Abstract
Clathrin-coated vesicles (CCVs) originating from the trans-Golgi network (TGN) provide a major transport pathway from the secretory system to endosomes/lysosomes. Herein we describe paralogous Sec14 domain-bearing proteins, clavesin 1/CRALBPL and clavesin 2, identified through a proteomic analysis of CCVs. Clavesins are enriched on CCVs and form a complex with clathrin heavy chain (CHC) and adaptor protein-1, major coat components of TGN-derived CCVs. The proteins co-localize with markers of endosomes and the TGN as well as with CHC and adaptor protein-1. A membrane mimic assay using the Sec14 domain of clavesin 1 reveals phosphatidylinositol 3,5-bisphosphate as a specific lipid partner. Phosphatidylinositol 3,5-bisphosphate is localized to late endosomes/lysosomes, and interestingly, isoform-specific knockdown of clavesins in neurons using lentiviral delivery of interfering RNA leads to enlargement of a lysosome-associated membrane protein 1-positive membrane compartment with no obvious influence on the CCV machinery at the TGN. Since clavesins are expressed exclusively in neurons, this new protein family appears to provide a unique neuron-specific regulation of late endosome/lysosome morphology.
Proteins entering the secretory pathway move through the Golgi apparatus to the trans-Golgi network (TGN)4 where they are sorted and packaged into carrier vesicles, including clathrin-coated vesicles (CCVs) for transport to their final destination (1). Adaptor protein-1 (AP-1), which is recruited to the TGN through dual interactions with Arf1 and phosphatidylinositol 4-phosphate (2), recruits clathrin to initiate CCV formation. AP-1 and clathrin form the membrane coat that shapes the vesicle and recruits an array of regulatory/accessory proteins, which control numerous aspects of CCV formation and function (3). Interactions with the clathrin·AP-1 coat complex also serve to recruit both transmembrane and cytosolic cargo to CCVs, allowing for their transport from the TGN to the endosomal network.
The specificity and function of intracellular compartments depends in part on the presence of distinct PtdIns species. For example, phosphatidylinositol 4-phosphate is found predominantly at the TGN and contributes to AP-1 recruitment, whereas phosphatidylinositol 3-phosphate recruits effectors, such as EEA1 (early endosome antigen 1), to early endosomes to mediate membrane fusion (4–9). Early endosomes subsequently transition into late endosomes/lysosomes, and during this process, phosphatidylinositol 3-phosphate is converted to PtdIns(3,5)P2 via the action of a phosphatidylinositol 3-phosphate 5-kinase named Fab1p in yeast and PIKfyve in mammals (10). PIKfyve is part of a protein complex nucleated by Vac14 (11, 12), and disruption of this complex leads to decreased PtdIns(3,5)P2 levels and the formation of enlarged cytoplasmic vacuoles of endosomal/lysosomal origin (10, 13, 14). Intriguingly, despite the fact that Vac14 is found in all tissues (15) and regulates a ubiquitous trafficking process (10), decreases in PtdIns(3,5)P2 levels resulting from Vac14 knock-out show massive neurodegeneration with little effect on other tissues (16). This neuron-specific effect has remained mysterious due to the lack of exclusively neuronal factors targeting PtdIns(3,5)P2.
The Sec14 domain is an evolutionarily ancient protein module that in humans is found in more than 45 proteins encoded by at least 25 genes (17). Mutations in several Sec14 proteins lead to human diseases, including neurodegeneration (18), yet most of the Sec14 proteins in mammals remain uncharacterized. The yeast protein Sec14p is the prototype for this module (19). Sec14p is essential for the transport of proteins from the Golgi (19), but more recently it was also found to be involved in the trafficking of protein cargo, specifically the mating factor receptor Ste3, from the plasma membrane via endosomes to the yeast vacuole (17). The yeast vacuole is the equivalent of the lysosome in mammalian cells. Yeast Sec14p is a phospholipid transfer protein that extracts phosphatidylinositol and phosphatidylcholine from membranes in vitro and regulates the metabolism of these lipids in cells (19, 20). Sec14p is composed of the Sec14 domain only, whereas the majority of Sec14 proteins in mammals contain a Sec14 domain in conjunction with other modules or protein/lipid binding domains (21, 22). In the few well characterized examples, these proteins function to integrate lipid binding/metabolism with other biological functions (21, 22).
In a subcellular proteomic analysis of CCVs from the brain, we identified eight novel open reading frames, including one encoding an Sec14 domain (23, 24). A subsequent bioinformatics analysis revealed a second, paralogous Sec14 protein. We now demonstrate that the proteins, which we have named clavesin (clathrin vesicle-associated Sec14 protein) 1 and 2, are neuron-specific proteins that function in the regulation of lysosome morphology.
EXPERIMENTAL PROCEDURES
Antibodies and cDNA Constructs
A rabbit polyclonal antibody that recognizes clavesin 1 and 2 was raised against a peptide from the C terminus of clavesin 1 (EKGENENTQPLLALD). Mouse monoclonal antibodies against various proteins were from the indicated commercial sources; CHC X22 (American Type Culture Collection), γ-adaptin (AP-1), α-adaptin (AP-2), CHC and GM130 (BD Transduction Laboratories), anti-FLAG M2 (Sigma), LAMP1 LY1C6 and EEA1 281.7 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), TGN38 2F7.1 (Abcam), and glial fibrillary acidic protein (Chemicon). Chicken MAP2 and sheep TGN46 polyclonal antibodies were from EnCor Biotechnology and Serotec, respectively. Rabbit anti-NECAP1 and endophilin 1 polyclonal antibodies were described previously (25, 26). Polyclonal antibodies against mannose 6-phosphate receptor 46 and 300 were a gift from Dr. Kurt von Figura. Alexa 594-, 633-, and 647-conjugated secondary antibodies were from Molecular Probes, Inc. Cy3-, Cy2-, and horseradish peroxidase-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories, Inc.
cDNA clones for human clavesin 1 (gi27503734) and clavesin 2 (gi115527290) were used as PCR templates to amplify DNA encoding full-length proteins. N-terminal GST, FLAG, and GFP tags were added by cloning the PCR products into pGEX-6P1 (Amersham Biosciences), pCMV-Tag2B (Stratagene), and pEGFP-C1 (BD Biosciences Clontech), respectively. Clavesin 1 and 2 truncation mutants were generated by PCR from full-length cDNA. Mutations of clavesin 1 resulting in a defective lipid binding were introduced into its cDNA using a QuikChange XL site-directed mutagenesis kit (Stratagene). Tandem FYVE domain (2×FYVE), derived from mouse hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) (27) was a gift of Dr. Fred Meunier. The enhanced green fluorescent protein from the pEGFP plasmid was replaced with mCherry. The enthoprotin C-terminal construct was previously described (23).
