Abstract
In skeletal muscle development, the genes and regulatory factors that govern the specification of myocytes are well described. Despite this knowledge, the mechanisms that regulate the coordinated assembly of myofiber proteins into the functional contractile unit or sarcomere remain undefined. Here we explored the hypothesis that modular domain proteins such as Bin1 coordinate protein interactions to promote sarcomere formation. We demonstrate that Bin1 facilitates sarcomere organization through protein-protein interactions as mediated by the Src homology 3 (SH3) domain. We observed a profound disorder in myofiber size and structural organization in a murine model expressing the Bin1 SH3 region. In addition, satellite cell-derived myogenesis was limited despite the accumulation of skeletal muscle-specific proteins. Our experiments revealed that the Bin1 SH3 domain formed transient protein complexes with both actin and myosin filaments and the pro-myogenic kinase Cdk5. Bin1 also associated with a Cdk5 phosphorylation domain of titin. Collectively, these observations suggest that Bin1 displays protein scaffold-like properties and binds with sarcomeric factors important in directing sarcomere protein assembly and myofiber maturation.
Skeletal muscle differentiation is a highly orchestrated phenomenon. The transition from cycling myoblasts to mature myofibers is dependent on a coordinated response involving up-regulation of muscle-specific transcription factors, engagement of a defined gene expression program, followed by an ordered assembly of muscle structural proteins to form the basic contractile units known as sarcomeres. The key molecular genetic features of this skeletal muscle differentiation program are well understood (1–3). Nevertheless, the regulatory networks that control and integrate sarcomeric assembly in developing myofibers remain comparatively unknown.
The sarcomere is composed of thick myosin and thin actin myofilaments together with the giant sarcomeric proteins titin and nebulin. The actin and myosin filaments are anchored at the Z-line and M-line, respectively. Titin has been coined a “molecular ruler” of the thick filament because it mediates an ordered and repetitive series of interactions with myosin and with several proteins at the Z- and M-lines that include the sarcomeric protein complex (4, 5). Similarly, nebulin has also been coined the molecular ruler of the thin filament for its ordered assembly of actin (6, 7), and recent evidence indicates that nebulin also mediates protein interactions of the sarcomere (reviewed in Refs. 4, 8).
The large number of protein interactions that initiate and establish the mature sarcomere implies that one or more protein structural motifs may be critical to the assembly process. Surprisingly, many of these proteins contain Src homology 3 domains (SH3),3 a well characterized protein-protein interaction domain (reviewed in Refs. 9, 10). Titin contains numerous SH3 domains, many of which affect its function. Similarly, nebulin function and incorporation into the mature sarcomere appear to be dependent on an endogenous SH3 domain (4, 11–13). Titin-associated proteins such as obscurin have SH3 motifs that appear to modulate the G-protein-coupled signal transduction pathways. Notably, a stretch of prolines representative of an SH3 binding region resides within the Rho guanine nucleotide exchange factor domain of obscurin (14, 15). These observations suggest that SH3 adaptor protein(s) play a pivotal role in the construction and stabilization of the sarcomere.
Once assembled, the sarcomere must be stabilized with other structures in the developing myofiber. Paramount among these components is the sarcolemma/t-tubule system. The sarcolemma is a highly specialized membrane with numerous involutions (t-tubules) that couple the external signal for contraction to the basic contractile unit, the sarcomere. As such, it is reasonable to hypothesize that sarcomere assembly and sarcolemmal biogenesis may be facilitated by an overlapping set of proteins. However, invoking such a model will be dependent on the identification of a modular protein that utilizes distinct domains to influence each of these disparate activities. Within this context, one candidate factor that has emerged is the tumor suppressor protein Bin1 (bridging integrator protein 1). Bin1 was initially characterized as a c-Myc interacting protein, capable of repressing c-Myc transcriptional activation (16, 17). Bin1 retains distinct modular features that include a mid-body c-Myc binding domain, a C-terminal SH3 domain with unique structural features not shared with SH3 regions of sequence-related proteins, and an N-terminal domain (referred to as the BAR domain) with sequence similarity to a larger family of synaptic vesicle/clathrin-interacting factors, exemplified by the neuron-enriched protein amphiphysin (18–20).
Bin1 has been implicated in regulating striated muscle function across a variety of model systems. Overexpression of Bin1 in a myoblast cell line inhibits cell growth and results in a more rapid onset of differentiation following growth factor withdrawal (21). Generation of mice with a null mutation in bin1 leads to a severe disruption in cardiomyocyte function through an undetermined mechanism (22). Null mutations of the Drosophila bin1 homologue have revealed that Bin1 is required for maintenance of excitation-contraction coupling in skeletal muscle (23, 24). Regulation of the contractile response was attributed to an ability of the Bin1 BAR domain to enhance sarcolemmal membrane curvature, influencing t-tubule assembly and maturation (25). Interestingly, a recent study has demonstrated that patients suffering from centronuclear myopathy have homozygous mutations in bin, at the regions encoding either the BAR or SH3 domain (26). A representative tissue culture model of the individual BAR domain or SH3 domain mutations displayed obvious defects in membrane tubulation events. This myopathy also displayed a phenotype consistent with sarcomere disorder, yet the significance of the Bin1 BAR and SH3 domains was not tested in this regard.
Here we explored the hypothesis that distinct modular domains of Bin1 separately influence the key events associated with skeletal muscle differentiation. Specifically, we propose that the Bin1 SH3 domain is a sarcomere-organizing protein. We demonstrate that transgenic overexpression of the Bin1 SH3 domain results in a profound perturbation of skeletal muscle ultrastructure, characterized by increased myofiber size with sarcomeric disorganization. This phenotypic outcome derives in part from a disruption in the endogenous interaction between the Bin1 SH3 domain and a number of contractile proteins, including sarcomeric actin and myosin. The sarcomere disruption in this model is also influenced by a loss in the endogenous interaction between the Bin1 SH3 domain and Cdk5, a pro-myogenic kinase that promotes sarcomeric assembly in part by phosphorylating a serine-responsive region of titin. Collectively, these observations suggest that Bin1 is a crucial adaptor protein that acts to promote skeletal muscle differentiation through domain-specific assembly of the mature sarcomere.
EXPERIMENTAL PROCEDURES
Transgenic Animals
The SH3 region (encoding amino acids 361–434) of the murine bin1 gene (gene identifier) was produced through PCR amplification. To direct systemic expression of the Bin1 SH3 construct, the sequence-verified PCR product was cloned in-frame into the pCAGGS expression vector containing a modified cytomegalovirus enhancer/promoter with a rabbit β-globin poly(A) (27, 28). Injection and derivation of transgenic mice were performed as described previously (29). A total of 12 founder lines were generated, of which 5 were subject to further characterization.
Histological Analyses and Electron Microscopy
Tissues were removed and fixed in 10% formalin for 4–5 days (skeletal muscle, heart, mammary gland, prostate, skin, liver, lung, brain). Fixed tissues were then embedded in paraffin, sectioned at 10 μm, stained, and counter-stained with hematoxylin and eosin. Muscle fiber diameters were assessed on hindlimb muscle groups from multiple founder lines, with a minimum of four individual muscles measured per group.
Ultrastructure examination of wild type and bin1SH3 gastrocnemius was performed as described previously (30). Longitudinal semi-thin (0.3–0.5 μm) sections were used to visualize the muscle ultrastructure under an accelerating voltage of 100 kV.
Whole Mount in Situ Hybridization
Embryos were collected between 8.5 and 12.5 days post-coitum (dpc) and prepared as described previously (31) A 5′-portion of the murine bin1 cDNA was used as a probe for whole mount in situ hybridizations. In situ detection of the pCAGGS transgene was performed with a riboprobe generated against the full-length rabbit β-globin poly(A) region. The myogenin riboprobe used was as described previously (32). Digoxigenin-labeled sense and antisense riboprobes were generated as per the manufacturer's instructions (Roche Applied Science). For in situ hybridization, murine embryos were treated as described (31).
Cell Culture
Skeletal myoblast c2c12 cells were cultured as described previously (33). All reagents for c2c12 culture were obtained from Invitrogen. Primary myoblast cell lines (muscle satellite cells) were isolated as described previously (33).
