization reaction, probably due to the occupation of uncleaved nicotinamide in the cyclization site.
FIGURE 2.
Structural features of cyclization reaction revealed from ADPRC complexes with substrate NAD and its analog N(2F-A)D. A, the active site structure of the E179GADPRC·NAD complex. Active site residues are shown as sticks with their carbon atoms in white; substrate NAD is also shown as sticks but with its carbon atoms in green. Polar interactions between NAD and active site residues are indicated as dashed cyan lines. Weighted Fo − Fc omit electron densities are shown as gray isomesh contoured at 2.5 σ. B, the active site structure of the WTADPRC·2F-ADPR·Nic complex. The color schemes for active site residues, shown in sticks, and the electron density map, contoured at 2.5 σ, are the same as in A. Nicotinamide, a leaving group from the cleavage of N(2F-A)D, is trapped in the active site and has obvious polar interaction with Asn-107 and hydrophobic interaction with Trp-140, respectively. The 2F-adenine terminus refolds toward the active site, forming a conformation suitable for cADPR formation by intramolecular linkage. C, comparison of the two states of the active site during nicotinamide cleavage reaction, from the initial state of substrate (NAD (gray)) binding to the state of the formation of the intermediate 2F-ADPR·Nic (pink). Residues Glu-98, Trp-140, Asn-107, and Phe-174 show slight but obvious conformational changes between transitions upon nicotinamide cleavage by ADPRC. Phe-174 shows hydrophobic interactions with nicotinamide and the 2F-adenine ring terminus of the ADPR intermediate. The Phe-174 phenyl ring coordinately rotates 10° in compensation for the dissociation of nicotinamide from 2F-ADPR.
ADPRC Favors Cyclization Reaction
To reveal the underlying mechanism for ADPRC in the cyclization reaction, we tested an NAD analog, N(2F-A)D (Fig. 1B), for its ability to form a complex with WTADPRC. In this analog, the addition of the fluorine atom at the 2-position of the adenine moiety apparently reduces the reactivity of the nitrogen at the 1-position, which is the site of cyclization. Consequently, N(2F-A)D is hydrolyzed by the WTADPRC to 2F-ADPR instead of being cyclized to 2F-cADPR (15). We determined the complex structure and refined it to 2.18 Å resolution. The electron densities clearly show two well separated species in the active site of WTADPRC (Fig. 2B). One is the cleaved nicotinamide, which is close to the Glu-98-Asn-107-Trp-140 triplet, and the other is a 2F-ADPR intermediate (2F-ADPRI). No electron density is between the C1′ atom of the ribose and nicotinamide, indicating that the nicotinamide is completely cleaved from the substrate. ADPRC is thus active even in the crystalline form and able to catalyze the hydrolysis of N(2F-A)D. Although the cleaved nicotinamide is still in the active site, the adenine moiety of 2F-ADPRI in the complex formed with the wild-type cyclase is now able to partially fold back toward the active site pocket. It is unexpected for nicotinamide being trapped in the active site after its cleavage and dissociation from 2F-ADPRI. In fact, the trapped nicotinamide in the active site has close interactions with Asn-107 and Trp-140, impeding the complete folding back of the 2F-adenine for cyclization. This would allow slow hydrolysis of N(2F-A)D to 2F-ADPR, consistent with the fact that in solution, the substrate is hydrolyzed by ADPRC instead of cyclized (15).
To find structural factors that contribute to the folding back of the adenine ring, we aligned and compared the E179GADPRC·NAD complex with the WTADPRC·2F-ADPRI·Nic complex (Fig. 2C). The alignment shows that Phe-174 rotates its side chain by 10° to coordinate with the folding of the 2F-adenine moiety. In the WTADPRC· 2F-ADPRI·Nic complex, Phe-174 interacts hydrophobically with the 2F-adenine moiety, whereas no such interactions are apparent in the E179GADPRC·NAD complex in which the adenine terminus is disordered (Fig. 2C). The differences suggest that Phe-174 is likely to be important in guiding the folding of the adenine moiety and thus should contribute to the cyclization preference of ADPRC, as will be described in more detail below.
