Abstract
The mechanisms by which mutant variants of Cu/Zn-superoxide dismutase (SOD1) cause familial amyotrophic lateral sclerosis are not clearly understood. Evidence to date suggests that altered conformations of amyotrophic lateral sclerosis mutant SOD1s trigger perturbations of cellular homeostasis that ultimately cause motor neuron degeneration. In this study we correlated the metal contents and disulfide bond status of purified wild-type (WT) and mutant SOD1 proteins to changes in electrophoretic mobility and surface hydrophobicity as detected by 1-anilinonaphthalene-8-sulfonic acid (ANS) fluorescence. As-isolated WT and mutant SOD1s were copper-deficient and exhibited mobilities that correlated with their expected negative charge. However, upon disulfide reduction and demetallation at physiological pH, both WT and mutant SOD1s underwent a conformational change that produced a slower mobility indicative of partial unfolding. Furthermore, although ANS did not bind appreciably to the WT holoenzyme, incubation of metal-deficient WT or mutant SOD1s with ANS increased the ANS fluorescence and shifted its peak toward shorter wavelengths. This increased interaction with ANS was greater for the mutant SOD1s and could be reversed by the addition of metal ions, especially Cu2+, even for SOD1 variants incapable of forming the disulfide bond. Overall, our findings support the notion that misfolding associated with metal deficiency may facilitate aberrant interactions of SOD1 with itself or with other cellular constituents and may thereby contribute to neuronal toxicity.
The sequence of events by which more than 100 mutations in the gene encoding Cu/Zn-superoxide dismutase (SOD1)3 cause familial forms of amyotrophic lateral sclerosis (ALS) is unknown. Studies of purified SOD1 proteins and cellular or rodent models of SOD1-linked ALS suggest that impaired metal ion binding or misfolding of mutant SOD1 proteins in the cellular environment may be related to their toxicity (1–10). Available evidence suggests that partially unfolded mutant SOD1 species could contribute to motor neuron death by promoting abnormal interactions that produce cellular dysfunction (11–16).
In previous studies we characterized physicochemical properties of 14 different biologically metallated ALS SOD1 mutants (17) and demonstrated altered thermal stabilities of these mutants compared with wild-type (WT) SOD1 (18). These “as-isolated” SOD1 proteins, which contain variable amounts of copper and zinc, were broadly grouped into two classes based on their ability to incorporate and retain metal ions with high affinity. WT-like SOD1 mutants retain the ability to bind copper and zinc ions and exhibit dismutase activity similar to the normal enzyme, whereas metal binding region (MBR) mutants are significantly deficient in copper and/or zinc (17, 19). We also observed that ALS-associated SOD1 mutants were more susceptible than the WT enzyme to reduction of the intrasubunit disulfide bond between Cys-57 and Cys-146 (20). The significance of these results is that even WT-like mutants, which exhibit a nearly normal backbone structure (21–23), may be vulnerable to destabilizing influences in vivo. Our group and others subsequently showed that the mutant SOD1 proteins share a susceptibility to increased hydrophobicity under conditions that reduce disulfide bonds and/or chelate metal ions (5) and that similar hydrophobic species exist in tissue lysates from mutant SOD1 transgenic mice (4–6). One consequence of such hydrophobic exposure could be the facilitation of abnormal interactions between the mutant enzymes and other cellular constituents (e.g. chaperones, mitochondrial components, or other targets), which might influence pathways leading to motor neuron death (15, 16, 24–27).
Accumulating evidence suggests that metal deficiency of SOD1 is an important factor that can influence SOD1 aggregation or neurotoxicity (4, 28–33), but the metal-deficient states of SOD1 that are most relevant to ALS remain unclear. Zinc-deficient, copper-replete SOD1 species, which can be produced in vitro by adding copper to SOD1 that has been stripped of its metal ions at acidic pH, were shown to be toxic to motor neurons in culture (28). However, it has not been shown that zinc-deficient, copper-replete SOD1 is produced in vivo as a consequence of ALS mutations, and loading of copper into SOD1 by the copper chaperone for SOD1 (CCS) is not required for toxicity (34, 35). Furthermore, the MBR mutants have a disrupted copper site and have been found to be severely deficient in both zinc and copper (17, 30), yet expression of these SOD1s still produces motor neuron disease (1, 2, 30, 34, 36, 37).
When recombinant human SOD1 was overexpressed in insect cells, we instead observed zinc-replete but copper-deficient species for most WT-like mutants, probably because the capacity of the copper-loading mechanism was exceeded (17). These preparations indicate that zinc can be efficiently incorporated into many WT-like mutants in vivo, and much of it is retained after purification. Furthermore, these copper-deficient biologically metallated proteins may be useful reagents to assess the influence of copper binding upon other properties of SOD1 mutants that may be relevant to their neurotoxicity.
We previously observed that reduction of the Cys-57—Cys-146 disulfide bond facilitates the ability of metal chelators to alter the electrophoretic mobility and to increase the hydrophobicity of SOD1 mutants (5). This is consistent with the known properties of this linkage to stabilize the dimeric interface, to orient Arg-143 via a hydrogen bond from the carbonyl oxygen of Cys-57 to Arg-143-NH2, and to prevent metal ion loss (38–40). However, it remains unclear whether the Cys-57—Cys-146 bond is required to prevent abnormal SOD1 hydrophobic exposure or whether the aberrant conformational change primarily results from metal ion loss. Ablation of the disulfide bond by the experimental (non-ALS) mutants C57S and C146S provides useful reagents to test the relative influence of the disulfide bond and copper binding upon SOD1 properties.
In this study we sought to correlate the consequences of copper deficiency, copper and zinc deficiency, and disulfide reduction upon the hydrodynamic behavior and surface hydrophobicity of WT and representative mutant SOD1 enzymes (Fig. 1A). We quantitated the metal contents of as-isolated SOD1 proteins, detected changes in conformation or metal occupancy using native PAGE to assess their electrophoretic mobility, a measure of global conformational change, and correlated these changes to hydrophobic exposure using 1-anilinonaphthalene-8-sulfonic acid (ANS), which is very sensitive to local conformational changes. ANS is a small amphipathic dye (Fig. 1B) that has been used as a sensitive probe to detect hydrophobic pockets on protein surfaces (41–44). Free ANS exhibits only weak fluorescence that is maximal near 520 nm, but when ANS binds to a hydrophobic site in a partially or fully folded protein, the fluorescence peak increases in amplitude and shifts to a shorter wavelength (42). ANS also has an anionic sulfonate group that can interact with cationic groups (e.g. Arg or Lys residues) through ion-pair formation which may be further strengthened by hydrophobic interactions (43–46).
FIGURE 1.
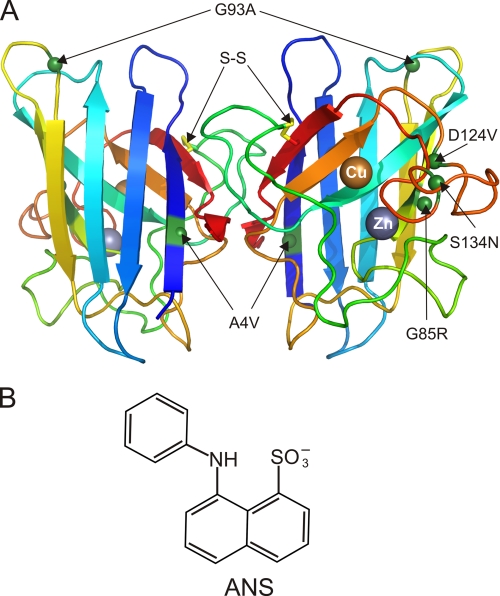
A, WT SOD1 structure showing the position of the C57-C146 intrasubunit disulfide bond (S–S, yellow), bound copper and zinc ions, and ALS mutant residues. The residues altered in A4V, G85R, G93A, D124V, and S134N SOD1s are indicated as green spheres. The backbone of the β-barrel core and the loops is shown in a rainbow color, from blue at the amino terminus to red at the carboxyl terminus. The figure was generated using PyMOL (84) and PDB entry 1HL5 (22). B, chemical structure of ANS fluorophore.
To evaluate further the importance of metal ion binding, we measured spectral changes related to the binding of cobalt and copper to the same SOD1 proteins. We observed that as-isolated WT-like mutants containing zinc could interact with copper ions to produce an electrophoretic mobility and decreased hydrophobicity resembling that of the fully metalated holo-WT SOD1. In contrast, we saw no evidence for copper binding to MBR mutants in a manner that alters their hydrodynamic properties or their hydrophobicity. Our data suggest that binding of both copper and zinc are important determinants of SOD1 conformation and that perturbation of such binding may be relevant to the ALS disease process.
EXPERIMENTAL PROCEDURES
Materials
EDTA was from Invitrogen; sodium phosphate (monobasic and dibasic), potassium phosphate (monobasic and dibasic), KCl, NaCl, glycine, and Tris base were from J. T. Baker Inc.; ANS was from Invitrogen; Tris(2-carboxyethyl)phosphine (TCEP), Coomassie Plus protein assay kit, BCA protein assay kit, and Gel Code blue were from Pierce; HEPES-free acid was from VWR; iodoacetamide, sodium acetate, native PAGE markers, trehalose, CoCl2, CuCl2, ZnCl2, and KOH were from Sigma; 1,4-dithiothreitol, SDS, HCl, methanol, and acetic acid were from EM Science; dual color SDS-PAGE markers, ChelexTM-100 resin, and bromphenol blue were from Bio-Rad. All solutions were prepared using Milli-Q (Millipore) ultrapurified water.
