Abstract
The anaerobic bacterium Aeromonas sobria is known to cause potentially lethal septic shock. We recently proposed that A. sobria serine protease (ASP) is a sepsis-related factor that induces vascular leakage, reductions in blood pressure via kinin release, and clotting via activation of prothrombin. ASP preferentially cleaves peptide bonds that follow dibasic amino acid residues, as do Kex2 (Saccharomyces cerevisiae serine protease) and furin, which are representative kexin family proteases. Here, we revealed the crystal structure of ASP at 1.65 Å resolution using the multiple isomorphous replacement method with anomalous scattering. Although the overall structure of ASP resembles that of Kex2, it has a unique extra occluding region close to its active site. Moreover, we found that a nicked ASP variant is cleaved within the occluding region. Nicked ASP shows a greater ability to cleave small peptide substrates than the native enzyme. On the other hand, the cleavage pattern for prekallikrein differs from that of ASP, suggesting the occluding region is important for substrate recognition. The extra occluding region of ASP is unique and could serve as a useful target to facilitate development of novel antisepsis drugs.
Aeromonas species are Gram-negative facultative anaerobic bacteria found ubiquitously in a variety of aquatic environments (1). The main syndrome caused by infection with Aeromonas is gastroenteritis (2, 3), although, in severe cases, sepsis is induced as a deuteropathy (4, 5). Two species, Aeromonas hydrophila and Aeromonas sobria, are associated with human disease (6, 7). Factors thought to play important roles in the pathogenesis include fimbrial and afimbrial adherence factors; a variety of exotoxins, including hemolysin, cytotonic enterotoxin, heat-stable enterotoxin, and heat-labile enterotoxin; and several secreted proteases and lipases (8–12). Recently, we purified a 65-kDa A. sobria serine protease (ASP)2 from the culture supernatant of A. sobria and found that the enzyme induces vascular leakage, reduces blood pressure through activation of the kallikrein/kinin system (13), promotes human plasma coagulation through activation of prothrombin (14), and causes the formation of pus and edema through the action of anaphylatoxin C5a (15). From these observations, we concluded that ASP mediates the induction of disseminated intravascular coagulation through α-thrombin production, which is a common and deadly consequence of sepsis (14).
ASP is a kexin-like serine protease belonging to the subtilisin family (subtilases) (16), which can be subdivided into six groups: the subtilisins, thermitases, proteinase K, lantibiotic peptidases, pyrolysins, and kexins. Among the kexins, the first identified was Kex2 (17), which is expressed by Saccharomyces cerevisiae; subsequently, the mammalian kexin-like protease furin was identified (18). Furin processes the precursors of biologically active peptides and proteins via limited proteolysis at paired basic amino acids to generate biologically active molecules (19). The domain structures of kexins and furins include a signal peptide, a partially conserved propeptide, a highly conserved subtilisin-like domain containing the characteristic Asp, His, and Ser catalytic residues, a relatively well conserved region called the P-domain, and a transmembrane domain followed by a cytoplasmic tail (20–22). Kex2 usually shows a high degree of specificity for cleavage after dibasic (P2-P1: Lys-Arg or Arg-Arg) or multiple basic residues (23). Among the kexins, which are nearly all eukaryotic and share a substantial degree of sequence homology (>40%), ASP is positioned as the most distant member of this family on the phylogenetic tree (16). The domain structure of ASP consists of the propeptide, the catalytic subtilisin-like domain, and the P-domain. For maturation of ASP, the first 24 residues of the propeptide are cleaved, and although a functional P-domain is reportedly necessary for maturation of the subtilisin domain in kexins (24, 25), the function of the P-domain in ASP remains unknown. Notably, in an earlier study of ASP expression, we found that for the maturation of the ASP subtilisin domain, another gene product, encoded by open reading frame 2, is required to serve as a chaperone in the periplasmic space (26).
Here, we report the crystal structure of wild-type ASP as a sepsis-related factor at 1.65 Å resolution. We found that ASP has a unique occluding region at the active site within the subtilisin domain and that a different form of ASP that is cleaved within the occluding region shows a different pattern of proteolysis from the native enzyme. Our findings suggested that the novel occluding region plays an important role in determining substrate specificity and that because it is unique, it could facilitate development of novel antisepsis drugs that have no inhibitory effect on furin-like human proteases.
EXPERIMENTAL PROCEDURES
Crystallization
We purified ASP from the culture supernatant of the A. sobria T94 strain transformed with pSA19-5528, including the asp and orf2 genes as described previously (12), and its sequence has been already registered in GenBank under the accession number AF253471. The sitting drop vapor diffusion method with polyethylene glycol (PEG) was used to prepare ASP crystals, which grew in 3–4 days at a PEG concentration of 10%. The droplets contained 10 mg/ml of ASP protease, 5% PEG 3000, and 50 mm CHES at a pH of 9.5. After reaching maximum dimensions of about 50 μm × 50 μm × 60 μm, the crystals were placed in mother liquor containing 40% trehalose and then briefly soaked and flash-vitrified in liquid nitrogen. Heavy atom derivatives were made by soaking the crystals in mother liquor containing a heavy atom compound (1 mm) for 24 h at 20 °C. ASP crystallizes in the space group P21 with unit cell parameter a = 49.8 Å, b = 112.0Å, c = 51.8 Å, and β = 110.7 and 42% solvent content.
