Abstract
There are three types of cell death; apoptosis, necrosis, and autophagy. The possibility that activation of the macroautophagy (autophagy) pathway may increase beta cell death is addressed in this study. Increased autophagy was present in pancreatic islets from Pdx1+/− mice with reduced insulin secretion and beta cell mass. Pdx1 expression was reduced in mouse insulinoma 6 (MIN6) cells by delivering small hairpin RNAs using a lentiviral vector. The MIN6 cells died after 7 days of Pdx1 deficiency, and autophagy was evident prior to the onset of cell death. Inhibition of autophagy prolonged cell survival and delayed cell death. Nutrient deprivation increased autophagy in MIN6 cells and mouse and human islets after starvation. Autophagy inhibition partly prevented amino acid starvation-induced MIN6 cell death. The in vivo effects of reduced autophagy were studied by crossing Pdx1+/− mice to Becn1+/− mice. After 1 week on a high fat diet, 4-week-old Pdx1+/− Becn1+/− mice showed normal glucose tolerance, preserved beta cell function, and increased beta cell mass compared with Pdx1+/− mice. This protective effect of reduced autophagy had worn off after 7 weeks on a high fat diet. Increased autophagy contributes to pancreatic beta cell death in Pdx1 deficiency and following nutrient deprivation. The role of autophagy should be considered in studies of pancreatic beta cell death and diabetes and as a target for novel therapeutic intervention.
Normal pancreatic beta cell function is essential for normal glucose tolerance, and abnormal beta cell function leads to glucose intolerance and diabetes. A progressive reduction in beta cell mass has been shown to occur in the evolution of diabetes (1). Thus understanding the mechanisms responsible for the reduction in beta cell mass is important for understanding the pathogenesis of diabetes and in developing novel approaches to prevention and treatment.
There are three types of cell death; apoptosis, necrosis, and autophagy (2). Previous studies have focused on apoptosis as the mechanism underlying beta cell death (1, 3–5). The possibility that activation of the macroautophagy (hereafter referred to as autophagy) pathway may increase beta cell death has not been systematically studied. Autophagy is a regulated lysosomal pathway leading to the degradation and recycling of long-lived proteins and organelles. During autophagy, cytoplasmic constituents are sequestered into autophagosomes with double membranes and fused to lysosomes (autolysosomes), where degradation occurs. Under certain circumstances such as in response to nutrient deprivation, autophagy may function as a pro-survival pathway by mediating cellular turnover of proteins and organelles (6–8). On the other hand, an increase in autophagy can cause autophagic cell death distinct from apoptosis (9, 10). It has been suggested that autophagy plays a key role in the turnover of insulin secretory granules and of mitochondria within the beta cell, thereby regulating insulin secretion (11, 12). Complete genetic ablation of Atg7 in beta cells resulted in degradation of islets and impaired glucose tolerance, suggesting that “basal autophagy” is important for maintenance of normal islet architecture and function (13, 14).
The present study was designed to determine whether activation of autophagy can contribute to pancreatic beta cell death that occurs with reduced expression of Pdx1 (pancreas duodenal homeobox 1). We chose to study Pdx1 deficiency because this homeodomain-containing transcription factor is essential for normal pancreatic beta cell function and survival. Complete deficiency of Pdx1 is associated with pancreatic agenesis, and partial deficiency leads to reduced insulin secretion and beta cell mass (15–19). Complementary studies were performed to determine whether nutrient deprivation leads to a similar increase in autophagy in the beta cell as it does in other cells and tissues (8).
EXPERIMENTAL PROCEDURES
Cell Culture
Mouse insulinoma MIN63 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 15% FBS, antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin), 1 mm sodium pyruvate, 10 mm HEPES, and 50 μm β-mercaptoethanol. For amino acid deprivation, the aforementioned media were changed to 5% FBS without amino acids. For experiments involving both amino acid and serum starvation, MIN6 cells were incubated in HBSS medium supplemented with 10 mm HEPES. Mouse fibroblastoid NIH 3T3 and HEK 293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with antibiotics plus 10% horse serum or 10% FBS, respectively. All of the cells were maintained at 37 °C in an atmosphere of 5% CO2/balance air and 100% humidity.
Western Blot
Attached MIN6 and NIH 3T3 cells and islets were lysed in cell lysis buffer containing 1% Triton X-100, 1 mm EDTA, 1 mm EGTA, 10 mm dithiothreitol, 1 mm Na3VO4, and complete protease inhibitor mixture. Equal amounts of protein were fractionated by SDS-PAGE, and blots were probed with antibodies against Pdx1 (sc-14664; Santa Cruz Biotechnology Inc., Santa Cruz, CA), Becn1 (3738; Cell Signaling Technologies, Beverly, MA), LC3 (NB100-2331; Novus Biologicals Inc., Littleton, CO), cleaved caspase-3 (9661; Cell Signaling Technologies), actin (A-2066; Sigma), and Atg5 (NB110-53818; Novus Biologicals).
Propidium Iodide/DAPI Cell Death Assay
For the last hour of incubation, 10 μg/ml propidium iodide (PI) and 20 μg/ml DAPI were added directly to the media. After this incubation, the MIN6 cells were washed three times with PBS and fixed with 3.7% formaldehyde for 15 min at 4 °C. Each condition reported represents >600 cells counted by randomized field selection. The percentage of cell death was calculated as the number of PI-stained nuclei over the total number of nuclei stained by DAPI.
Isolation and Culture of Primary Pancreatic Islets
Mouse islets were isolated by using collagenase and filtration as previously described (3), and human islets were provided by the Human Islet Isolation Core at Washington University in St. Louis. The islets were cultured in RPMI 1640 medium with antibiotics, 10% FBS, pH 7.4, with NaOH at 37 °C and 5% CO2.
Quantification of mRNA Levels
RNA isolation, first strand cDNA synthesis, and TaqMan gene expression assays were performed as previously described (20). The RNA level was normalized to the amount of Hmbs mRNA in the same sample. Applied Biosystems (Foster City, CA) TaqMan assay numbers were: Hmbs, Mm00660262_g1; and Pdx1, Mm00435565_m1.
Lentivirus-mediated shRNA Expression
The pLKO.1-puro lentivirus vector was generously provided by Dr. Sheila Stewart of Washington University Medical School (St. Louis, MO). Potential shRNA targets in the murine Pdx1 mRNA (GenBankTM accession number NM_008814) were identified using the Dharmacon on-line siDESIGN tool. Following a BLAST search of the NCBI data base, one potential target was identified that lacks significant homology to non-Pdx1 gene products. Pdx1 target sequence 5′-CAGTGAGGAGCAGTACTAC-3′ or control sequence 5′-ACTACCGTTGTTATAGGTG-3′ was subcloned into the AgeI/EcoRI restriction site of pLKO-1-puro. shRNA targeting mouse Becn1 and Atg5 were obtained from Sigma. Recombinant lentiviral particles were prepared by transfecting HEK 293T cells with the appropriate pLKO.1-puro plasmid plus pHR′CVM8.2 delta R and pCMV-VSV-G plasmids. Lentivirus was added to the medium on days 1 and 2 followed by 2 μg/ml puromycin selections on days 4, 5, and 7. The media including autophagy inhibitor were changed on days 3, 4, 5, and 7 (supplemental Fig. S1).