SOD1 Protein Purification
Human WT or ALS-related mutant SOD1 enzymes (A4V, G85R, G93A, D124V, D125H, and S134N) containing biologically incorporated metal ions were isolated from a baculoviral expression system as described previously (17) but without supplementation of the culture medium with additional copper and zinc. Proteins expressed in insect cells were purified by solubility in 60% NH4(SO4)2 followed by sequential hydrophobic interaction and ion exchange gradient chromatography at physiological pH values as described (17). Protein concentrations were determined spectroscopically using a dimeric molar extinction coefficient of 10,800 m−1 cm−1 at 280 nm (47) and also by a modified Bradford assay (Coomassie PlusTM, Pierce). EDTA (0.1 mm) was included in the isolation buffer to inhibit proteases and also to remove any loosely bound metals before metal content analysis. Non-ALS mutant SOD1s lacking the disulfide bond (C57S and C146S) were expressed in the EGy118 strain of Saccharomyces cerevisiae and purified as described previously (48).
Metal Contents and Chelation
The metal contents of all the SOD1 proteins were assessed using inductively coupled plasma mass spectrometry (Agilent 7500ce) and an octopole reaction system utilizing both hydrogen and helium gas for polyatomic interference removal at the UCLA Department of Chemistry and Biochemistry Elemental Analysis and Speciation Facility. The fully metallated holoenzyme is expected to contain two equivalents each of copper and zinc per dimer. ApoWT SOD1, lacking both copper and zinc, was prepared using a demetallation protocol modified from Lyons et al., (49). Briefly, WT SOD1 was dialyzed against 100 mm acetate buffer (pH 3.8) in the presence of 10 mm EDTA for 48 h at 4 °C with 3 changes against 1 liter of buffer every 10–12 h. The protein was then dialyzed overnight at 4 °C against 100 mm acetate buffer (pH 5.5) containing 1 mm EDTA. The buffers used for further dialysis were treated with ChelexTM 100 resin (Bio-Rad) to remove any contaminating metal ions. SOD1 was equilibrated with 2 changes of a buffer containing 20 mm HEPES (pH 7.2) plus 150 mm KCl. After dialysis, the protein was concentrated by centrifugal filtration (Amicon Ultra, Millipore). Metal content analysis was performed as above following the demetallation procedure.
PAGE
Purified SOD1 proteins were separated by gel electrophoresis under native, non-reducing, fully denaturing, or partially denaturing conditions as described previously (5, 17, 20). For native PAGE, proteins were equilibrated for 30 min with the non-denaturing sample buffer (62 mm Tris (pH 6.8), 10% glycerol, 0.01% bromphenol blue) before running on 15% gels (unless otherwise indicated) using ice-cold Tris-glycine buffer (25 mm Tris, 192 mm glycine (pH 8.3)). For non-reducing SDS-PAGE and analysis of disulfide bond status, samples were treated with 5 mm iodoacetamide for 1 h at 25 °C before the addition of non-reducing sample buffer (62 mm Tris (pH 6.8) 10% glycerol, 2% SDS, 0.01% bromphenol blue). Samples were boiled for 3 min and then loaded on 15% gels.
Fluorescence
Steady-state fluorescence spectra were collected using a T-format Spex Fluorolog-3 (Instruments S. A. Inc., Edison, NJ) outfitted with Glan-Thompson polarizers at the Univ. of Massachusetts Medical School Dept. of Biochemistry and Molecular Pharmacology. Samples were excited with vertically polarized light at 370 nm and detected at the optimal angle (54.7°) to eliminate polarization bias from the monochromators and detectors. Bandwidths were 5 and 10 nm for excitation and emission, respectively. Spectra were collected 3 times using a 1-nm step size and a 0.5-s integration time at 25 °C and then averaged. Except for the ANS titration, all experiments were performed at pH 7.2 using 10 μm ANS with 10 μm dimeric SOD1 either in phosphate buffer (20 mm potassium phosphate (pH 7.2) + 150 mm KCl + 200 mm trehalose) or in HEPES buffer (20 mm HEPES (pH 7.2) + 150 mm KCl + 200 mm trehalose). ANS was incubated with proteins for 1 h at 25 °C before collecting the spectra.
ANS Titration
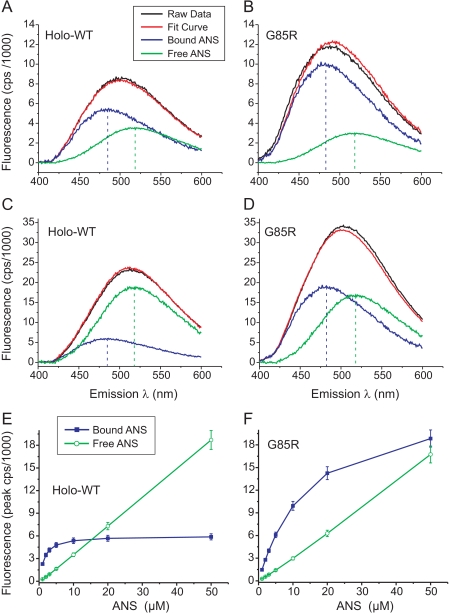
Increasing amounts of ANS (0, 0.5, 1, 2, 3, 5, 10, 20, 50, and 100 μm) were added to 10 μm dimeric SOD1 proteins (holo-WT or G85R), and the mixtures were equilibrated at 25 °C for 10 min before acquiring each spectrum. Titration of ANS into the buffer without protein yielded a fluorescence peak at 518 ± 1.5 nm for free ANS that was independent of the ANS concentration, and these curves provided a constraint for subsequent fitting. ANS titration spectra were fitted globally to a single site binding model ([P] + [ANS] ↔ [P/ANS]) using a Levenberg-Marquardt nonlinear least-squares algorithm implemented in Savuka Version 6.2 (50, 51). ANS fluorescence values at emission wavelengths from 400–600 nm were fitted simultaneously with the Kd as a global variable parameter. The free ANS fluorescence intensity at a given wavelength was a local parameter fixed to the experimentally determined free ANS fluorescence spectrum. A global scaling parameter for the free ANS spectrum was used to account for differences in the system gain and small (<5%) variations in the determination of the ANS concentration. The bound ANS fluorescence intensity at each wavelength (for 201 values using 1-nm increments) was treated as a local variable parameter to be optimized during the fit. In total, 203 parameters (201 local and 2 global) were simultaneously fitted to 201 binding curves. This analysis yielded a Kd value for ANS binding and allowed us to deconvolute the spectra for bound versus free ANS fluorescence. 10 μm ANS was found to be optimal to ensure detection of weak binding to holo-WT SOD1 yet avoid excessive fluorescence from free ANS (Fig. 3).
FIGURE 3.
Titration of ANS binding to holo-WT and G85R mutant SOD1s. 10 μm concentrations of dimeric SOD1 proteins (holo-WT or G85R) were incubated with increasing amounts of ANS (0, 0.5, 1, 2, 3, 5, 10, 20, 50, and 100 μm), and fluorescence spectra were obtained upon excitation at 370 nm. The spectra after subtraction of fluorescence in the absence of ANS are shown as solid black traces in panels A and B (10 μm ANS) and C and D (50 μm ANS) and are not shown for the other ANS concentrations. For each SOD1 protein, all nine ANS spectra were fitted simultaneously to a single-site binding model (see “Experimental Procedures”). For each concentration of ANS, the fit curve (red traces) was deconvolved into fluorescence attributable to bound ANS (blue traces) versus free ANS (green traces). The vertical dashed lines indicate λmax for ANS bound to holo-WT (A and C, 484 nm), ANS bound to G85R (B and D, 482 nm), and free ANS (A–D, 518 nm). The peak fluorescence for bound versus free ANS as a function of ANS concentration is summarized for holo-WT SOD1 (E) and G85R SOD1 (F). Error bars indicate ±S.D. Metal contents for the SOD1 proteins are as indicated in Fig. 2.
Metal Ion Titrations
Titrations of the SOD1 proteins with solutions of copper and cobalt chloride were monitored by UV-visible spectroscopy using a Hewlett Packard 8453 spectrophotometer (Wilmington, DE). Five scans were averaged for each spectrum using a Hewlett Packard UV-visible Chemstation (Version A.06.04) and were analyzed using Origin (MicroCal) software. A volume of 150 μl of 0.15–0.25 mm dimeric SOD1 was titrated with 2 eq/dimer of a 10 mm metal chloride solution. All titrations were in HEPES buffer (20 mm HEPES (pH 7.2) + 150 mm KCl). After the addition and mixing of metal ions, the protein solutions were centrifuged at 20,000 × g for 5 min at 4 °C before spectral observations and were maintained at 4 °C between successive scans.
RESULTS
We purified the WT and five familial ALS-associated human SOD1 mutants (A4V, G85R, G93A, D124V, and S134N) from an insect cell expression system (17). These mutants (Fig. 1A) were selected because (i) they are located in distinct regions of the enzyme, (ii) many of their physicochemical and structural properties have been characterized, and (iii) they include variants shown to produce an ALS phenotype in transgenic rodent models (A4V, G85R, and G93A) (1, 7, 52). A4V perturbs the packing within the β-barrel interior near the dimer interface (53). G85R disrupts conserved hydrogen bonding and alters the metal binding loop IV conformation (54), G93A perturbs a tight turn at one pole of the β-barrel and increases the mobility of loops III and V (55), D124V is expected to prevent hydrogen bonding important for metal ion coordination, and S134N in the electrostatic loop VII also decreases metal binding (3, 17). A4V and G93A belong to a subset of ALS mutants that exhibit several “WT-like” properties, including relatively preserved metal binding and enzymatic activity, whereas G85R, D124V, and S134N are “metal binding region” mutants that behave similarly to the WT apoprotein with regard to decreased enzymatic activity and altered structure of loops IV and VII (17, 19, 55).
