Abstract
This study identified the peptide-binding motif of HLA-DRB1*1401 (DR1401). First, peptides containing DR1401 restricted epitopes were identified using tetramer guided epitope mapping. Among these, an influenza B peptide was selected for the motif study. After confirming the binding register for this peptide using a set of arginine substitutions, binding affinities were determined for 33 peptides derived from this influenza B sequence with single amino acid substitutions. The DR1401 peptide binding motif was deduced from the relative binding affinities of these peptides and confirmed by structural modeling. Pocket 1 demonstrated a preference for aliphatic anchor residues and methionine. Pocket 4 accommodated methionine and aliphatic residues, but also allowed some polar and charged amino acids. Pocket 6 preferred basic residues but also allowed some polar and aliphatic amino acids. Pocket 9 preferred aliphatic and aromatic amino acids and tolerated some polar residues but excluded all charged residues. Together these preferences define a distinct set of peptides that can be presented by DR1401. The resulting motif was used to verify T cell epitopes within the novel antigenic peptides identified by tetramer guided epitope mapping and within peptides from published reports that contain putative DR1401 epitopes.
Keywords: HLA-DR14, peptide motif, binding assay, Class II MHC, structural modeling
Introduction
T cell mediated immunity relies on the presentation of immunogenic peptides by MHC molecules on the surface of antigen presenting cells. In the case of MHC class II, peptides of variable length are bound specifically via interactions with distinct pockets within the peptide binding cleft. Among the hundreds of different class II alleles, these peptide binding pockets include polymorphic residues, such that each allele accommodates a specific set of residues for a given binding pocket, thereby determining its binding affinity for peptide sequences (1). As such, each class II allele binds and presents a distinct set of peptides based on the specific amino acid residues that comprise its peptide binding pockets. The peptide binding motifs for a number of class II alleles have been defined (2–4), but many remain unknown. Among these, DR1401 is of considerable interest because of its association with HLA driven HIV escape (5), protection from type I diabetes (6), and susceptibility to psoriasis vulgaris (7) in certain populations. However, remarkably little has been reported about the peptide binding preferences or epitopes recognized by this allele. The objective of this study was to identify the peptide binding motif of DR1401 using an in vitro peptide competition assay complimented by a structural modeling approach. The resulting peptide binding preferences were used to verify T cell epitopes within antigenic peptides that were identified by tetramer guided epitope mapping and within peptides from published reports that contain putative DR1401 epitopes.
Methods
Peptides
Panels of 20-mer peptides with overlapping sequences spanning the Influenza A/Puerto Rico/8/34 Nucleoprotein (Flu A-PR NP), Influenza B/Hong Kong/330/2001 Hemagglutinin (Flu B HA), Influenza A/Panama/2007/99 Hemagglutinin (Flu A-PA HA), Influenza A/New Caledonia/20/99 Hemagglutinin (Flu A-NC HA), and Influenza A/Wyoming/3/03 Hemagglutinin (Flu A-WY HA) proteins were synthesized on polyethylene pins with 9-fluorenylmethoxycarbonyl chemistry by Mimotopes (Clayton, Australia). Similarly, a panel of 13-mer peptides with sequences based on the Flu B HA 116-128 sequence (RIRLSNHNVINAE) was synthesized by Mimotopes. The Biotinylated reference peptide p53 193-204 (LIRVEGNLRVE) was synthesized using an Applied Biosystems 433A Peptide Synthesizer (Foster City, CA). This peptide contains a DR1401 restricted epitope (8). For flexibility, two Fmoc-6-aminohexanoic acid spacers were added between the n-terminal biotin label and the remainder of the peptide sequence. Each peptide was dissolved in DMSO at 20 mg/ml and subsequently diluted as needed.