Phase Determination and Model Building
Initial phases were determined by the method of the multiple isomorphous replacement with anomalous scattering method using one native data set and three derivative data sets (thimerosal and mersalyl acid) using R-AXIS VII and FR-E at 100 K. The initial multiple isomorphous replacement with anomalous scattering phasing (three mercury derivatives) was carried out at 2.0 Å using SOLVE (27) and gave a figure-of-merit of 0.34 and a score of 24.9. The quality of the electron density map was excellent. After solvent flattening, the chain was traced automatically using RESOLVE (28), and 79.6% (478 amino acids) of the model was constructed. Several cycles of manual building using XTALVIEW (29) and refinement using CNS (30) were then performed. At this stage, another high resolution data set (1.65 Å) was obtained using synchrotron radiation at Photon Factory AR NW12 (Table 1) at 100 K, and data processing was carried out using HKL2000 (31). The initial model was then subjected to refinement and iterative model building against the high resolution data. Finally, we subjected TLS refinement (41) using REFMAC5 (32) and manual refinement using MolProbity (33) and Coot (34). At this stage, we found that our previous report about the asp nucleotide sequence might be partially wrong. To resolve this suspicion, we carefully reconfirmed the sequence. Consequently, we found that four residues at positions 70 (His), 114 (Gly), 176 (His), and 179 (Arg) must be revised because of our wrong nucleotide sequence determination. These amino acid residues were instead Gln, Ala, Gln, and Gly, respectively. We have already revised the sequence on GenBank (accession number AF253471). The final R factor and Rfree were 16.8% and 20.1%, respectively. The final model included residues 3–595, three calcium ions, and 700 water molecules (supplemental Fig. S1). Two N-terminal and five C-terminal residues were missing from the electron density map.
TABLE 1.
Data collection and refinement statistics
| Native 1 | Thiomerosal 01 (anomalous data) | Thiomerosal 02 | Mersalyl acid (anomalous data) | Native 2 | |
|---|---|---|---|---|---|
| Data collection | |||||
| Radiation source | R-AXIS VII/FR-E | PF-AR NW12 | |||
| Wavelength (Å) | 1.5418 | 1.5418 | 1.5418 | 1.5418 | 1.00 |
| Resolution range (Å) | 50.0–1.9 | 50.0–2.0 | 50.0–2.0 | 50.0–2.43 | 50.0–1.65 |
| Highest resolution shell (Å) | 2.0–1.9 | 2.11–2.00 | 2.11–2.00 | 2.52–2.43 | 1.74–1.65 |
| No. of unique reflections | 42,545 | 61,942 | 31,397 | 20,913 | 62,466 (8734) |
| Redundancy | 2.9 (2.1) | 1.4 (1.4) | 2.6 (2.3) | 6.1 (5.8) | 3.7 (3.4) |
| Completeness (%) | 97.1 (82.5) | 84.6 (70.8) | 83.9 (77.9) | 99.6 (98.8) | 98.7 (94.9) |
| Rsym (%)a | 7.5 (29.3) | 8.4 (30.0) | 8.7 (30.0) | 5.7 (17.1) | 3.9 (13.7) |
| I/σ(I) | 7.1 (1.9) | 4.8 (1.5) | 6.7 (1.9) | 9.4 (2.3) | 22.5 (6.2) |
| Rpimb | 3.7 (12.3) | ||||
| Rrim (Rmeas)c | 7.0 (22.8) | ||||
| MIRASd phasing FOM | 0.34 | ||||
| SOLVE score | 24.91 | ||||
| Refinement | |||||
| Resolution (Å) | 1.65 | ||||
| Rcryst (%)e | 16.8 | ||||
| Rfree (%)f | 20.1 | ||||
| No. of calcium ions | 3 | ||||
| No. of water molecules | 700 | ||||
| r.m.s.d.f from ideal bond length (Å) | 0.014 | ||||
| r.m.s.d. from ideal bond angles | 1.571° | ||||
| Ramachandran plot | |||||
| Residues in most favorable regions (%) | 87.5 | ||||
| Residues in additional allowed regions (%) | 11.9 | ||||
| Residues in generously allowed regions (%) | 0.4 | ||||
| Residues in disallowed regions (%) | 0.2 | ||||
| Space group P21/unit cell parameters | |||||
| Native 1 | a = 50.7, b = 112.3, c = 54.0 Å; α = γ = 90°, β = 113.0° | ||||
| Thiomerosal 01 | a = 50.4, b = 112.1, c = 53.5 Å; α = γ = 90°, β = 112.6° | ||||
| Thiomerosal 02 | a = 50.7, b = 112.4, c = 53.9 Å; α = γ = 90°, β = 112.9° | ||||
| Mersalyl acid | a = 50.6, b = 112.4, c = 53.8 Å; α = γ = 90°, β = 112.8° | ||||
| Native 2 | a = 49.7, b = 112.0, c = 51.8 Å; α = γ = 90°, β = 110.7° | ||||
aRsym = ΣhklΣi=1N|〈Ihkl〉 − Iihkl|/ΣhklΣi=1NIihkl.
bRpim = Σhkl(1/N − 1)1/2Σi=1N|〈Ihkl〉 − Iihkl|/ΣhklΣi=1NIihkl where N is the multiplicity of the observed reflection.
cRrim (Rmeas) = Σhkl(N/N − 1)1/2Σi=1N|〈Ihkl〉 − Iihkl|/ΣhklΣi=1NIihkl where N is the multiplicity of the observed reflection.
d MIRAS, multiple isomorphous replacement method with anomalous scattering; r.m.s.d., root mean square deviation.
e Rwork = Σh‖Fobs| − |Fcalc‖/Σh|Fobs|.
f Rfree was calculated with randomly selected reflections (5%).
Model of ASP in Complex with Substrate
A model of ASP in complex with a substrate was created by referring to the crystal structure of Kex2 with the inhibitor Ac-Arg-Glu-Lys-boroArg peptidyl boronic acid bound (35). First, ASP was superimposed to the structure of Kex2 with inhibitor (Protein Data Bank code 1R64). Second, the inhibitor of Kex2 and ASP was extracted. The amino acid was swapped from arginine to lysine in P1, and finally, the torsion angel of this P1 lysine and P3 glutamate was modified to contact with Arg-566.