Immunocytochemistry
Immunolocalization of skeletal muscle differentiation markers in cultured cells was performed as described previously (33). Cells were fixed in 4% paraformaldehyde and stained with anti-myosin heavy chain MF20 hybridoma and visualized by counterstaining with a fluorescein isothiocyanate-conjugated secondary antibody.
Adenoviral Constructs and Infection
Adenoviral expression vectors were generated using the AdEasy adenoviral vector system (Stratagene). The AdHA-SH3-hrGFP-2 construct was generated using the bin1 SH3 region (exons 15 and 16). The KSP region of titin (cDNA gift from Siegfried Labeit, Mannheim, Germany) was used to generate the AdHA-TiKSP-GFP.
Flow Cytometry/Cell Viability Assays
Annexin-V-propidium iodide (PI) analyses of apoptotic and necrotic cells were performed using the annexin-V-FLUOS staining kit (Roche Applied Science) on a Beckman-Coulter ALTRA flow cytometer.
Cell growth assays were evaluated in both cycling cells and in cells after 2 days in low serum media (differentiation media) using a 5-bromo-2-deoxyuridine cell proliferation assay kit (Roche Applied Science). Myoblast fusion was evaluated by determining the number of myosin heavy chain-positive myotubes containing two or more nuclei relative to the total number of myosin heavy chain-positive myotubes.
Immunoblot Analysis
Cell and tissue protein lysates were prepared in modified radioimmunoprecipitation assay (RIPA) buffer (33). Immunoblots assays were performed as described previously (33) using antibodies against myogenin, p38, MEF2C, skeletal α-actin and myosin, Cdk5, and Bin199D (all from Santa Cruz Biotechnology), myosin heavy chain MF20 hybridoma, and M-cadherin and HA tag (Sigma).
Size Exclusion Chromatography
Lysates from c2c12 cultured cells were prepared as above, and 800–1000 μg of total cell lysate was loaded onto an equilibrated Superose 6HR 10/30 column. Samples were run in 0.05 m phosphate buffer with 0.15 m NaCl using an ÄKTA 10 Explorer FPLC.
Co-immunoprecipitation, GST Pulldown, and Kinase Activity Assays
Co-immunoprecipitation assays were performed as described previously (33). GST-pulldown assays were performed using recombinant GST and GST-SH3 purified on glutathione beads with 150 μg of total protein from tissue culture lysates. Protein kinase analyses were performed as described previously (33).
Mass Spectrometry
A two-dimensional nano-liquid chromatography MS/MS approach was used to identify Bin1 SH3-interacting proteins. Following GST pulldown assays, the tagged protein complexes were subjected to on-bead tryptic digests, and peptides were bound to a cation exchange column on a CapLC capillary LC system (Waters). Peptides were eluted with a gradient ammonium acetate solution (2, 5, 10, 25, 50 100, and 200 mm) and separated through a C18 reverse phase column followed by electrospray ionization and quadrupole/time-of-flight MS/MS on a Q-TOF Ultima hybrid mass spectrometer. Peptide MS/MS spectra were searched against the NCBInr data base using the MASCOT searching algorithm.
RESULTS
The modular domain structure of Bin1 indicated that this protein may serve as an ideal adaptor protein for the assembly and maintenance of the mature myofiber. However, the post-natal distribution of Bin1 has been reported to be ubiquitous with an enriched expression pattern in neural and cardiac tissues and in adult skeletal muscle (16, 18, 34). Therefore, to address the function of Bin1 in skeletal muscle as a probable maturation regulatory factor, we examined its distribution during murine embryogenesis using an antisense riboprobe specific to the murine bin1 cDNA (16).
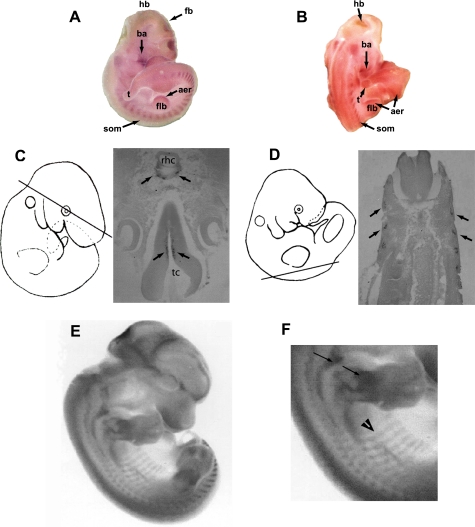
bin1 expression was not detectable in developing embryos prior to 8.5 dpc. By 10–10.5 dpc, bin1 transcripts were readily visible within the branchial arches, somites, and the ependymal region of the developing forebrain and hindbrain (Fig. 1, A–D). Somites represent the tissue reservoirs from which most skeletal muscle is derived. The myogenic stem cells originating from the somites provide the myoblast pool from which all trunk and limb musculature arises, whereas the branchial arch myoblasts contribute to components of head musculature (35). Histological sections of embryos at 10.5 dpc confirmed that bin1 expression was concentrated in the differentiated regions of the somite (myotome portion) and ependymal layers of the developing brain (Fig. 1, C and D). In addition, embryos revealed elevated bin1 expression in both the epicardial region of ventricular trabeculae and within the developing optic cup (data not shown), although the predominant expression appeared to be within the skeletal muscle primordia. At later stages of embryogenesis (12.5 dpc), the expression of bin1 remained elevated within tissues of the skeletal muscle lineage, including the developing pre-muscle masses of the limbs, head, and scapular regions (Fig. 1E). However, closer examination of these embryos revealed a divergence in the skeletal muscle expression pattern of bin1 in that the highest expression was found in regions where differentiation had recently initiated such as in limb and scapular muscle groups (Fig. 1, E and F). Conversely, bin1 expression declined in areas of more established muscle development, such as the primordia of body wall muscle at 12.5 dpc (Fig. 1F).
FIGURE 1.
Developmental expression of bin1. A, bin1 is accumulated within the somites (som), branchial arches (ba), and regions within the developing hindbrain (hb) and forebrain (fb) at 10.5 dpc. t, tailbud; aer, apical ectodermal ridge; flb, forelimb bud. B, larger concentration of bin1 is detected in regions of differentiating skeletal muscle at 10.5 dpc, including the somites (som) and branchial arches (ba). Limited staining is also observed in the apical ectodermal ridge (aer) of the developing forelimb bud (flb) and in regions of the hindbrain (hb). C, transverse sections of the embryo at 10 dpc reveal that bin1 expression is limited to the ependymal layers of the developing hindbrain/rhombencephalon (rhc) (arrows) and forebrain/telencephalon (tc) (arrows). D, bin1 expression in 10.5 dpc embryos is restricted to a somite subdomain (arrow) in which myogenic differentiation has been initiated. E and F, enhanced expression of bin1 in early stage muscle differentiation at 12.5 dpc. F, bin1 expression is elevated in the developing forelimb and shoulder girdle muscles (arrows). bin1 expression declines in areas of more established muscle development such as those within the body-wall (arrowhead).
The enriched expression of bin1 in regions specific to skeletal muscle development suggested a site-specific function for this protein. Interestingly, accumulation of differentially spliced bin1 variants in neural, cardiac, testicular, and other tissues may also suggest site-specific functions of bin1 (reviewed in Ref. 20). Indeed, other members of the BAR domain family have been assigned a variety of cellular functions, including transcription, apoptosis, endocytosis, tumor suppression, and cell growth control (reviewed in Ref. 20). Conceivably, such divergent roles for Bin1 may be explained by invoking a model whereby separable domains of the Bin1 protein manage domain-specific cellular activities. Therefore, to directly test this hypothesis, we created a transgenic strain that overexpressed the SH3 domain of Bin1. The SH3 domain remains the most conserved feature of all the Bin1 splice variants (36). Moreover, the Bin1 SH3 domain appears to be structurally unique across a wide variety of SH3 motifs suggesting a distinct or limited function for this region of the Bin1 protein (19, 36, 37). As such, we anticipated that the overexpression of this domain would disrupt endogenous Bin1 interactions and thereby impair Bin1 function with little to no effect on other SH3-containing proteins. We utilized a ubiquitously expressed transgene (pCAGGS expression vector) as a targeted delivery system for overexpression of the Bin1 SH3 domain (27) (Fig. 2A). The enhancer promoter combination of this transgene has frequently demonstrated a global expression pattern in transgenic mice (28). Importantly, the use of a ubiquitous promoter allowed us to address whether the SH3 domain of Bin1 performed tissue-specific functions or whether this region of the Bin1 protein conveyed non-muscle cellular functions, in addition to the anticipated role in skeletal muscle.