Detection of Autophagy by Immunostaining
The presence of autophagosomes in MIN6 cells or islets was evaluated using immunostaining. MIN6 cells were fixed with 3.7% formaldehyde at room temperature for 60 min followed by immunostaining with anti-LC3 antibody (NB100-2331; Novus Biologicals). For immunostaining of islets, the mice were anesthetized and perfused through the left ventricle with 3.7% formaldehyde. The pancreas was isolated and then fixed at room temperature overnight. To quantitate the extent of autophagy, the number of LC3 punctae in each cell was counted in five independent visual fields from at least two independent experiments. Bafilomycin A1 (BafA1) is known as a strong inhibitor of the vacuolar type H+-ATPase and thereby inhibits the final step of lysosomal digestion in autophagy (21). To determine whether reduced Pdx1 expression in MIN6 cells increases autophagic flux, 1 nm BafA1 (Sigma) was added to the medium 4 h prior to the end of the treatment.
Electron Microscopy
MIN6 cells were infected with the shPdx1 or control vector, and islets were isolated from 3-week-old male Pdx1+/− or Pdx1+/+ mice followed by fixation with modified Karnovsky's fixative containing 3% glutaraldehyde and 1% paraformaldehyde in sodium cacodylate buffer, pH 7.4, overnight. The cells or islets were then rinsed in sodium cacodylate buffer followed by post-fixation in cacodylate-buffered 1% OsO4, for 1 h, dehydrated in graded ethanol with a final dehydration in propylene oxide, and embedded in EMbed-812 (Electron Microscopy Sciences, Hatfield, PA). Tissue blocks were sectioned at ninety nanometers thick, post-stained with uranyl acetate and Venable's lead citrate, and viewed with a JEOL model 1200EX electron microscope (JEOL, Tokyo, Japan). Digital images were acquired using the AMT Advantage HR (Advanced Microscopy Techniques, Danvers, MA) high definition CCD, 1.3 megapixel transmission electron microscopy camera.
In Vivo Characterization of Mice
The Pdx1+/− mice have been previously described (16) and were kindly provided by Dr. Helena Edlund (University of Umea, Umea, Sweden). Becn1+/− mice (22) were kindly provided by Dr. Beth Levine of University of Texas Southwestern Medical Center (Dallas, TX). For high fat diet, male mice were fed food containing 42% fat (Harlan Laboratories, Inc., Indianapolis, IN) from 3 weeks of age and provided with water ad libitum. Wild type littermates were used as controls. Intraperitoneal glucose tolerance tests were performed after a 4 h fast (2 g of dextrose/kg of body weight). Autophagic flux was evaluated by intraperitoneal administration of BafA1 (Sigma) at 0.3 mg/kg for 24 h. For morphometric analysis, the sections were immunostained with antibody against cleaved caspase-3 (9661; Cell Signaling Technologies) or Ki-67 (Zymed Laboratories Inc./Invitrogen). Pancreatic area and beta cell area were each estimated using the intensity thresholding function of the integrated morphometry package in MetaMorph as previously described (3). All of the experiments in this study using animal protocols were approved by the Washington University Animal Studies Committee.
Statistical Analysis
Statistical analyses were performed by Student's unpaired t test. The differences were considered significant when p < 0.05. The results are presented as the means ± S.E.
RESULTS
Reduced Pdx1 Expression Leads to Autophagy in MIN6 Cells
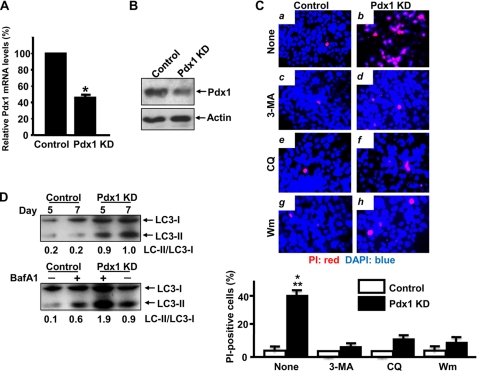
To determine whether reduced Pdx1 expression affects autophagy, we developed a lentivirus-based system to deliver an shRNA construct to MIN6 cells a mouse insulinoma cell line to knock down-expression of Pdx1. Infection of MIN6 cells with lentivirus encoding this construct significantly reduced Pdx1 mRNA levels to 52 ± 3% of control on day 5 (Fig. 1A). A decrease in Pdx1 protein levels was also observed on day 6 (Fig. 1B), and an increase in cell death was observed on day 7 as assessed by PI staining, which stains dying and dead cells (Fig. 1C, panels a and b). Infection of MIN6 cells with lentivirus containing a control construct did not cause cell death, and the Pdx1 KD construct did not cause death in mouse fibroblastoid NIH 3T3 cells that do not express Pdx1 (data not shown). Thus the increase in cell death after lentiviral infection is due to a decrease in Pdx1 expression and not nonspecific cellular effect(s) of viral infection.
FIGURE 1.
Pdx1 deficiency induces MIN6 cell death accompanied by increased autophagy. A, MIN6 cells were infected with a lentiviral vector that drives expression of an shRNA targeting Pdx1 transcript (Pdx1 KD) or control lentiviral vector. Five days after infection, Pdx1 mRNA levels assayed by real time reverse transcription-PCR were reduced to 52 ± 3% of control values (n = 3) (*, p < 0.0001). B, six days after infection, Pdx1 protein levels assayed by Western blot were reduced in the Pdx1 KD MIN6 cells compared with control (n = 3). C, following infection with the control or Pdx1 KD vector, MIN6 cells received no treatment (panels a and b), 1 mm 3-MA (panels c and d), 0.3 μm chloroquine (CQ) (panels e and f), or 30 nm wortmannin (Wm) (panels g and h). PI (red)/DAPI (blue) staining revealed that all three treatments prevented the Pdx1 KD-induced cell death on day 7. 40 ± 3% of Pdx1 KD stained with PI, and this was reduced (n = 3). Bottom panel, control versus Pdx1 KD (*, p < 0.001), Pdx1 KD versus Pdx1 KD with 3-MA (**, p < 0.001), Pdx1 KD versus Pdx1 KD with chloroquine (**, p < 0.001), and Pdx1 KD versus Pdx1 KD with wortmannin (**, p < 0.001). D, in the upper panel, LC3-II levels in MIN6 cells infected with a control or Pdx1 KD lentiviral construct were measured on days 5 and 7. LC3-II levels were increased in Pdx1 KD MIN6 cells on day 5 before cell death became evident and on day 7 when death of these cells was evident (n = 3). The lower panel shows LC3-II levels in control or Pdx1 KD MIN6 cells cultured in the absence or presence of 1 nm BafA1 on day 7. LC3-II levels were increased in the presence of BafA1 in Pdx1 KD MIN6 cells, indicating increased autophagic flux in Pdx1 KD MIN6 (n = 3). The intensity of LC3-I and LC3-II bans was quantified, and the calculated LC3-II/LC3-I ratio is indicated.