Purification of Nicked ASP
To purify ASP, A. sobria strain T94 harboring pSA19–5528 was cultured in Luria broth supplemented with chloramphenicol (50 μg/ml) for 20 h at 37 °C with shaking (12). After cultivation, the cells were removed from the culture fluid by centrifugation, and ammonium sulfate was added to the supernatant to 50% saturation. The resultant precipitate was removed by centrifugation (18,000 × g for 30 min), and ammonium sulfate was added to 60% saturation. The solution was kept at 4 °C for 15 h, after which the precipitate was collected by centrifugation (18,000 × g for 45 min), dissolved in 1 mm phosphate buffer (pH 7.2), and dialyzed against the same buffer. The resultant protein solution served as the crude sample.
The crude sample was loaded onto a hydroxyapatite column (Seikagaku Kogyo Co., Tokyo, Japan) equilibrated with 1 mm phosphate buffer (pH 7.2). After washing the column with the same buffer, the adsorbed material was eluted first with a linear gradient of 1 to 120 mm phosphate buffer (pH 7.2) and with a gradient of 120–600 mm phosphate buffer (pH 7.2). The proteolytic activity in each fraction was assessed by cleavage of azocasein (Wako Chemical Co., Tokyo, Japan). Fractions showing activity were collected, concentrated 20-fold by ultrafiltration (Vivaspin 20 Polyethersulfone 30,000 molecular weight cut off; Vivascience, Inc., Göttingen, Germany), and diluted 3-fold with water to lower the phosphate concentration. The sample was loaded onto a hydroxyapatite column in an HPLC system (Bio-Scale CHT2-I column; Bio-Rad Laboratories). After washing the column, the adsorbed material was eluted with a linear gradient of 1–100 mm phosphate buffer.
Mass Spectrometry
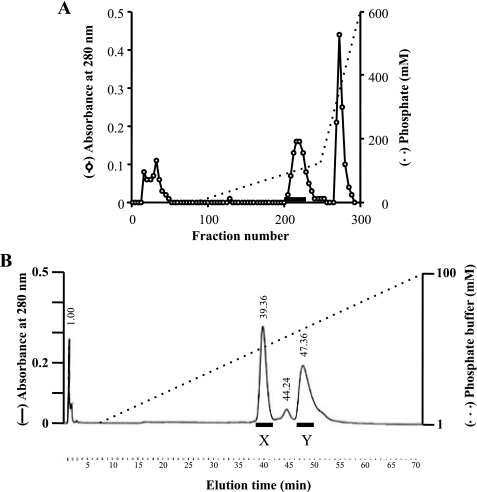
A matrix-assisted laser desorption ionization type mass spectrometer (Kompact MALDI 4 tDE; Shimadzu, Kyoto, Japan) was used for determination of the molecular mass of ASP. A sample from fraction Y of the hydroxyapatite column chromatography (see Fig. 4B) was mixed with 3,5-dimethoxy-4-hydroxy-cinnamic acid (10 mg/ml in 50% acetonitrile containing 0.05% trifluoroacetic acid). After drying, the sample was dissolved in distilled water, and the molecular mass of ASP was measured using the spectrometer according to the manufacturer's instructions.
FIGURE 4.
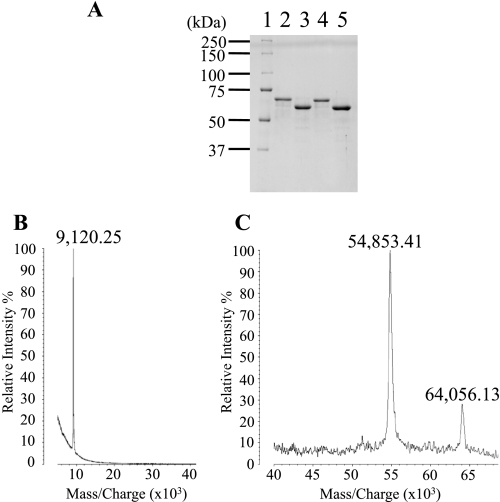
Purification of two types of ASP. A, the crude sample was loaded onto a hydroxyapatite column equilibrated with 1 mm phosphate buffer (pH 7.2). After washing the column, the adsorbed material was eluted first with a linear gradient of 1–120 mm phosphate buffer and then with a gradient of 120 to 600 mm phosphate buffer. B, fractions presenting proteolytic activity are indicated by the horizontal bar in A. The active fractions from A were collected and concentrated 20×, after which the sample was diluted 3-fold with water and loaded onto a hydroxyapatite column in a HPLC system. After washing, the adsorbed material was eluted with a linear gradient of 1–100 mm phosphate buffer. Two peaks, designated fractions X and Y, expressed proteolytic activity.