WT and mutant SOD1s were expressed in Sf21 insect cells and purified as previously described (17), except here we did not supplement the culture medium with additional copper and zinc. We verified the purity of the proteins by SDS-PAGE and by electrospray mass spectrometry (17, 20). These preparations of SOD1 proteins were generally deficient in copper and variably deficient in zinc (Fig. 2A), as measured by inductively coupled plasma mass spectrometry.
FIGURE 2.
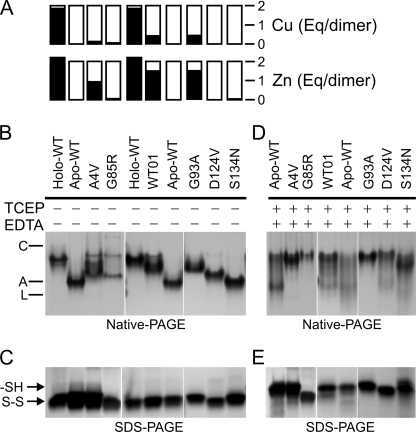
Metal contents and electrophoretic mobilities of purified SOD1s. A, copper and zinc contents by inductively coupled plasma mass spectrometry, where fully metallated holo-SOD1 is expected to contain 2 eq of each metal per dimer. Holo-WT, 1.85 copper, 2.2 zinc; apoWT, 0.02 copper, 0.04 zinc; as-isolated WT01, 0.57 copper, 1.69 zinc; A4V, 0.16 copper, 0.97 zinc; G85R, 0.09 copper, 0.09 zinc; G93A, 0.48 copper, 1.58 zinc; D124V, 0.01 copper, 0.03 zinc; S134N, 0.0 copper, 0.08 zinc. B, native PAGE of SOD1 proteins (10 μg/lane) after incubation at 25 °C for 16–18 h without exposure to TCEP or EDTA. Markers were carbonic anhydrase (C), albumin (A), and lactalbumin (L). C, non-reducing SDS-PAGE of WT and mutant SOD1 proteins from B after tagging free SH groups with iodoacetamide (5 μg/lane). Arrows indicate the position of the reduced (SH) and oxidized (S-S) SOD1s (6). D, native PAGE of SOD1 proteins (10 μg/lane) after incubation at 25 °C for 16–18 h with 20 mm TCEP and 1 mm EDTA. E, non-reducing SDS-PAGE after iodoacetamide tagging of SOD1 proteins treated with 20 mm TCEP and 1 mm EDTA. Panels B and D were from a single gel, as were panels C and E.
Disulfide Reduction and Metal Deficiency of WT and Mutant SOD1s Impeded Electrophoretic Mobility
The migration of proteins during native PAGE is influenced by contributions from net charge and hydrodynamic volume, thus enabling it to detect both conformational changes and differences in non-covalent binding events. For SOD1 in particular, native PAGE can differentially resolve metallated species, including those produced upon expression of these proteins in insect cells (17, 20). We first compared the mobilities of fully metallated SOD1 holoprotein from human erythrocytes (holo-SOD1), the SOD1 apoprotein lacking both copper and zinc (apoSOD1), and the as-isolated WT (WT01) and mutant (A4V, G93A, G85R, D124V, S134N) SOD1 preparations under native conditions (Fig. 2B). The mobility of holo-WT SOD1 was slower than that of disulfide-oxidized apoWT SOD1, as expected because although both proteins are dimeric (48), apoSOD1 has a greater net negative charge. As-isolated WT01, A4V, and G93A SOD1s migrated as species with mobilities intermediate between those of apo- and holo-SOD1, consistent with their copper deficiency and the presence of differentially metallated species (Fig. 2B). Metal binding region mutants D124V and S134N, which were deficient in both copper and zinc, migrated near the apoWT protein, with D124V migrating slower because the mutation caused additional loss of a negative charge. Also consistent with this, a major component of the metal-deficient G85R mutant preparation migrated similarly to D124V on native PAGE (Fig. 2B).
Migration of SOD1 during native PAGE is expected to be sensitive to conformational changes that may occur upon either disulfide reduction or loss of bound metal ions or both. Therefore, to determine the status of the disulfide bond in our preparations by an independent method, we treated the proteins with iodoacetamide to tag free thiol groups and then separated them by non-reducing SDS-PAGE as described (6). The results of Fig. 2C confirmed that the disulfide bond was intact for all of our as-isolated SOD1s.
Our previous study showed that exposure to disulfide-reducing agents slows the mobility of ALS mutants during native PAGE and can expose the normally buried Cys-6 residue at physiological pH (20). Furthermore, the combination of a reducing agent and a metal chelator was most effective to unmask an aberrantly increased surface hydrophobicity of ALS SOD1 mutants (5), suggesting that disulfide reduction facilitated the extraction of bound metals from the SOD1s and promoted either SOD1 monomerization or partial unfolding. In the present study we, therefore, exposed the as-isolated SOD1 proteins to 20 mm TCEP and 1 mm EDTA to determine whether PAGE could detect conformational changes that might correlate with increased hydrophobicity under these conditions. In Fig. 2D, the electrophoretic patterns for the metal-deficient G85R, D124V, and S134N mutants shifted toward a slower migration. Because these mutants already lacked detectable metal binding before exposure to the chelator, the mobility shift was most consistent with a conformational change attributable to disulfide reduction and known formation of unfolded apomonomers under these conditions (20, 48, 56), which would increase the hydrodynamic volume and decrease the mobility. It has also been shown by ultracentrifugation as well as size exclusion chromatography that in the absence of bound metals the disulfide bond is required to maintain the dimeric state (48, 57). We, therefore, expected that metal-deficient, disulfide-reduced SOD1s (whether WT or mutant) would be monomeric, with disordered loops and a partially open β-barrel, as was observed by solution NMR for a monomeric apoSOD1 mutant (58). Consistent with this, the A4V and G93A mutants treated with TCEP and EDTA also migrated at the same rate as metal-deficient SOD1 mutants (Fig. 2D), suggesting that they were converted to apo species that were either monomeric or partially unfolded to a similar extent (59). Both apoWT and the zinc-containing WT01 SOD1 also converted to these slowly migrating species, although these proteins appeared more resistant than A4V, G85R, and G93A SOD1s. Iodoacetamide tagging followed by non-reducing SDS-PAGE of the treated samples from Fig. 2D confirmed that the disulfide bond was reduced for all of the proteins (Fig. 2E). Overall, these results indicated that treatment of WT and mutant SOD1s with TCEP + EDTA produced conformational changes that were consistent with disulfide reduction and demetallation of the enzymes.
ANS Binding to SOD1 Mutants
Normally folded SOD1 is a highly water-soluble enzyme with hydrophilic surface properties. To determine whether the slowed migration we observed for ALS mutants during PAGE (a measure of global conformational change) correlated with a change in surface hydrophobicity, we measured the fluorescence properties of ANS binding to the SOD1 proteins. ANS is a hydrophobic fluorophore that is very sensitive to local conformational changes. We performed titrations of 0.5–100 μm ANS in the presence of 10 μm (dimeric concentration) of holo-WT or G85R SOD1s to identify an optimal ANS concentration that would allow us to detect protein-bound ANS fluorescence while minimizing the signal contributed by free ANS (Fig. 3). We observed that the peak fluorescence attributable to ANS bound to holo-WT SOD1 saturated by 10 μm ANS, whereas the peak fluorescence of free ANS continued to increase linearly with ANS concentration (Fig. 3E). For the G85R mutant, however, the fluorescence attributable to bound ANS was greater compared with WT at >5 μm ANS (Fig. 3F). Although saturation of ANS binding to the G85R SOD1 occurred above 50 μm ANS, the signal attributable to free ANS also increased at high ANS concentrations. We considered 10 μm ANS to be a reasonable ANS concentration for the assay because it ensured sensitivity to detect ANS binding even to the holo-WT enzyme and specificity to detect the blue shift in the fluorescence peak upon binding.
In Fig. 4A we observed that the fluorescence emission spectrum for a solution containing 10 μm holo-WT SOD1 and 10 μm ANS (solid black curve) was nearly identical to the spectrum for ANS in buffer alone (dotted curve) and exhibited a λmax at 507 nm. This suggested only negligible ANS binding to the fully metallated SOD1 enzyme. In contrast, the as-isolated WT01 SOD1 showed a shift in the fluorescence peak to 472 nm and a 7.7-fold increase in the magnitude of the peak fluorescence compared with holo-WT (after subtracting the fluorescence signal of ANS in the buffer without protein). This was consistent with binding of ANS to one or more accessible sites in copper-deficient SOD1. Pretreatment of WT01 SOD1 with 20 mm TCEP and 1 mm EDTA did not alter the fluorescence spectrum (Fig. 4A, dashed line), indicating that disulfide reduction had no further detectable effect upon ANS binding to the copper-deficient WT SOD1.
FIGURE 4.