To determine whether autophagy is present in MIN6 cells with reduced Pdx1 expression, Western blots of microtubule-associated protein 1 LC3 were performed on Pdx1 KD MIN6 cells at two time points: 48 h before cell death became microscopically evident (on day 5) and on day 7 when cell death was evident. LC3-II levels were increased at both time points (Fig. 1D, upper panel). In addition, we treated Pdx1 KD MIN6 cells with BafA1, which inhibits the final step of lysosomal digestion in autophagy to determine whether reduced Pdx1 expression in MIN6 cells increases autophagic flux. Reduced Pdx1 expression in the presence of 1 nm BafA1 resulted in accumulation of LC3-II in comparison with the absence of BafA1 in Pdx1 KD MIN6 cells on day 7 (Fig. 1D, lower panel), indicating that autophagic flux is increased in Pdx1-reduced MIN6 cells. Immunostaining revealed a diffuse pattern of LC3 fluorescence in control cells, whereas LC3 developed the punctate appearance characteristic of autophagy (23) in the Pdx1 KD MIN6 cells on day 5. This effect was blocked by the autophagy inhibitor 1 mm 3-methyladenine (3-MA) (Fig. 2A). Quantitative morphometric analysis revealed a significant increase in LC3 punctae in the Pdx1 KD MIN6 cells compared with control MIN6 cells. Exposure of the cells to 1 mm 3-MA blocked this effect and reduced the number of LC3 punctae to levels seen in cells exposed to lentivirus containing a control vector (Fig. 2B). Thus 1 mm 3-MA prevents activation of autophagy but does not inhibit basal autophagy. Transmission electron microscopy provided additional morphological evidence for autophagy in the Pdx1 KD MIN6 cells. Autophagosomes and autolysosomes were evident 5 days after Pdx1 KD but not in MIN6 cells infected with lentivirus containing a control vector (Fig. 2C). All of these findings indicate that autophagy is increased in Pdx1-reduced MIN6 cells.
FIGURE 2.
Inhibition of autophagy in Pdx1-reduced MIN6 cells. A and B, 5 days after infection with the shPdx1 or control vector in the absence or presence of 1 mm 3-MA, MIN6 cells were fixed and stained for LC3. A representative area (box) from Pdx1 KD MIN6 cells was magnified (High mag). The number of LC3 punctae per cell increased from 8.61 ± 1.54 in control cells to 27.0 ± 3.14 in Pdx1 KD cells (*, p < 0.001), and this increase was prevented by treatment with 1 mm 3-MA (7.86 ± 3.87) (*, p < 0.001). C, representative electron micrographs of Pdx1 KD or control MIN6 cells on day 5. Pdx1 KD cells showed the presence of autophagosomes and autolysosomes (arrows) with cytoplasmic contents ranging from granular cytoplasm with occasional organelles to well formed autophagosomes containing degenerating organelles. Autophagosomes and autolysosomes were rarely seen in control MIN6 cells. The scale bars represent 2 μm. D, following infection with a control or Pdx1 KD vector, MIN6 cells were exposed to 1 mm 3-MA, 10 μm DEVD-CHO, or no treatment. On day 9, 63 ± 11% of Pdx1 KD cells stained positive for PI, whereas only 19 ± 12% of DEVD-CHO-treated Pdx1 KD cells and 53 ± 16% of 3-MA-treated Pdx1 KD cells stained for PI (n = 3). Pdx1 KD versus Pdx1 KD with DEVD-CHO on day 9 (*, p < 0.001). E, cleaved caspase-3 levels in 1 mm 3-MA or 10 μm DEVD-CHO treated Pdx1 KD MIN6 cells. Increased cleaved caspase-3 levels were observed in 3-MA-treated Pdx1 KD cells on day 9 compared with day 5 and day 7, whereas cleaved caspase-3 was not detected in DEVD-CHO-treated Pdx1 KD cells (n = 4).
Autophagy Inhibition Enhances Survival of Pdx1-reduced MIN6 Cells
MIN6 cells with reduced Pdx1 expression were exposed to agents that inhibit formation of autolysosomes by different mechanisms (24). One mm 3-MA, 0.3 μm chloroquine, and 30 nm wortmannin almost completely prevented Pdx1 KD-induced MIN6 cell death on day 7 (Fig. 1C). Optimal concentrations were 5–10 times lower than the concentrations used in previous studies (24) (data not shown). To determine the relative roles of apoptosis and autophagy on the observed cell death, MIN6 cells with reduced Pdx1 expression were treated with 3-MA (1 mm) or caspase-3 inhibitor DEVD-CHO (10 μm). Pdx1 KD MIN6 cells treated with DEVD-CHO showed nearly complete inhibition of Pdx1 KD-induced cell death compared with 1 mm 3-MA-treated Pdx1 KD MIN6 cells on day 9 (Fig. 2D). A progressive increase in cleaved caspase-3 levels was observed in 3-MA-treated Pdx1 KD MIN6 cells on days 5, 7, and 9, respectively (Fig. 2E). This increase in cleaved caspase-3 over time was completely prevented in DEVD-CHO-treated Pdx1 KD MIN6 cells. These results indicate that 3-MA delays Pdx1 KD-induced MIN6 cell death, but 3-MA-treated Pdx1 KD MIN6 cells finally die by caspase-3-dependent cell death. Thus in the temporal evolution of the cell death induced by reduced Pdx1 expression, an increase in autophagy appears to occur early, and this is followed by an increase in apoptosis.
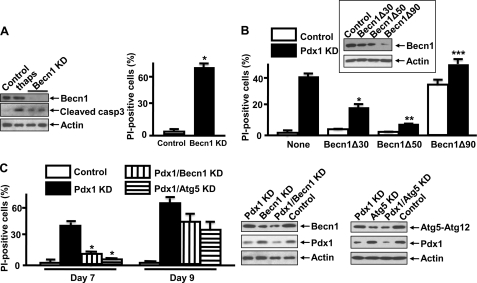
Effect of Becn1 and Atg5 Knockdown
Because the autophagy inhibitors used in this study may have nonspecific effects (24), we also studied the effects of knocking down expression of Becn1 or Atg5 in MIN6 cells. Becn1 and Atg5 are key mediators of autophagosome formation (25, 26). Becn1 is the mammalian orthologue of the yeast Atg6/Vps30 gene and a regulator of the class III phosphoinositide 3-kinase complex involved in autophagosome formation (25). Nearly complete Becn1 KD in MIN6 cells lead to an increase in cleaved caspase-3, and this was associated with a substantial increase in the death of the cells, with 68 ± 3% of cells taking up PI indicative of cell death (Fig. 3A). Different shRNA constructs were next utilized to reduce Becn1 expression in Pdx1 KD MIN6 cells by 30% (Becn1Δ30), 50% (Becn1Δ50), and 90% (Becn1Δ90) (Fig. 3B, inset). Reducing Becn1 expression in Pdx1-deficient cells reduced MIN6 cell death as judged by the percentage of cells that take up PI from 41 ± 2% in control cells to 18 ± 2 and 7 ± 1% in cells in which Becn1 expression had been reduced by 30 and 50%, respectively (*, p < 0.01; **, p < 0.001). On the other hand when Becn1 expression was reduced by 90%, the percentage of cells taking up PI increased to 55 ± 4% (***, p < 0.001) (Fig. 3B). These results suggest that a minimum basal level of autophagy is required for MIN6 cell survival and are consistent with the results reported in the beta cell-specific Atg7 knock-out mouse showing that a complete absence of Atg7 is associated with the activation of caspase-3 in islets and an increase in beta cell death (13, 14). Based on these results, the Becn1Δ50 construct was used to inhibit activated autophagy in subsequent experiments. Inhibition of autophagy by shBecn1 or shAtg5 prevented Pdx1 KD-induced MIN6 cell death on day 7, but ultimately cells started dying on day 9 (Fig. 3C), consistent with the result obtained with 1 mm 3-MA-treated Pdx1 KD MIN6 cells (Fig. 2D).