The Proteolytic Activity of ASP
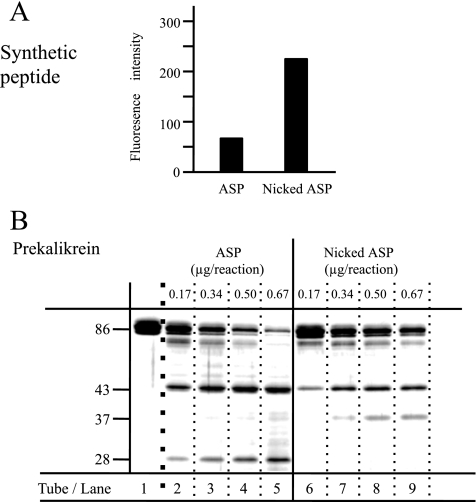
We assessed the proteolytic activity of ASP based on its ability to cleave Boc-Glu-Lys-Lys-4-methylylcoumaryl-7-amide (MCA) (Peptide Institute, Inc., Osaka, Japan). The reaction was started by adding 50 μl of protease solution containing ASP or nicked ASP to 700 μl of solution containing the peptide at a concentration of 1.1 mm. The concentration of protease in the reaction mixture was 0.493 ng/μl, and the solvent used was 10 mm sodium phosphate buffer (pH 7.5). After incubation at 37 °C for 30 min, the reaction was stopped by adding 750 μl of acetic acid (130 mm). The fluorescence of the solution (λex = 340 nm and λem = 440 nm) was measured to assess the amount of 7-amino-4-methyl-coumarin formed. To test the ability of nicked ASP to cleave prekallikrein B1 (PK) (EMD Biosciences, Inc., La Jolla, CA), 5 μg of PK were incubated for 30 min at 37 °C with various amounts of nicked ASP (0.17 μg, 0.34 μg, 0.50 μg, and 0.67 μg) in 15 μl of 10 mm sodium phosphate buffer (pH 7.5). ASP was not added to the sample in lane 1 (see Fig. 6B). After incubation, sample buffer containing SDS and 2-mercaptoethanol was added to each solution, which was heated at 100 °C for 5 min. The samples were separated by SDS-PAGE, and the gel was stained with Coomassie Brilliant Blue.
FIGURE 6.
Proteolytic activity of ASP. Boc-Glu-Lys-Lys-MCA and PK were incubated with ASP or nicked ASP. A, the proteolytic activity of ASP or nicked ASP was measured using a fluorogenic synthetic peptide substrate, (Boc)-Glu-Lys-Lys-MCA. B, samples of PK (5 μg) were incubated for 30 min at 37 °C in 15 μl of solution containing various amounts of ASP (0.17 μg, 0.34 μg, 0.50 μg, or 0.67 μg). ASP was not added to the sample for lane 1. After incubation, these samples were separated by SDS-PAGE. Molecular masses are calibrated on the side.
RESULTS
Overview of the Structure of ASP
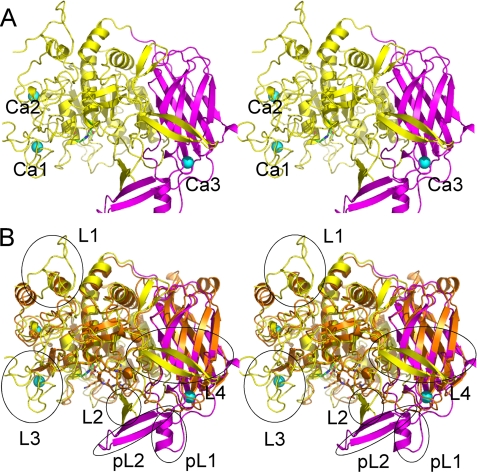
Following the x-ray crystallography carried out as described above, the final structure obtained consisted of residues 3–595 of a protein with the sequence of ASP, three Ca2+ ions, and 700 water molecules. The ASP molecule consists of two regions, an N-terminal region extending from Gly-3 to Pro-431 and forming the subtilisin domain and a C-terminal region extending from Leu-432 to His-595 and forming the P-domain (see Fig. 2A). As we predicted from the sequence, the structure of ASP is similar to those of Kex2 and furin. Superimposition of subtilisin, furin, and Kex2 gave overall Cα root mean square deviations of 2.5 Å, 2.2 Å, and 2.1 Å for 245, 445, and 452 amino acids, respectively. When the structure of ASP was superimposed on that of Kex2, a prominent difference was seen in the protruding regions marked by circles in Fig. 2B, although the two structures overlapped fairly well in the subtilisin domain and P-domain.
FIGURE 2.
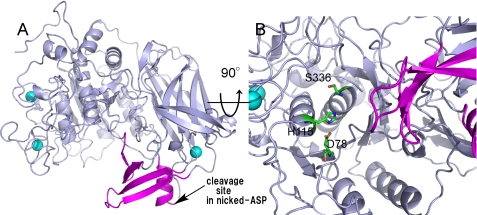
Overall structure of ASP. A, stereo ribbon representation of the overall structure of ASP. The domain architecture is shown in different colors: the N-terminal subtilisin domain (amino acids 3–431) is shown in yellow, and the C-terminal P-domain (amino acids 432–595) is shown in purple. Three bound Ca2+ ions are shown as greenish cyan spheres. B, superimposed stereo view of ASP (yellow and purple as in A) and Kex2 (orange). The four loops in the N-terminal domain of ASP are labeled L1, L2, L3, and L4, whereas the two parts of the unique extra occluding region in the C-terminal domain are labeled as pL1 and pL2.
Structure of the Subtilisin Domain
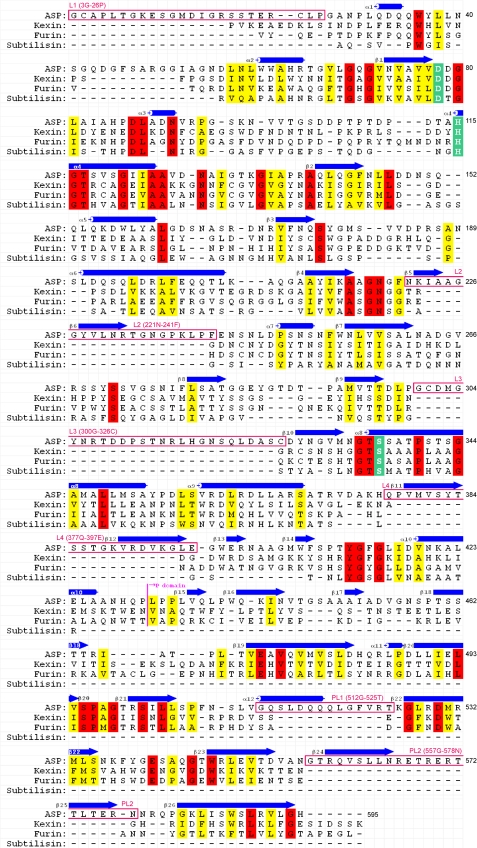
The core of the subtilisin domain consists of 10 helices (α1–α10) and twelve β-chains (β1–β10 and β13–β14) (Fig. 1). Superimposition of ASP on Kex2 (Fig. 2B) revealed that the fold of the N-terminal domain of ASP resembles that of the catalytic domain of Kex2, which was previously shown to be similar to those of subtilisin and other subtilisin-related proteases (35). The Kex2 catalytic site contains the catalytic Asp, His, and Ser residues characteristic of subtilisins, and the corresponding residues are present in ASP as Asp-78, His-115, and Ser-336 (Fig. 3B). On the other hand, four loops (L) protrude from the N-terminal subtilisin domain of ASP: Gly-3–Pro-26 (L1), Asn-221–Phe-241 (L2), Gly-300–Cys-326 (L3), and Gln-377–Glu-397 (L4). L1, L2, and L3 have random coil structure, whereas L4 forms a β-hairpin that protrudes toward the P-domain (Fig. 2B). Moreover, two disulfide bridges are formed between Cys-4 and Cys-24 in L1 and between Cys-301 and Cys-326 in L3, which stabilize those loops.