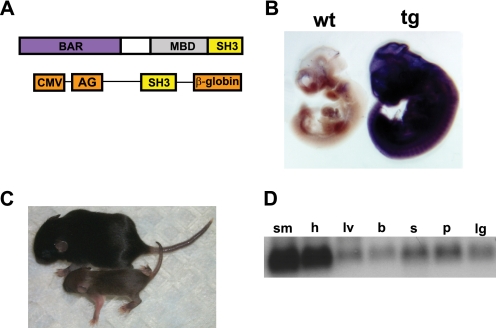
FIGURE 2.
Transgenic overexpression of the Bin1 SH3 domain. A, schematic representation of Bin1 showing the relative positions of the Bin1-Amphiphysin-RSV (BAR) domain, Myc-binding domain (MBD), and the Src homology 3 (SH3) domain. A schematic representation of the pCAGGS bin1SH3 transgene used to generate bin1SH3 mice is also shown. The SH3 domain is flanked 5′ with the sequences from murine cytomegalovirus (CMV) and rabbit AG (AG) promoter and 3′ with the rabbit β-globin poly(A) tail. B, bin1SH3 expression is ubiquitously high in the transgenic (tg) embryo (left) and absent in the wild type (wt) embryo (right) at 10.5 dpc. C, wild type mouse (top) is shown relative to a smaller bin1SH3 transgenic littermate at post-embryonic day 14. D, bin1SH3 transgene was highly expressed in adult skeletal muscle (sm) and heart (h). lv, liver; b, brain; s, skin; p, prostate, lg, lung.
We performed numerous rounds of pronuclear injections to collect and analyze founder embryos (transient transgenics) that might display profound changes in vivo and create founder lines for breeding to homozygosity. As expected, a ubiquitous expression of Bin1 SH3 gene product was detected in embryos at 10.5 days with a riboprobe designed against the β-globin poly(A) sequence from the pCAGGS targeting construct (Fig. 2B). Transgenic mice expressing the SH3 region (bin1SH3) were characteristically smaller than littermate controls (Fig. 2C). Despite the ubiquitous expression of bin1SH3 in young pups, we observed an enriched accumulation of the Bin1 SH3 transgene in both the heart and skeletal muscles of adult mice. The pronounced expression of the SH3 domain in skeletal muscle may indicate a bias for expression of the transgene/enhancer combination in this tissue type. Nevertheless, robust expression was noted in several other cell types.
To investigate putative pathological effects caused by the overexpression of the Bin1 SH3 domain, we performed an extensive histologic examination of the Bin1 SH3 gene product in mice. The loss of Bin1 function has been associated with malignant transformation in a variety of tissues and cells (38–42). In addition, homozygous ablation of bin1 resulted in lung and mammary carcinoma as well as severe cardiomyopathy (22, 43, 44), suggesting that altered expression of the Bin1 SH3 domain may result in pleitrophic pathology. We inspected the progeny of five independent bin1SH3 transgenic lines (>200 mice) during the natural lifespan of these animals. Overt tumor formation was never observed in any progeny even up to 1 year of age. Moreover, with the exception of skeletal muscle, histologic inspection of target organs in aged mice showed normal cellular architecture with no evidence of hyperplasia despite the robust expression of bin1SH3 (Fig. 3A). For example, sections of heart, lung, liver, prostate, and skin revealed no overt alteration in cellular architecture or in gross morphology (Fig. 3A).
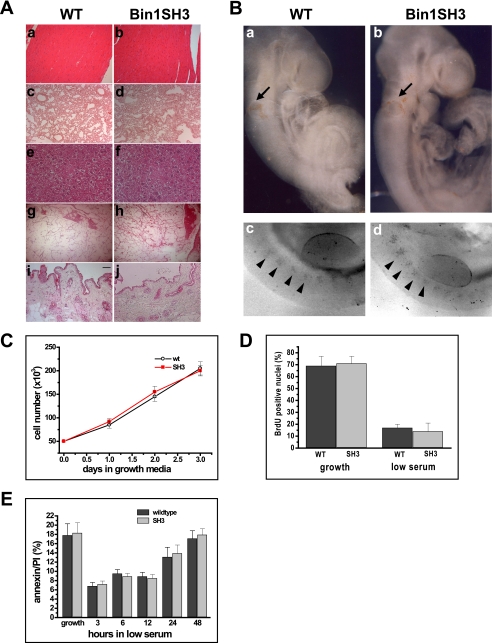
FIGURE 3.
Pathological examination of the bin1SH3 genotype. A, hematoxylin/eosin staining of the heart (panels a and b), lung (panels c and d), liver (panels e and f), prostate (panels g and h), and skin (panels i and j) showed no anomalies in cell morphology and tissue density in wild type (WT) and bin1SH3 mice. Bar represents 100 μm. B, whole mount tunnel assays for apoptosis show no differences in cell death at 9.5 dpc between wild type (panel a) and bin1SH3 (panel b) embryos at the otic vesicles (arrows). A comparable degree of apoptosis is also observed within the somites (arrowheads) and along the apical ectodermal ridge in both wild type (panel c) and bin1SH3 (panel d) embryos. C, growth kinetics of wild type (wt) and bin1SH3 myoblasts held in growth medium were similar. n = 9 animals per culture for each wild type and bin1SH3. D, cells were incubated in growth medium, and cell proliferation was examined using 5-bromo-2-deoxyuridine (BrdU). n = 9 animals per culture for each wild type (WT) and bin1SH3. E, annexin-V/propidium iodide (PI) apoptosis analyses of wild type and bin1SH3 myoblasts during incubation in low serum media. Both wild type and bin1SH3 myoblasts had comparable levels of apoptosis at the indicated time points. n = 9 animals per culture for each wild type and bin1SH3.
As noted above, the absence of pathology in these tissues is in contrast to studies demonstrating tumor formation with bin1 disruption. Bin1 and its homologue in yeast have also been implicated as integral signal components in apoptotic programs (38, 42, 45–47). To further evaluate the consequences of Bin1 SH3 overexpression on cellular homeostasis, we performed comparative whole mount terminal dUTP nick-end labeling assays on both founder and bin1SH3 homozygous animals. Terminal dUTP nick-end labeling positive regions were noted in the otic vesicles, somites, and apical ectodermal ridge in all embryos examined. However, the degree of apoptosis associated during development did not vary between the bin1SH3 and wild type littermates (Fig. 3B). These results suggest that the overexpression of the SH3 domain of Bin1 does not impede the normal apoptotic process that has been previously characterized in other models of Bin1 perturbation (26, 39, 43, 44, 47, 48).
Myoblasts isolated from the skeletal muscles of both wild type and bin1SH3 mice were plated at equal concentrations in high serum conditions. Differences in the growth rates of either culture were not observed (Fig. 3C). Additionally, the degree of DNA replication was comparable between wild type and SH3 overexpressing myoblasts in both growth and differentiation conditions (Fig. 3D). To address potential alterations in apoptosis, we employed a comparative fluorescence-activated cell sorter analysis using propidium iodide (PI) staining, coupled with annexin V during low serum induction of differentiation. During the differentiation time course, late apoptotic events (PI + annexinV/PI co-stained cells) appeared in a relatively similar frequency in both wild type and bin1SH3 cultures (Fig. 3E). Taken together, our results indicate that the bin1SH3 transgene does not impact cellular apoptosis and has no demonstrable effect on cell cycle kinetics.
Histologic examination of the skeletal musculature from bin1SH3 mice revealed significant differences in the myofiber structure compared with wild type littermates. For example, myofibers from bin1SH3 muscle had visibly increased diameters and distorted myofiber structure (Fig. 4, A and B). We employed a transverse tubule stain using potassium ferricyanide (30) to examine the ultrastructural organization of skeletal muscle from wild type and bin1SH3 mice. The I-band and z-disk from transgenic muscles were strikingly wider and appeared more disseminated than myofibers from wild type counterparts (Fig. 4, G and H). Closer comparison exposed a distinctive misalignment of the z-disk in the transgenic muscles (Fig. 4H, arrowheads). Importantly, the t-tubule-specific stain did not reveal any overt differences in the t-tubule structure of wild type and bin1SH3 mice suggesting that the SH3 domain mediates a sarcomere-specific function.