Subcellular Fractionation
Adult rat tissues and various cell lines were homogenized in buffer A (10 mm HEPES, pH 7.4, 0.83 mm benzamidine, 0.23 mm phenylmethylsulfonyl fluoride, 0.5 μg/ml aprotinin, and 0.5 μg/ml leupeptin) and centrifuged at 800 × g for 5 min. The protein content of the postnuclear supernatants was determined, and equal protein aliquots were analyzed by SDS-PAGE and Western blot. In other cases, the postnuclear supernatant was further centrifuged at 12,000 × g to create a P2, and the supernatant of this spin was centrifuged at 200,000 × g to create a microsomal pellet (P3) and a cytosolic fraction (S3). CCVs were isolated from rat brain, as described previously (23).
Protein Binding Assays
Postnuclear supernatants were prepared from brain or transfected HEK293 cells as described above. Triton X-100 was added to 1% final, and the extracts were spun at 265,000 × g for 30 min. The soluble supernatants were incubated with GST fusion proteins purified from Escherichia coli BL21 cells and prebound to glutathione-Sepharose beads (Amersham Biosciences). Samples were incubated overnight at 4 °C and washed three times with buffer A containing 1% Triton X-100. Proteins specifically bound to the beads were subjected to Western blot.
Primary Rat Hippocampal Cultures
Embryonic day 18–19 rat hippocampal neurons were prepared as described (28). Cells were fed every 7 days with Neurobasal medium supplemented with B-27, N-2, l-glutamine (500 μm), and penicillin/streptomycin (100 units/ml) (Invitrogen) and were processed for immunofluorescence analysis or Western blot.
Immunofluorescence
For most immunofluorescence experiments, hippocampal neurons plated on poly-l-lysine-coated coverslips were washed in phosphate buffered saline (PBS; 20 mm NaH2PO4, 0.9% NaCl, pH 7.4) and fixed in 4% paraformaldehyde for 10 min. Cells were then washed in PBS, permeabilized in PBS with 0.2% Triton X-100, and incubated in blocking buffer (PBS with 1% bovine serum albumin and 0.02% Triton X-100). Following this block, coverslips were incubated with primary antibodies in blocking buffer for 2 h at room temperature. Following this incubation, cells were washed in PBS with 0.02% Triton X-100, incubated for 1 h at room temperature with fluorescent secondary antibodies, and washed, and the coverslips were mounted on glass slides with DAKO (cytomation fluorescence mounting medium; Dakocytomation) and imaged on a Zeiss 510 confocal microscope. For quantification of lysosomes, raw images were converted to binary, particles larger than 0.1 μm2 were extracted, and the area of the extracted particles was measured using ImageJ software (National Institutes of Health). Data were analyzed statistically using JMP software (SAS Institute, Inc.). For quantification of intensity of CHC and AP-1 at the TGN, total fluorescence in the regions of interest was measured using ImageJ software.
Knockdown
MicroRNA (miRNA) sequences were designed to deplete rat clavesin 1 (gi109476027) and clavesin 2 (gi109460582) using Invitrogen's RNAi Designer. The sequences target clavesin 1 and clavesin 2 mRNA starting at nucleotides 489 (clavesin 1 sequence 1), 956 (clavesin 1 sequence 2), 1087 (clavesin 2 sequence 1), and 1186 (clavesin 2 sequence 2) and are TGATGATCATGTCTCTGACCT, AAAGCGAGCAGGAAAGCTGTC, TTCAGTGCCTGCTTGATGCCA, and ACCAGTGTATACCTGCTCTGA, respectively. A nontargeting miRNA was used for controls, and its sequence is AATTCTCCGAACGTGTCACGT. Oligonucleotides encoding the miRNA sequences were cloned into pcDNA 6.2-GW/EmGFP-miR plasmid (Invitrogen), and lentivirus was produced as described (29). Virus vectors were generally transduced at 7 days in vitro (DIV), and neurons were processed for immunofluorescence or Western blot at 14–21 DIV.
Lipid Binding
Lipid binding was performed in 10 mm HEPES with 100 mm NaCl essentially as described (30).
RESULTS
Identification of Clavesin 1 and 2
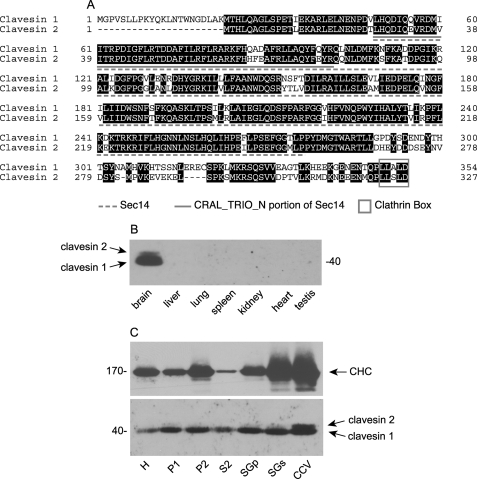
Clavesin 1 was originally identified as a novel open reading frame in a proteomic analysis of rat brain CCVs (23), and a data base search revealed a clavesin 1 paralogue, clavesin 2. The proteins consist of an Sec14 domain flanked by a short stretch of residues on the N-terminal side and an ∼75 residue C-terminal region ending in a putative clathrin box, a motif involved in clathrin binding (Fig. 1A). The proteins also contain a CRAL_TRIO_N domain (Fig. 1A). Although this module is often annotated as a separate entity, it is a component of the globular Sec14 domain (21). Clavesin 1 was independently isolated from a cDNA library based on the fact that it contains a CRAL_TRIO_N domain (31). Due to homology of the protein with a cellular retinaldehyde-binding protein (CRALBP) it was called CRALBPL (CRALBP-like) (31). Reverse transcription-PCR analysis indicated that the mRNA for CRALBPL/clavesin 1 was expressed in the brain, but neither expression nor localization of the endogenous protein was examined (31). We thus sought to generate antibodies against the clavesins. Our first attempts to generate isoform-specific antibodies were unsuccessful. However, we were successful in generating a polyclonal antibody against a peptide from the C terminus of clavesin 1 that recognizes recombinant clavesin 1 and 2 (supplemental Fig. S1). Western blots of brain extracts reveal a doublet with a strong band at 40 kDa and a weaker band that migrates slightly slower (Fig. 1B). Isoform-specific knockdown in cultured hippocampal neurons indicates that the lower and upper bands are clavesin 1 and 2, respectively (see Fig. 9A). Clavesins are detected in the brain but are undetectable, even with extended exposure in non-neuronal tissues or in various cell lines (Fig. 1B and supplemental Fig. S2; data not shown). Both clavesin isoforms are enriched on CCVs, although their fractionation pattern does not precisely parallel the clathrin heavy chain (CHC) (Fig. 1C), indicating that clavesins are not exclusively CCV proteins. Thus, clavesins are brain-specific proteins associated with CCVs.