FIGURE 1.
Structure-based sequence alignment of ASP, kexin, furin, and subtilisin. Identical and similar residues are shown in red and yellow, respectively. The catalytic Asp, His, and Ser residues are shown in green. Secondary structures and extra regions are also shown. The residues are numbered (3–595) from the amino-terminal residue of ASP.
FIGURE 3.
Structure of extra occluding region of ASP. A, the extra occluding region consisting of L2, pL1, and pL2 is shown in purple. Other regions and Ca2+ ions are shown in gray and greenish cyan, respectively. The cleavage site between residues 519 and 520 of nicked ASP is indicated by the arrow. B, a close-up view of the catalytic site is shown. The color scheme is the same as in A, except for the catalytic triad (green).
Structure of the P-domain
The C-terminal region of Kex2 is referred to as the P-domain. The core of the P-domain in ASP consists of a jelly roll-like fold with eight β-strands (β16, β18–β23, and β26) (Fig. 1). As illustrated in Fig. 2B, the structure of the ASP P-domain is similar to that of Kex2, although there are specific structures that form a unique occluding region (Fig. 3A) in ASP (Fig. 2B) that is not seen in Kex2. This extra occluding region is comprised of two parts, pL1 (Gly-521–Thr-525, β5, β6, and α12) and pL2 (Gly-557–Asn-578, β25, and β25), and is particularly interesting because it is situated close to the catalytic triad Asp-78, His-115, and Ser-336 (Fig. 3B).
Three Ca2+ Binding Sites
Three Ca2+ binding sites were assigned to ASP based on electron density, counter charges, and coordination. Two of these sites (Ca1 and Ca2) are situated in the N-terminal domain, and the third site (Ca3) is situated in the C-terminal domain (Fig. 2A). In Kex2, by contrast, two Ca2+ binding sites are located in the vicinity of the catalytic site, and a third is at a position corresponding Ca2 in ASP. The two Ca2+ ions in the vicinity of the catalytic site are indispensable for the enzymatic activity of Kex2 (35), which makes it noteworthy that ASP contains no Ca2+ binding sites near its catalytic site.
Analysis of Nicked ASP Using Mass Spectrometry
In earlier studies, we purified ASP through a series of successive column chromatography steps using a DEAE-Sephadex A-25 column, a hydroxyapatite column, and a gel filtration column in an HPLC system (12). Because that method was both laborious and time-consuming, we have developed an improved method that entailed expressing recombinant ASP in A. sobria T94, a strain we found to produce large amounts of ASP. After cultivation of the ASP-expressing cells, the culture supernatant was collected after centrifugation and salted out with 50% saturated ammonium sulfate. The precipitate was then collected by centrifugation but showed little proteolytic activity. We therefore presumed that the majority of ASP remained in the supernatant. To collect ASP, this time we added ammonium sulfate to 60% saturation, and the precipitate generated was again collected by centrifugation. This material showed strong proteolytic activity and was used as the crude ASP sample.
The crude sample was dialyzed against 1 mm phosphate buffer and separated by hydroxyapatite column chromatography. The fractions eluted with ∼90 mm phosphate showed proteolytic activity (Fig. 4A) and were thus separated further on a hydroxyapatite column in an HPLC system (Fig. 4B). Two peaks, eluted with 43 and 62 mm phosphate, respectively, showed proteolytic activity. These peaks were designated fractions X and Y (Fig. 4B) and were analyzed by SDS-PAGE (Fig. 5A). The analysis showed the molecular mass of the protein in fraction X to be ∼64,000 Da (Fig. 5A, lane 2). Because the theoretical value of full-length ASP is 64,210.61 Da, the protease eluted in peak X was thought to be full-length ASP.
FIGURE 5.
Analysis of nicked ASP using mass spectrometry. A, SDS-PAGE (10% polyacrylamide gel) of the purified samples from fractions X (lane 2) and Y (lane 3) in Fig. 4B. Similarly, we also obtained the purified samples from two fractions corresponding to X (lane 4) and Y (lane 5) using the mutant strain producing ASP(H115A). Numbers along the side indicate molecular masses in kDa. Lane 1, molecular mass standards. B and C, a sample of lane 3 in A was analyzed by a matrix-assisted laser desorption ionization type mass spectrometer following the method described in the text. The mass values of each peak are shown on the figure.