DR1401 Protein and Tetramers
Recombinant DR1401 protein was produced as previously described (9). Briefly, soluble DR1401 was purified from insect cell culture supernatants by affinity chromatography. For the preparation of MHC class II tetramers, DR1401 protein was biotinylated at a sequence-specific site using biotin ligase (Avidity, Denver, CO). The biotinylated monomer was loaded with 0.2 mg/ml of peptide by incubating at 37°C for 72 hours in the presence of 2.5 mg/ml n-octyl-β-D-glucopyranoside and 1 mM Pefabloc SC (Sigma-Aldrich, St. Louis, MO). Peptide loaded monomers were subsequently conjugated as tetramers using R-PE streptavidin (Biosource International, Camarillo, CA) at a molar ratio of 8 to1. For peptide binding studies, DR1401 protein was left unbiotinylated and dialyzed directly into phosphate storage buffer.
Tetramer Guided Epitope Mapping
The Tetramer guided Epitope Mapping procedure was conducted as previously described (10) for each protein. PBMC were isolated from the blood of vaccinated healthy DR1401 subjects by ficoll underlay. Subsequently, CD4+ T cells were isolated using the Miltenyi CD4+ T cell isolation kit. Cells recovered from the CD4− fraction were incubated in 48 well plates (3 × 106 cells per well) for 1 hour and washed, leaving adherent cells as APC. After adding 2 million CD4+ T cells per well, each well was stimulated with a pool of five consecutive peptides spanning the Flu A-PR NP, Flu B HA, Flu A-PA HA, Flu A-NC HA and Flu A-WY HA protein sequences. After 14 days, 100 μl of resuspended cells were stained with pooled peptide PE-conjugated tetramers for 60 min at 37°C. Subsequently, cells were stained with CD4-PerCP (BD Biosciences), CD3-FITC and CD25-APC mAbs (eBioscience) and analyzed by flow cytometry. Cells from pools that gave positive staining were analyzed again with the corresponding individual peptide tetramers to identify each antigenic peptide.
Peptide binding competition
Various concentrations of each test peptide were incubated in competition with 0.1 μM biotinylated p53 peptide in wells coated with recombinant HLA-DR1401 protein as previously described (11). After washing, residual biotin-p53 peptide was labeled using europium-conjugated streptavidin (Perkin Elmer) and quantified using a Victor2 D time resolved fluorometer (Perkin Elmer). Peptide binding curves were simulated by non-linear regression with Prism software (Version 4.03, GraphPad Software Inc.) using a sigmoidal dose response curve. IC50 binding values were calculated from the resulting curves as the peptide concentration needed for 50% inhibition of reference peptide binding. Relative binding affinity (RBA) values were calculated as the IC50 value of the substituted peptide divided by the IC50 value of the non-substituted peptide. The ratio of two IC50 values reflects the fold difference in peptide binding affinity.
Molecular modeling
Models of DR1401 with the Flu B HA 116-128 peptide and its variants were prepared on a Silicon Graphics Indigo 2 work station using the program Insight II, version 2000 (Accelrys, San Diego, CA, USA), essentially as previously described (12). Energy minimization at was performed at pH 5.4, the ambient pH for binding studies. The crystal structure of HLA-DRB1*0301 in complex with the CLIP peptide (13) was used as the base molecule for all simulation studies. Figures were drawn with the aid of WebLabViewer version 3.5 and DSViewer Pro version 6.0, of Accelrys.