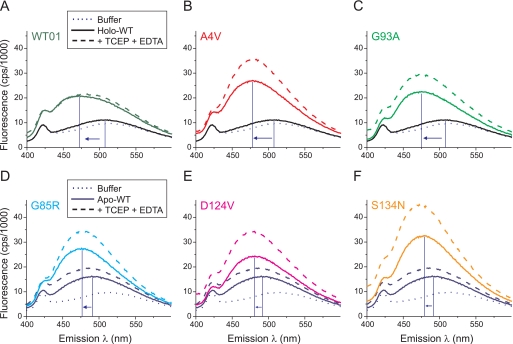
SOD1 mutants exhibited increased intensity and blue shift of ANS fluorescence compared with WT SOD1. WT and mutant SOD1s with metal contents as indicated in Fig. 2A were incubated for 16–18 h at 25 °C with or without 20 mm TCEP + 1 mm EDTA. Each protein (10 μm) was then incubated with 10 μm ANS in phosphate buffer (pH 7.2) for 1 h at 25 °C before collecting fluorescence spectra elicited by excitation at 370 nm. A–C, as-isolated, copper-deficient WT01, A4V, and G93A SOD1s (solid colored lines) showed increased ANS fluorescence and blue shift (arrows) compared with holo-WT SOD1 (solid black line) or ANS in buffer without protein (dotted line). Incubation with TCEP + EDTA (dashed lines) increased the ANS fluorescence of A4V and G93A mutants but not that of WT01 SOD1. D–F, as-isolated metal-deficient mutants G85R, D124V, and S134N (solid colored lines) exhibited increased ANS fluorescence and blue shift (arrows) compared with apoWT SOD1 (solid dark blue line). Incubation with TCEP + EDTA (dashed lines) increased the ANS fluorescence of the mutants more than that of apoWT SOD1.
For two ALS mutant preparations containing zinc but lacking appreciable copper (A4V and G93A), we observed a similar shift of λmax (to 478 and 475 nm, respectively) and an increase in fluorescence (by 11.9- and 8.9-fold, respectively, after subtraction of ANS in buffer alone) compared with the holo-WT SOD1. Treatment of A4V or G93A proteins with TCEP + EDTA produced a 48 and 51% increase, respectively, in the peak fluorescence compared with untreated proteins, consistent with a greater vulnerability to hydrophobic exposure for these mutants compared with WT01 SOD1 (Fig. 4, A–C). Despite a similar slowing of global mobility for the WT01, A4V, and G93A proteins under these conditions (Fig. 2D), the increased ANS fluorescence for the mutants suggested that the effects of these substitutions on the backbone structure (53, 55) or other localized consequences could be distinguished by the ANS probe under solution conditions.
Our preparation of WT SOD1 from which copper and zinc were removed (apoWT) showed a small shift in λmax to 491 nm and 4.5-fold increase in fluorescence compared with holo-WT SOD1 after subtraction of ANS in buffer alone (compare Figs. 4, A and D, solid lines). Upon treatment of apoWT SOD1 with TCEP + EDTA, the fluorescence increased by 1.5-fold, and λmax shifted to 487 nm (Fig. 4D, dashed line). Under these conditions we expected the disulfide-reduced apoWT SOD1 to monomerize (48, 57) and to exhibit disorder of loops IV and VII (22, 39, 40). When disulfide-intact species of the three metal binding region SOD1 mutants (G85R, D124V, and S134N) were analyzed under the same conditions, we observed a larger shift in λmax (to 476, 481, and 477 nm, respectively) and greater ANS fluorescence (by 2.7-, 2.3-, and 3.5-fold, respectively) compared with that of the apoWT protein (Fig. 4, D–F, solid lines). Upon treatment of each mutant with TCEP + EDTA, the relative fluorescence increased by 1.4-, 1.7-, and 1.5-fold, respectively (Fig. 4, D–F, dashed lines). Overall, these results suggested that (i) both WT-like and metal binding region ALS mutants perturb the conformation of SOD1 to favor interaction with ANS and (ii) ALS mutant species are especially vulnerable to abnormal ANS binding under conditions of disulfide reduction or metal deficiency.
Titration of Copper and Cobalt into SOD1s at Physiological pH
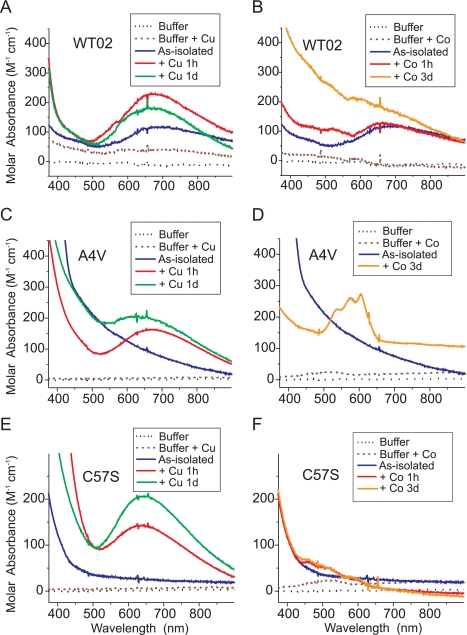
The results of Fig. 4 suggested that impaired metal ion binding in the context of an ALS mutation may contribute to exposure of hydrophobic residues capable of binding ANS. We hypothesized that if exogenously added copper or zinc can bind to the appropriate sites in WT or a subset of mutant SOD1s at physiological pH, then such binding might minimize the structural aberration that produced abnormally increased ANS fluorescence. Because previous metal titrations into SOD1 were performed using apoproteins at pH 5.5 (49, 60–62), we used UV-visible spectroscopy to test whether metals added to as-isolated WT and mutant SOD1s at pH 7.2 could bind to their correct sites (Fig. 5). Binding of Cu2+ to the copper site in SOD1 produces a peak absorbance near 670 nm (60). Because Zn2+ is spectroscopically silent, we measured the binding of Co2+, which has similar chemical properties and produces characteristic spectra at 500–650 nm when bound to the zinc site in SOD1 (49, 60).
FIGURE 5.
UV-visible absorption spectra for as-isolated and metal-substituted SOD1 derivatives. UV-visible spectra of as-isolated WT02 SOD1 and WT02 incubated for 1 h or 1 day (d) at 4 °C with 2 eq/dimer of Cu2+ (A) or 2 eq/dimer of Co2+ (B). As-isolated A4V SOD1 and A4V incubated as above with 2 eq/dimer of Cu2+ (C) or 2 eq/dimer of Co2+ (D). As-isolated C57S SOD1 and C57S incubated as above with 2 eq/dimer of Cu2+ (E) or 2 eq/dimer of Co2+ (F). All proteins were 150–250 μm in buffer containing 20 mm HEPES (pH 7.2) + 150 mm KCl. The metal contents (eq/dimer) of as-isolated SOD1 proteins were: WT02, 0.44 copper and 1.42 zinc; A4V, 0.16 copper, 0.97 zinc; C57S, 0.03 copper, 2.25 zinc.
We observed that as-isolated WT02 SOD1 (1.42 eq/dimer of zinc and 0.44 eq/dimer of copper) readily bound additional Cu2+ in the copper site, as evidenced by augmentation of the absorbance at 670 nm (Fig. 5A). If appreciable Cu2+ had instead bound to the zinc site, the absorbance peak would have been near 810 nm (49, 60, 62). In Fig. 5B, we observed that this WT SOD1 preparation did not appear to bind appreciable Co2+, as evidenced by the lack of signature absorbance peaks in the range of 500–650 nm. This suggested that the zinc site was nearly fully occupied by Zn2+, which Co2+ could not displace.
Fig. 5C shows that the as-isolated A4V SOD1 (0.97 eq/dimer of zinc and 0.16 eq/dimer of copper) was capable of binding added Cu2+ to the copper site at pH 7.2, similar to WT SOD1. An increase in absorbance at 500 nm was observed over 24 h that suggested possible copper shifting to the zinc site or other changes occurring at the copper site (60). Furthermore, this preparation of A4V bound Co2+, as evidenced by the absorbance peaks at 500–650 nm (Fig. 5D), which was consistent with incomplete occupancy of the zinc site in the as-isolated preparation.
Because the intrasubunit disulfide bond between Cys-57 and Cys-146 may affect the ability to load Cu2+ into SOD1, we next tested whether genetic ablation of the disulfide linkage might affect metal binding. Two human SOD1 mutants that prevent disulfide bond formation (C57S and C146S) were expressed in yeast and purified (48). For C57S (2.25 eq/dimer of zinc and 0.03 eq/dimer of copper), we detected binding of additional Cu2+ to the copper site (Fig. 5E). For this mutant, though, the absorbance peak was shifted to 635 nm, indicating a more square planar copper coordination geometry compared with WT SOD1. Only minimal Co2+ binding was observed (Fig. 5F). These results suggested that this preparation of C57S could accommodate Cu2+ to the copper site and maintained high Zn2+ occupancy of the zinc site, as was also observed for C146S SOD1 (not shown). Furthermore, no high affinity copper binding was detected for G85R SOD1 (not shown).