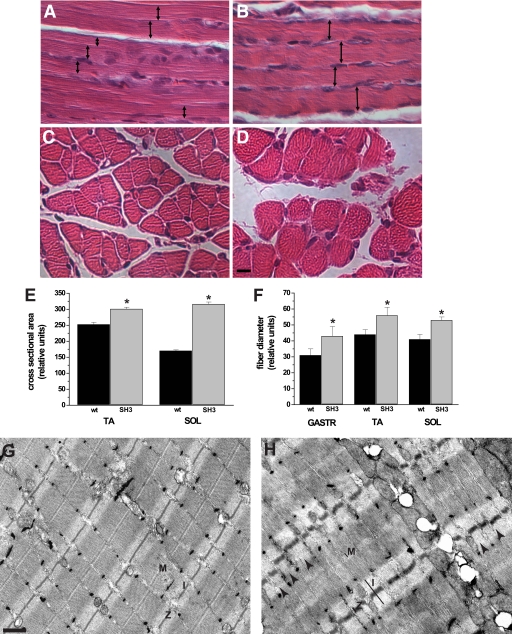
FIGURE 4.
Loss of Bin1 SH3 function results in an unorganized myofibril arrangement. A and B, longitudinal section of hematoxylin/eosin-stained soleus muscle from post-embryonic day 10, wild type, and bin1SH3 mice. The widths of myofibers from the bin1SH3 mice are markedly larger (double-headed arrows). C and D, cross-sectional fiber diameters of the soleus from bin1SH3 mice are also noticeably larger than wild type. Scale bar represents 100 μm. E, cross-sectional area of tibialis anterior (TA) and soleus (SOL) muscles from wild type (wt) and bin1SH3 (SH3) mice were compared. n = 6 muscles per group with at least 60–80 fibers per muscle were counted (*, significantly different, p < 0.001). F, relative fiber diameters of the gastrocnemius (GASTR), tibialis anterior (TA), and soleus (SOL) of p10 wild type (wt) and bin1SH3 (SH3) mice were compared. n = 6 per group (*, significantly different, p < 0.001). G and H, transmission electron microscopy showing the ultrastructure of the gastrocnemius from wild type (G) and bin1SH3 (H) muscle. The M-bands (M), z-disks (Z), and I-bands (I) from bin1SH3 muscle are distended, and the z-disks are periodically misaligned (arrows). Scale bar represents 500 nm.
At a gross morphology level, the tibialis anterior and soleus muscles from bin1SH3 mice were significantly larger in cross-sectional area than in wild type littermates (Fig. 4E). Additionally, pronounced enlargements in individual fiber diameters were evident in the gastrocnemius, tibialis anterior, and soleus muscles of bin1SH3 mice (Fig. 4, C, D, and F). These muscles represent tissues with a broad metabolic range and cellular physiology, suggesting that Bin1 exerted a dominant effect on skeletal muscle cyto-structure independent of metabolic or fiber-type specificity.
Given the early expression pattern of bin1, it is reasonable to assume that the Bin1 SH3 domain may affect sarcomere structure indirectly by altering myogenic gene activation and protein accumulation. Therefore, to discriminate direct versus indirect effects of Bin1 on myofiber maturation, we examined the expression of key myogenic regulatory factors. In 12.5 dpc embryos, whole mount in situ analysis revealed significant accumulation of myogenin message within the somites and at the forelimb buds of wild type litters (Fig. 5A, panels a and c). Although myogenin expression appeared to be somewhat attenuated in bin1SH3 embryos, the overall expression pattern was conserved (Fig. 5A, panels b and d).
FIGURE 5.
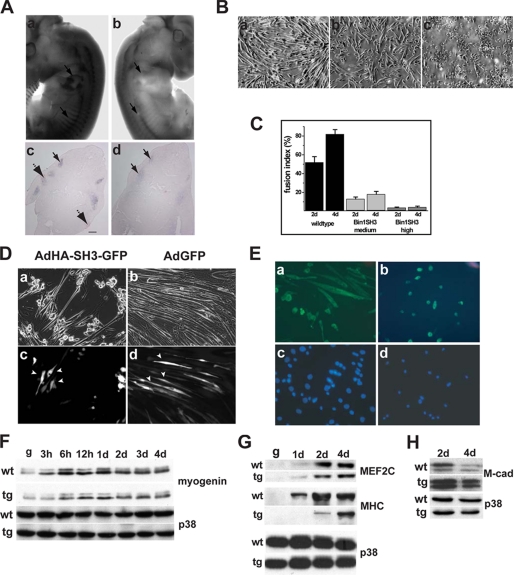
Bin1 SH3 domain affects myoblast fusion. A, whole mount in situ hybridization of a wild type 12-dpc embryo (panel a) demonstrates significant accumulation of myogenin message within the somites and at the forelimb buds (arrows). Similar areas of active myogenesis in the bin1SH3 embryo (panel b) have noticeably absent levels of myogenin message. Cross-sectional analysis of a wild type embryo (panel c) confirms the presence of myogenin message within the somites (solid arrow) and forelimb buds (broken arrow). A similar analysis of a bin1SH3 embryo (panel d) shows a very low accumulation of myogenin message within the somites and nondetectable levels of myogenin at the forelimb buds. Scale bar represents 100 μm. B, primary myoblasts isolated from wild type (panel a) muscle differentiate into elongated myotubes after 48 h. A marked reduction in myotube pattern was noted in myoblasts isolated from a low expressing bin1SH3 muscle (panel b). Myoblasts isolated from a high expressing bin1SH3 mice (panel c) failed to elongate and fuse. C, higher percentage of cells containing two or more nuclei within a myosin heavy chain positive cell was observed in wild type myoblasts compared with both myoblasts from low and high expressing bin1SH3 mice following incubation in low serum media. D, c2c12 myoblasts were infected with AdHA-SH3-GFP (left panel) or AdGFP control (right panel) and incubated with low serum for 4 days. Bright field micrographs (panels a and b) were compared with GFP fluorescence (panels c and d). SH3GFP-positive cells (panel c showing cells indicated by arrowheads) remained unicellular and did not fuse, whereas SH3-GFP-negative cells were elongated. Both GFP-positive and GFP-negative cells were elongated and visibly differentiated (panel b compared with panel d). Arrows in panel d show GFP-infected cells that were elongated. E, myosin heavy chain expression in wild type (panels a and c) and bin1SH3 (panels b and d) myoblasts after 2 days in low serum medium. Myoblasts from Bin1 SH3-expressing mice show an accumulation of myosin heavy chain but do not elongate and fuse to form mature myotubes. Nuclei were visualized using 4′,6-diamidino-2-phenylindole staining (panels c and d). F, myogenin levels remained relatively similar in both wild type (wt) and transgenic (tg) myoblasts after early and prolonged low serum exposure. Loading was assessed using p38α. d, day. G, MEF2C was also present in both wild type and transgenic myoblasts after 4 days in low serum media. Myosin heavy chain accumulated more slowly and to a lesser extent in transgenic myoblasts. H, after 4 days in low serum conditions, wild type M-cad levels decreased indicating that these cells had fused. M-cad levels remained elevated in transgenic myoblasts at 4 days.
To further investigate the effect of the Bin1 SH3 domain on skeletal myogenesis, we isolated satellite cells from the hindlimbs of wild type and bin1SH3 mice. These cells displayed comparable growth kinetics during proliferation. However, induction of differentiation resulted in strikingly disparate morphologies between wild type and transgenic myoblast cultures. After 4 days in low serum, wild type myoblasts underwent robust differentiation and displayed elongated and arrayed extensions containing multiple nuclei (Fig. 5, B, panel a, and C). In contrast, high expressing bin1SH3 myoblasts displayed an appropriate increase in myosin heavy chain expression, yet did not form ordered myotube structures (Fig. 5, B, panel c, and C). Interestingly, myoblasts from a transgenic line with moderate expression of bin1SH3 did not form stable myotubes to the same degree as the wild type cells, but the myogenic phenotype had progressed further than the bin1SH3 high expressing cells (Fig. 5, B, panel b, and C). The limitation in myotube structure and patterning was not a simple temporal delay because extended low serum exposure (≥7 days) did not result in robust myotubes in transgenic cultures (data not shown). These experiments confirmed our observations that bin1SH3 expression affects the morphological characteristics of myoblast differentiation in a dose-dependent manner.