Results
Tetramer Guided Epitope Mapping of DR1401 restricted epitopes
The Tetramer guided Epitope Mapping approach was applied to identify DR1401 restricted epitopes within the Influenza A/Puerto Rico/8/34 Nucleoprotein (Flu A-PR NP), Influenza B/Hong Kong/330/2001 Hemagglutinin (Flu B HA), Influenza A/Panama/2007/99 Hemagglutinin (Flu A-PA HA), Influenza A/New Caledonia/20/99 Hemagglutinin (Flu A-NC HA), and Influenza A/Wyoming/3/03 Hemagglutinin (Flu A-WY HA) proteins. Panels of overlapping peptides (20 amino acids long with a 12 residue overlap) were synthesized for each protein and divided into pools as described in Materials and Methods. CD4+ T cells from DR1401 subjects were stimulated with pooled peptides and analyzed using the corresponding pooled peptide tetramers after two weeks in culture. Pools that were tetramer positive were analyzed using individual peptide tetramers to reveal the antigenic peptides within each pool. Representative results for pooled peptide tetramer analysis and subsequent individual peptide tetramer analysis are shown in Figure 1. For Flu B HA, positive staining was observed for 3 peptide pools (Fig. 1A) and for 5 individual peptides within these pools (Fig. 1B). Using this approach, similar analysis was completed for the remaining proteins. Overall, 11 peptides were identified that contain DR1401 restricted epitopes. Among these peptides, the core region of the Flu B HA 113-132 peptide, residues 116-128, was chosen as a study peptide to determine the binding preferences of DR1401.
Figure 1.
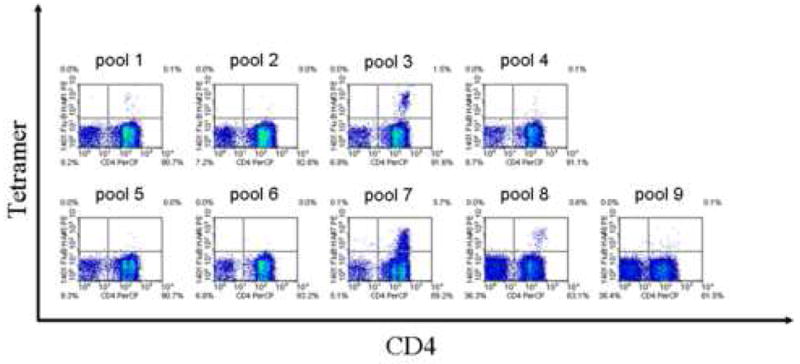
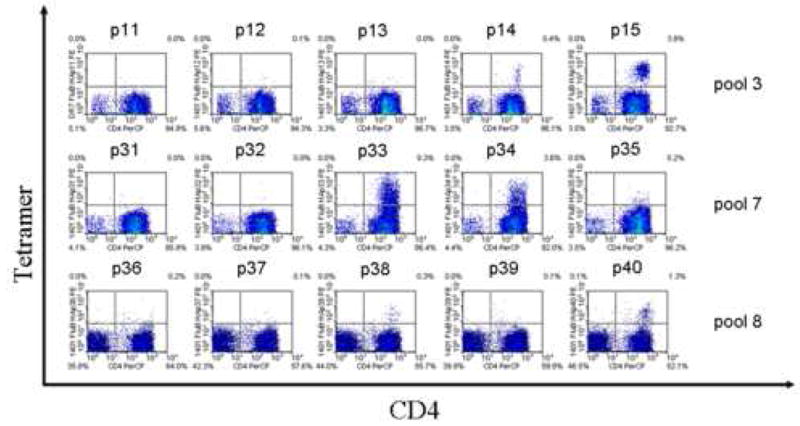
Tetramer Guided Epitope Mapping of Infuenza B epitopes. (A) Pool Mapping: T cells from a DR1401 donor were stimulated with overlapping peptide mixtures spanning Flu B HA and stained using pooled peptide loaded tetramers after two weeks. Peptide pools 3, 7, and 8 gave positive staining. (B) Individual peptide mapping: T cells from the tetramer positive panels shown in A were stained again using tetramers loaded with individual peptide from the corresponding peptide pools. Peptides p14, p15, p33, p34, and p40 were identified as peptides containing DR1401 restricted epitopes.