FIGURE 3.
Effects of Becn1 and Atg5 KD in Pdx1-reduced MIN6 cells. A, Becn1 and cleaved caspase-3 levels were measured by Western blot 11 days after infection with the control or shBecn1 vector. Nearly complete suppression of Becn1 to inhibit basal autophagy induced cleaved caspase-3. MIN6 cells exposed to 1 μm thapsigargin for 24 h were used as a positive control for cleaved caspase-3 (n = 2). PI-positive cells were increased from 3 ± 1 to 67 ± 4% in Becn1 KD MIN6 cells (*, p < 0.0001). B, Becn1 protein levels 7 days after coinfection with shPdx1 and shBecn1Δ30, shBecn1Δ50, or shBecn1Δ90 are shown in the inset (n = 3). Becn1Δ30 and Becn1Δ50 reduced the proportion of Pdx1 KD cells staining positive for PI from 41 ± 2 to 18 ± 2% (*, p < 0.01) and 7 ± 1% (**, p < 0.001), respectively. Becn1 KD by 90% (Becn1Δ90) increased the proportion of Pdx1 KD MIN6 cells staining with PI to 55 ± 4% (***, p < 0.001) (n = 3). C, following infection with a Pdx1 KD vector, MIN6 cells were exposed to lentivirus containing control, shBecn1, or shAtg5 vectors. On day 7, 39 ± 3% of Pdx1 KD cells stained positive for PI, whereas only 12 ± 2 and 7 ± 1% of Pdx1/Becn1 KD cells and Pdx1/Atg5 KD cells, respectively, stained with PI. On day 9, the proportion of PI-positive Pdx1/Becn1 KD cells and Pdx1/Atg5 KD cells increased to 44 ± 4 and 33 ± 6%, respectively (n = 3). Pdx1 KD versus Pdx1/Becn1 KD (*, p < 0.001) and Pdx1/Atg5 KD (*, p < 0.001) on day 7. The right panels show Western blots of Pdx1, Becn1, and Atg5 from the aforementioned experiments on day 7 (n = 3).
Activation of Autophagy in Pdx1+/− Beta Cells
To determine whether reduced Pdx1 expression is associated with autophagy in vivo, we studied Pdx1+/− mouse islets. Pdx1+/− beta cells demonstrated the punctate staining for LC3 that is characteristic of autophagy (Fig. 4A). Quantitative morphometric analysis of LC3 images revealed a significant increase in the number of LC3-positive punctae in Pdx1+/− beta cells compared with Pdx1+/+ controls (Fig. 4B). The levels of LC3-II and the LC-II/LC-I ratio were increased in Pdx1+/− islets compared with Pdx1+/+ islets by Western blot analysis (Fig. 4C), indicating that autophagy is increased in islets from Pdx1+/− mice. To gain further insight into the pathological changes present in Pdx1+/− islets, the ultrastructure of beta cells was examined by transmission electron microscopy. Beta cells could easily be distinguished from non-beta cells because of the presence of a small electron-dense core surrounded by a large halo (27) as shown in normal beta cells from Pdx1+/+ islets (Fig. 4D, panel a). Autophagosomes and autolysosomes accumulated in Pdx1+/− beta cells (Fig. 4D, panels b–f), a finding rarely seen in Pdx1+/+ beta cells. We also observed occasional apoptotic bodies in Pdx1+/− beta cells (Fig. 4D, panels e and f) but not in Pdx1+/+ beta cells. Next we treated mice with BafA1 to determine whether reduced Pdx1 expression in beta cells increases autophagic flux in vivo. BafA1 (0.3 mg/kg) resulted in an increase in the number of LC3-positive punctae in Pdx1+/− beta cells compared with mice treated with vehicle controls after 24 h (Fig. 4E), suggesting that autophagic flux is increased in Pdx1+/− beta cells. Together, these results suggest that autophagy is activated in Pdx1+/− beta cells in vivo.
FIGURE 4.
Autophagy is increased in Pdx1+/− beta cells. A, immunostaining for LC3 (green), insulin (red), and DAPI (blue) in islets from 11-week-old male Pdx1+/− and Pdx1+/+ mice. A representative area from an islet was magnified (High mag) in gray from each inset. Pdx1+/− beta cells revealed more frequent LC3 punctae formation compared with Pdx1+/+ beta cells. B, the number of LC3 punctae/beta cell was increased from 1.49 ± 0.11 in Pdx1+/+ beta cells to 9.38 ± 1.64 in Pdx1+/− beta cells (*, p < 0.0001). C, Western blots of LC3 in islets from 3-week-old male Pdx1+/− mice showed an increased LC3-II and LC3-I/LC3-II ratio compared with Pdx1+/+ islets (n = 3). D, pancreatic islets isolated from 3-week-old male Pdx1+/+ (panel a) and Pdx1+/− (panels b–f) mice analyzed by transmission electron microscopy. Pdx1+/+ islets showed numerous granules surrounded by halos in beta cells (panel a) in the absence of autophagosomes, autolysosomes, and apoptotic bodies. Beta cells from Pdx1+/− mouse islets contained autophagosomes and autolysosomes (arrows), which are shown at several magnifications (panel b–f). A representative area from the beta cell was magnified from each inset. Note that rough endoplasmic reticulum is visible in autophagosomes in panel d from a Pdx1+/− beta cell. Panels e and f show several apoptotic bodies (asterisks) with autophagosomes and autolysosomes (arrows) from Pdx1+/− mouse islets. The scale bars represent 2 μm. E, the effect of 0.3 mg/kg BafA1 on autophagic flux was studied by LC3 staining. BafA1 treatment increased in LC3-positive punctae in Pdx1+/− beta cells from 8.85 ± 3.56 to 17.31 ± 2.26, indicating increased autophagic flux in Pdx1+/− compared with Pdx1+/+ beta cells (*, p < 0.01).