Fig. 2C also shows that after the N-glycosidic bond breaks, the Glu-98-Asn-107-Trp-140 triplet appears to pull the nicotinamide away from the remaining 2F-ADPRI C1′ atom in a dissociative manner, which can contribute to the cleavage of the N-glycosidic bond. On the other hand, the diphosphate of the substrates aligns quite well in both structures, indicating that there is only minimal movement in this portion of the molecule during the reaction. The conformational changes of the Glu-98-Asn-107-Trp-140 triplet result in a slight enlargement of the active site, which can be particularly important for the release of the cleaved nicotinamide and the subsequent entry of the adenine moiety into the active site during cyclization of NAD. The triplet thus has a similar functional role as has been described previously for the corresponding residues in CD38 (28).
E179GADPRC·N1-cIDPR Complex Reveals a Novel Binding Site
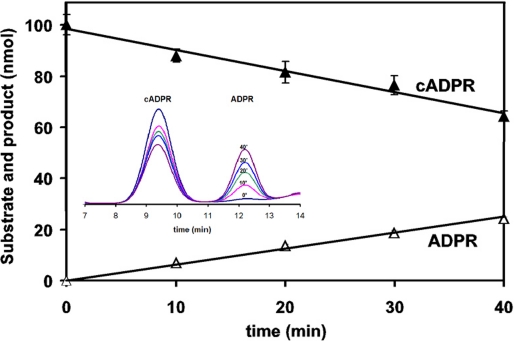
ADPRC cyclizes NAD to produce cADPR, a product that is released from the enzyme. There has been no report that ADPRC can form a complex with cADPR, nor has there been any investigation about whether it can hydrolyze cADPR. The prior effort in obtaining an ADPRC·cADPR complex was not successful (17), either because the cyclase has no affinity for cADPR or because the enzyme hydrolyzes it during crystallization. We thus tested whether the cyclase can hydrolyze cADPR. This turns out to be the case, as shown in Fig. 3. Incubation of cADPR with the ADPRC resulted in its progressive hydrolysis to ADPR, as analyzed by HPLC.
FIGURE 3.
ADPRC catalyzes the hydrolysis of cADPR in a progressive way. ADPRC (5 μg/ml) was incubated with 1 mm cADPR in 25 mm Tris-HCl, pH 8, for 10–40 min. The ADPR product was analyzed by HPLC.
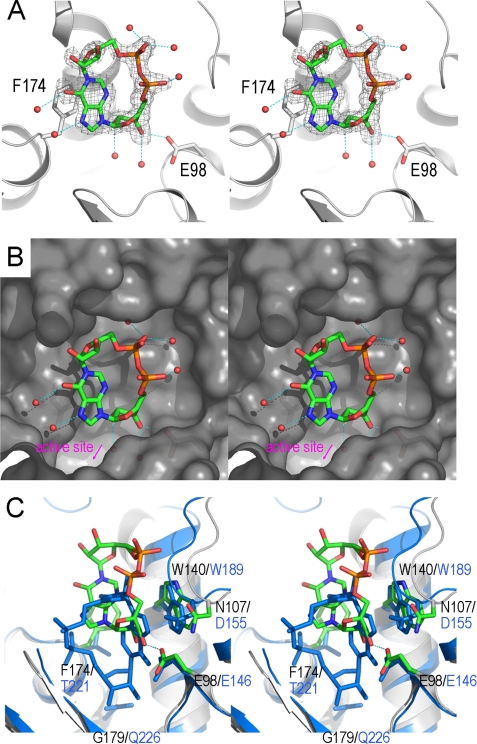
We therefore used an analog of cADPR, N1-cIDPR (20), for complex formation with the E179GADPRC mutant. This analog has previously been used to obtain the CD38·N1-cIDPR complex (28). N1-cIDPR was infused into preformed crystals of E179GADPRC by a soaking procedure similar to that used in obtaining the CD38·N1-cIDPR complex. Fig. 4A shows that N1-cIDPR can indeed bind to ADPRC. The electron density unambiguously defines the entity of N1-cIDPR in a cleft close to the active site. The hypoxanthine moiety of N1-cIDPR interacts hydrophobically with the phenyl ring of Phe-174, whereas the hypoxanthine ribose 3′-OH group forms an H-bond with the carboxylate group of Glu-98. Although both residues (Phe-174 and Glu-98) are part of the pocket, the N1-cIDPR binding site is away from the active site, as shown in Fig. 4B. A number of bound water molecules are observed to be recruited for N1-cIDPR binding.