On the other hand, the molecular mass of peak Y was estimated to be ∼58,000 Da (Fig. 5A, lane 3). To determine the nature of the protein in fraction Y, we transferred the band from the gel to a nitrocellulose membrane and determined the amino acid sequence at the N terminus. We found the sequence to be NEGCAPLT, which is identical to that of full-length ASP (16), suggesting the protein in peak Y is an ASP variant whose N-terminal region remains intact but whose C-terminal region is truncated. The amount of the intact form (peak X) was ∼1.3-fold larger than that of the nicked form (peak Y). We think that appearance of the nicking form (peak Y) is not due to the purification procedure because both proteins had been already observed in the culture supernatant before purification.
For a more detailed analysis of the protein eluted in fraction Y, we next subjected the fraction to MALDI-TOF MS (AXIMA-CFR; Shimazu Biotech, Kyoto, Japan). Three peaks were observed with m/z values of 64,056.13, 54,853.41, and 9,120.25, respectively (Figs. 5B and 2C). We determined that the molecular mass of the protease in lane 3 of Fig. 5A was 54,853.41 Da and that the protein extended from residues 1 to 519 of the mature ASP. (The theoretical molecular mass of this region is 54,881.17 Da.) Thus, the cleavage appears to have occurred between residues 519 and 520 of ASP. It also appears that the carboxyl-terminal peptide generated by the cleavage is bound to the main body of the protein, which would form the nicked ASP eluted in fraction Y during hydroxyapatite column chromatography (Fig. 4B). However, SDS-PAGE would be expected to denature the protein and detach the carboxyl peptide from the main body (Fig. 5A, lane 3).
The molecular mass of the peptide that was detached from the main body of the protein was estimated to be 9,120.25 Da (Fig. 5B), which would account for the region 520–598. (The theoretical valve for this fragment is 9,104.19 Da.) A peptide of that size would have been too small to detect by SDS-PAGE under the conditions used, so it is not surprising that no corresponding band was observed on the gel (Fig. 5A). During the MALDI-TOF MS experiment, the components of the majority of nicked ASP disassociated; however, a small portion of the nicked ASP likely remained in the associated form to yield a peak at 64,056.13 (Fig. 5B). From these results, we conclude that ASP was cleaved between residues 519 and 520 and that the carboxyl-terminal peptide generated by the cleavage was bound to the main body of the protein to give the nicked form of ASP. The cleavage site is shown in Fig. 3A.
Is such a nicking process autocatalytic? To answer this question, we carried out another study using the mutant strain producing ASP(H115A), whose histidine residue at position 115 in the active site was substituted with Ala. The ASP(H115A) molecules were secreted into the milieu as seen in ASP-producing cells. Although ASP(H115A) did not show any proteolytic activity, the nicked form was still observed. As shown in Fig. 5A (lanes 4 and 5), we could isolate both intact and nicked forms from the culture supernatant of the ASP(H115A)-producing cells. Moreover, the amino acid residues around the cleavage site of the nicked ASP were composed of neutral amino acid residues, although ASP preferentially cleaves the peptide bond that follows two basic amino acid residues (36). Based on these results, we consider that the nicking must be proceeded via proteolytic action by any bacterial protease other than ASP.
Proteolytic Activity of Nicked ASP
We previously showed that ASP preferentially cleaves peptide bonds that follow two basic residues, one of which is Lys, and that the synthetic peptide substrate Boc-Glu-Lys-Lys-MCA, as well as PK, are efficiently cleaved by ASP (36). To determine the proteolytic activity of nicked ASP, we initially incubated the synthetic peptide with nicked ASP, and we compared the activity of the enzyme with that of native ASP. We found that the amount of coumarin generated from the synthetic peptide by the action of nicked ASP was higher than that generated by intact ASP (Fig. 6A), indicating nicked ASP has a greater ability to cleave the small synthetic peptide than the native enzyme.
To further examine the ability of the nicked ASP to hydrolyze a protein substrate, we incubated nicked ASP with PK and again compared its activity with that of native ASP (Fig. 6B). In tubes 6, 7, 8, and 9, we incubated 5 μg of PK with 0.17 μg, 0.34 μg, 0.50 μg, and 0.67 μg of nicked ASP, respectively, whereas PK was incubated with native enzyme in tubes 2, 3, 4, and 5. After incubation for 30 min at 37 °C, the samples were subjected to SDS-PAGE analysis. Both full-length ASP and nicked ASP hydrolyzed PK in a dose-dependent manner, but the SDS-PAGE pattern of the sample incubated with the full-length ASP was different from that of nicked ASP. Whereas a substantial amount of apparently undigested PK (86 kDa) remained, even after incubation with 0.67 μg of the nicked ASP, almost all the PK was cleaved after treatment with the same amount of native ASP (Fig. 6B, lanes 9 and 5). Moreover, a novel band appeared at 37 kDa after incubation with nicked ASP, which suggests that substrate recognition by nicked ASP differs from that by native ASP.
DISCUSSION
In this report, we determined the tertiary structure of ASP, which is considered to be a member of the subtilase family of enzymes characterized by the catalytic Asp, His, and Ser triad. In ASP, these residues are confirmed to be located at positions 78, 115, and 336, respectively (Fig. 3B). In addition, the substrate specificity of ASP resembles that of Kex2 and furin, which are representative of this group of proteases. Given that ASP cleaves the peptide bond that follows two basic amino acid residues, we predicted that the structure of the catalytic site of ASP would resemble those of Kex2 and furin, which was confirmed by our analysis of the crystal structure of ASP. Nonetheless, there are some important differences in the structures of these proteases.
The amino acid sequences of Kex2 and furin share 38% identity, and their tertiary structures are similar. The crystal structures of Kex2 and furin have a common fold, and the catalytic residues and the residues involved in P1 recognition and active site Ca2+ binding are completely conserved (16, 35, 37, 38). There is a small difference in the amino acid residues involved in the P2 and P4 recognition sites, however; because of this difference, the substrate specificity of Kex2 differs somewhat from that of furin. Kex2 recognizes Arg or Lys at P2, and, at P4, it has a dual specificity for basic and branched chain aliphatic amino acids (16). Consequently, the suitable substrate sequence for Kex2 is thought to be Arg(aliphatic residue)-X-Arg(Lys)-Arg. By contrast, furin is less selective at P2, and the interaction between furin and the substrate relies more heavily on interactions at P1 and P4. Finally, the furin cleavage site is known to be the amino-terminal peptide bond following Arg-X-X-Arg (37, 39, 40).