Inhibition of Autophagy in Pdx1+/− Mice
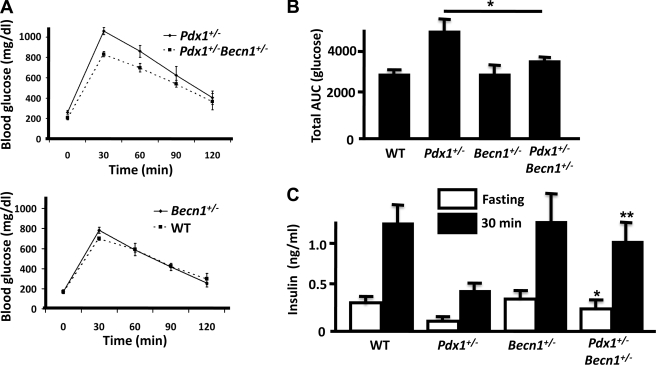
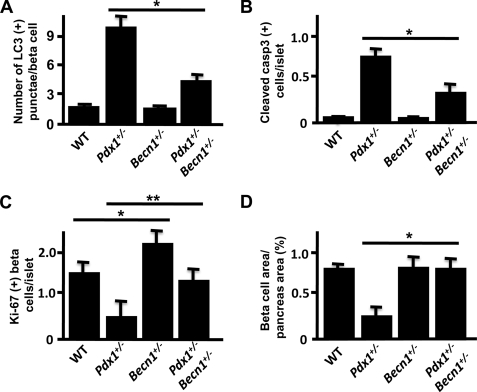
To study the functional significance of increased autophagy in Pdx1+/− beta cells, Pdx1+/− mice were crossed to Becn1+/− mice to inhibit autophagy in Pdx1+/− mice. Because a high fat diet increases autophagy in beta cells (13), we put these mice on a high fat diet from 3 weeks of age. After 1 week on the high fat diet, Pdx1+/− Becn1+/− mice showed significantly lower blood glucose concentrations after the intraperitoneal administration of glucose compared with Pdx1+/− mice. Becn1+/− and wild type mice had similar glucose concentrations (Fig. 5, A and B). This was accompanied by significant increases in both fasting and 30-min insulin levels after glucose administration in Pdx1+/− Becn1+/− mice compared with Pdx1+/− mice (Fig. 5C). Next we evaluated whether autophagy and apoptosis were altered in Pdx1+/− Becn1+/− mice. LC3-positive punctae were decreased in Pdx1+/− Becn1+/− beta cells compared with beta cells from Pdx1+/− mice, but LC3 punctae formation was not completely inhibited (Fig. 6A), suggesting persistence of basal autophagy in Pdx1+/− Becn1+/− beta cells. We have previously reported increased caspase-3-dependent apoptosis in Pdx1+/− islets accompanied by reduced beta cell mass (3). Cleaved caspase-3-positive cells in islets were significantly decreased in Pdx1+/− Becn1+/− mice compared with Pdx1+/− mice (Fig. 6B), indicating that inhibition of autophagy in Pdx1+/− islets prevents caspase-3 activation. Beta cell proliferation was studied by staining with Ki-67. Ki-67-positive beta cells were increased in Pdx1+/− Becn1+/− mice and Becn1+/− mice compared with Pdx1+/− mice and wild type mice, respectively (Fig. 6C). As a reflection of beta cell mass, the area occupied by beta cells within multiple pancreatic sections was quantified and expressed as a percentage of the pancreas area. As shown in Fig. 6D, the beta cell area/pancreas area was significantly greater in Pdx1+/− Becn1+/− mice than in Pdx1+/− mice and was similar to the levels found in wild type and Becn1+/− mice. Thus after 1 week on a high fat diet, when compared with Pdx1+/− mice, Pdx1+/− Becn1+/− mice demonstrate inhibition of autophagy, reduced activation of caspase-3, and increased proliferation of beta cells This results in increased insulin secretion and preserved glucose tolerance and beta cell mass.
FIGURE 5.
Comparison of Pdx1+/− and Pdx1+/−Becn1+/− mice after 1 week on a high fat diet. A, blood glucose concentrations during intraperitoneal glucose tolerance tests after 1 week on a high fat diet in 4-week-old Pdx1+/− and Pdx1+/− Becn1+/− mice (n = 4) (top panel) and Becn1+/− (n = 4) and WT mice (n = 3) (lower panel). B, total areas under the glucose curves (AUC) from A. The AUC was reduced in Pdx1+/− Becn1+/− mice compared with Pdx1+/− mice (*, p < 0.05) and was similar to the AUC in the WT and Becn1+/− mice. C, fasting and 30-min serum insulin levels measured during intraperitoneal glucose tolerance tests in Pdx1+/− Becn1+/− mice (n = 4) were significantly greater than in Pdx1+/− mice (n = 4) (*, p < 0.05; **, p < 0.01) and similar to the levels obtained in WT (n = 4) and Becn1+/− mice (n = 4).
FIGURE 6.
Beta cell autophagy, apoptosis, proliferation and mass in pancreatic islets from mice after 1 week on a high fat diet. These analyses were conducted in the mouse groups depicted in Fig. 5. A, the number of LC3 punctae was decreased from 10.19 ± 3.36 punctae in Pdx1+/− beta cells to 4.33 ± 2.34 punctae in Pdx1+/− Becn1+/− beta cells (*, p < 0.001). The number of LC3 punctae in beta cells from WT and Becn1+/− mice were 2.54 ± 0.21 and 2.30 ± 0.22 punctae (p = ns), respectively. B, the number of cells staining positive for cleaved caspase-3 was significantly reduced in islets from Pdx1+/− Becn1+/− mice compared with Pdx1+/− mice (*, p < 0.01). The percentage of cells staining positive for cleaved caspase-3 was similar in islets from Becn1+/− and WT mice (n = 3). C, the number of Ki-67-positive beta cells per islet was increased in Pdx1+/− Becn1+/− and Becn1+/− mice compared with Pdx1+/− and WT mice, respectively (*, Pdx1+/− Becn1+/− versus Pdx1+/−, p < 0.05; **, Becn1+/− versus WT, p < 0.01) (n = 3). D, beta cell area at 1 week after high fat diet was estimated using insulin immunoreactivity as described under “Experimental Procedures” and normalized to the total pancreas area. Five or six sections were analyzed from each animal (n = 4). Pdx1+/− Becn1+/− beta cell area/pancreas area (0.76 ± 0.09%) was preserved compared with that of Pdx1+/− (0.29 ± 0.05%) (*, p < 0.01) and comparable with that seen in WT (0.76 ± 0.03%) and Becn1+/− mice (0.78 ± 0.09%).
To determine the durability of these effects, these experiments were repeated in animals maintained on a high fat diet for 7 weeks. At this stage Pdx1+/− Becn1+/− and Pdx1+/− mice demonstrated similar levels of hyperglycemia (supplemental Fig. S2, A and B), similar impairment of insulin secretion (supplemental Fig. S2C), similar increases in cleaved caspase-3 levels (supplemental Fig. S3A), and similar reductions in beta cell area/pancreas area (supplemental Fig. S3B). On the other hand at 7 weeks on a high fat diet, proliferation was still increased in Pdx1+/− Becn1+/− mice compared with Pdx1+/− mice (supplemental Fig. S3C), and the number of LC3-positive punctae was reduced (supplemental Fig. S3D), suggesting that autophagy is still inhibited in Pdx1+/− Becn1+/− beta cells at this stage. These results indicate that if the activation of autophagy that occurs in Pdx1+/− beta cells is prevented, the onset of cell death is delayed but ultimately occurs because of activation of caspase-3-dependent apoptosis.