Binding Register of the study peptide
The binding register of the study peptide, Flu B HA 116-128, was confirmed using a set of arginine substituted peptides. Replacement of anchor residues with this basic amino acid has been previously shown to disrupt peptide binding (14). As shown in Figure 2, arginine substitution at residues 117, 120, and 125 reversed peptide binding, suggesting that these are primary anchor residues for binding to DR1401. The spacing of these residues establishes 117I as the P1 anchor, 120S as the P4 anchor, and 125I as the P9 anchor. Substitution at the remaining positions had only minor influences on binding.
Figure 2.
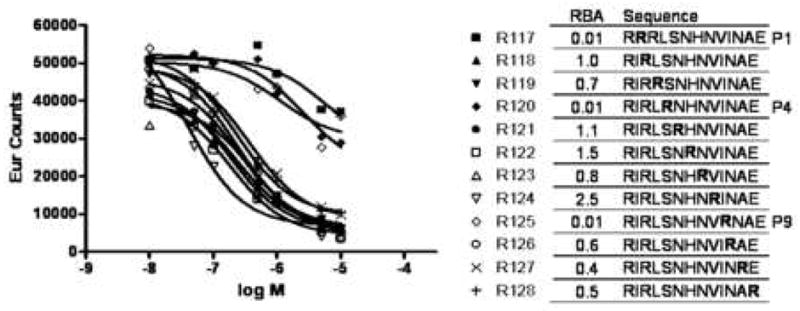
Binding register of the test peptide. Peptide binding curves (left panel) and relative binding affinity (right panel) for arginine substituted versions of the Influenza B 116-128 hemaglutinin peptide are shown. The binding curves indicate europium counts (residual reference peptide) versus test peptide concentration. Relative binding affinity (RBA) values were calculated as the IC50 value of the substituted peptide divided by the IC50 value of the un-substituted peptide.
Pocket Binding Preferences
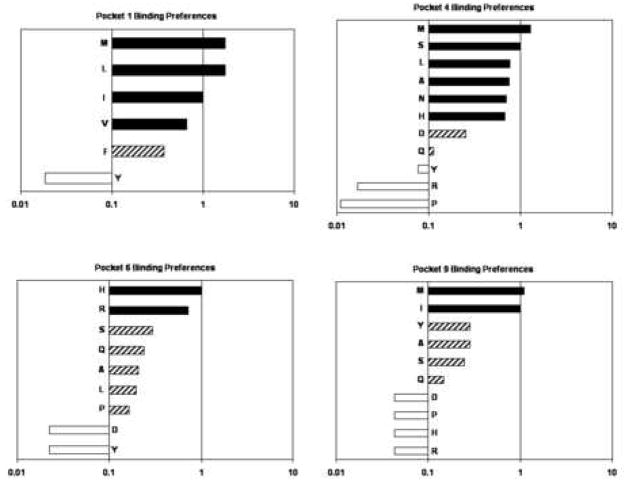
To determine the amino acid preferences for the binding pockets of DR1401, a panel of Flu B HA 116-128 derived peptides with single amino acid substitutions was synthesized. These substitutions represented obligatory anchor residues (15) for pocket 1 and general classes of amino acids for pockets 4, 6, and 9. Each of these peptides was bound (at various concentrations) to recombinant DR1401 protein in competition with biotinylated p53 reference peptide. The sequences of these 33 peptides and their relative binding affinities (RBA) are summarized in Table I. The observed binding preferences for each pocket are summarized in Figure 2. Pocket 1 preferred Met, Leu, Ile, and Val but also tolerated Phe. Pocket 4 preferred Met, Ser, Leu, Ala, Asn, and His but also tolerated Asp and Gln. Pocket 6 preferred His and Arg but also tolerated Ser, Gln, Ala, Leu, and Pro. Pocket 9 preferred Met and Ile but also tolerated Tyr, Ala, Ser and Gln. For this analysis, values within 2.5-fold of the reference peptide (RBA ≥ 0.4) were preferred, values within 10-fold of the reference peptide (RBA ≥ 0.1) were tolerated, and values less than 10-fold of the reference peptide (RBA ≤ 0.1) were not tolerated.