FIGURE 1.
Identification and CCV enrichment of clavesin proteins. A, alignment of clavesin 1 and 2. Identical amino acids are shaded black. The dashed underline indicates the Sec14 domain, and the solid underline indicates the CRAL_TRIO_N portion of the Sec14 domain. The box denotes a clathrin box. B, the level of clavesin 1 and 2 in the indicated tissues was determined by Western blot. C, rat brain subcellular fractions from a preparation leading to highly enriched CCVs were blotted with clavesin 1/2 and CHC antibodies. H, homogenate; P, pellet; S, supernatant; SGp, sucrose gradient pellet; SGs, sucrose gradient supernatant.
FIGURE 9.
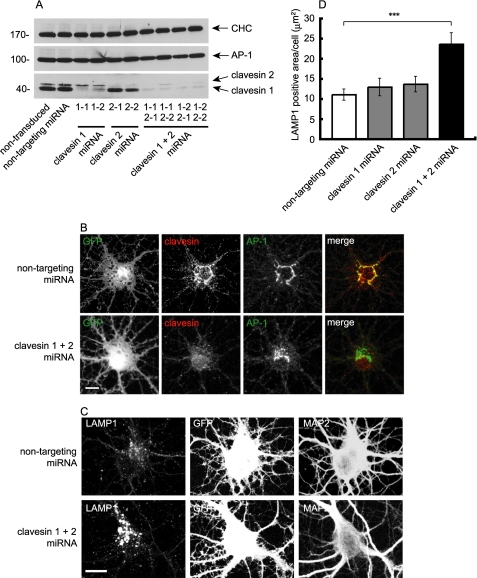
Knockdown of clavesins in neurons alters lysosome morphology. A, hippocampal neurons at 4 DIV were transduced with lentivirus encoding a non-targeting miRNA or with lentivirus encoding two different miRNAs targeting clavesin 1 (1-1, 1-2), two different miRNAs targeting clavesin 2 (2-1, 2-2), or different combinations of the clavesin 1 and 2 miRNAs, as indicated. At 12 DIV, lysates were prepared from the cultures, and equal protein aliquots were processed for Western blots with antibodies against CHC, AP-1, or clavesin 1/2. B, hippocampal neurons at 4 DIV were transduced with lentivirus encoding clavesin 1-1 and 2-1 miRNAs. At 21 DIV, cells were processed for indirect immunofluorescence with polyclonal antibody against clavesin (red) and monoclonal antibody against AP-1. The AP-1 signal was processed in the CY5 channel (633) and was pseudocolored green for the purpose of merging with the red clavesin signal. The lentivirus also drives expression of GFP (leftmost panels). Bar, 10 μm. C, hippocampal neurons at 7 DIV were transduced with lentivirus encoding non-targeting miRNA, clavesin 1-1 miRNA, clavesin 2-1 miRNA, or both clavesin miRNAs. At 14 DIV, cells were processed for indirect immunofluorescence with antibodies against LAMP1 (red) and MAP2 (blue). The lentivirus also drives expression of GFP. Bar, 10 μm. D, the total LAMP1-positive area was quantified. Each bar represents the mean ± S.E. with n = 40. ***, a significance of p < 0.001 using Welch's t test.
Clavesins are Localized in Part to the TGN
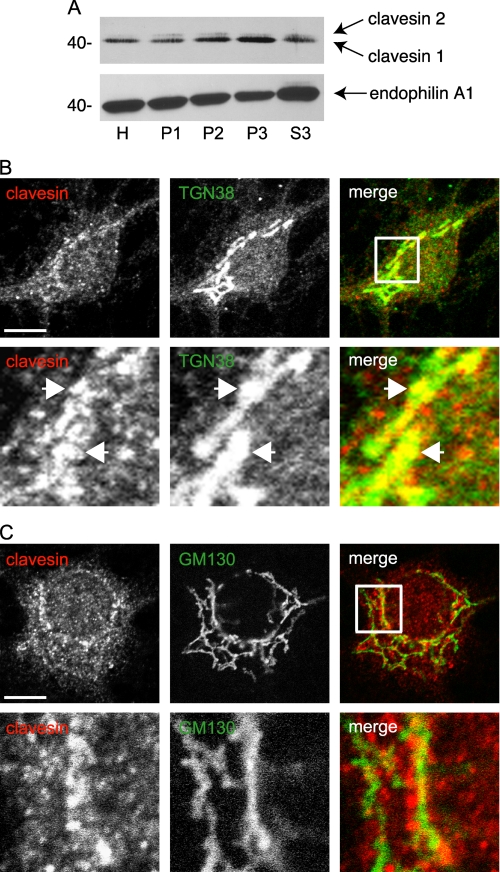
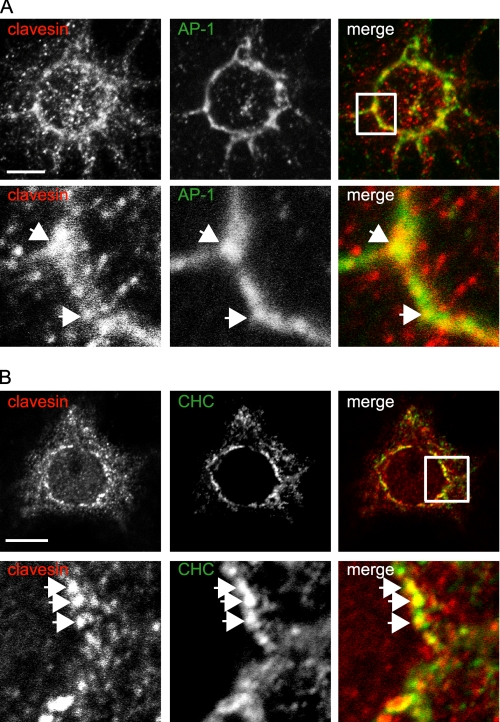
In hippocampal cultures at 21 DIV, clavesins are detected by immunofluorescence in MAP2-positive neurons but not glial fibrillary acidic protein-positive astrocytes (supplemental Fig. S3). Neuronal staining is punctate and prominent in cell bodies with enrichment in a perinuclear compartment (supplemental Fig. S3). Subcellular fractionation of brain lysates by differential centrifugation reveals that clavesins are most prominent in the pellet fractions (Fig. 2A). Clavesin 1 is enriched in the P3 microsomal fraction, whereas clavesin 2 is more evenly distributed between membrane fractions (Fig. 2A). There is less of the clavesins in the S3 cytosolic fraction, which is enriched in endophilin A1. Thus, the clavesin puncta detected by immunofluorescence probably represent membrane-associated proteins. Clavesin puncta in the perinuclear region partially overlap with TGN38 (Fig. 2B), which is highly enriched at the TGN at steady-state, whereas little co-localization is seen with the cis-Golgi protein GM130 (Fig. 2C). Clavesins also partially co-localize with AP-1 and CHC (Fig. 3, A and B). Strongly diminished immunofluorescence following treatment with inhibitory RNAs targeting the clavesins indicates the specificity of the antibody (see Fig. 9B). However, there is some weak immunofluorescence seen in nuclei (Fig. 2, B and C, for example), and this is not lost with inhibitory RNA treatment, suggesting that it represents a cross-reactivity of the antibody. Together, our data demonstrate that clavesins are neuron-specific, membrane-associated proteins that co-localize with clathrin coat components on the TGN.