Decreased Interaction of ANS with SOD1 upon Metal Loading
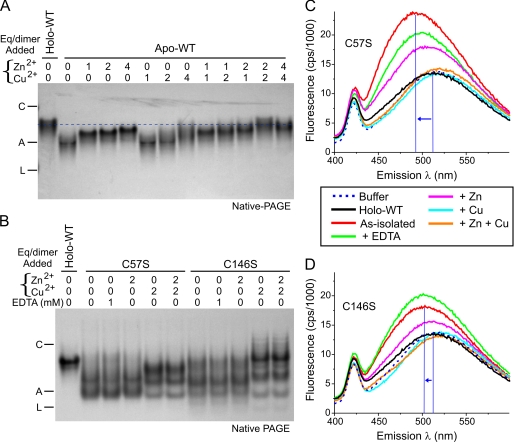
The results of Fig. 5 suggested that copper and zinc ions added at pH 7.2 can occupy their appropriate sites in as-isolated WT SOD1 and some of the ALS mutants. We hypothesized that repletion of metal ions may alter the SOD1 electrophoretic mobility in a manner that correlates with decreased abnormal hydrophobic exposure. To define the effects of metallation upon SOD1 mobility during native PAGE, we first titrated zinc and copper ions into apoWT SOD1 at pH 7.2 (Fig. 6A). ApoWT SOD1 treated with only 1.0 eq/dimer of Zn2+ showed an electrophoretic mobility intermediate between the untreated apoWT and the holo-WT protein (Fig. 6A, first through third lanes), as was expected for the decreased net negative charge. The addition of 2.0 or 4.0 eq/dimer of Zn2+ to apoWT SOD1 caused only a modest additional slowing of mobility, consistent with previous studies showing that binding of the first Zn2+ to apoSOD1 is a major thermodynamic event (63) that increases stability of the protein (18, 48, 56, 64). The zinc loading was also consistent with calorimetry of bovine SOD1 in which addition of 4.0 eq/dimer of Zn2+ increased the melting temperature (Tm) of the enzyme to 79 °C, which still remained lower than the Tm of 96 °C for bovine holo-WT SOD1 (65). In the absence of prior zinc loading, however, exposure of apoWT SOD1 to either 1.0 or 2.0 eq/dimer of Cu2+ did not substantially alter the SOD1 mobility (Fig. 6A, sixth and seventh lanes). This was consistent with studies suggesting that Zn2+ binding may be a prerequisite for efficient copper loading into SOD1 (66). We observed that prior loading of apoSOD1 with 2.0 eq/dimer of Zn2+ followed by the addition of 2.0 eq/dimer of Cu2+ was most effective to produce a species with an electrophoretic mobility that closely resembled holo-WT SOD1 (Fig. 6A, compare first and twelfth lanes).
FIGURE 6.
Metal ion binding to SOD1 proteins slowed the electrophoretic mobility and decreased ANS fluorescence. A, apoWT SOD1 was incubated with Zn2+ (0–4 eq/dimer) for 1 h followed by Cu2+ (0–4 eq/dimer) for 24 h at 25 °C. The mobility for these species during native PAGE (18% gel, 5 μg of protein/lane) was compared with that of holo-WT SOD1 (dashed line). Markers were carbonic anhydrase (C), albumin (A), and lactalbumin (L). B, as-isolated C57S and C146S SOD1 mutants incapable of forming the native disulfide bond were incubated with 0–2 eq/dimer of Zn2+ or 1 mm EDTA for 1 h followed by 0–2 eq/dimer of Cu2+ for 24 h at 25 °C. Electrophoretic mobility was visualized by native PAGE (15% gel, 5 μg/lane) and compared with that of holo-WT SOD1. The metal contents (eq/dimer) of as-isolated SOD1 proteins were: C57S, 0.03 copper, 2.25 zinc; C146S, 0.19 copper, 1.96 zinc. C, ANS fluorescence was measured as in Fig. 4. The ANS spectrum for as-isolated C57S mutant SOD1 (10 μm) was increased and shifted toward shorter wavelengths (blue arrow) compared with holo-WT SOD1. C57S SOD1 that was treated with 2 eq/dimer Zn2+ or Cu2+ or 1 mm EDTA as in B showed a decrease in ANS fluorescence and a shift toward longer wavelengths. D, as-isolated C146S mutant SOD1 was treated as in C before collecting fluorescence spectra.
We next tested whether two experimental (non-ALS) SOD1 mutants, C57S and C146S, which prevent disulfide bond formation (48), also alter the electrophoretic mobility upon addition of metal ions. Metal analysis by inductively coupled plasma mass spectrometry of these as-isolated preparations showed that they were fully zinc-loaded (2.0–2.3 eq/dimer) but copper-deficient (0.03–0.19 eq/dimer). For as-isolated C57S, we observed two main electrophoretic species on native PAGE with substantially accelerated migration compared with holo-WT SOD1 (Fig. 6B, first and second lanes). Our C57S preparation was zinc-replete and thereby expected to be dimeric and well folded (48); the migration differences in Fig. 6B, therefore, depend mainly on charge differences caused by differential metal occupancy. Exposure to 1 mm EDTA did not alter the electrophoretic pattern, supporting the likelihood that the metal ions present were tightly bound. The addition of an extra 2.0 eq/dimer of Zn2+ also did not change the mobility of the observed species, consistent with full zinc-site occupancy for these proteins. In contrast, upon the addition of 2.0 eq/dimer of Cu2+ to as-isolated C57S, the fastest migrating species was decreased in abundance, and a new species with slower mobility appeared (Fig. 6B, fifth lane), consistent with copper binding to the copper site as observed in the spectra of Fig. 5E. Similar findings were obtained for the C146S SOD1 mutant (Fig. 6B, seventh through eleventh lanes). Overall, these results suggested that Cu2+ can bind efficiently to zinc-loaded SOD1 and can alter its mobility during native PAGE without the requirement of an intact disulfide bond.
The effects of metal ion loading to affect SOD1 electrophoretic mobility raised the possibility that zinc or copper binding might alter the SOD1 surface hydrophobicity. We next measured ANS fluorescence of the as-isolated C57S (Fig. 6C) and C146S (Fig. 6D) SOD1s before and after treatment with EDTA, Zn2+, Cu2+, or Zn2+ + Cu2+ ions. Cu2+ is a paramagnetic ion known to quench the fluorescence from tryptophan and tyrosine residues (67, 68). As it is not known if the paramagnetic effects of copper can similarly quench ANS fluorescence, we measured the effect of up to 15-fold higher concentration of Cu2+ ions in the buffer (up to 300 μm) and observed that copper did not quench or decrease ANS fluorescence (data not shown). Furthermore, ANS has been successfully used as a hydrophobic fluorophore for other experiments that employed paramagnetic ions such as copper and iron (69–71).
In Fig. 6C, ANS incubated with holo-WT SOD1 (which contained 2.2 zinc and 1.85 copper per dimer) showed minimal fluorescence above the background observed in the absence of protein, indicating no significant ANS binding to a hydrophobic site. In contrast, ANS incubated with the copper-deficient as-isolated C57S SOD1 showed increased fluorescence intensity and a shift of the λmax from 513 to 493 nm (Fig. 6C), consistent with increased hydrophobic exposure compared with holo-WT SOD1. Exposure to 1 mm EDTA slightly decreased the ANS fluorescence of C57S SOD1. The addition of an extra 2.0 eq/dimer of Zn2+ to the as-isolated C57S SOD1 (which already contained 2.3 eq/dimer of Zn2+) produced a decrease in ANS fluorescence and a shift of λmax to 506 nm. Moreover, the most effective treatment to abrogate the ANS binding was the addition of 2.0 eq/dimer of Cu2+ to C57S SOD1, which produced a fluorescence spectrum very similar to holo-WT SOD1 (Fig. 6C). We obtained similar results using the as-isolated C146S mutant, which was less completely copper-deficient and contained 0.19 eq/dimer Cu2+; for this mutant, EDTA slightly increased the ANS fluorescence, possibly by chelating Cu2+ that was loosely bound (Fig. 6D). Overall, these findings demonstrated that incompletely metallated SOD1 exhibits hydrophobic properties, as detected by ANS fluorescence that can be fully abrogated by loading with a total of 2.0 Zn2+ + 2.0 Cu2+ per dimer and that such hydrophobic masking does not require the presence of the intrasubunit disulfide bond.