To further determine SH3-specific effects, we employed the c2c12 myoblast cell line. These cells were infected with adenovirus expressing either GFP (Ad-IRES-GFP) or the SH3 domain of Bin1 (AdGFP-IRES-SH3), maintained in low serum media for 4 days, and then examined for GFP expression (Fig. 5D). GFP-positive cells that also expressed the Bin1 SH3 domain remained largely mononuclear and did not efficiently form myotubes, whereas uninfected cells (GFP-negative) differentiated normally. This was in contrast to c2c12 cells infected with adenovirus-GFP in which both GFP-positive and GFP-negative cells consistently formed robust multinucleated myotubes (Fig. 5D, right panels). Trypan blue dye exclusion analyses did not indicate cellular toxicity with either Ad-IRES-GFP or AdGFP-IRES-SH3 (<3% of total infected cells) through a range of adenoviral infections (0–500 infectious particles/cell) (supplemental Fig. 1). These experiments verified that structural deficiencies in myotube formation are attributable to the SH3 domain of Bin1.
The apparent lack of myotube formation after low serum incubation in bin1SH3 cultures prompted a more thorough examination of muscle-specific transcription factors in these cells. After 2 days in low serum media, both wild type and binSH3 myoblasts showed early accumulation of the terminal differentiation markers such as myosin heavy chain, myogenin, and MEF2C (Fig. 5, E–G), further confirming our previous immunocytochemical results and suggesting that the Bin SH3 domain did not impact the differentiation program per se (Fig. 5, F and G).
Myoblast fusion is a distinct feature of muscle cells. The trans-membrane glycoprotein muscle cadherin (M-cadherin) is a member of the cadherin family of adhesion receptor molecules and has a role in myoblast fusion. M-cadherin is normally expressed at high levels during myoblast differentiation and rapidly declines following myoblast fusion (49–51). The lack of elongated, multinucleated myotubes observed in our ex vivo cultures of wild type and bin1SH3 myoblasts prompted us to compare the pattern of M-cadherin protein in these cells. A notable level of M-cadherin was present in both cultures after 2 days following induction of muscle differentiation. However, M-cadherin levels declined after 4 days in wild type cells but remained substantially elevated in bin1SH3 cells (Fig. 5H). These results suggested that at least in cultured cells, M-cadherin is improperly regulated in bin1SH3 myoblasts. Intriguingly, fusion did not appear to be perturbed in bin1SH3 muscles in vivo despite the presence of disrupted sarcomeric structures. Although we cannot directly account for this inconsistency, it would seem likely that other myoblast fusion-related factors play a role in vivo (52). Therefore, we cannot irrefutably conclude that M-cadherin regulation is directly influenced by Bin1. However, the significance of Bin1 in prompting adhesion-dependent signals that influence cell fate has been noted previously (53).
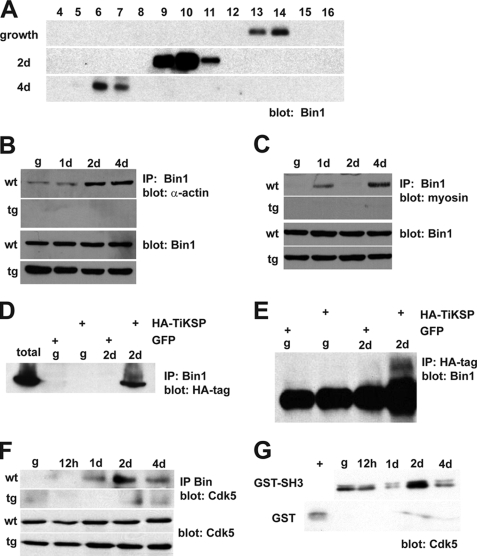
The modular structure of Bin1 implicates its participation in protein-protein interactions. Furthermore, the phenotypic outcome from overexpression of the Bin1 SH3 domain in both cultured cells and transgenic animals suggested that Bin1 associates with protein(s) critical for assembly of the muscle sarcomere, at least in part through the SH3 region. In a developmental context, it is conceivable that a protein involved in myo-structural assembly would form transient protein complexes with myocyte proteins. To begin to examine Bin1-dependent protein interactions during myogenesis, we analyzed the native size of Bin1 in differentiating c2c12 cells using gel filtration chromatography. With a Bin1-specific antibody probed against a series of gel filtration fractions, we observed a shift in the elution of Bin1 protein by immunoblot analyses (Fig. 6A). In these experiments, elution at a lower volume (fraction number) indicated a larger size in the native protein. Bin1 was observed in fractions 13–14 from lysates derived from cycling myoblasts. After 2 and 4 days of low serum induction of differentiation, the prospective Bin1 complex was found in fractions 9–11 and 6–7, respectively, signifying a substantial increase in the size of Bin1. Interestingly, Bin1 did not seem to undergo dynamic structural changes in bin1SH3 myoblasts. In an earlier study, alternative splicing led to higher molecular weight isoforms of Bin1 in differentiated c2c12 cells (∼65-kDa growth to 68–70-kDa differentiation) (21). In our experiments, we found similar isoforms in 4- day differentiated cultures using the same anti-Bin1 antibody (99D). However, the elution profile from these gel filtration experiments still designated Bin1 as a large molecular weight protein of variable size. Taken together these data suggest that the size of Bin1 and presumably Bin1-associated protein complexes increased in mass during early c2c12 differentiation. Moreover, a deviation in the native size of Bin1 (∼67–440 kDa during growth to >700 kDa during differentiation) suggested that Bin1 engages transient protein interactions during the process of myoblast differentiation.
FIGURE 6.
Bin1 is found in sarcomere-associated protein complexes. A, shift in the elution profile of Bin1 was observed in differentiating myoblasts indicating an increase in the molecular weight of Bin1 and Bin1-associated protein complexes (fraction = 1 ml). Bin1 associates with sarcomeric actin (B) and myosin (C) in wild type (wt) but not in bin1SH3 (tg) myoblasts. Bin1-myosin and -actin interactions occurred in a temporal fashion throughout the 4-day differentiation time course. d, day; IP, immunoprecipitation. D, c2c12 cells expressing either AdHA-TiKSP or AdGFP were collected at growth (g) or after 2 days in low serum media (differentiation). Lysates were immunoprecipitated with an anti-Bin1 antibody and immunoblotted with an anti-HA tag antibody. A 16-kDa HA-TiKSP fragment was detected after 2 days of differentiation. The reverse immunoprecipitation-immunoblot was performed in E. A 57-kDa Bin1 band was observed close to the heavy IgG band after 2 days in low serum media. F, Bin1 was immunoprecipitated from wild type (wt) and bin1SH3 (tg) cells, and samples were analyzed for the presence of Cdk5. G, c2c12 cells were collected throughout a 4-day differentiation time course, and lysates were incubated with either GST-SH3 or GST and then immunoblotted for Cdk5. + indicates positive control using whole cell lysate.
Clearly, the dynamic range in the molecular weight of Bin1 is attributable in part to the SH3 interaction domain. In a preliminary LC-MS/MS screen of Bin1 SH3-interacting proteins, we identified skeletal muscle myosin and sarcomeric actin as associated proteins within a Bin1-SH3 complex. To further define the scope of myofilament protein interactions with Bin1, a temporal analysis of the actin-Bin1 association was examined in wild type and bin1SH3 cultured myoblasts. Using an α-sarcomeric actin-specific antibody, we observed a positive Bin1-actin interaction in differentiating wild type myoblasts but not in bin1SH3 transgenic myoblasts (Fig. 6B). Transient interactions between actin and Bin1 occurred at a 4-day post low serum induction of differentiation. A similar analysis was undertaken to verify the degree of Bin1-myosin interaction. As with actin, a myosin-Bin1 interaction was only evident in differentiated wild type myoblasts. However, the most pronounced degree of myosin interaction occurred after 1 day and again after 4 days of differentiation (Fig. 6C). These results further corroborate the LC-MS/MS findings and suggest that the Bin1 SH3 domain interacts with actin and myosin in a temporal manner during myoblast differentiation.