Table I.
Relative Binding of Substituted Flu B Peptides
| Sequence | Substitution | RBA |
|---|---|---|
| RIRLSNHNVIN | Unmodified | 1.0 |
| RLRLSNHNVIN | P1 I-->L | 1.7 |
| RYRLSNHNVIN | P1 I-->Y | 0.02 |
| RFRLSNHNVIN | P1 I-->F | 0.4 |
| RVRLSNHNVIN | P1 I-->V | 0.7 |
| RMRLSNHNVIN | P1 I-->M | 1.8 |
| RIRLYNHNVIN | P4 S-->Y | 0.1 |
| RIRLLNHNVIN | P4 S-->L | 0.8 |
| RIRLPNHNVIN | P4 S-->P | 0.01 |
| RIRLANHNVIN | P4 S-->A | 0.7 |
| RIRLRNHNVIN | P4 S-->R | 0.02 |
| RIRLHNHNVIN | P4 S-->H | 0.7 |
| RIRLDNHNVIN | P4 S-->D | 0.3 |
| RIRLQNHNVIN | P4 S-->Q | 0.1 |
| RIRLNNHNVIN | P4 S-->N | 0.7 |
| RIRLMNHNVIN | P4 S-->M | 1.3 |
| RIRLSNYNVIN | P6 H-->Y | 0.02 |
| RIRLSNLNVIN | P6 H-->L | 0.2 |
| RIRLSNPNVIN | P6 H-->P | 0.2 |
| RIRLSNANVIN | P6 H-->A | 0.2 |
| RIRLSNRNVIN | P6 H-->R | 0.7 |
| RIRLSNDNVIN | P6 H-->D | 0.02 |
| RIRLSNQNVIN | P6 H-->Q | 0.2 |
| RIRLSNSNVIN | P6 H-->S | 0.3 |
| RIRLSNHNVYN | P9 I-->Y | 0.3 |
| RIRLSNHNVPN | P9 I-->P | 0.04 |
| RIRLSNHNVAN | P9 I-->A | 0.3 |
| RIRLSNHNVRN | P9 I-->R | 0.04 |
| RIRLSNHNVHN | P9 I-->H | 0.04 |