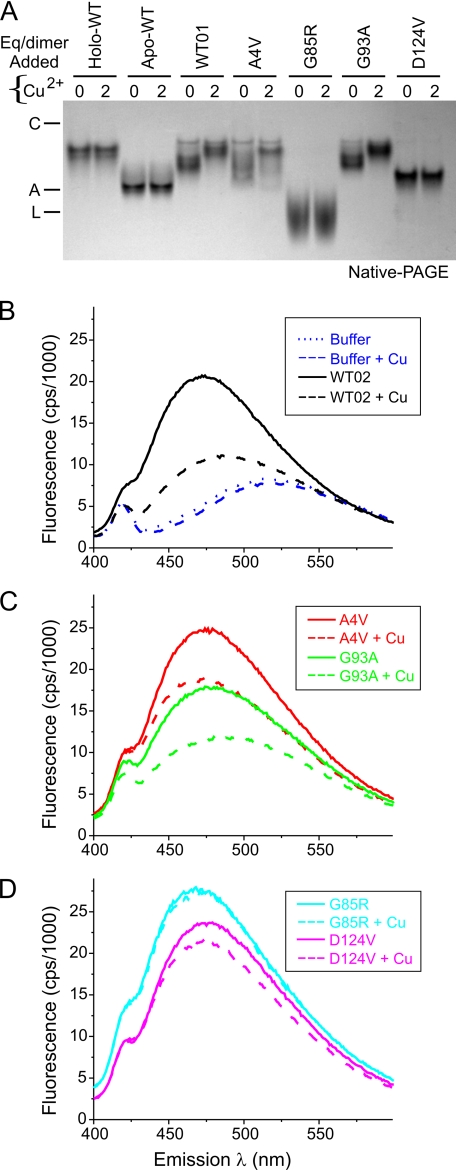
The as-isolated WT and ALS mutant SOD1s either contained significant zinc but little copper (WT, A4V, and G93A) or were deficient in both zinc and copper (G85R, D124V, and S134N). For these proteins we correlated changes in mobility on native PAGE and changes in ANS fluorescence that resulted upon exposure to 2.0 eq/dimer of Cu2+ (Fig. 7). We measured the background ANS fluorescence of buffer with and without copper and found no change in fluorescence intensity (Fig. 7B). When copper was added to WT, A4V, and G93A SOD1s that contained zinc and could accept copper, the mobility was slowed (Fig. 7A), and the ANS fluorescence decreased for WT (Fig. 7B) and for the A4V or G93A mutants (Fig. 7C). When 2.0 eq/dimer of Cu2+ was added to G85R and D124V, which each contained <0.1 eq/dimer of Zn2+, no change in the mobility (Fig. 7A) and only a small decrease in ANS fluorescence (Fig. 7D) were observed. Similarly, apoWT SOD1 did not show any change in electrophoretic mobility when copper was added in the absence of prior zinc loading (Fig. 7A). Overall, these results supported the hypothesis that full occupancy of both zinc and copper sites may be required to minimize ANS fluorescence and hydrophobic exposure of mutant SOD1s.
FIGURE 7.
Altered electrophoretic mobility of as-isolated WT-like SOD1 mutants upon incubation with Cu2+ correlated with decreased ANS fluorescence. A, native PAGE of SOD1 proteins (5 μg/lane) after incubation with 0–2 eq/dimer of Cu2+ for 24 h at 25 °C. As-isolated WT, A4V, and G93A SOD1s exhibited slower mobility after exposure to Cu2+, whereas MBR mutants G85R and D124V were unaffected. The metal contents are shown in Fig. 2A. Markers were carbonic anhydrase (C), albumin (A), and lactalbumin (L). B, ANS fluorescence of as-isolated WT SOD1 incubated with 0–2 eq/dimer of Cu2+ for 24 h at 25 °C. For this preparation of WT SOD1 (WT02), the starting metal content (eq/dimer) was 0.44 copper and 1.42 zinc. The addition of Cu2+ decreased the ANS fluorescence in a manner that correlated with the slowing of mobility observed for more highly metallated SOD1 in A. ANS and SOD1 were 10 μm each in HEPES buffer (pH 7.2, see “Experimental Procedures”). C, ANS fluorescence decreased in parallel with the slowed mobility for the as-isolated WT-like mutants A4V and G93A after incubation with 2 eq/dimer of Cu2+ for 24 h at 25 °C. D, ANS fluorescence of MBR mutants G85R and D124V was minimally affected by incubation with Cu2+.
DISCUSSION
A central unanswered question regarding ALS pathogenesis related to SOD1 mutants is, How do so many subtle and distinct perturbations of the enzyme produce a strikingly similar downstream toxicity to motor neurons? The properties of ALS-related SOD1 mutants may be altered directly by each mutant residue or more globally as a consequence of changes in disulfide bond status or metal ion binding in vivo. Novel findings of the present study are (i) as-isolated SOD1 mutants capable of incorporating exogenous copper, zinc, or cobalt ions appear to bind metals to the correct sites at pH 7.2, (ii) metal occupancy of both the zinc and copper sites of SOD1 can prevent abnormal hydrophobic exposure as detected by ANS fluorescence, (iii) an intact disulfide bond between Cys-57 and Cys-146 is not required to prevent hydrophobicity so long as the metal sites are occupied, and (iv) the requirement of zinc binding to organize loops IV (residues 49–84) and VII (residues 121–142) and to preorganize the copper binding site may explain why metal binding region mutants remain highly hydrophobic even in the presence of available copper.
Previous studies have characterized human SOD1 structural changes in solution as a function of metal binding, disulfide bond formation, dimerization, and the presence of disease-related or experimental mutations (38, 39, 48, 55–58, 72–76). Zinc binding to apoSOD1 decreases the mobility of the zinc binding (loop IV) and electrostatic (loop VII) regions and also preorganizes the copper binding site (58, 74). Consistent with this, the crystal structures for apoWT and metal-deficient mutant SOD1s (e.g. H46R, G85R, D124V, and S134N) show a structured β-barrel core with disorder of loops IV and VII (3, 22, 54, 77). The importance of zinc binding to order loops IV and VII has been suggested by molecular dynamics simulations (78) and isothermal titration calorimetry (63); furthermore, bound copper is insufficient to order the loops in the absence of zinc binding (79). The dimer interface region of loop IV is tethered to the β-barrel by the disulfide linkage between Cys-57 and Cys-146, and either zinc acquisition or disulfide oxidation can convert disulfide-reduced apoSOD1 monomers to a dimeric form (48, 56). Low metal occupancy or loss of metals because of oxidative damage may also contribute to SOD1 destabilization and aggregation (29, 31).
These considerations have led to the suggestion that diverse ALS mutants may all share a tendency to increase the population of metal-deficient or disulfide-reduced SOD1 species as a consequence of perturbed folding or post-translational processing (12, 19, 57, 80). Our data showing a uniform slowing of electrophoretic mobility for disulfide-reduced, metal-depleted SOD1 species (Fig. 2D) demonstrate that ALS mutants and WT SOD1 can acquire similar conformational properties. Because mutant SOD1 species also exhibit increased hydrophobic exposure under similar conditions (5) and in ALS tissues (5, 6), it is important to define the relative importance of the disulfide linkage and metal binding to SOD1 hydrophobicity. In this study we used spectroscopic methods and a hydrophobic probe (ANS) to determine that the abnormal increase in SOD1 hydrophobicity correlated best with impaired SOD1 metal binding rather than with absence of the disulfide bond.
The zinc binding affinity was previously shown to be decreased for A4V, A4T, L38V, and I113T mutants compared with WT SOD1 under conditions of 2 m urea (81). Furthermore, mislocalization of metals occurred when zinc and copper ions were added at pH 5.5 to mutant SOD1s (A4V, L38V, and G93A) that had been previously stripped of metals at acid pH (62). Given this evidence for perturbation of the metal binding properties for reasonably well folded ALS mutants, it was surprising that the as-isolated A4V SOD1 could accept copper and cobalt ions to the appropriate sites at pH 7.2 (Fig. 5, C and D). The absorbance maximum at 673 nm for Cu2+ bound to both WT and A4V SOD1s after 1 h was indicative of the normal distorted tetragonal coordination geometry favorable to redox cycling of the active site copper. In contrast, binding of Cu2+ to the experimental mutant C57S produced an absorbance maximum that was shifted to 635 nm (Fig. 5E). This suggests that in the absence of the disulfide bond, bound Cu2+ adopts a more square planar coordination similar to that observed for the yeast H48C SOD1 mutant (82) or the ALS mutant H48Q (17). The implication is that disulfide bond formation may contribute to the “strain” induced by the protein upon the copper coordination geometry that enables efficient cycling between Cu2+ and Cu+ states.
Also surprising was the effectiveness of copper binding to decrease the hydrophobicity of as-isolated WT, A4V, G93A, and the experimental mutants C57S and C146S. Although zinc binding is clearly important for ordering loops IV and VII of SOD1, our current findings indicate that copper deficiency also produces a significant conformational perturbation, as copper loading was required for ablation of ANS fluorescence and for restoring electrophoretic mobility (Figs. 6 and 7). Although incubation with excess zinc also decreased hydrophobicity of C57S and C146S, zinc was not as effective as copper. Zn2+ coordination to the copper site would be expected to favor a more tetrahedral geometry, similar to Cu+ binding. Because of its completely filled d shell, Zn2+ is not affected by the ligand field stabilization energy that can influence Cu2+ coordination. In contrast to the zinc-containing mutants, metal binding region mutants that do not bind Zn2+ with high affinity (G85R and D124V) retained their abnormal mobility and ANS fluorescence despite incubation with Cu2+ (Fig. 7). Overall, these results indicate that full metal occupancy is not only critical for SOD1 extreme thermal stability (18, 31, 65) but is also more important than disulfide bond status to prevent abnormal hydrophobic exposure.
Both the binding and fluorescence properties of ANS are influenced strongly by local environmental variables, which include not only hydrophobicity but also neighboring charged groups and the solvent polarity and viscosity (43, 44, 83). Interaction of positively charged arginine or lysine side chains with the sulfonate group of ANS (Fig. 1B), for instance, may contribute to ANS binding and the charge transfer processes that define the ANS fluorescence emission maximum (43, 44, 83). We were surprised to observe that compared with apoWT SOD1, the partially metalated WT01 SOD1 produced an increase in ANS fluorescence and blue shift of the peak (Fig. 4, A and D). This could be caused either by formation of a local hydrophobic pocket or by a change in the position of neighboring charged residues upon zinc binding. ANS can, therefore, act as a sensitive probe to measure subtle changes in the local environment of the protein.