FIGURE 4.
Divergent binding of ADPRC and CD38 with a cADPR analog, N1-cIDPR. A, the structure of the binding site of the E179GADPRC·N1-cIDPR complex. The color scheme for residues involved in N1-cIDPR binding is the same as in Fig. 2A. Residues Glu-98 and Phe-174 have polar and non-polar interactions with N1-cIDPR, respectively, and play a major role in N1-cIDPR binding. Red spheres are water molecules that participate in N1-cIDPR binding through hydrogen-bonding interactions. Electron density for N1-cIDPR is shown as gray isomesh contoured at 2.5 σ. B, surface representation, colored in gray, of the N1-cIDPR binding site with N1-cIDPR (sticks) and water molecules (spheres) in the binding cleft. C, N1-cIDPR binds to a site in ADPRC that is different from CD38. In CD38 (aquamarine), N1-cIDPR binds deep into the active site, whereas in ADPRC, the site is farther away from the catalytic residue (6.1Å) and in an upside-down and left-right conformation.
Although N1-cIDPR can bind to the active site ADPRC, the binding is different from that in CD38 shown previously (28). Fig. 4C compares E179GADPRC·N1-cIDPR and E226QCD38·N1-cIDPR. It can be seen that N1- cIDPR binds to ADPRC in an upside-down and left-right orientation as compared with its position in CD38. In ADPRC, the adenine ribose of N1-cIDPR points to the catalytic residue Glu-179, whereas in CD38, it is the northern ribose that points to the catalytic residue Glu-226. Because of the hydrophobic interaction between Phe-174 and the hypoxanthine moiety, N1-cIDPR does not interact with residues Arg-170, Ser-78, and Phe-175. However, in the E179GADPRC·NAD complex, these residues are involved in substrate recognition and binding (Fig. 2A). We have also determined the WTADPRC·N1-cIDPR complex and observed the same binding of N1-cIDPR as in the E179G mutant (data not shown), indicating that, unlike the linear NAD·ADPRC complex described above, the catalytic residue Glu-179 has minimal ability to recruit N1-cIDPR into the active site.
Another notable difference between the N1-cIDPR binding sites in CD38 and ADPRC is that the former is much deeper into the active site pocket (Fig. 4C). This is most likely because of the strong hydrophobic stacking between Phe-174 in ADPRC with the hypoxanthine moiety of N1-cIDPR, preventing the cyclic nucleotide from entering deeper into the pocket. In CD38, the residue corresponding to Phe-174 (in ADPRC) is Thr-221. The absence of the benzyl ring on Thr-221 may greatly reduce the stacking interaction and allows N1-cIDPR to reach deeper into the active site in both the wild-type CD38 and its mutant E226QCD38 (28) (Fig. 4C).
CD38 Favors the NAD Hydrolyzation Reaction
The structural results described above indicate the preference of ADPRC for the cyclization reaction. It is then of interest to determine the structural mechanism of the preference of CD38 in the hydrolyzation reaction. We thus tested two substrate analogs of NAD, ara-2′F-NAD and ribo-2′F-NAD. We have previously shown that ara-2′F-NMN forms a covalent intermediate with CD38 (29). This is also true for the ara-2′F-NAD, as shown in Fig. 5A. Soaking CD38 crystals with the substrate resulted in the cleavage of the nicotinamide, and the remaining nucleotide formed a covalent adduct between the nicotinamide ribose C1′ and the carboxylate group of the catalytic residue, Glu-226, as clearly shown by the observed electron densities (Fig. 5A). The formation of the covalent adduct fixes one end at the C1′ position of the nicotinamide ribose and allows structural evaluation of the folding ability of the other end, the adenine terminus, which is observed to fold back and enter the active site (Fig. 5A). This is similar to that described above in the 2F-ADPR·ADPRC complex (Fig. 2B). Two water molecules are close to the C1′ carbon, but neither of them can hydrolyze the covalent adduct, indicating that the linkage is stable and resistant to further hydrolysis by the bound water. The diphosphate group forms a number of H-bonds with Trp-125, Ser-126, Arg-127, and Phe-222. There is an H-bond between the adenine ribose and Thr-221, suggesting a role of this residue in stabilizing and guiding the adenine ring terminus into the active site. Although the adenine ring is folded back into the active site, its conformation does not favor the cyclizing N-1-C1′ linkage. The reason is that, in the conformation shown in Fig. 5A, it is the N-7 atom, not the N-1, on adenine moiety that is close to the C1′ of the terminal ribose.