At the sarcomere, actin and myosin are in close proximity with the giant myofilament protein titin. Titin has been extensively characterized and is known to contain several key features required for muscle force generation (54, 55). Titin scaffolds numerous myofibrillar proteins, including α-actinin, myomesin, telethonin, components of the thick filament, and several signaling proteins (5, 56). The loss of functional titin gives rise to disordered actin-myosin arrangement and ultimate loss of myofilament assembly. Interestingly, a region important for muscle development and myoblast differentiation was identified in an earlier study of titin structure (57). In this study, a series of lysine-serine-proline (KSP) repeats was identified within the C-terminal region of titin localized at the M-line of the sarcomere. We expressed this 28-residue titin KSP region as an HA-tagged protein (HA-TiKSP) in myoblasts and examined its interaction with Bin1 using both Bin1-immunoprecipitated lysates and HA tag-immunoprecipitated lysates. From these experiments, we observed an HA-TiKSP-Bin1 interaction in myoblasts following 2 days of differentiation (Fig. 6, D and E). However in cycling myoblasts, this interaction was not evident.
KSP is a recognized binding motif for the proline-directed serine-threonine kinase Cdk5, a member of the cyclin-dependent kinase family. Given the apparent Bin1-TiKSP interaction, we hypothesized that Bin1 behaves as a molecular scaffold to mediate phosphorylation at the titin-KSP region by Cdk5 during myogenesis. Therefore, we examined the extent of Bin1-Cdk5 association in wild type and Bin1 SH3-overexpressing myoblasts. Cdk5 was present in Bin1-immunoprecipitated lysates from wild type cells after 1, 2, and 4 days of differentiation (Fig. 6F). However, under similar conditions Cdk5 was absent in bin1SH3 myoblasts. To determine whether this interaction was mediated by the SH3 domain of Bin1, we conducted a series binding assays from myoblast lysates. Interestingly, Cdk5 was present at growth and after differentiation, although the most robust association was observed after 2 days of differentiation (Fig. 6G). Taken together, our results suggest that Bin1 associates with Cdk5 in a temporal manner. The degree of association may be important for mediating myogenically dependent phosphorylation of titin within the KSP region.
DISCUSSION
Although the molecular genetic mechanisms of muscle differentiation and specification have been elucidated, the assembly of a functional myofiber remains to be defined. Here we demonstrate that the SH3 domain of Bin1 mediates the assembly and organization of the skeletal muscle sarcomere through associations with actin and myosin filaments. Transgenic overexpression of the Bin1 SH3 domain resulted in mis-expression of skeletal muscle-specific proteins with consequent disruption of muscle fiber size and ultrastructural organization. We propose that SH3-mediated interactions with actin, myosin, and Cdk5 allow Bin1 to direct the assembly and organization of the skeletal muscle sarcomere.
Bin1 is a protein with diverse cellular functions. Early reports classed Bin1 as a Myc-interacting protein with tumor suppressor properties. and more recent experiments have verified this function (16, 43, 44, 48, 58). In these studies, high expression of Bin1 was identified in skeletal muscle found near actin filaments (16, 18, 34). Localization of Bin1 at the t-tubules of striated muscle implicated it as a modulator of membrane curvature that was mediated through interactions of the BAR domain (25, 34). To date, the role of the SH3 domain has remained a major gap in the understanding of Bin1 function. Our observations suggest that the Bin1 SH3 domain provides a skeletal muscle-specific role for an otherwise ubiquitous protein that maintains a variety of cellular functions.
In this study, we anticipated that endogenous SH3-mediated Bin1 functions would be suppressed by dominant expression of the SH3 transgene. In eukaryotes, SH3 domains share a significant degree of sequence similarity and mediate protein interactions by binding proline-rich sequences with a core PXXP motif (where X represents any amino acid) (59, 60). Despite this generality, there are numerous examples of SH3 domains that bind to alternate sequence motifs on target proteins (61–65). Although the atomic resolution of the Bin1 SH3 region with either actin or myosin was not determined in this study, it is likely that these proteins associate through noncanonical SH3-ligand recognition mechanisms. In our experiments, we demonstrate that the SH3 domain of Bin1 interacts with both actin and myosin and that these interactions occur in a temporally sensitive fashion at early stages of myoblast differentiation.
During myogenesis, sufficient and sequential activation of muscle-specific genes is a requisite for assembling the muscle cyto-architecture. The enlarged fiber diameter and irregular ultrastructure of Bin1 SH3 murine skeletal muscle clearly indicated defects in muscle sarcomere organization. Remarkably, the net accumulation of muscle-specific proteins in the SH3 expressing animals was not affected. Despite an adequate expression of myogenic proteins, the functional components are seemingly unable to assemble into ordered units of the myofibril structure. To date, knowledge of sarcomere assembly has been generally limited to large proteins of the sarcomere complex such as obscurin (14, 15, 66–72).
In our experiments, we demonstrate that endogenous Bin1 associates with the C-terminal KSP region of titin that is highly phosphorylated during skeletal muscle differentiation (57, 73). KSP is a substrate-binding motif for cyclin-dependent kinase 5 (Cdk5) (74–76). In addition to interactions with the titin-KSP region, we also demonstrate Bin1 interactions with Cdk5. This association is more prominent during the early phases of muscle differentiation, and importantly, this interaction is attenuated in muscle lysates from Bin1 SH3 animals.
The in vivo phosphorylation of titin, as directed by Cdk5, is required for proper myofibril arrangement and sarcomere organization. These proteins form a core scaffold binding region and anchor with other proteins in a multicomponent complex. The Bin1-Cdk5 and Bin1-titin interactions that we demonstrate in vitro are consistent with the hypothesis that Bin1 acts as a scaffold protein. However, further experimentation will be required to confirm that Bin1 bridges Cdk5 to titin either through direct or indirect interactions. From the evidence presented in this study, the role of Bin1 appears not to form part of the functional sarcomere but rather to mediate interactions with sarcomeric proteins to ensure efficient assembly and structure of the sarcomere itself.
The sarcomere assembly activity of Bin1 may be complementary to other functions of this adaptor protein. For example, the BAR domain of Bin1 has been implicated in transverse-tubule (T-tubule) biogenesis (25). T-tubules are invaginations of the sarcolemma that occur on either side of the Z-line and penetrate deep into the myocyte to mediate Na+/K+ dynamics to couple muscle excitation and contraction. Accordingly, Bin1 has been localized to the T-tubular system where the BAR domain binds phosphatidylinositol 4,5-bisphosphate (19, 25, 34). Expression of the BAR domain with the 15-amino acid exon10 sequence in non-muscle cells induced both morphological and biochemical features similar to membrane tubules (25). Interestingly, exon10 of Bin1 has been implicated in regulating Bin1 SH3 protein-protein interactions during myogenesis (19). In these experiments, exon10 associated with the SH3 domain and masked its affinity for PXXP ligands. However, binding of phosphoinositides with exon10, as would occur at T-tubules, unmasked the Bin1 SH3 domain. In our experiments, we demonstrate that the SH3 domain alone can influence myogenesis without notable effects on T-tubule morphology. It is conceivable that the localization of Bin1 to T-tubules as directed by the BAR domain and the interaction of exon10 with phosphoinositides permits the Bin1 SH3 domain to participate in further protein-protein interactions that mediate sarcomere organization. In its entirety, these results may further verify the multifunctional and dynamic nature of Bin1.
Our data suggest that Bin1 is retained within an unusually large complex during differentiation. Whether the Bin1-ligand interactions occur as multicomponent complexes or as individual protein-ligand interactions during muscle differentiation remains to be determined. Nonetheless, it is clear that Bin1 behaves as a transient and dynamic adaptor that complexes with sarcomeric proteins at select points during myofilament assembly. Such transient interactions with myofilament proteins have also been demonstrated for the tubulin-associated RING/B-box protein MURF2 during sarcomere assembly (77–79).
Structurally, SH3 domains have a common helical fold, an antiparallel β-sheet region, and variable Arg-Thr (RT) and N-Src loops (reviewed in Refs. 10, 80, 81). NMR spectral analysis and crystal resolution of the Bin1 SH3 structure indicated features that were very unique among SH3 domains that may enhance Bin1 SH3 binding specificity (36, 37). Accordingly, we observed unambiguous anomalies in only the skeletal musculature of Bin1 SH3-expressing animals. Interestingly, homozygous deletion of Bin1 gave rise to substantial cardiomyopathy (22), an effect not observed in this study where only the SH3 domain was targeted. Furthermore, we did not observe the previously reported tumorigenic or cell cycle effects associated with the disruption of Bin1 activity (43, 44, 48). The compartmentalization and spatial distribution of binding partners are likely to contribute to the function and specificity of the Bin1 SH3 as is the case for other proteins with modular interaction domains (reviewed in Refs. 10, 59, 82).