Our observation that Cu2+ binding inhibits abnormal ANS fluorescence (Figs. 5–7) suggests that associated conformational changes upon copper site occupancy can mask either the ANS binding or its fluorescence emission. The conformation of Arg-143 is critical for SOD1 enzymatic activity and is properly constrained in the dimeric holoenzyme but not in monomeric (38) or disulfide-reduced Empty, Zinc-SOD1 (38). It is noteworthy that Val-118, which is buried in the SOD1 holoenzyme, becomes exposed in a hydrophobic pocket near Arg-143 in disulfide-reduced Empty, Zinc-SOD1 (39). Further definitive studies are in progress to map the locations of ANS interaction with different SOD1 species. Overall, these findings suggest that failure of SOD1 mutant species to acquire or retain metal ions, especially copper, may influence the observed hydrophobic exposure and contribute to abnormal interactions with other proteins or lipids in vivo.
Acknowledgments
We thank Hongru Zhou and Laura Baldassari for assistance with the protein purification. Dr. Jill Zitzewitz and Can Kayatekin contributed insightful discussions during the course of the project.
This work was supported, in whole or in part, by National Institutes of Health Grants NS44170 (to L. J. H.), NS049134 (to J. S. V.), NS39112 (to P. J. H.), and GM54836 (to C. R. M.) from NINDS. This work was also supported by grants from the ALS Association (to A. T. and L. J. H.), the ALS Therapy Alliance (to A. T. and L. J. H.), and The Angel Fund (to A. T.).
- SOD1
- Cu/Zn-superoxide dismutase
- ALS
- amyotrophic lateral sclerosis
- WT
- wild type
- ANS
- 1-anilinonaphthalene-8-sulfonic acid
- MBR
- metal binding region
- TCEP
- Tris(2-carboxyethyl)phosphine.
REFERENCES
- 1.Bruijn L. I., Becher M. W., Lee M. K., Anderson K. L., Jenkins N. A., Copeland N. G., Sisodia S. S., Rothstein J. D., Borchelt D. R., Price D. L., Cleveland D. W. (1997) Neuron 18, 327–338 [DOI] [PubMed] [Google Scholar]
- 2.Wang J., Xu G., Borchelt D. R. (2002) Neurobiol. Dis. 9, 139–148 [DOI] [PubMed] [Google Scholar]
- 3.Elam J. S., Taylor A. B., Strange R., Antonyuk S., Doucette P. A., Rodriguez J. A., Hasnain S. S., Hayward L. J., Valentine J. S., Yeates T. O., Hart P. J. (2003) Nat. Struct. Biol. 10, 461–467 [DOI] [PubMed] [Google Scholar]
- 4.Jonsson P. A., Ernhill K., Andersen P. M., Bergemalm D., Brännström T., Gredal O., Nilsson P., Marklund S. L. (2004) Brain 127, 73–88 [DOI] [PubMed] [Google Scholar]
- 5.Tiwari A., Xu Z., Hayward L. J. (2005) J. Biol. Chem. 280, 29771–29779 [DOI] [PubMed] [Google Scholar]
- 6.Jonsson P. A., Graffmo K. S., Andersen P. M., Brännström T., Lindberg M., Oliveberg M., Marklund S. L. (2006) Brain 129, 451–464 [DOI] [PubMed] [Google Scholar]
- 7.Deng H. X., Shi Y., Furukawa Y., Zhai H., Fu R., Liu E., Gorrie G. H., Khan M. S., Hung W. Y., Bigio E. H., Lukas T., Dal Canto M. C., O'Halloran T. V., Siddique T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7142–7147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaarsma D., Teuling E., Haasdijk E. D., De Zeeuw C. I., Hoogenraad C. C. (2008) J. Neurosci. 28, 2075–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaw B. F., Lelie H. L., Durazo A., Nersissian A. M., Xu G., Chan P. K., Gralla E. B., Tiwari A., Hayward L. J., Borchelt D. R., Valentine J. S., Whitelegge J. P. (2008) J. Biol. Chem. 283, 8340–8350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banci L., Bertini I., Boca M., Girotto S., Martinelli M., Valentine J. S., Vieru M. (2008) PLoS ONE 3, e1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruijn L. I., Miller T. M., Cleveland D. W. (2004) Annu. Rev. Neurosci. 27, 723–749 [DOI] [PubMed] [Google Scholar]
- 12.Hart P. J. (2006) Curr. Opin. Chem. Biol. 10, 131–138 [DOI] [PubMed] [Google Scholar]
- 13.Pasinelli P., Brown R. H. (2006) Nat. Rev. Neurosci. 7, 710–723 [DOI] [PubMed] [Google Scholar]
- 14.Kabashi E., Durham H. D. (2006) Biochim. Biophys. Acta 1762, 1038–1050 [DOI] [PubMed] [Google Scholar]
- 15.Zhang F., Ström A. L., Fukada K., Lee S., Hayward L. J., Zhu H. (2007) J. Biol. Chem. 282, 16691–16699 [DOI] [PubMed] [Google Scholar]
- 16.Gal J., Ström A. L., Kilty R., Zhang F., Zhu H. (2007) J. Biol. Chem. 282, 11068–11077 [DOI] [PubMed] [Google Scholar]
- 17.Hayward L. J., Rodriguez J. A., Kim J. W., Tiwari A., Goto J. J., Cabelli D. E., Valentine J. S., Brown R. H., Jr. (2002) J. Biol. Chem. 277, 15923–15931 [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez J. A., Valentine J. S., Eggers D. K., Roe J. A., Tiwari A., Brown R. H., Jr., Hayward L. J. (2002) J. Biol. Chem. 277, 15932–15937 [DOI] [PubMed] [Google Scholar]
- 19.Tiwari A., Hayward L. J. (2005) Neurodegener. Dis. 2, 115–127 [DOI] [PubMed] [Google Scholar]
- 20.Tiwari A., Hayward L. J. (2003) J. Biol. Chem. 278, 5984–5992 [DOI] [PubMed] [Google Scholar]
- 21.Hart P. J., Liu H., Pellegrini M., Nersissian A. M., Gralla E. B., Valentine J. S., Eisenberg D. (1998) Protein Sci. 7, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strange R. W., Antonyuk S., Hough M. A., Doucette P. A., Rodriguez J. A., Hart P. J., Hayward L. J., Valentine J. S., Hasnain S. S. (2003) J. Mol. Biol. 328, 877–891 [DOI] [PubMed] [Google Scholar]
- 23.Valentine J. S., Doucette P. A., Zittin Potter S. (2005) Annu. Rev. Biochem. 74, 563–593 [DOI] [PubMed] [Google Scholar]
- 24.Shinder G. A., Lacourse M. C., Minotti S., Durham H. D. (2001) J. Biol. Chem. 276, 12791–12796 [DOI] [PubMed] [Google Scholar]
- 25.Liu J., Lillo C., Jonsson P. A., Vande Velde C., Ward C. M., Miller T. M., Subramaniam J. R., Rothstein J. D., Marklund S., Andersen P. M., Brännström T., Gredal O., Wong P. C., Williams D. S., Cleveland D. W. (2004) Neuron 43, 5–17 [DOI] [PubMed] [Google Scholar]
- 26.Pasinelli P., Belford M. E., Lennon N., Bacskai B. J., Hyman B. T., Trotti D., Brown R. H., Jr. (2004) Neuron 43, 19–30 [DOI] [PubMed] [Google Scholar]
- 27.Harraz M. M., Marden J. J., Zhou W., Zhang Y., Williams A., Sharov V. S., Nelson K., Luo M., Paulson H., Schöneich C., Engelhardt J. F. (2008) J. Clin. Invest. 118, 659–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Estévez A. G., Crow J. P., Sampson J. B., Reiter C., Zhuang Y., Richardson G. J., Tarpey M. M., Barbeito L., Beckman J. S. (1999) Science 286, 2498–2500 [DOI] [PubMed] [Google Scholar]
- 29.Rakhit R., Cunningham P., Furtos-Matei A., Dahan S., Qi X. F., Crow J. P., Cashman N. R., Kondejewski L. H., Chakrabartty A. (2002) J. Biol. Chem. 277, 47551–47556 [DOI] [PubMed] [Google Scholar]
- 30.Wang J., Xu G., Gonzales V., Coonfield M., Fromholt D., Copeland N. G., Jenkins N. A., Borchelt D. R. (2002) Neurobiol. Dis. 10, 128–138 [DOI] [PubMed] [Google Scholar]
- 31.Stathopulos P. B., Rumfeldt J. A., Scholz G. A., Irani R. A., Frey H. E., Hallewell R. A., Lepock J. R., Meiering E. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7021–7026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rakhit R., Crow J. P., Lepock J. R., Kondejewski L. H., Cashman N. R., Chakrabartty A. (2004) J. Biol. Chem. 279, 15499–15504 [DOI] [PubMed] [Google Scholar]
- 33.Stathopulos P. B., Rumfeldt J. A., Karbassi F., Siddall C. A., Lepock J. R., Meiering E. M. (2006) J. Biol. Chem. 281, 6184–6193 [DOI] [PubMed] [Google Scholar]
- 34.Subramaniam J. R., Lyons W. E., Liu J., Bartnikas T. B., Rothstein J., Price D. L., Cleveland D. W., Gitlin J. D., Wong P. C. (2002) Nat. Neurosci. 5, 301–307 [DOI] [PubMed] [Google Scholar]
- 35.Wang J., Slunt H., Gonzales V., Fromholt D., Coonfield M., Copeland N. G., Jenkins N. A., Borchelt D. R. (2003) Hum. Mol. Genet. 12, 2753–2764 [DOI] [PubMed] [Google Scholar]