FIGURE 5.
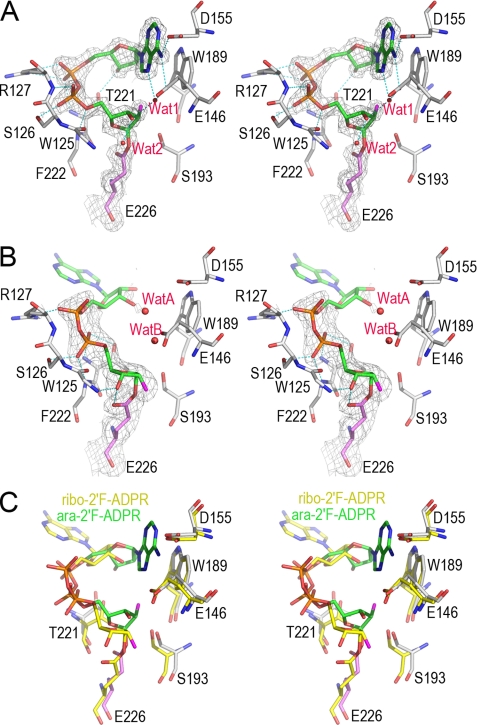
The structural features of hydrolyzation reaction in CD38 revealed from two covalent reaction intermediates. A, the active site structure of the WTCD38·ara-2′F-ADPR complex. The active site residues are shown as sticks. Covalent adduct ara-2′F-ADPR is also shown as sticks with its carbon atoms in green. Weighted 2Fo − Fc omit electron density is shown as gray isomesh contoured at 1.2 σ covering both ara-2′F-ADPR and Glu-226. Two water molecules trapped in the active site are shown as red spheres. After cleavage and removal of nicotinamide from substrate ara-2′F-NAD, the remaining part forms a covalent adduct with the catalytic residue Glu-226, resulting in inhibition of the enzyme. After the release of the nicotinamide moiety, the adenine ring terminus can partially enter the active site, but it cannot cyclize with the terminal ribose because of the stability of the covalent intermediate and the incorrect orientation of the adenine ring, with the N1-cyclizing site pointing away. Two water molecules are present but cannot hydrolyze the intermediate, indicating its stability. B, the active site structure of the WTCD38·ribo-2′F-ADPR complex. The drawing and color scheme are the same as in A. C, structural comparison of ara-2′F-ADPR (green carbon) and ribo-2′F-ADPR (yellow carbon) complexed with CD38. The alignment shows that after the release of the nicotinamide, the adenine moiety is not able to either enter the catalytic site or enter in a wrong orientation. Both conformations of the adenine moiety favor the hydrolyzation reaction to take place.
The adenine moiety has hydrophobic interaction with Trp-189 and H-bonds with Glu-146, Asp-155, as well as nearby bound water. The presence of trapped water in the active site shows that water enters the active site more readily than the adenine ring, which needs to fold back to form cADPR. This is consistent with the fact that CD38 produces mostly ADPR from NAD through a hydrolyzation reaction, instead of cADPR through a cyclization reaction.