Although Bin1 does not form part of the functional sarcomere itself, we propose that Bin1 serves a role in sarcomere assembly by mediating interactions with sarcomeric proteins. This is in conjunction with the other functions of Bin1 as imposed by its modular protein regions, namely the Myc-binding domain and the BAR domain. Our data also suggest that Bin1 is retained within an unusually large protein complex during myogenesis. Taken together, these observations and the results of this study suggest that myofiber assembly is dependent on transient protein interactions that guide and position sarcomeric proteins at the appropriate time and place.
Supplementary Material
Acknowledgments
We thank Kim Balazsi and Beata Pekalska for skilled technical assistance and Drs. Jeffrey Dilworth and David Picketts for helpful discussions.
This work was supported in part by grants from the Canadian Institutes of Health Research (to L. A. M. and R. K.) and the Muscular Dystrophy Association (to L. A. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Fig. 1.
- SH3
- Src homology 3
- HA
- hemagglutinin
- dpc
- days post-coitum
- GFP
- green fluorescent protein
- MS/MS
- tandem mass spectrometry
- LC
- liquid chromatography
- PI
- propidium iodide.
REFERENCES
- 1.Sabourin L. A., Rudnicki M. A. (2000) Clin. Genet. 57, 16–25 [DOI] [PubMed] [Google Scholar]
- 2.Buckingham M., Bajard L., Chang T., Daubas P., Hadchouel J., Meilhac S., Montarras D., Rocancourt D., Relaix F. (2003) J. Anat. 202, 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao P., Hoffman E. P. (2004) Dev. Dyn. 229, 380–392 [DOI] [PubMed] [Google Scholar]
- 4.Au Y. (2004) Cell. Mol. Life Sci. 61, 3016–3033 [DOI] [PubMed] [Google Scholar]
- 5.Lange S., Ehler E., Gautel M. (2006) Trends Cell Biol. 16, 11–18 [DOI] [PubMed] [Google Scholar]
- 6.Labeit S., Gibson T., Lakey A., Leonard K., Zeviani M., Knight P., Wardale J., Trinick J. (1991) FEBS Lett. 282, 313–316 [DOI] [PubMed] [Google Scholar]
- 7.McElhinny A. S., Schwach C., Valichnac M., Mount-Patrick S., Gregorio C. C. (2005) J. Cell Biol. 170, 947–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinz A., Wang K. (2001) Biochemistry 40, 7903–7913 [DOI] [PubMed] [Google Scholar]
- 9.Kaneko T., Li L., Li S. S. (2008) Front. Biosci. 13, 4938–4952 [DOI] [PubMed] [Google Scholar]
- 10.Li S. S. (2005) Biochem. J. 390, 641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma K., Forbes J. G., Gutierrez-Cruz G., Wang K. (2006) J. Biol. Chem. 281, 27539–27556 [DOI] [PubMed] [Google Scholar]
- 12.Ma K., Wang K. (2002) FEBS Lett. 532, 273–278 [DOI] [PubMed] [Google Scholar]
- 13.Politou A. S., Millevoi S., Gautel M., Kolmerer B., Pastore A. (1998) J. Mol. Biol. 276, 189–202 [DOI] [PubMed] [Google Scholar]
- 14.Young P., Ehler E., Gautel M. (2001) J. Cell Biol. 154, 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bang M. L., Centner T., Fornoff F., Geach A. J., Gotthardt M., McNabb M., Witt C. C., Labeit D., Gregorio C. C., Granzier H., Labeit S. (2001) Circ. Res. 89, 1065–1072 [DOI] [PubMed] [Google Scholar]
- 16.Sakamuro D., Elliott K. J., Wechsler-Reya R., Prendergast G. C. (1996) Nat. Genet. 14, 69–77 [DOI] [PubMed] [Google Scholar]
- 17.Elliott K., Sakamuro D., Basu A., Du W., Wunner W., Staller P., Gaubatz S., Zhang H., Prochownik E., Eilers M., Prendergast G. C. (1999) Oncogene 18, 3564–3573 [DOI] [PubMed] [Google Scholar]
- 18.Wechsler-Reya R., Sakamuro D., Zhang J., Duhadaway J., Prendergast G. C. (1997) J. Biol. Chem. 272, 31453–31458 [DOI] [PubMed] [Google Scholar]
- 19.Kojima C., Hashimoto A., Yabuta I., Hirose M., Hashimoto S., Kanaho Y., Sumimoto H., Ikegami T., Sabe H. (2004) EMBO J. 23, 4413–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren G., Vajjhala P., Lee J. S., Winsor B., Munn A. L. (2006) Microbiol. Mol. Biol. Rev. 70, 37–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wechsler-Reya R. J., Elliott K. J., Prendergast G. C. (1998) Mol. Cell. Biol. 18, 566–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller A. J., Baker J. F., DuHadaway J. B., Ge K., Farmer G., Donover P. S., Meade R., Reid C., Grzanna R., Roach A. H., Shah N., Soler A. P., Prendergast G. C. (2003) Mol. Cell. Biol. 23, 4295–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Razzaq A., Robinson I. M., McMahon H. T., Skepper J. N., Su Y., Zelhof A. C., Jackson A. P., Gay N. J., O'Kane C. J. (2001) Genes Dev. 15, 2967–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zelhof A. C., Bao H., Hardy R. W., Razzaq A., Zhang B., Doe C. Q. (2001) Development 128, 5005–5015 [DOI] [PubMed] [Google Scholar]
- 25.Lee E., Marcucci M., Daniell L., Pypaert M., Weisz O. A., Ochoa G. C., Farsad K., Wenk M. R., De Camilli P. (2002) Science 297, 1193–1196 [DOI] [PubMed] [Google Scholar]
- 26.Nicot A. S., Toussaint A., Tosch V., Kretz C., Wallgren-Pettersson C., Iwarsson E., Kingston H., Garnier J. M., Biancalana V., Oldfors A., Mandel J. L., Laporte J. (2007) Nat. Genet. 39, 1134–1139 [DOI] [PubMed] [Google Scholar]
- 27.Niwa H., Yamamura K., Miyazaki J. (1991) Gene 108, 193–199 [DOI] [PubMed] [Google Scholar]
- 28.Lobe C. G., Koop K. E., Kreppner W., Lomeli H., Gertsenstein M., Nagy A. (1999) Dev. Biol. 208, 281–292 [DOI] [PubMed] [Google Scholar]
- 29.Guy L. G., Kothary R., DeRepentigny Y., Delvoye N., Ellis J., Wall L. (1996) EMBO J. 15, 3713–3721 [PMC free article] [PubMed] [Google Scholar]
- 30.Franzini-Armstrong C. (1991) Dev. Biol. 146, 353–363 [DOI] [PubMed] [Google Scholar]
- 31.Sassoon D., Lyons G., Wright W. E., Lin V., Lassar A., Weintraub H., Buckingham M. (1989) Nature 341, 303–307 [DOI] [PubMed] [Google Scholar]
- 32.Wilkinson D. G., Nieto M. A. (1993) Methods Enzymol. 225, 361–373 [DOI] [PubMed] [Google Scholar]
- 33.Fernando P., Kelly J. F., Balazsi K., Slack R. S., Megeney L. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11025–11030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butler M. H., David C., Ochoa G. C., Freyberg Z., Daniell L., Grabs D., Cremona O., De Camilli P. (1997) J. Cell Biol. 137, 1355–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buckingham M. (2006) Curr. Opin. Genet. Dev. 16, 525–532 [DOI] [PubMed] [Google Scholar]
- 36.Owen D. J., Wigge P., Vallis Y., Moore J. D., Evans P. R., McMahon H. T. (1998) EMBO J. 17, 5273–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pineda-Lucena A., Ho C. S., Mao D. Y., Sheng Y., Laister R. C., Muhandiram R., Lu Y., Seet B. T., Katz S., Szyperski T., Penn L. Z., Arrowsmith C. H. (2005) J. Mol. Biol. 351, 182–194 [DOI] [PubMed] [Google Scholar]