Regulation of Autophagy in Beta Cells by Nutrient Deprivation
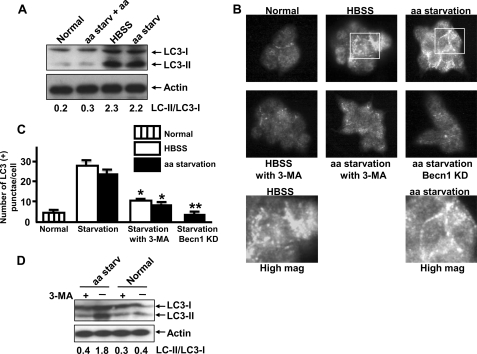
Because an increase in autophagy is a major adaptive response to nutrient deprivation, we determined whether nutrient deprivation increases autophagy in beta cells as in other cell lines and organ systems (8, 28). MIN6 cells incubated for 48 h in HBSS medium (deficient both in amino acids and serum) or amino acid-starved medium demonstrated an increase in LC3-II levels compared with normal medium, and the addition of amino acids in amino acid-deprived medium inhibited starvation-induced LC3-II (Fig. 7A). In response to nutrient deprivation, the punctate pattern of LC3 staining became evident (Fig. 7B). Autophagosome formation induced by nutrient deprivation was significantly inhibited by 1 mm 3-MA or Becn1 KD (Fig. 7C). An increase in LC3-II was also observed in human islets incubated in medium deficient in amino acid for 48 h compared with normal medium, and this increase was inhibited by 1 mm 3-MA (Fig. 7D).
FIGURE 7.
Nutrient deprivation induces autophagy in MIN6 cells. A, MIN6 cells were cultured for 48 h in normal medium, amino acid-starved (aa starv) medium plus amino acids, HBSS medium that does not contain either amino acids or serum, or amino acid-starved medium. LC3-II levels were increased in HBSS medium and amino acid-starved medium, and the addition of amino acids in amino acid-starved medium prevented the increase in LC3-II levels as assessed by Western blot (n = 3). B, MIN6 cells were cultured in normal medium, HBSS medium, or amino acid-starved medium with or without 1 mm 3-MA for 24 h. Six days after infection with a lentivirus containing shBecn1, MIN6 cells were cultured without amino acid for 24 h. C, LC3 punctae were increased from 4.31 ± 1.37 punctae/cell in normal medium to 28.0 ± 3.51 punctae/cell in HBSS medium and 26.4 ± 3.77 punctae/cell in normal medium without amino acids. The number of punctae/cell was significantly decreased to 9.87 ± 0.27 in HBSS medium with 3-MA, 8.74 ± 2.17 in amino acid-starved medium with 3-MA, and 3.21 ± 1.47 in amino acid-starved Becn1 KD MIN6 cells. D, human islets were incubated for 48 h with or without amino acids in the presence or absence of 1 mm 3-MA. Amino acid starvation increased LC3-II levels, and this was prevented by3-MA (n = 3).
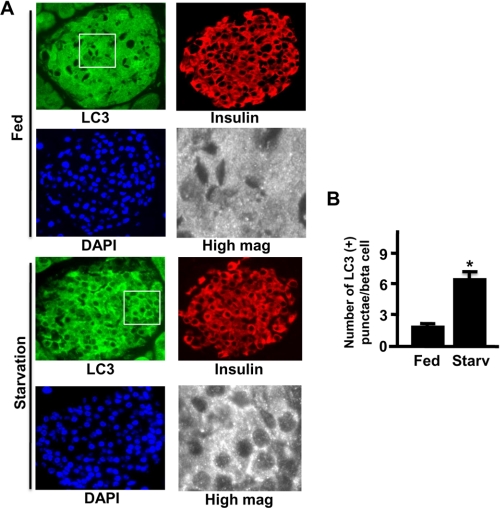
Recently, Ebato et al. (13) reported that starvation induces autophagy in INS1 beta cells, and overnight fasting does not induce autophagy in islets in vivo. Thus we starved mice for 24 h and stained islets with LC3. The LC3 signal was distributed diffusely in the cytoplasm of islet beta cells from fed animals, whereas 24-h starvation induced punctate staining of LC3 (Fig. 8A). Quantitative morphometric analysis of LC3 images revealed a significant increase in the number of LC3-positive punctae in beta cells from animals starved for 24 h compared with animals in the fed state (Fig. 8B). Taken together, these results indicated that starvation induces autophagy in the beta cell both in vitro and in vivo.
FIGURE 8.
Starvation induces autophagy in beta cells in vivo. A, immunostaining for LC3 (green), insulin (red), and DAPI (blue) in islets from fed or 24-h fasted mice. A representative area from an islet was magnified (High mag) in gray from each inset. Starvation increased the number of LC3-positive punctae/beta cell in fasted compared with fed mice. B, LC3 punctae were increased from 2.36 ± 0.21 punctae in fed mice to 6.18 ± 1.24 punctae in starved mice (*, p < 0.01) (n = 3).
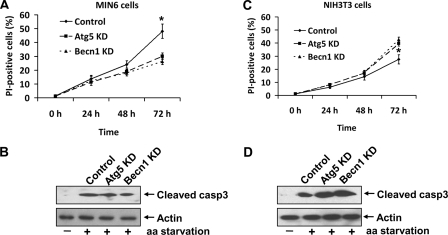
Effect of Autophagy Inhibition on Starvation-induced Cell Death
Inhibition of autophagy using Atg7 small interfering RNA prevented cell death during starvation in neurons, whereas it increased cell death in fibroblasts (29). Thus we investigated whether the increase in autophagy induced by amino acid deprivation improves cell survival or leads to cell death in MIN6 cells. A progressive increase in MIN6 cell death was observed during a 72-h incubation in medium lacking amino acids. In MIN6 cells, KD of Atg5 or Becn1 reduced but did not eliminate the increase in cell death induced by amino acid deprivation (Fig. 9A), suggesting that the increase in cell death is partially mediated by autophagy in MIN6 cells. Cleaved caspase-3 levels were also increased in amino acid-deprived MIN6 cells (Fig. 9B), indicating that apoptosis is also present. In contrast to the result in MIN6 cells, KD of Atg5 or Becn1 in amino acid-deprived fibroblastoid NIH 3T3 cells lead to an increase in cell death and cleaved caspase-3 levels compared with control (Fig. 9, C and D). Thus the effects of autophagy inhibition on amino acid starvation-induced cell death differ in different cell types.
FIGURE 9.
Amino acid deprivation-induced cell death in MIN6 and NIH 3T3 cells. A, PI-positive cells were measured in control, Atg5, or Becn1 KD MIN6 cells incubated for 72 h in amino acid (aa)-deprived medium. 48 ± 5% of cells incubated in amino acid-starved medium stained positive for PI, and this was decreased to 30 ± 2 and 26 ± 2% in Atg5 KD and Becn1 KD MIN6 cells, respectively (*, p < 0.01) (n = 3). B, representative Western blots of cleaved caspase-3 from the cells at 72 h described for A. Cleaved caspase-3 levels were increased in MIN6 cells maintained in amino acid-deprived medium compared with normal medium. Note that Atg5 KD and Becn1 KD did not affect the level of cleaved caspase-3 in amino acid-starved MIN6 cells (n = 3). C, in NIH 3T3 cells, PI-positive cells were increased to 28 ± 3% when cells were incubated in amino acid-deprived medium for 72 h, and this was further increased to 40 ± 2 and 42 ± 2% in Atg5 KD and Becn1 KD NIH 3T3 cells, respectively (*, p < 0.05) (n = 3). D, amino acid deprivation increased cleaved caspase-3 levels in NIH 3T3 cells, and this was further elevated in Atg5 KD and Becn1 KD NIH 3T3 cells at 72 h (n = 3).