FIGURE 2.
Clavesins are membrane-associated proteins that localize to the TGN. A, rat brain homogenates (H) were subjected to differential centrifugation, leading to pellets (P1–P3) with increasingly smaller membranes as well as a cytosolic supernatant fraction (S3). Equal protein aliquots of the fractions were blotted with antibodies against clavesins or endophilin 1. Hippocampal neurons at 21 DIV were processed by indirect immunofluorescence with polyclonal antibody against clavesin (red) and monoclonal antibodies against TGN38 (B) and GM130 (C), each in green. In each case, the lower panels are from the area indicated by the white box in the merge images. The arrows point to co-localizing structures. Bars, 10 and 2 μm for the low and high power images, respectively.
FIGURE 3.
Clavesins co-localize with clathrin coat components. Hippocampal neurons at 21 DIV were processed by indirect immunofluorescence with polyclonal antibody against clavesin (red) and monoclonal antibodies against AP-1 (A) and CHC (B), each in green. In each case, the lower panels are from the area indicated by the white box in the merge images. The arrows point to co-localizing structures. Bars, 10 and 2 μm for the low and high power images, respectively.
Clavesins Localize in Part to Endosomes
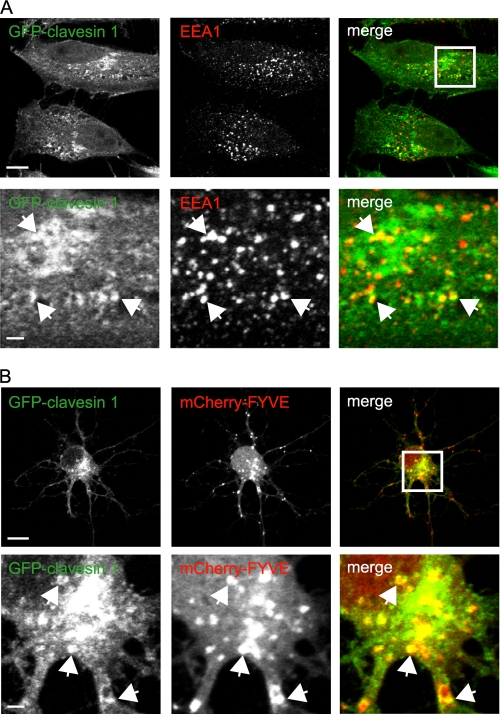
In addition to the TGN localization, we noticed that clavesins also displayed a more peripherally distributed punctate staining in neuronal cell bodies (Figs. 2, B and C, and 3). Since clavesins are associated with TGN-derived CCVs that deliver cargo to endosomes, it was reasonable to hypothesize that clavesins would also localize in part to endosomes. To test this possibility, we transfected HeLa cells with GFP-clavesin 1 and examined its localization in relationship to the early endosome marker EEA1 (Fig. 4A). Interestingly, in addition to a juxtanuclear pool corresponding to the TGN, there was a more peripheral punctate pool that showed prominent co-localization with EEA1, indicating the presence of an endosomal pool of clavesin 1 (Fig. 4A; see also Fig. 7). Similar results were seen with GFP-clavesin 2 (data not shown). Unfortunately, the EEA1 antibody did not work for immunofluorescence in neurons. Thus, we co-transfected hippocampal cultures with GFP-clavesin 1 and a construct encoding tandem copies of the PtdIns 3-phosphate-binding FYVE domain of Hrs fused to mCherry, a marker of early endosomes (27). To allow for increased transfection efficiency, cells were transfected at 6 DIV and imaged at 7 DIV. GFP-clavesin 1 displayed juxtanuclear staining corresponding to the TGN (Fig. 4B; also see Fig. 6). At 7 DIV, the TGN has a more restricted juxtanuclear pattern (Figs. 4 and 6) than at 21 DIV (Figs. 2 and 3). In addition to the TGN staining, a pool of GFP-clavesin 1 co-localizes with mCherry-FYVE, indicating endosomal localization (Fig. 4B). Thus, clavesins are localized to endosomes in addition to the TGN.
FIGURE 4.
Clavesins localize to endosomes. A, HeLa cells were transfected with plasmid encoding GFP-clavesin 1 and subsequently processed by indirect immunofluorescence with antibody against EEA1 (red). The lower panels are from the area indicated by the white box in the merge image. Bars, 10 and 2 μm for the low and high power images, respectively. B, hippocampal neurons at 6 DIV were transfected with plasmids encoding mCherry-tagged tandem FYVE domains of Hrs (mCherry-FYVE) (red) and GFP-clavesin 1 (green) and were imaged at 7 DIV. The lower panels are from the area indicated by the white box in the merge image. Bars, 10 and 2 μm for the low and high power images, respectively.
FIGURE 7.
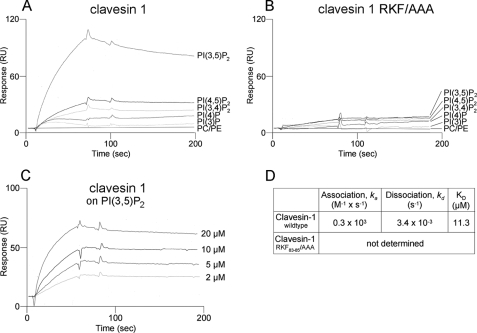
Clavesins bind to PtdIns(3,5)P2. A, liposomes composed of 80% phosphatidylcholine, 20% phosphatidylethanolamine (PC/PE) and liposomes in which 10% phosphatidylcholine was substituted for the indicated PtdIns were probed for binding of recombinant clavesin 1 using a surface plasmon resonance biosensor. Binding to PtdIns(3,5)P2 was most significant. B, a triple point mutation in the Sec14 domain (RFK/AAA mutant) abolished binding, indicating that clavesin 1 binding is specific, although it exhibits a relatively low affinity. C and D, a series of injections with varying concentrations of clavesin-1 were used to calculate a rate constant of 11.3 μm for clavesin 1 binding to PtdIns(3,5)P2.