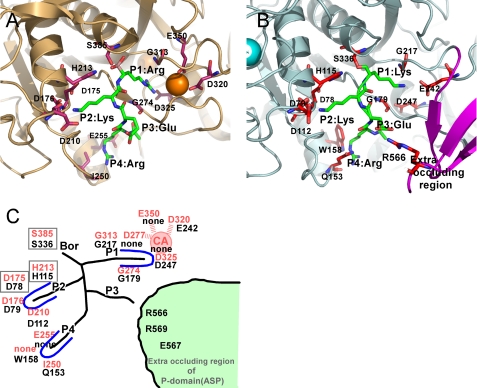
ASP preferentially cleaves the peptide bond that follows two Lys residues (36). A schematic representation of the binding of the peptide Arg-Glu-Lys-Lys to ASP is illustrated in Fig. 7B. For comparison, the binding of the peptide Arg-Glu-Lys-Arg to Kex2 is depicted in Fig. 7A, and the two binding modes are summarized in Fig. 7C, where residues depicted in red and black are from Kex2 and ASP, respectively. The surface of the catalytic site has a strong negative charge. In Kex2, the S1 site contains four negatively charged residues (Asp-277, Asp-320, Asp-325, and Glu-350), along with a buried Ca2+ cation, which are involved in the selection of the amino acid residue at P1 (e.g. Arg). However, neither the corresponding residues nor the Ca2+ are present in ASP. The S1 site seems to have large space because of a lack of calcium ion and Glu-242, located at the bottom of the S1 site, could function to select the substrate at the S1 site. As a result, Lys becomes the favored at P1 for ASP. The residues involved in determining substrate specificity at the Kex2 S2 site (Asp-176 and Asp-210) are conserved in ASP (Asp-79 and Asp-112) (Fig. 7C). It is therefore reasonable that ASP requires the presence of a Lys residue at P2. In the Kex2 S4 site, Glu-255 and Ile-250 are reportedly involved in the selection of basic and branched chain aliphatic amino acids at P4 of the substrate. No amino acid corresponding to Glu-255 is present in ASP. Trp-158 and Gln-153 of ASP may be involved in substrate selection, and it is noteworthy that Arg-566, Arg-569, and Glu-567, located in the novel occluding region in the P-domain, probably also affect recognition of protein substrates by ASP.
FIGURE 7.
Structural comparison of the subsites in ASP and Kex2. A, close-up view of catalytic site of S. cerevisiae Kex2 with a bound inhibitor (Protein Data Bank code 1R64), which is shown in light orange. One Ca2+ ion bound to Kex2 is shown as orange spheres. The important residues making up the subsites are shown as orange sticks. B, close-up view of the catalytic site of ASP with a bound substrate model (Lys-Lys-Glu). The important residues that make up the subsites are shown as red sticks. The Ca2+ ion bound to ASP is shown as greenish cyan spheres. The extra occluding region is shown in purple. C, schematic representation of the subsites of Kex2 and ASP. The site of Kex2 and ASP are labeled in red and black letters.
We eventually purified a stable form of nicked ASP and were able show a difference between the proteolytic activities of nicked ASP and the native enzyme. The cleavage site in nicked ASP is just behind the region formed by Arg-566, Arg-569, and Glu-567 in the extra occluding region. Against PK, nicked ASP showed a different proteolytic pattern, and against a small substrate, nicked ASP showed much greater activity than native ASP. These observations suggest that the cleavage is affected by the difference in the structure and/or flexibility of the extra occluding region, which ultimately results in a difference in the pattern of proteolysis. Although this occluding region, which is unique to ASP, appears to be important for determining substrate specificity with protein substrates like PK, with other proteins it may act as a steric obstacle. Although it is an open question as to why nicked ASP facilitates activity against low molecular mass substrates, we suggest that the greater flexibility of nicked ASP may facilitate activity. Lastly, we would like to stress that ASP possesses unique features that distinguish it from the human protease. This unique occluding region identified in ASP could facilitate development of novel antisepsis drugs that have no inhibitory effect on other furin-like human proteases.
Supplementary Material
Acknowledgments
We thank Drs. N. Igarashi, N. Matsugaki, and S. Wakatsuki for help in data collection at KEK AR NW12. We thank two anonymous reviewers for positive advice for this manuscript.
This work was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and by the Academic Frontier Promotion Program of MEXT of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The atomic coordinates and structure factors (code 3HJR) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) AF253471.
- ASP
- Aeromonas sobria serine protease
- MCA
- methylylcoumaryl-7-amide
- PEG
- polyethylene glycol
- HPLC
- high performance liquid chromatography
- MALDI-TOF MS
- matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- PK
- prekallikrein
- L
- loop
- CHES
- N-cyclohexyl-2-aminoethanesulfonic acid.