- 36.Nagai M., Aoki M., Miyoshi I., Kato M., Pasinelli P., Kasai N., Brown R. H., Jr., Itoyama Y. (2001) J. Neurosci. 21, 9246–9254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J., Caruano-Yzermans A., Rodriguez A., Scheurmann J. P., Slunt H. H., Cao X., Gitlin J., Hart P. J., Borchelt D. R. (2007) J. Biol. Chem. 282, 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banci L., Bertini I., Cramaro F., Del Conte R., Rosato A., Viezzoli M. S. (2000) Biochemistry 39, 9108–9118 [DOI] [PubMed] [Google Scholar]
- 39.Banci L., Bertini I., Cantini F., D'Amelio N., Gaggelli E. (2006) J. Biol. Chem. 281, 2333–2337 [DOI] [PubMed] [Google Scholar]
- 40.Hörnberg A., Logan D. T., Marklund S. L., Oliveberg M. (2007) J. Mol. Biol. 365, 333–342 [DOI] [PubMed] [Google Scholar]
- 41.Slavík J. (1982) Biochim. Biophys. Acta 694, 1–25 [DOI] [PubMed] [Google Scholar]
- 42.Matulis D., Baumann C. G., Bloomfield V. A., Lovrien R. E. (1999) Biopolymers 49, 451–458 [DOI] [PubMed] [Google Scholar]
- 43.Gasymov O. K., Glasgow B. J. (2007) Biochim. Biophys. Acta 1774, 403–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hawe A., Sutter M., Jiskoot W. (2008) Pharm. Res. 25, 1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ichimura S., Zama M. (1977) Biopolymers 16, 1449–1464 [DOI] [PubMed] [Google Scholar]
- 46.Matulis D., Lovrien R. (1998) Biophys. J. 74, 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goto J. J., Gralla E. B., Valentine J. S., Cabelli D. E. (1998) J. Biol. Chem. 273, 30104–30109 [DOI] [PubMed] [Google Scholar]
- 48.Doucette P. A., Whitson L. J., Cao X., Schirf V., Demeler B., Valentine J. S., Hansen J. C., Hart P. J. (2004) J. Biol. Chem. 279, 54558–54566 [DOI] [PubMed] [Google Scholar]
- 49.Lyons T. J., Nersissian A., Goto J. J., Zhu H., Gralla E. B., Valentine J. S. (1998) J. Biol. Inorg. Chem. 3, 650–662 [Google Scholar]
- 50.Zitzewitz J. A., Bilsel O., Luo J., Jones B. E., Matthews C. R. (1995) Biochemistry 34, 12812–12819 [DOI] [PubMed] [Google Scholar]
- 51.Bilsel O., Zitzewitz J. A., Bowers K. E., Matthews C. R. (1999) Biochemistry 38, 1018–1029 [DOI] [PubMed] [Google Scholar]
- 52.Gurney M. E., Pu H., Chiu A. Y., Dal Canto M. C., Polchow C. Y., Alexander D. D., Caliendo J., Hentati A., Kwon Y. W., Deng H. X. (1994) Science 264, 1772–1775 [DOI] [PubMed] [Google Scholar]
- 53.Hough M. A., Grossmann J. G., Antonyuk S. V., Strange R. W., Doucette P. A., Rodriguez J. A., Whitson L. J., Hart P. J., Hayward L. J., Valentine J. S., Hasnain S. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5976–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao X., Antonyuk S. V., Seetharaman S. V., Whitson L. J., Taylor A. B., Holloway S. P., Strange R. W., Doucette P. A., Valentine J. S., Tiwari A., Hayward L. J., Padua S., Cohlberg J. A., Hasnain S. S., Hart P. J. (2008) J. Biol. Chem. 283, 16169–16177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shipp E. L., Cantini F., Bertini I., Valentine J. S., Banci L. (2003) Biochemistry 42, 1890–1899 [DOI] [PubMed] [Google Scholar]
- 56.Arnesano F., Banci L., Bertini I., Martinelli M., Furukawa Y., O'Halloran T. V. (2004) J. Biol. Chem. 279, 47998–48003 [DOI] [PubMed] [Google Scholar]
- 57.Furukawa Y., O'Halloran T. V. (2005) J. Biol. Chem. 280, 17266–17274 [DOI] [PubMed] [Google Scholar]
- 58.Banci L., Bertini I., Cramaro F., Del Conte R., Viezzoli M. S. (2003) Biochemistry 42, 9543–9553 [DOI] [PubMed] [Google Scholar]
- 59.Shaw B. F., Durazo A., Nersissian A. M., Whitelegge J. P., Faull K. F., Valentine J. S. (2006) J. Biol. Chem. 281, 18167–18176 [DOI] [PubMed] [Google Scholar]
- 60.Lyons T. J., Liu H., Goto J. J., Nersissian A., Roe J. A., Graden J. A., Café C., Ellerby L. M., Bredesen D. E., Gralla E. B., Valentine J. S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12240–12244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lyons T. J., Nersissian A., Huang H., Yeom H., Nishida C. R., Graden J. A., Gralla E. B., Valentine J. S. (2000) J. Biol. Inorg. Chem. 5, 189–203 [DOI] [PubMed] [Google Scholar]
- 62.Goto J. J., Zhu H., Sanchez R. J., Nersissian A., Gralla E. B., Valentine J. S., Cabelli D. E. (2000) J. Biol. Chem. 275, 1007–1014 [DOI] [PubMed] [Google Scholar]
- 63.Potter S. Z., Zhu H., Shaw B. F., Rodriguez J. A., Doucette P. A., Sohn S. H., Durazo A., Faull K. F., Gralla E. B., Nersissian A. M., Valentine J. S. (2007) J. Am. Chem. Soc. 129, 4575–4583 [DOI] [PubMed] [Google Scholar]
- 64.Rodriguez J. A., Shaw B. F., Durazo A., Sohn S. H., Doucette P. A., Nersissian A. M., Faull K. F., Eggers D. K., Tiwari A., Hayward L. J., Valentine J. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10516–10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roe J. A., Butler A., Scholler D. M., Valentine J. S., Marky L., Breslauer K. J. (1988) Biochemistry 27, 950–958 [DOI] [PubMed] [Google Scholar]
- 66.Furukawa Y., Torres A. S., O'Halloran T. V. (2004) EMBO J. 23, 2872–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lehrer S. S. (1969) J. Biol. Chem. 244, 3613–3617 [PubMed] [Google Scholar]
- 68.Goldberg M., Pecht I. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 4684–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schäfer B. W., Fritschy J. M., Murmann P., Troxler H., Durussel I., Heizmann C. W., Cox J. A. (2000) J. Biol. Chem. 275, 30623–30630 [DOI] [PubMed] [Google Scholar]
- 70.Uversky V. N., Li J., Fink A. L. (2001) J. Biol. Chem. 276, 44284–44296 [DOI] [PubMed] [Google Scholar]
- 71.Ricchelli F., Drago D., Filippi B., Tognon G., Zatta P. (2005) Cell. Mol. Life Sci. 62, 1724–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Banci L., Benedetto M., Bertini I., Del Conte R., Piccioli M., Viezzoli M. S. (1998) Biochemistry 37, 11780–11791 [DOI] [PubMed] [Google Scholar]
- 73.Banci L., Bertini I., Cramaro F., Del Conte R., Viezzoli M. S. (2002) Eur J Biochem. 269, 1905–1915 [DOI] [PubMed] [Google Scholar]
- 74.Banci L., Bertini I., Cantini F., D'Onofrio M., Viezzoli M. S. (2002) Protein Sci. 11, 2479–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Banci L., Bertini I., D'Amelio N., Gaggelli E., Libralesso E., Matecko I., Turano P., Valentine J. S. (2005) J. Biol. Chem. 280, 35815–35821 [DOI] [PubMed] [Google Scholar]
- 76.Banci L., Bertini I., D'Amelio N., Libralesso E., Turano P., Valentine J. S. (2007) Biochemistry 46, 9953–9962 [DOI] [PubMed] [Google Scholar]
- 77.Elam J. S., Malek K., Rodriguez J. A., Doucette P. A., Taylor A. B., Hayward L. J., Cabelli D. E., Valentine J. S., Hart P. J. (2003) J. Biol. Chem. 278, 21032–21039 [DOI] [PubMed] [Google Scholar]
- 78.Strange R. W., Yong C. W., Smith W., Hasnain S. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10040–10044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roberts B. R., Tainer J. A., Getzoff E. D., Malencik D. A., Anderson S. R., Bomben V. C., Meyers K. R., Karplus P. A., Beckman J. S. (2007) J. Mol. Biol. 373, 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shaw B. F., Valentine J. S. (2007) Trends Biochem. Sci. 32, 78–85 [DOI] [PubMed] [Google Scholar]
- 81.Crow J. P., Sampson J. B., Zhuang Y., Thompson J. A., Beckman J. S. (1997) J. Neurochem. 69, 1936–1944 [DOI] [PubMed] [Google Scholar]
- 82.Hart P. J., Balbirnie M. M., Ogihara N. L., Nersissian A. M., Weiss M. S., Valentine J. S., Eisenberg D. (1999) Biochemistry 38, 2167–2178 [DOI] [PubMed] [Google Scholar]
- 83.Schonbrunn E., Eschenburg S., Luger K., Kabsch W., Amrhein N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6345–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.DeLano W. (2002) De Lano, W. L. (2002)The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]