FIGURE 6.
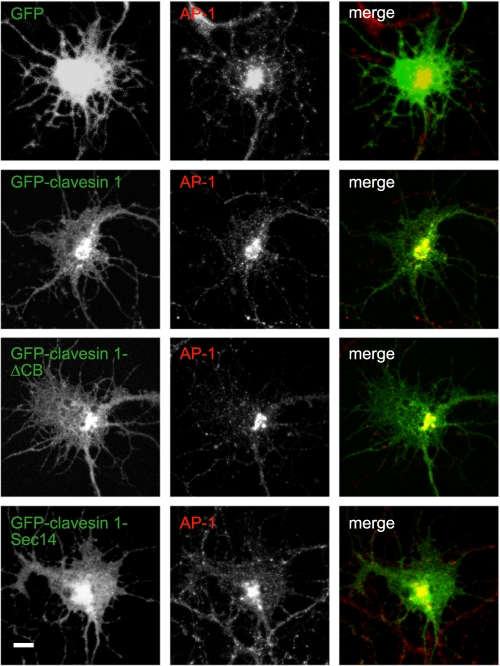
The Sec14 domain targets clavesin to the membrane. Hippocampal neurons at 6 DIV transfected with plasmids encoding GFP, GFP-clavesin 1, GFP-clavesin 1 lacking the clathrin box (ΔCB), or GFP-clavesin 1 Sec14 domain, as indicated, were processed by indirect immunofluorescence with antibody against AP-1 at 7 DIV. Bar, 2 μm.
Clavesins Are in a Complex with CHC and AP-1
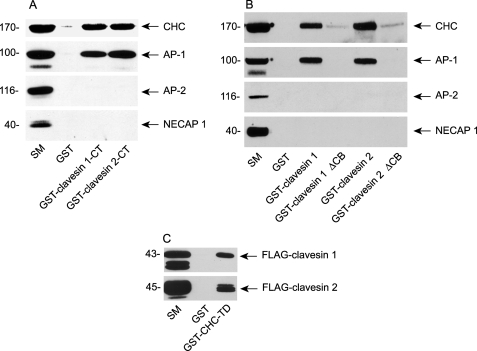
Clavesin 1 and 2 end with the sequence LLALD and LLSLD, respectively (Fig. 1A). These sequences match the consensus (θ-θ-X-θ-Ac, where θ represents a bulky hydrophobic residue, and Ac is an acidic residue) for a type I clathrin box (32, 33). GST-tagged clavesin 1 or 2 C-terminal constructs (residues 298–354 and 276–327, respectively) affinity-selected CHC from brain extracts (Fig. 5A). Similar results were seen with full-length proteins, and deletion of the clathrin box strongly reduced CHC binding (Fig. 5B). Clathrin box motifs interacted with the N-terminal domain of the CHC, and a GST-terminal domain construct (GST-CHC-TD) affinity-selected clavesin 1 and 2 from transfected cell lysates (Fig. 5C). AP-1 was also affinity-selected with GST-clavesin 1 and 2, and this interaction depended on residues in the clathrin box (Fig. 5, A and B). Clavesins did not bind the plasma membrane clathrin adaptor protein-2 (AP-2) (Fig. 5, A and B). The mechanism of AP-1 binding is unknown, but it may be indirect through CHC. An important future avenue will be to determine the selectivity of clavesins for AP-1 versus AP-2.
FIGURE 5.
Clavesins form a complex with AP-1 and CHC. A, GST fusion proteins encoding clavesin 1 or 2 C-terminal regions or GST alone were precoupled to glutathione-Sepharose and incubated with soluble brain extracts. Affinity-selected proteins were blotted with the indicated antibodies. The starting material (SM) is an aliquot of extract equal to one-tenth of that added to the fusion proteins. B, as in A except using GST-full-length clavesin 1 and 2 and full-length constructs lacking the clathrin box (ΔCB). C, a GST fusion protein encoding the N-terminal domain of the CHC (GST-CHC-TD) or GST alone was precoupled to glutathione-Sepharose and incubated with soluble extracts from HEK-293 cells transfected with FLAG-tagged clavesin 1 or 2. Affinity-selected proteins were blotted with an anti-FLAG antibody.
The Sec14 Domain Targets Clavesins to Membranes
We next sought to identify the region of the clavesins responsible for their membrane targeting. Whereas GFP alone transfected into hippocampal cultures was found throughout neurons, GFP-clavesin 1 was co-localized with AP-1 in the juxtanuclear area (Fig. 6). Interestingly, GFP-clavesin 1 lacking the clathrin box showed a similar AP-1 co-localization as the full-length protein, and in fact the Sec14 domain alone appeared to be sufficient for guiding localization (Fig. 6). Similar results were observed for clavesin 2 in neurons and for both clavesin 1 and 2 ectopically expressed in HeLa cells (data not shown).
The Sec14 Domain of Clavesin 1 Binds PtdIns(3,5)P2
Sec14 domains generally function in lipid binding (21, 22). Preliminary studies with PIP strips indicated that the Sec14 domain of clavesin 1 and 2 bound to phosphorylated forms of PtdIns but not PtdIns alone or other phospholipids, such as phosphatidylcholine or phosphatidylserine (data not shown). We thus examined Sec14 domain binding to PtdIns species using liposomes of known composition, captured on biosensor chips. This membrane mimic assay allows for real time detection of lipid binding with affinity measurements (30). Intriguingly, the Sec14 domain of clavesin 1 showed specific binding to PtdIns(3,5)P2 (Fig. 7A). Much weaker binding was seen with PtdIns 4,5-bisphosphate and PtdIns 3,4-bisphosphate, and binding of PtdIns 4-phosphate or PtdIns 3-phosphate was only marginally above the background seen with the phosphoglycerolipids (Fig. 7A). To further demonstrate the specificity of these interactions, we sought to design a phospholipid-binding mutant. PTP-MEG2 is a Sec14 domain-bearing protein phosphatase, and point mutations have been identified that disrupt PtdIns binding to the Sec14 domain (34, 35). These residues are conserved in the Sec14 domain of clavesin 1 and 2, and we thus made a triple point mutation in clavesin 1 (R83A/K84A/F85A). The RKF/AAA triple mutant Sec14 domain was expressed as well as the wild-type version but displayed abrogated binding to PtdIns(3,5)P2 (Fig. 7B), confirming the specificity of the interaction. A concentration series was performed for the binding of the Sec14 domain to PtdIns(3,5)P2 (Fig. 7C), revealing a Kd of 11.3 μm (Fig. 7D).