| RIRLSNHNVDN | P9 I-->D | 0.04 |
| RIRLSNHNVQN | P9 I-->Q | 0.1 |
| RIRLSNHNVSN | P9 I-->S | 0.2 |
| RIRLSNHNVMN | P9 I-->M | 1.1 |
Modeling analysis of the DR1401 peptide binding motif
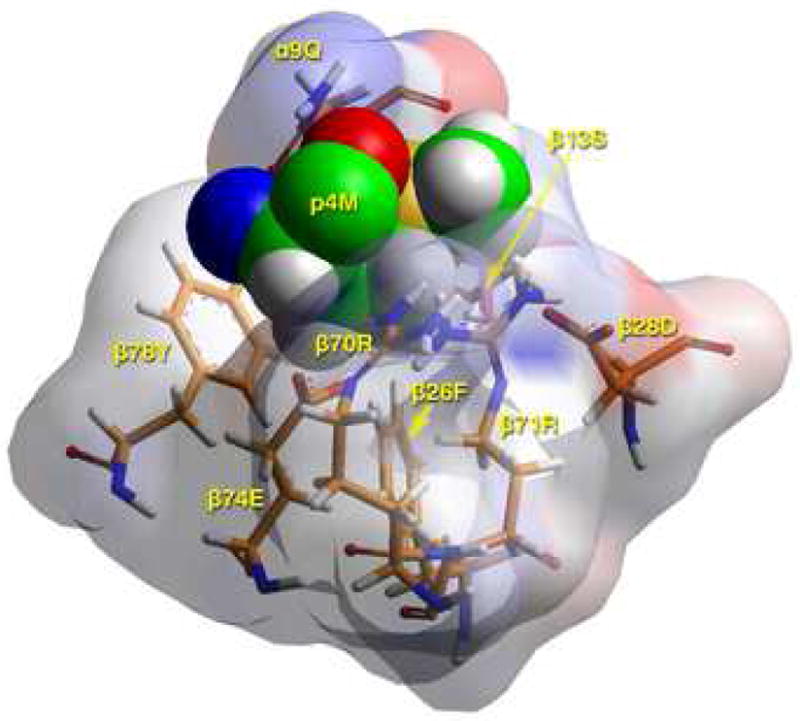
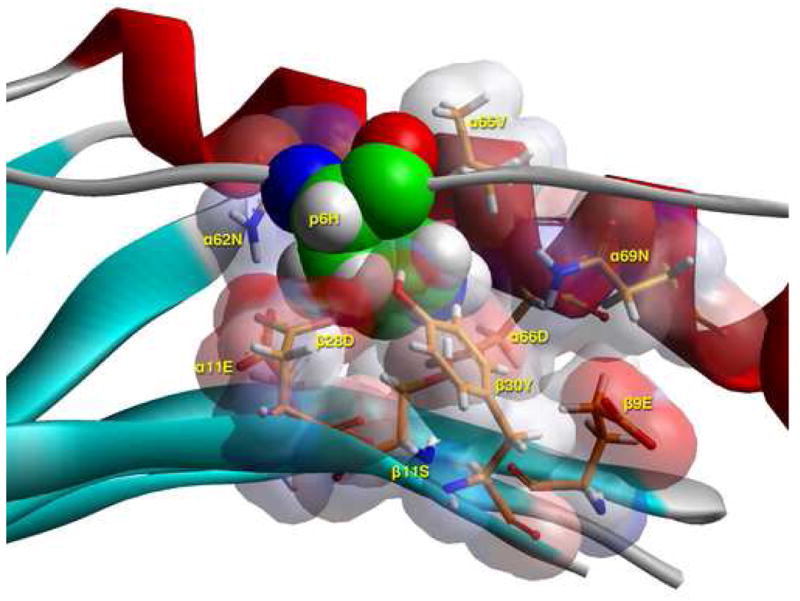
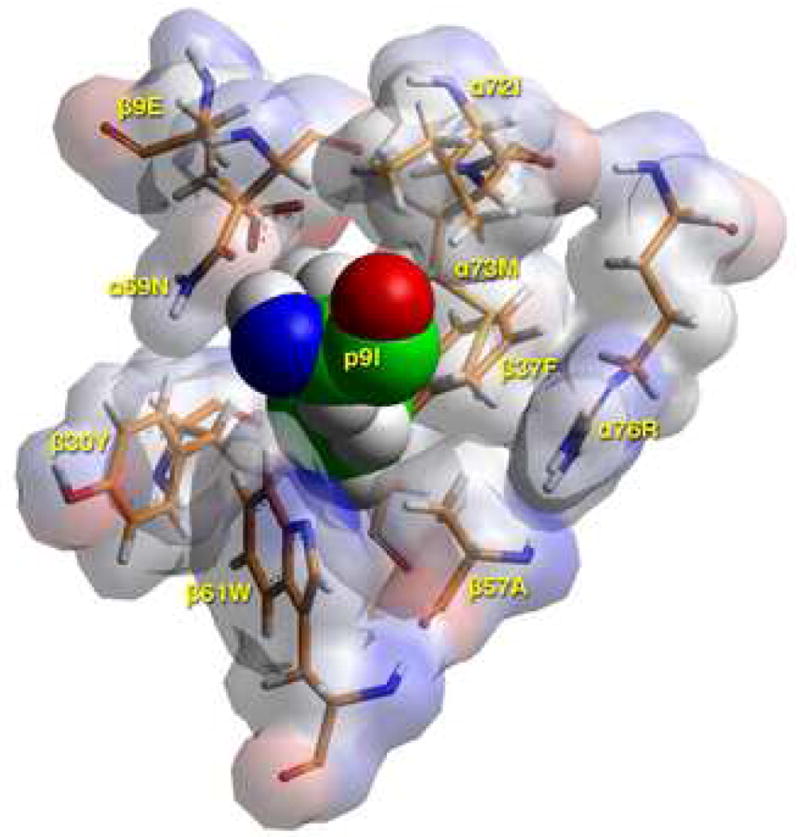
Models of the Flu B HA 116-128 peptide bound to DR1401 were created based on the crystal structure of DR0301 in complex with CLIP peptide (13) after energy minimization at pH 5.4, as described in Materials and Methods. As shown in Figure 4A, the Flu B HA peptide binds with geometry consistent with a high affinity interaction. This view highlights several DR14 residues that appear to have important interactions with the antigenic peptide (shown in stick form). This modeling confirmed that the peptide binds in a register with 117I in pocket 1, 120S in pocket 4, 122H in pocket 6, and 125I in pocket 9. Figure 4B shows a TCR view of pocket 4 of the bound study peptide. The arrangement of the various αβ residues in this pocket