- 38.Tajiri T., Liu X., Thompson P. M., Tanaka S., Suita S., Zhao H., Maris J. M., Prendergast G. C., Hogarty M. D. (2003) Clin. Cancer Res. 9, 3345–3355 [PubMed] [Google Scholar]
- 39.Ge K., Minhas F., Duhadaway J., Mao N. C., Wilson D., Buccafusca R., Sakamuro D., Nelson P., Malkowicz S. B., Tomaszewski J., Prendergast G. C. (2000) Int. J. Cancer 86, 155–161 [DOI] [PubMed] [Google Scholar]
- 40.Hogarty M. D., Liu X., Thompson P. M., White P. S., Sulman E. P., Maris J. M., Brodeur G. M. (2000) Med. Pediatr. Oncol. 35, 559–562 [DOI] [PubMed] [Google Scholar]
- 41.Galderisi U., Di Bernardo G., Cipollaro M., Jori F. P., Piegari E., Cascino A., Peluso G., Melone M. A. (1999) J. Cell. Biochem. 74, 313–322 [PubMed] [Google Scholar]
- 42.Ge K., DuHadaway J., Du W., Herlyn M., Rodeck U., Prendergast G. C. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9689–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang M. Y., Boulden J., Katz J. B., Wang L., Meyer T. J., Soler A. P., Muller A. J., Prendergast G. C. (2007) Cancer Res. 67, 7605–7612 [DOI] [PubMed] [Google Scholar]
- 44.Chang M. Y., Boulden J., Sutanto-Ward E., Duhadaway J. B., Soler A. P., Muller A. J., Prendergast G. C. (2007) Cancer Res. 67, 100–107 [DOI] [PubMed] [Google Scholar]
- 45.Ramalingam A., Farmer G. E., Stamato T. D., Prendergast G. C. (2007) Cell Cycle 6, 1914–1918 [DOI] [PubMed] [Google Scholar]
- 46.Telfer J. F., Urquhart J., Crouch D. H. (2005) Cell. Signal. 17, 701–708 [DOI] [PubMed] [Google Scholar]
- 47.Elliott K., Ge K., Du W., Prendergast G. C. (2000) Oncogene 19, 4669–4684 [DOI] [PubMed] [Google Scholar]
- 48.Muller A. J., DuHadaway J. B., Donover P. S., Sutanto-Ward E., Prendergast G. C. (2004) Cancer Biol. Ther. 3, 1236–1242 [DOI] [PubMed] [Google Scholar]
- 49.Charrasse S., Comunale F., Fortier M., Portales-Casamar E., Debant A., Gauthier-Rouvière C. (2007) Mol. Biol. Cell 18, 1734–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wróbel E., Brzóska E., Moraczewski J. (2007) Eur. J. Cell Biol. 86, 99–109 [DOI] [PubMed] [Google Scholar]
- 51.Zeschnigk M., Kozian D., Kuch C., Schmoll M., Starzinski-Powitz A. (1995) J. Cell Sci. 108, 2973–2981 [DOI] [PubMed] [Google Scholar]
- 52.Hollnagel A., Grund C., Franke W. W., Arnold H. H. (2002) Mol. Cell. Biol. 22, 4760–4770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duhadaway J., Rowe F., Elliott K., Mao N. C., Prendergast G. C. (1999) Cell Adhes. Commun. 7, 99–110 [DOI] [PubMed] [Google Scholar]
- 54.Fukuda N., Granzier H. L., Ishiwata S., Kurihara S. (2008) J. Physiol. Sci. 58, 151–159 [DOI] [PubMed] [Google Scholar]
- 55.Granzier H. L., Labeit S. (2005) Adv. Protein Chem. 71, 89–119 [DOI] [PubMed] [Google Scholar]
- 56.Clark K. A., McElhinny A. S., Beckerle M. C., Gregorio C. C. (2002) Annu. Rev. Cell Dev. Biol. 18, 637–706 [DOI] [PubMed] [Google Scholar]
- 57.Gautel M., Leonard K., Labeit S. (1993) EMBO J. 12, 3827–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DuHadaway J. B., Sakamuro D., Ewert D. L., Prendergast G. C. (2001) Cancer Res. 61, 3151–3156 [PubMed] [Google Scholar]
- 59.Mayer B. J. (2001) J. Cell Sci. 114, 1253–1263 [DOI] [PubMed] [Google Scholar]
- 60.Kay B. K., Williamson M. P., Sudol M. (2000) FASEB J. 14, 231–241 [PubMed] [Google Scholar]
- 61.Moncalián G., Cárdenes N., Deribe Y. L., Spínola-Amilibia M., Dikic I., Bravo J. (2006) J. Biol. Chem. 281, 38845–38853 [DOI] [PubMed] [Google Scholar]
- 62.Tian L., Chen L., McClafferty H., Sailer C. A., Ruth P., Knaus H. G., Shipston M. J. (2006) FASEB J. 20, 2588–2590 [DOI] [PubMed] [Google Scholar]
- 63.Jia C. Y., Nie J., Wu C., Li C., Li S. S. (2005) Mol. Cell. Proteomics 4, 1155–1166 [DOI] [PubMed] [Google Scholar]
- 64.Kami K., Takeya R., Sumimoto H., Kohda D. (2002) EMBO J. 21, 4268–4276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barnett P., Bottger G., Klein A. T., Tabak H. F., Distel B. (2000) EMBO J. 19, 6382–6391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Armani A., Galli S., Giacomello E., Bagnato P., Barone V., Rossi D., Sorrentino V. (2006) Exp. Cell Res. 312, 3546–3558 [DOI] [PubMed] [Google Scholar]
- 67.Kontrogianni-Konstantopoulos A., Catino D. H., Strong J. C., Sutter S., Borisov A. B., Pumplin D. W., Russell M. W., Bloch R. J. (2006) FASEB J. 20, 2102–2111 [DOI] [PubMed] [Google Scholar]
- 68.Bagnato P., Barone V., Giacomello E., Rossi D., Sorrentino V. (2003) J. Cell Biol. 160, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kontrogianni-Konstantopoulos A., Jones E. M., Van Rossum D. B., Bloch R. J. (2003) Mol. Biol. Cell 14, 1138–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bowman A. L., Catino D. H., Strong J. C., Randall W. R., Kontrogianni-Konstantopoulos A., Bloch R. J. (2008) Mol. Biol. Cell. 19, 3782–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bang M. L., Gregorio C., Labeit S. (2002) J. Struct. Biol. 137, 119–127 [DOI] [PubMed] [Google Scholar]
- 72.Nave R., Fürst D. O., Weber K. (1990) FEBS Lett. 269, 163–166 [DOI] [PubMed] [Google Scholar]
- 73.Maruyama K., Endo T., Kume H., Kawamura Y., Kanzawa N., Kimura S., Kawashima S., Maruyama K. (1994) J. Biochem. 115, 147–149 [DOI] [PubMed] [Google Scholar]
- 74.Sharma P., Steinbach P. J., Sharma M., Amin N. D., Barchi J. J., Jr., Pant H. C. (1999) J. Biol. Chem. 274, 9600–9606 [DOI] [PubMed] [Google Scholar]
- 75.Shetty K. T., Link W. T., Pant H. C. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 6844–6848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hellmich M. R., Pant H. C., Wada E., Battey J. F. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10867–10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Spencer J. A., Eliazer S., Ilaria R. L., Jr., Richardson J. A., Olson E. N. (2000) J. Cell Biol. 150, 771–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pizon V., Iakovenko A., Van Der Ven P. F., Kelly R., Fatu C., Fürst D. O., Karsenti E., Gautel M. (2002) J. Cell Sci. 115, 4469–4482 [DOI] [PubMed] [Google Scholar]
- 79.McElhinny A. S., Perry C. N., Witt C. C., Labeit S., Gregorio C. C. (2004) J. Cell Sci. 117, 3175–3188 [DOI] [PubMed] [Google Scholar]
- 80.Yu H., Rosen M. K., Shin T. B., Seidel-Dugan C., Brugge J. S., Schreiber S. L. (1992) Science 258, 1665–1668 [DOI] [PubMed] [Google Scholar]
- 81.Feng S., Kasahara C., Rickles R. J., Schreiber S. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 12408–12415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ladbury J. E., Arold S. (2000) Chem. Biol. 7, R3–R8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.