PtdIns Binding Is Not Required for the Localization of Clavesins
The Sec14 domain of the clavesins is sufficient for membrane targeting (Fig. 6), suggesting that PtdIns binding may guide the proteins to membranes. However, this seemed somewhat unlikely, given the modest affinity for PtdIns(3,5)P2. To test if lipid binding was important for localization, we transfected cells with GFP-tagged clavesin 1 containing the triple mutation. This construct showed a similar localization pattern in HeLa cells as the wild-type protein, revealing co-localization with AP-1 at the TGN and endosomes (Fig. 8). Similar results were seen in neurons (data not shown). Moreover, wild-type Sec14 domain alone (lacking the clathrin-binding motif) showed a similar localization pattern as the Sec14 domain alone with the triple mutation (supplemental Fig. S4). In fact, PTP-MEG2 mutated for PtdIns binding also shows normal Sec14 domain-dependent localization to secretory vesicles (35). Thus, whereas the Sec14 domain appears sufficient for membrane targeting, the phospholipid binding is not required. Future studies will be directed to identifying the interacting factors that target clavesins via their Sec14 domains to membranes.
FIGURE 8.
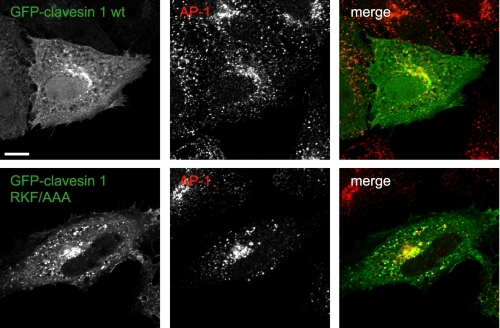
Membrane localization of clavesin is PtdIns-independent. HeLa cells were transfected with plasmids encoding GFP-clavesin 1 or GFP-clavesin 1 containing a triple point mutation in the Sec14 domain that eliminates PtdIns binding (RFK/AAA). The cells were subsequently processed by indirect immunofluorescence with antibody against AP-1 (red). Bar, 10 μm.
Clavesin Knockdown Disrupts the Morphology of LAMP1-positive Late Endosomes/Lysosomes
The TGN and endosomal localization of the CCV-associated clavesins together with the binding to PtdIns(3,5)P2, which is involved in the formation/maturation of late endosomes/lysosomes, suggests that clavesins function in trafficking between the TGN and the endosomal system and/or in the transition of early endosomes to late endosomes/lysosomes. To explore these possibilities, we took a loss-of-function approach using a lentiviral system that allows for the efficient delivery of inhibitory miRNAs into postmitotic cells, including neurons, and examined the steady-state localization of the mannose 6-phosphate receptor and LAMP1. The mannose 6-phosphate receptor uses CCVs to transport lysosomal hydrolases from the TGN to endosomes (36–38). The hydrolases are eventually targeted to lysosomes, whereas the receptor returns to the TGN (39). Thus, alterations in the localization of the mannose 6-phosphate receptor are indicative of disruptions in the TGN/endosomal trafficking cycle. In contrast, LAMP1 is a resident late endosome/lysosomal protein that is delivered in a clathrin-independent manner from the TGN to early endosomes, which then transition into late endosomes/lysosomes (40, 41). Cultured hippocampal neurons were transduced with a control virus encoding a non-targeting sequence or with viruses encoding two sequences specific for clavesin 1 and two sequences specific for clavesin 2. In addition, the clavesin 1 and 2 miRNA targeting viruses were used in varying combinations (Fig. 9A). The viruses were highly effective in allowing for isoform-specific knockdown in neurons (Fig. 9A). We were also able to efficiently knock down both isoforms using various combinations of isoform-specific viruses (Fig. 9A). Immunofluorescence revealed a greatly reduced signal in knockdown neurons compared with controls (Fig. 9B). Thus, we are able to effect efficient clavesin knockdown. However, we were unable to detect any change in the localization of either the 46- or 300-kDa isoform of the mannose 6-phosphate receptor (supplemental Fig. S5). Moreover, although there was a slight reduction in the recruitment of both CHC and AP-1 to the TGN, neither effect was significant (supplemental Fig. S6), nor was there any influence of clavesin knockdown on TGN morphology (data not shown). In contrast, LAMP1 staining was dramatically altered following clavesin knockdown (Fig. 9C). Single knockdown of either clavesin 1 or 2 led to a slight and not significant enlargement of LAMP1-positive late endosomes/lysosomes that became highly significant when both clavesins were depleted (Fig. 9, C and D). Thus, clavesin function is required for the normal morphology of LAMP1-positive late endosomes/lysosomes in neurons.
DISCUSSION
Subcellular proteomics has proven to be an extremely versatile approach toward the identification of new protein components of organelles (42). Our proteomic analysis of CCVs isolated from rat brain yielded eight proteins that at the time of the analysis had not been described at the protein level (23, 24). Six of these proteins have now been linked to clathrin-mediated membrane trafficking, including AAK1 (adaptor-associated kinase 1) (43), NECAP1 and -2 (44, 45), CVAK104 (coated vesicle-associated kinase of 104 kDa)/Scyl-like 2 (46), FENS1 (FYVE domain endosomal protein 1) (47), and enthoprotin/epsinR (23). These proteins function in trafficking at various subcellular locations. AAK1 and NECAP1 and -2 interact with AP-2 and are involved in the formation of cargo-laden CCVs at the plasma membrane during endocytosis (43, 45). Interestingly, like clavesins, FENS1, CVAK104, and enthoprotin/epsinR are localized to membranes of the TGN and/or endosomes and are involved in trafficking at these compartments (23, 46–50). Both FENS1 and enthoprotin/epsinR interact with PtdIns through specific lipid-binding modules, and both CVAK104 and enthoprotin/epsinR bind to AP-1 (23, 46–52). Thus, clavesins join an expanding repertoire of CCV-associated proteins that provide an interface between the clathrin machinery and PtdIns.