We have also solved the crystal structure of WTCD38 complexed with ribo-2′F-NAD (Fig. 5B), a stereo isomer that is identical to ara-2′F-NAD except that the 2′-position, where fluorine is attached, is in the opposite configuration (Fig. 1C). Like ara-2′F-NAD, it also forms a covalent adduct with the catalytic residue Glu-226. The electron density for the nicotinamide ribose and diphosphate are well defined, but there is no electron density covering the adenine ribose and the adenine terminus, showing them to be disordered. Also seen in this complex, the nicotinamide ribose 3′-OH group forms H-bonds with Glu-226 and the Trp-125 main chain nitrogen atom. The diphosphate group has five H-bonds to residues Thr-221, Phe-222, Ser-126, and Arg-127. Again, water was observed to be trapped in the active site with the closest distance of only 3.6 Å to C1′ atom of the nicotinamide ribose.
Superimposition of ara-2′F-ADPR on ribo-2′F-ADPR indicates that no matter which NAD analog was used, the conformation of the adenine terminus does not favor the cyclization reaction (Fig. 5C). In the ara-2′F-ADPR complex, the adenine terminus can fold back into the active site, but the adenine ring is in an improper orientation with the N1-cyclization site pointing away from the C1′ of the ribose. In the ribo-2′F-ADPR complex, the adenine terminus remains disordered out of the active site with only water in the active site. Moreover, it is water, instead of the adenine terminus, closer to C1′ of the covalent adducts. With NAD as substrate, the proposed reaction intermediate will be non-covalent, which is more reactive than the covalent adduct (29). Water will then be ready to hydrolyze the intermediate to form ADPR. The catalysis of CD38 thus strongly favors hydrolyzation instead of cyclization with NAD as the substrate.
Mutational Conversion of the ADPRC to an CD38-like NAD Hydrolase
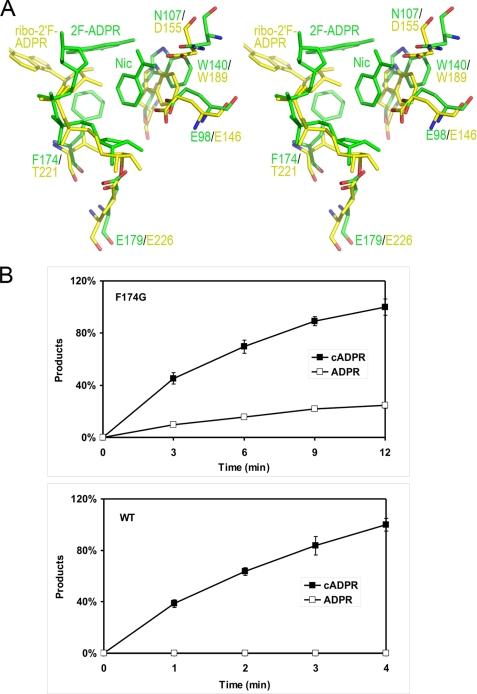
From the above analysis, it is clear that the cyclization ability of either ADPRC or CD38 depends on the correct folding of the adenine terminus of the substrate into the active site after nicotinamide cleavage. It is thus of interest to compare the structural features of the active sites of the two enzymes. In Fig. 6A, the WTADPRC·2F-ADPR·Nic complex is superimposed with the WTCD38·ribo-2′F-ADPR complex. In both complexes, the first step of the reaction, e.g. the N-glycosidic bond cleavage, has finished. The comparison shows structural differences in the positioning of some key resides in their active sites, including Glu-98 (Glu-146 in CD38), Asn-107 (Asp-155 in CD38), Trp-140 (Trp-189 in CD38), and Phe-174 (Thr-221 in CD38). Of particular interest is Phe-174 in ADPRC, which is more bulky than the corresponding residue, Thr-221, in CD38 and appears to have hydrophobic interaction with Trp-140. More importantly, Phe-174 in the WTADPRC·2F-ADPR·Nic complex also shows hydrophobic interaction with the 2F-adenine moiety of 2F-ADPR, which is likely to be important in guiding the folding of adenine ring into the active site. Indeed, when comparing the complexes of unfolded and folded substrates, as shown in Fig. 2C, the phenyl ring of the Phe-174 residue is shown to rotate 10° to promote the folding of the adenine moiety. A corollary of this interpretation is that when changing Phe-174 to a less bulky and/or hydrophobic residue, the cyclization activity of the enzyme would be reduced, whereas its hydrolytic activity should be enhanced. This is the case in CD38, which has a less hydrophobic threonine at the corresponding position and thus high hydrolytic activity. To further confirm the importance of Phe-174 in the cyclization reaction, we removed its hydrophobic character by mutating the residue to a glycine. As shown in Fig. 6B, indeed, the mutation bestows the F174G mutant with high NAD-hydrolyzing activity, producing about 30% of its catalytic products as ADPR, whereas none is observed with the wild-type ADPRC.