DISCUSSION
The present study addressed the role of autophagy in beta cell survival and death. The possibility that an increase in autophagy could play a role in the pathogenesis of diabetes has received little attention, and it is not known whether autophagy contributes to pancreatic beta cell death and reduced beta cell mass. Marsh et al. (11) have suggested that the rate of microautophagy within the insulin secretory granule determines the rate of turnover of these granules, and this is an important mechanism for ensuring that the pool of insulin secretory granules is adequate to meet the demands of the body for insulin. Mice with beta cell-specific knock-out of Atg7 showed degradation of islets and impaired glucose-induced insulin secretion because of abnormal turnover and function of cellular organelles (13, 14). Beta cells in db/db and C57BL/6 mice that had been fed a high fat diet showed autophagosome formation, probably because of increased insulin resistance caused by high calorie intake or obesity (13). Autophagy has been noted in high glucose-induced ubiquitination of proteins into cytoplasmic aggregates in a diabetic model (30). Evidence of autophagy induced by high glucose was assessed by transfecting GFP-LC3 in INS1 cells. Because transfected GFP-LC3 associates with protein aggregates, and this aggregation is thought to be independent of autophagy (31), at present it is not clear whether high glucose induces autophagy in beta cells.
Pdx1 deficiency was selected as the primary experimental model for our studies because normal expression of this homeodomain transcription factor is essential for beta cell development, survival and function. Humans with a single mutated Pdx1 allele develop a non-insulin-dependent form of diabetes (18). Mice heterozygous for a null mutation in Pdx1 as well as animals in which the Pdx1 gene has been disrupted specifically in beta cells develop hyperglycemia caused by defective insulin secretion (15–17, 19), and Pdx1 plays an important role in the pro-survival actions of insulin signaling in the beta cell (4).
Our results have documented an important role for autophagy in regulating the cellular response to reduced Pdx1 expression in vivo in Pdx1-deficient mice and in vitro in insulin-secreting MIN6 cells. Pdx1+/− mice develop hyperglycemia soon after birth, particularly on a high fat diet, and have reduced insulin secretion and pancreatic beta cell mass. Autophagy is clearly increased in these animals with an increase in LC3 staining and the presence of autophagosomes and autolysosomes under electron microscopy. When autophagy was reduced in Pdx1+/− mice by crossing them with Becn1+/− mice, the onset of hyperglycemia was delayed in the resulting Pdx1+/− Becn1+/− offspring, and these animals demonstrated preserved insulin secretion and beta cell mass and increased beta cell proliferation compared with Pdx1+/− mice. The protective effects on beta cell function of reduced autophagy in the Pdx1+/− Becn1+/− mice were not durable, and after 7 weeks on a high fat diet the Pdx1+/− Becn1+/− and Pdx1+/− had similar levels of hyperglycemia, impairment of insulin secretion, and reduction in beta cell mass. The reduction in autophagy delayed but did not prevent the development of caspase-3-dependent apoptosis in the Pdx1+/− Becn1+/− mice, and this was ultimately responsible for the development of pancreatic beta cell death and reduced beta cell mass. Our data also showed reduced caspase-3-dependent apoptosis and increased beta cell proliferation in Pdx1+/− Becn1+/− mice compared with Pdx1+/− mice at 1 week after starting a high fat diet. However, Pdx1+/− Becn1+/− mice ultimately developed glucose intolerance and reduced beta cell mass presumably caused by increased caspase-3-dependent apoptosis.
Autophagy was also observed in the Pdx1-reduced MIN6 cells including autophagosomes seen under the electron microscope and the characteristic increase in punctate LC3 staining and LC3-II levels. The effects of inhibiting autophagy were explored by exposing the Pdx1-reduced MIN6 cells to the autophagy inhibitor 3-MA, Becn1, or Atg5 KD construct. This delayed the onset of cell death but did not prevent the cells from dying, and cell death was associated with increases in activated caspase-3 levels. Thus autophagy is an integral part of the beta cell response to reduced Pdx1, and it modulates the onset and timing of beta cell death.
Nutrient deprivation is a classic stimulus of autophagy (8). GFP-LC3 transgenic mice exhibited an increase in LC3 punctae in starved pancreatic exocrine cells. However, low levels of expression of GFP-LC3 in the transgenic mice prevented any conclusions being drawn about changes in pancreatic islets (28). Experiments were therefore performed to determine whether the same was true in insulin-secreting beta cells. An increase in autophagy was observed in MIN6 cells incubated in nutrient-deprived medium, and this effect was inhibited by exposure of the cells to 3-MA and reduced Becn1 expression as previously reported in other cells (32, 33). Autophagy was observed in pancreatic beta cells from mice that had been starved for 24 h and in beta cells deprived of nutrients in vitro. Autophagy was also activated in human islets deprived of amino acids.
The predominant view of autophagy is that it represents an adaptive response to promote cell survival in response to various stressors such as nutrient deprivation. Our results suggest that this process plays a more complex role in the insulin-secreting beta cell. We have demonstrated an improvement in the survival of Pdx1-reduced islets and insulin-secreting cells and nutrient-deprived cells if autophagy is inhibited by genetic (Becn1 and Atg5 knockdown) manipulations, suggesting that at least under these circumstances autophagy promotes cell death. The relationship between autophagy and apoptosis is the topic of ongoing debate. These two processes have been reported to coexist in the same cell (32, 33) and our results demonstrate that the two processes clearly coexist in insulin-secreting beta cells. Under the experimental conditions studied (reduced Pdx1 and nutrient deprivation), autophagy was increased before apoptosis. Inhibition of autophagy delayed the onset of cell death but did not prevent apoptosis. Near total inhibition of basal autophagy in MIN6 cells induces cell death apparently mediated by apoptosis in light of the increase in cleaved caspase-3 level. Thus, whereas autophagy may modulate the development of cell death, in the systems we have studied apoptosis is the critical process responsible for cell death. Inhibition of autophagy does not prevent eventual apoptosis, and near total inhibition of basal autophagy may actually induce caspase-3-dependent apoptosis.
In summary, an increase in autophagy is an integral component of the response of insulin-secreting beta cells to reduced Pdx1 expression and nutrient deprivation. Autophagy is evident prior to the onset of cell death induced by these conditions and inhibition of autophagy delays but does not prevent cell death, which is mediated primarily by apoptosis. The potential role of autophagy should be considered in future studies of pancreatic beta cell death in diabetes and in attempts to develop novel agents that target this process for prevention and treatment of the disorder.