Given that clavesins are associated with TGN-derived CCVs and that they form a complex with CHC and AP-1, it seemed possible that they could be involved in the formation of CCVs at the TGN membrane, specifically that they interact with the TGN membrane through the Sec14 domain and serve a bridging function in the recruitment of CHC·AP-1 complexes. However, we saw no significant change in the localization of AP-1 or CHC at the TGN following clavesin knockdown. Moreover, we saw no alterations in the localization of mannose 6-phosphate receptors, inconsistent with a role for clavesins in clathrin-mediated trafficking from the TGN to endosomes or recycling of mannose 6-phosphate receptors from endosomes back to the TGN. However, clavesin knockdown in hippocampal neurons did cause a significant enlargement of a LAMP1-positive compartment. LAMP1 traffics from the TGN to early endosomes before progression into late endosomes/lysosomes (40). Given the lack of influence of clavesin knockdown on TGN to endosome trafficking, these data suggest that clavesins function in a pathway between early endosomes and mature lysosomes. In this scenario, clavesins use CCVs as a mechanism to reach the endosomal system. LAMP1 itself traffics from the TGN to early endosomes using a clathrin-independent pathway (41). Thus, clavesins will not encounter LAMP1-positive membranes until early endosomes, further supporting the possibility that the influence of clavesins on LAMP1 occurs after early endosomes. We do not at present understand the mechanism of clavesin function or the means by which clavesin proteins influence the morphology of the LAMP1-positive compartment. However, given that the Sec14 domain binds PtdIns(3,5)P2 and that decreased levels of this inositol phospholipid lead to the formation of enlarged cytoplasmic LAMP2-positive vacuoles (10, 11, 13, 14, 53), it is interesting to speculate that clavesin knockdown could cause decreased PtdIns(3,5)P2 levels. Unfortunately, our attempts to assay PtdIns(3,5)P2 in knockdown versus control neurons following loading with [3H]myo-inositol were frustrated by the low level of this PtdIns species (54) and limitations in the material available from knockdown neuronal cultures. This will clearly be an important avenue for future exploration.
The ingls (infantile gliosis) mouse arose as a spontaneous mutation at The Jackson Laboratory. The mice show enlargement of brain ventricles and gliosis (astrocyte proliferation) as a result of extensive and severe neuronal degeneration (11). It was recently demonstrated that ingls results from a mutation in Vac14 that disrupts the Vac14-PIKfyve complex and leads to decreased PtdIns(3,5)P2 levels (11). It is not understood why alterations in the level of a lipid involved in “housekeeping” trafficking pathways would lead to selective neuronal loss (11). However, these data imply the presence of neuron-specific PtdIns(3,5)P2 effectors that are important for cell survival. To the best of our knowledge, clavesin 1 is the first neuron-specific PtdIns(3,5)P2-binding protein. Moreover, it is interesting that clavesins function in trafficking to lysosomes. The importance of lysosomes for cell survival is underscored not only by their ability to degrade damaged macromolecules but also by the fact that compromised lysosomal function can lead to apoptotic cell death (55). Neurons are particularly sensitive to lysosomal dysfunction, and alterations in lysosomal function are the underlying cause of numerous neurodegenerative diseases (55). These data imply that neurons have unique mechanisms for regulation of lysosomal function and/or formation/maintenance, probably involving PtdIns(3,5)P2. An important avenue for the future will be to determine the mechanism of clavesin function in LAMP1 trafficking in neurons.
Supplementary Material
Acknowledgments
We are grateful to Lucia Rameh for assistance with high pressure liquid chromatography analysis of labeled inositol phospholipids. We thank Jacynthe Philie for expert technical assistance, Dr. Fred Meunier for the tandem FYVE construct, and Dr. Kurt von Figura for mannose 6-phosphate receptor-specific antibodies.
This work was supported by Canadian Institutes of Health Research (CIHR) Grant MOP-62684 (to P. S. M.).
The nucleotide sequence reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) BK006899–BK006900.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- TGN
- trans-Golgi network
- CCV
- clathrin-coated vesicle
- AP
- adaptor protein
- PtdIns
- phosphatidylinositol
- PtdIns(3,5)P2
- phosphatidylinositol 3,5-bisphosphate
- GST
- glutathione S-transferase
- GFP
- green fluorescent protein
- PBS
- phosphate-buffered saline
- DIV
- days in vitro
- miRNA
- microRNA
- CHC
- clathrin heavy chain.
REFERENCES
- 1.Donaldson J. G., McPherson P. S. (2009) EMBO Rep. 10, 132–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y. J., Wang J., Sun H. Q., Martinez M., Sun Y. X., Macia E., Kirchhausen T., Albanesi J. P., Roth M. G., Yin H. L. (2003) Cell 114, 299–310 [DOI] [PubMed] [Google Scholar]
- 3.Ritter B., McPherson P. S. (2004) Top. Curr. Genet. 10, 9–37 [Google Scholar]
- 4.Gillooly D. J., Raiborg C., Stenmark H. (2003) Histochem. Cell Biol. 120, 445–453 [DOI] [PubMed] [Google Scholar]
- 5.Burd C. G., Emr S. D. (1998) Mol. Cell 2, 157–162 [DOI] [PubMed] [Google Scholar]
- 6.Patki V., Lawe D. C., Corvera S., Virbasius J. V., Chawla A. (1998) Nature 394, 433–444 [DOI] [PubMed] [Google Scholar]
- 7.Gaullier J. M., Simonsen A., D'Arrigo A., Bremnes B., Stenmark H., Aasland R. (1998) Nature 394, 432–433 [DOI] [PubMed] [Google Scholar]
- 8.Simonsen A., Lippé R., Christoforidis S., Gaullier J. M., Brech A., Callaghan J., Toh B. H., Murphy C., Zerial M., Stenmark H. (1998) Nature 394, 494–498 [DOI] [PubMed] [Google Scholar]
- 9.Mills I. G., Jones A. T., Clague M. J. (1998) Curr. Biol. 16, 881–884 [DOI] [PubMed] [Google Scholar]
- 10.Dove S. K., Dong K., Kobayashi T., Williams F. K., Michell R. H. (2009) Biochem. J. 419, 1–13 [DOI] [PubMed] [Google Scholar]