FIGURE 6.
Conversion of the ADPRC to an NADase by a single residue mutation. A, residue Phe-174 is critical for cyclization reaction. Comparison of the complexes of WTADPRC·2F-ADPR·nicotinamide (colored in green) and CD38·ribo-2′F-ADPR (colored in yellow) shows that Phe-174 in ADPRC may help direct the adenine ring into the active site correctly for cyclization, whereas the corresponding residue Thr-221 in CD38 disfavors the entry process. Also, the steric repulsion of the bulky phenyl ring of Trp-140 (in ADPRC) by other residues results in a bigger active site that favors the easy entry of the adenine ring during the cyclization reaction. B, mutation of Phe-174 to Gly-174 turns ADPRC into a CD38-like hydrolase. The WTADPRC plasmid was converted to the F174G mutant by site-directed mutagenesis and transfected into P. pastoris; the mutant form of the protein was expressed and purified. Both WTADPRC (0.1 μg/ml) and F174G (1 μg/ml) were incubated with 0.1 mm NAD in Tris-HCl, pH 8, for the times indicated, and the products were analyzed by HPLC. Under these conditions, the WTADPR produces only cADPR, whereas the F174G mutant produces both cADPR and ADPR. n = 4 for each time point.
Acknowledgments
The crystallographic data were collected at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation and NIGMS National Institutes of Health under award DMR-0225180. MacCHESS is supported by National Institutes of Health Grant RR01646.
This work was supported, in whole or in part, by National Institutes of Health Grant GM061568 (to H. C. L. and Q. H.). This work was also supported by Hong Kong General Research Fund grants from the National Science Foundation of China/Research Grant Council of Hong Kong (NSFC/RGC) Joint Research Scheme (to Q. H. and H. C. L.), Project Grant 084068 from the Wellcome Trust (to B. V. L. P.), and the Dreyfus Foundation New Faculty Award (to H. L.).
- cADPR
- cyclic ADP-ribose
- ADPR
- adenosine diphosphate ribose
- ADPRC
- ADP-ribosyl cyclases, N(2F-A)D, nicotinamide 2-fluoro-adenine dinucleotide
- Nic
- nicotinamide
- N1-cIDPR
- N1-cyclic inosine diphosphate ribose
- ara-2′F-NAD
- arabinosyl-2′-fluoro-deoxy-nicotinamide adenine dinucleotide
- ara-2′F-ADPR
- arabinosyl-2′-fluoro-deoxy-adenosine diphosphate ribose
- ribo-2′F-NAD
- ribosyl-2′-fluoro-deoxy-nicotinamide adenine dinucleotide
- ribo-2′F-ADPR
- ribosyl-2′-fluoro-deoxy-adenosine diphosphate ribose
- 2F-ADPRI
- 2F-ADPR intermediate
- WT
- wild type
- HPLC
- high pressure liquid chromatography
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Lee H. C. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 317–345 [DOI] [PubMed] [Google Scholar]
- 2.Schuber F., Lund F. E. (2004) Curr. Mol. Med. 4, 249–261 [DOI] [PubMed] [Google Scholar]
- 3.Malavasi F., Deaglio S., Funaro A., Ferrero E., Horenstein A. L., Ortolan E., Vaisitti T., Aydin S. (2008) Physiol. Rev. 88, 841–886 [DOI] [PubMed] [Google Scholar]
- 4.Lee H. C., Aarhus R. (1991) Cell Regul. 2, 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.States D. J., Walseth T. F., Lee H. C. (1992) Trends Biochem. Sci. 17, 495. [DOI] [PubMed] [Google Scholar]