REFERENCES
- 1.Jones B. L., Wilcox M. H. (1995) J. Antimicrob. Chemother. 35, 453–461 [DOI] [PubMed] [Google Scholar]
- 2.Janda J. M., Abbott S. L. (1998) Clin. Infect. Dis. 27, 332–344 [DOI] [PubMed] [Google Scholar]
- 3.Janda J. M., Brenden R. (1987) J. Infect. Dis. 155, 589–591 [DOI] [PubMed] [Google Scholar]
- 4.Moses A. E., Leibergal M., Rahav G., Perouansky M., Or R., Shapiro M. (1995) Eur. J. Clin. Microbiol. Infect. Dis. 14, 237–240 [DOI] [PubMed] [Google Scholar]
- 5.Brink A. J., Giannakopoulos E., Viljoen H. G. (1998) S. Afr. Med. J. 88, 1011–1012 [PubMed] [Google Scholar]
- 6.Seshadri R., Joseph S. W., Chopra A. K., Sha J., Shaw J., Graf J., Haft D., Wu M., Ren Q., Rosovitz M. J., Madupu R., Tallon L., Kim M., Jin S., Vuong H., Stine O. C., Ali A., Horneman A. J., Heidelberg J. F. (2006) J. Bacteriol. 188, 8272–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamada S., Matsushita S., Dejsirilert S., Kudoh Y. (1997) Epidemiol. Infect. 119, 121–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuang Y. C., Chiou S. F., Su J. H., Wu M. L., Chang M. C. (1997) Microbiology 143, 803–812 [DOI] [PubMed] [Google Scholar]
- 9.Fujii Y., Nomura T., Kanzawa H., Kameyama M., Yamanaka H., Akita M., Setsu K., Okamoto K. (1998) Microbiol. Immunol. 42, 703–714 [DOI] [PubMed] [Google Scholar]
- 10.Galindo C. L., Fadl A. A., Sha J., Gutierrez C., Jr., Popov V. L., Boldogh I., Aggarwal B. B., Chopra A. K. (2004) J. Biol. Chem. 279, 37597–37612 [DOI] [PubMed] [Google Scholar]
- 11.Nomura T., Fujii Y., Okamoto K. (1999) Microbiol. Immunol. 43, 29–38 [DOI] [PubMed] [Google Scholar]
- 12.Okamoto K., Nomura T., Hamada M., Fukuda T., Noguchi Y., Fujii Y. (2000) Microbiol. Immunol. 44, 787–798 [DOI] [PubMed] [Google Scholar]
- 13.Imamura T., Kobayashi H., Khan R., Nitta H., Okamoto K. (2006) J. Immunol. 177, 8723–8729 [DOI] [PubMed] [Google Scholar]
- 14.Nitta H., Kobayashi H., Irie A., Baba H., Okamoto K., Imamura T. (2007) FEBS Lett. 581, 5935–5939 [DOI] [PubMed] [Google Scholar]
- 15.Nitta H., Imamura T., Wada Y., Irie A., Kobayashi H., Okamoto K., Baba H. (2008) J. Immunol. 181, 3602–3608 [DOI] [PubMed] [Google Scholar]
- 16.Siezen R. J., Leunissen J. A. (1997) Protein Sci. 6, 501–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leibowitz M. J., Wickner R. B. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 2061–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roebroek A. J., Schalken J. A., Bussemakers M. J., van Heerikhuizen H., Onnekink C., Debruyne F. M., Bloemers H. P., Van de Ven W. J. (1986) Mol. Biol. Rep. 11, 117–125 [DOI] [PubMed] [Google Scholar]
- 19.Hosaka M., Nagahama M., Kim W. S., Watanabe T., Hatsuzawa K., Ikemizu J., Murakami K., Nakayama K. (1991) J. Biol. Chem. 266, 12127–12130 [PubMed] [Google Scholar]
- 20.Fuller R. S., Brake A., Thorner J. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 1434–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van de Ven W. J., Creemers J. W., Roebroek A. J. (1991) Enzyme 45, 257–270 [DOI] [PubMed] [Google Scholar]
- 22.van den Ouweland A. M., van Duijnhoven H. L., Keizer G. D., Dorssers L. C., Van de Ven W. J. (1990) Nucleic Acids Res. 18, 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuller R. S., Sterne R. E., Thorner J. (1988) Annu. Rev. Physiol. 50, 345–362 [DOI] [PubMed] [Google Scholar]
- 24.Zhou A., Martin S., Lipkind G., LaMendola J., Steiner D. F. (1998) J. Biol. Chem. 273, 11107–11114 [DOI] [PubMed] [Google Scholar]
- 25.Gluschankof P., Fuller R. S. (1994) EMBO J. 13, 2280–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nomura T., Fujii Y., Yamanaka H., Kobayashi H., Okamoto K. (2002) J. Bacteriol. 184, 7058–7061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1937–1940 [DOI] [PubMed] [Google Scholar]
- 29.McRee D. E. (1999) J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 30.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 32.The CCP4 suite: Programs for Protein Crystallography (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 33.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35.Holyoak T., Wilson M. A., Fenn T. D., Kettner C. A., Petsko G. A., Fuller R. S., Ringe D. (2003) Biochemistry 42, 6709–6718 [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi H., Takahashi E., Oguma K., Fujii Y., Yamanaka H., Negishi T., Arimoto-Kobayashi S., Tsuji T., Okamoto K. (2006) FEMS Microbiol. Lett. 256, 165–170 [DOI] [PubMed] [Google Scholar]
- 37.Henrich S., Cameron A., Bourenkov G. P., Kiefersauer R., Huber R., Lindberg I., Bode W., Than M. E. (2003) Nat. Struct. Biol. 10, 520–526 [DOI] [PubMed] [Google Scholar]
- 38.Holyoak T., Kettner C. A., Petsko G. A., Fuller R. S., Ringe D. (2004) Biochemistry 43, 2412–2421 [DOI] [PubMed] [Google Scholar]
- 39.Krysan D. J., Rockwell N. C., Fuller R. S. (1999) J. Biol. Chem. 274, 23229–23234 [DOI] [PubMed] [Google Scholar]
- 40.Rockwell N. C., Krysan D. J., Komiyama T., Fuller R. S. (2002) Chem. Rev. 102, 4525–4548 [DOI] [PubMed] [Google Scholar]
- 41.Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta. Cryst. D57, 122–133 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.