- 11.Jin N., Chow C. Y., Liu L., Zolov S. N., Bronson R., Davisson M., Petersen J. L., Zhang Y., Park S., Duex J. E., Goldowitz D., Meisler M. H., Weisman L. S. (2008) EMBO J. 27, 3221–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michell R. H., Dove S. K. (2009) EMBO J. 28, 86–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutherford A. C., Traer C., Wassmer T., Pattni K., Bujny M. V., Carlton J. G., Stenmark H., Cullen P. J. (2006) J. Cell Sci. 119, 3944–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jefferies H. B., Cooke F. T., Jat P., Boucheron C., Koizumi T., Hayakawa M., Kaizawa H., Ohishi T., Workman P., Waterfield M. D., Parker P. J. (2008) EMBO Rep. 9, 164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemaire J. F., McPherson P. S. (2006) FEBS Lett. 580, 6948–6954 [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Zolov S. N., Chow C. Y., Slutsky S. G., Richardson S. C., Piper R. C., Yang B., Nau J. J., Westrick R. J., Morrison S. J., Meisler M. H., Weisman L. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17518–17523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curwin A. J., Fairn G. D., McMaster C. R. (2009) J. Biol. Chem. 284, 7364–7375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips S. E., Vincent P., Rizzieri K. E., Schaaf G., Bankaitis V. A., Gaucher E. A. (2006) Crit. Rev. Biochem. Mol. Biol. 41, 21–49 [DOI] [PubMed] [Google Scholar]
- 19.Bankaitis V. A., Aitken J. R., Cleves A. E., Dowhan W. (1990) Nature 347, 561–562 [DOI] [PubMed] [Google Scholar]
- 20.Patton-Vogt J. L., Griac P., Sreenivas A., Bruno V., Dowd S., Swede M. J., Henry S. A. (1997) J. Biol. Chem. 272, 20873–20883 [DOI] [PubMed] [Google Scholar]
- 21.Saito K., Tautz L., Mustelin T. (2007) Biochim. Biophys. Acta 1771, 719–726 [DOI] [PubMed] [Google Scholar]
- 22.Mousley C. J., Tyeryar K. R., Vincent-Pope P., Bankaitis V. A. (2007) Biochim. Biophys. Acta. 1771, 727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wasiak S., Legendre-Guillemin V., Puertollano R., Blondeau F., Girard M., de Heuvel E., Boismenu D., Bell A. W., Bonifacino J. S., McPherson P. S. (2002) J. Cell Biol. 158, 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blondeau F., Ritter B., Allaire P. D., Wasiak S., Girard M., Hussain N. K., Angers A., Legendre-Guillemin V., Roy L., Boismenu D., Kearney R. E., Bell A. W., Bergeron J. J., McPherson P. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3833–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritter B., Denisov A. Y., Philie J., Deprez C., Tung E. C., Gehring K., McPherson P. S. (2004) EMBO J. 23, 3701–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Micheva K. D., Kay B. K., McPherson P. S. (1997) J. Biol. Chem. 272, 27239–27245 [DOI] [PubMed] [Google Scholar]
- 27.Wen P. J., Osborne S. L., Morrow I. C., Parton R. G., Domin J., Meunier F. A. (2008) Mol. Biol. Cell 19, 5593–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burman J. L., Wasiak S., Ritter B., de Heuvel E., McPherson P. S. (2005) FEBS Lett. 579, 2177–2184 [DOI] [PubMed] [Google Scholar]
- 29.Thomas S., Ritter B., Verbich D., Sanson C., Bourbonnière L., McKinney R. A., McPherson P. S. (2009) J. Biol. Chem. 284, 12410–12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Höning S., Ricotta D., Krauss M., Späte K., Spolaore B., Motley A., Robinson M., Robinson C., Haucke V., Owen D. J. (2005) Mol. Cell 18, 519–531 [DOI] [PubMed] [Google Scholar]
- 31.Kong Y. H., Ye G. M., Qu K., Pan W. Q., Liu X. H., Wan B., Guo J. H., Yu L. (2006) Biotechnol. Lett. 28, 1327–1333 [DOI] [PubMed] [Google Scholar]
- 32.Ramjaun A. R., McPherson P. S. (1998) J. Neurochem. 70, 2369–2376 [DOI] [PubMed] [Google Scholar]
- 33.ter Haar E., Harrison S. C., Kirchhausen T. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruger J. M., Fukushima T., Cherepanov V., Borregaard N., Loeve C., Shek C., Sharma K., Tanswell A. K., Chow C. W., Downey G. P. (2002) J. Biol. Chem. 277, 2620–2628 [DOI] [PubMed] [Google Scholar]
- 35.Huynh H., Wang X., Li W., Bottini N., Williams S., Nika K., Ishihara H., Godzik A., Mustelin T. (2003) J. Immunol. 171, 6661–6671 [DOI] [PubMed] [Google Scholar]
- 36.Hille-Rehfeld A. (1995) Biochim. Biophys. Acta 1241, 177–194 [DOI] [PubMed] [Google Scholar]
- 37.Kornfeld S. (1992) Annu. Rev. Biochem. 61, 307–330 [DOI] [PubMed] [Google Scholar]
- 38.Kornfeld S., Mellman I. (1989) Annu. Rev. Cell Biol. 5, 483–525 [DOI] [PubMed] [Google Scholar]
- 39.Seaman M. N. (2007) J. Cell Sci. 120, 2378–2389 [DOI] [PubMed] [Google Scholar]
- 40.Cook N. R., Row P. E., Davidson H. W. (2004) Traffic 5, 685–699 [DOI] [PubMed] [Google Scholar]
- 41.Karlsson K., Carlsson S. R. (1998) J. Biol. Chem. 273, 18966–18973 [DOI] [PubMed] [Google Scholar]
- 42.Au C. E., Bell A. W., Gilchrist A., Hiding J., Nilsson T., Bergeron J. J. (2007) Curr. Opin. Cell Biol. 19, 376–385 [DOI] [PubMed] [Google Scholar]
- 43.Conner S. D., Schmid S. L. (2002) J. Cell Biol. 156, 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritter B., Philie J., Girard M., Tung E. C., Blondeau F., McPherson P. S. (2003) EMBO Rep. 4, 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ritter B., Denisov A. Y., Philie J., Allaire P. D., Legendre-Guillemin V., Zylbergold P., Gehring K., McPherson P. S. (2007) EMBO J. 26, 4066–4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conner S. D., Schmid S. L. (2005) J. Biol. Chem. 280, 21539–21544 [DOI] [PubMed] [Google Scholar]
- 47.Ridley S. H., Ktistakis N., Davidson K., Anderson K. E., Manifava M., Ellson C. D., Lipp P., Bootman M., Coadwell J., Nazarian A., Erdjument-Bromage H., Tempst P., Cooper M. A., Thuring J. W., Lim Z. Y., Holmes A. B., Stephens L. R., Hawkins P. T. (2001) J. Cell Sci. 114, 3991–4000 [DOI] [PubMed] [Google Scholar]
- 48.Hirst J., Motley A., Harasaki K., Peak Chew S. Y., Robinson M. S. (2003) Mol. Biol. Cell 14, 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalthoff C., Groos S., Kohl R., Mahrhold S., Ungewickell E. J. (2002) Mol. Biol. Cell 13, 4060–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mills I. G., Praefcke G. J., Vallis Y., Peter B. J., Olesen L. E., Gallop J. L., Butler P. J., Evans P. R., McMahon H. T. (2003) J. Cell Biol. 160, 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Düwel M., Ungewickell E. J. (2006) Mol. Biol. Cell 17, 4513–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borner G. H., Rana A. A., Forster R., Harbour M., Smith J. C., Robinson M. S. (2007) Traffic 8, 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ikonomov O. C., Sbrissa D., Shisheva A. (2006) Am. J. Physiol. Cell Physiol. 291, C393–C404 [DOI] [PubMed] [Google Scholar]
- 54.Bonangelino C. J., Nau J. J., Duex J. E., Brinkman M., Wurmser A. E., Gary J. D., Emr S. D., Weisman L. S. (2002) J. Cell Biol. 156, 1015–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pivtoraiko V. N., Stone S. L., Roth K. A., Shacka J. J. (2009) Antioxid. Redox Signal. 11, 421–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.