Supplementary Material
Acknowledgments
We thank Dr. Sheila Stewart for generous assistance in setting up the lentivirus shRNA expression system, Dr. Beth Levine for providing the Becn1+/− mice, and Dr. Helena Edlund for providing the Pdx1+/− mice.
This work was supported by the Radioimmunoassay and Morphology Cores of the Diabetes Research and Training Center of Washington University Grants P60 DK-20579 and DK-31842 (to K. S. P.), Washington University Digestive Diseases Research Center Morphology Core Grant 5P30 DK052574, Clinical and Translational Science Award to Washington University Grant UL1RR024992, and the Blum Kovler Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- MIN6
- mouse insulinoma 6
- KD
- knockdown
- LC3
- microtubule-associated protein 1 light chain 3
- 3-MA
- 3-methyladenine
- shRNA
- small hairpin RNA
- FBS
- fetal bovine serum
- HBSS
- Hanks' balanced salt solution
- DAPI
- 4′,6-diamidino-2-phenylindole
- PI
- propidium iodide
- BafA1
- bafilomycin A1
- CHO
- Chinese hamster ovary
- GFP
- green fluorescent protein
- WT
- wild type.
REFERENCES
- 1.Butler P. C., Meier J. J., Butler A. E., Bhushan A. (2007) Nat. Clin. Pract. Endocrinol. Metab. 3, 758–768 [DOI] [PubMed] [Google Scholar]
- 2.Scarlatti F., Granata R., Meijer A. J., Codogno P. (2009) Cell Death Differ. 16, 12–20 [DOI] [PubMed] [Google Scholar]
- 3.Johnson J. D., Ahmed N. T., Luciani D. S., Han Z., Tran H., Fujita J., Misler S., Edlund H., Polonsky K. S. (2003) J. Clin. Invest. 111, 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson J. D., Bernal-Mizrachi E., Alejandro E. U., Han Z., Kalynyak T. B., Li H., Beith J. L., Gross J., Warnock G. L., Townsend R. R., Permutt M. A., Polonsky K. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19575–19580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y., Cao X., Li L. X., Brubaker P. L., Edlund H., Drucker D. J. (2005) Diabetes 54, 482–491 [DOI] [PubMed] [Google Scholar]
- 6.Klionsky D. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 931–937 [DOI] [PubMed] [Google Scholar]
- 7.Levine B., Kroemer G. (2008) Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mortimore G. E., Pösö A. R. (1987) Annu. Rev. Nutr. 7, 539–564 [DOI] [PubMed] [Google Scholar]
- 9.Shimizu S., Kanaseki T., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C. B., Tsujimoto Y. (2004) Nat. Cell Biol. 6, 1221–1228 [DOI] [PubMed] [Google Scholar]
- 10.Yu L., Alva A., Su H., Dutt P., Freundt E., Welsh S., Baehrecke E. H., Lenardo M. J. (2004) Science 304, 1500–1502 [DOI] [PubMed] [Google Scholar]
- 11.Marsh B. J., Soden C., Alarcón C., Wicksteed B. L., Yaekura K., Costin A. J., Morgan G. P., Rhodes C. J. (2007) Mol. Endocrinol. 21, 2255–2269 [DOI] [PubMed] [Google Scholar]
- 12.Twig G., Elorza A., Molina A. J., Mohamed H., Wikstrom J. D., Walzer G., Stiles L., Haigh S. E., Katz S., Las G., Alroy J., Wu M., Py B. F., Yuan J., Deeney J. T., Corkey B. E., Shirihai O. S. (2008) EMBO J. 27, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebato C., Uchida T., Arakawa M., Komatsu M., Ueno T., Komiya K., Azuma K., Hirose T., Tanaka K., Kominami E., Kawamori R., Fujitani Y., Watada H. (2008) Cell Metab. 8, 325–332 [DOI] [PubMed] [Google Scholar]
- 14.Jung H. S., Chung K. W., Won Kim J., Kim J., Komatsu M., Tanaka K., Nguyen Y. H., Kang T. M., Yoon K. H., Kim J. W., Jeong Y. T., Han M. S., Lee M. K., Kim K. W., Shin J., Lee M. S. (2008) Cell Metab. 8, 318–324 [DOI] [PubMed] [Google Scholar]
- 15.Ahlgren U., Jonsson J., Jonsson L., Simu K., Edlund H. (1998) Genes Dev. 12, 1763–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonsson J., Carlsson L., Edlund T., Edlund H. (1994) Nature 371, 606–609 [DOI] [PubMed] [Google Scholar]
- 17.McKinnon C. M., Docherty K. (2001) Diabetologia 44, 1203–1214 [DOI] [PubMed] [Google Scholar]
- 18.Stoffers D. A., Ferrer J., Clarke W. L., Habener J. F. (1997) Nat. Genet. 17, 138–139 [DOI] [PubMed] [Google Scholar]
- 19.Wang H., Hagenfeldt-Johansson K., Otten L. A., Gauthier B. R., Herrera P. L., Wollheim C. B. (2002) Diabetes 51, S333–S342 [DOI] [PubMed] [Google Scholar]
- 20.Althage M. C., Ford E. L., Wang S., Tso P., Polonsky K. S., Wice B. M. (2008) J. Biol. Chem. 283, 18365–18376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., Tashiro Y. (1998) Cell Struct. Funct. 23, 33–42 [DOI] [PubMed] [Google Scholar]
- 22.Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E. L., Mizushima N., Ohsumi Y., Cattoretti G., Levine B. (2003) J. Clin. Invest. 112, 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clavé C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di, Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Dröge W., Dron M., Dunn W. A., Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fésüs L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., González-Estévez C., Gorski S., Gottlieb R. A., Häussinger D., He Y. W., Heidenreich K., Hill J. A., Høyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jäättelä M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovács A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., López-Otín C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Meléndez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Münz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nürnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D. C., Ryan K. M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P. O., Seleverstov O., Settleman J., Shacka J. J., Shapiro I. M., Sibirny A., Silva-Zacarin E. C., Simon H. U., Simone C., Simonsen A., Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Tallóczy Z., Tanaka K., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcátegui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 26.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 27.Greider M. H., Howell S. L., Lacy P. E. (1969) J. Cell Biol. 41, 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. (2004) Mol. Biol. Cell 15, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du L., Hickey R. W., Bayir H., Watkins S. C., Tyurin V. A., Guo F., Kochanek P. M., Jenkins L. W., Ren J., Gibson G., Chu C. T., Kagan V. E., Clark R. S. (2009) J. Biol. Chem. 284, 2383–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaniuk N. A., Kiraly M., Bates H., Vranic M., Volchuk A., Brumell J. H. (2007) Diabetes 56, 930–939 [DOI] [PubMed] [Google Scholar]
- 31.Shvets E., Elazar Z. (2008) Autophagy 4, 1054–1056 [DOI] [PubMed] [Google Scholar]
- 32.Hwang S. O., Lee G. M. (2008) Biotechnol. Bioeng. 15, 678–685 [DOI] [PubMed] [Google Scholar]
- 33.Sadasivan S., Waghray A., Larner S. F., Dunn W. A., Jr., Hayes R. L., Wang K. K. (2006) Apoptosis 11, 1573–1582 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.