- 6.Howard M., Grimaldi J. C., Bazan J. F., Lund F. E., Santos-Argumedo L., Parkhouse R. M., Walseth T. F., Lee H. C. (1993) Science 262, 1056–1059 [DOI] [PubMed] [Google Scholar]
- 7.Lee H. C., Graeff R., Walseth T. F. (1995) Biochimie 77, 345–355 [DOI] [PubMed] [Google Scholar]
- 8.Hirata Y., Kimura N., Sato K., Ohsugi Y., Takasawa S., Okamoto H., Ishikawa J., Kaisho T., Ishihara K., Hirano T. (1994) FEBS Lett. 356, 244–248 [DOI] [PubMed] [Google Scholar]
- 9.Adebanjo O. A., Anandatheerthavarada H. K., Koval A. P., Moonga B. S., Biswas G., Sun L., Sodam B. R., Bevis P. J., Huang C. L., Epstein S., Lai F. A., Avadhani N. G., Zaidi M. (1999) Nat. Cell Biol. 1, 409–414 [DOI] [PubMed] [Google Scholar]
- 10.Barbosa M. T., Soares S. M., Novak C. M., Sinclair D., Levine J. A., Aksoy P., Chini E. N. (2007) FASEB J. 21, 3629–3639 [DOI] [PubMed] [Google Scholar]
- 11.Lee H. C. (2006) Mol. Med. 12, 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrero E., Saccucci F., Malavasi F. (1999) Immunogenetics 49, 597–604 [DOI] [PubMed] [Google Scholar]
- 13.Cakir-Kiefer C., Muller-Steffner H., Schuber F. (2000) Biochem. J. 349, 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauve A. A., Munshi C., Lee H. C., Schramm V. L. (1998) Biochemistry 37, 13239–13249 [DOI] [PubMed] [Google Scholar]
- 15.Zhang B., Muller-Steffner H., Schuber F., Potter B. V. (2007) Biochemistry 46, 4100–4109 [DOI] [PubMed] [Google Scholar]
- 16.Graeff R., Munshi C., Aarhus R., Johns M., Lee H. C. (2001) J. Biol. Chem. 276, 12169–12173 [DOI] [PubMed] [Google Scholar]
- 17.Munshi C., Thiel D. J., Mathews I. I., Aarhus R., Walseth T. F., Lee H. C. (1999) J. Biol. Chem. 274, 30770–30777 [DOI] [PubMed] [Google Scholar]
- 18.Munshi C. B., Fryxell K. B., Lee H. C., Branton W. D. (1997) Methods Enzymol. 280, 318–330 [DOI] [PubMed] [Google Scholar]
- 19.Sleath P. R., Handlon A. L., Oppenheimer N. J. (1991) J. Org. Chem. 56, 3608–3613 [Google Scholar]
- 20.Wagner G. K., Guse A. H., Potter B. V. L. (2005) J. Org. Chem. 70, 4810–4819 [DOI] [PubMed] [Google Scholar]
- 21.Liu Q., Kriksunov I. A., Graeff R., Munshi C., Lee H. C., Hao Q. (2005) Structure 13, 1331–1339 [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z., Minor W. (1997) Macromolecular Crystallogr. Pt A 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 23.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 24.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25.Schüttelkopf A. W., van Aalten D. M. F. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 26.Morris R. J., Perrakis A., Lamzin V. S. (2003) Macromolecular Crystallogr. D 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 27.Munshi C., Lee H. C. (1997) Protein Expr. Purif. 11, 104–110 [DOI] [PubMed] [Google Scholar]
- 28.Liu Q., Kriksunov I. A., Moreau C., Graeff R., Potter B. V., Lee H. C., Hao Q. (2007) J. Biol. Chem. 282, 24825–24832 [DOI] [PubMed] [Google Scholar]
- 29.Liu Q., Kriksunov I. A., Jiang H., Graeff R., Lin H., Lee H. C., Hao Q. (2008) Chem. Biol. 15, 1068–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]