promotes the anchoring of both small and bulky aliphatic residues, while allowing polar and some charged amino acids. Figure 4C depicts a side view of pocket 6 of the peptide-MHC complex. The arrangement of αβ residues in this pocket permits the anchoring of a wide variety of residues. However, the presence of four acidic residues promotes the binding of positively charged residues. Figure 4D shows a TCR view of pocket 9 of the complex. The particular arrangement of αβ residues in this pocket promotes the anchoring of large aliphatic and aromatic amino acids while excluding both acidic and basic amino acids.
Figure 4.
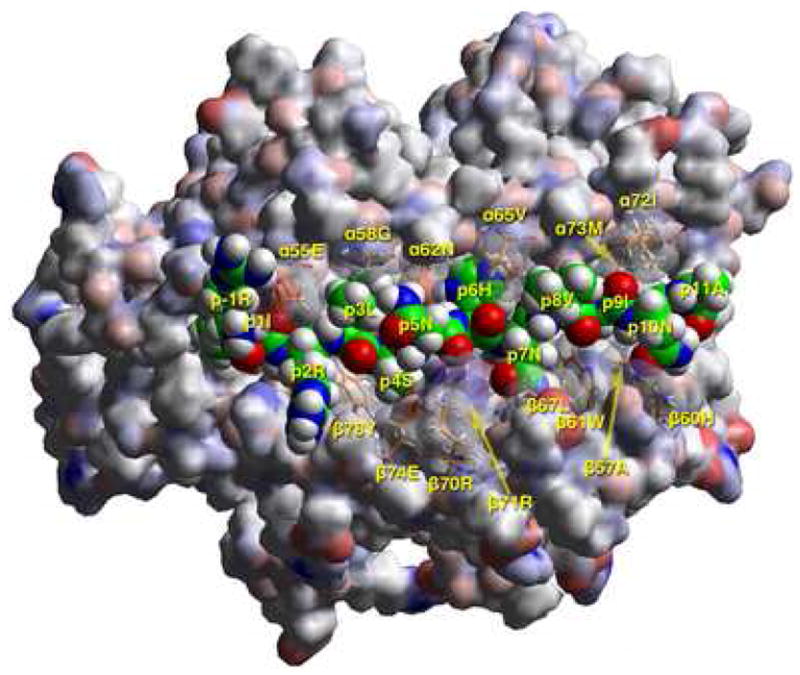
A) T cell receptor view of the antigenic peptide (RIRLSNHNVINA, anchors in bold) in the groove of DRB1*1401 (DR1401), after energy minimization at pH 5.4 based on the crystal structure of HLA-DRB1*0301 in complex with the CLIP peptide (13). The αlβl domain of the DR14 molecule is depicted in van der Waals surface representation, with the surface atomic charges color-coded (blue, positive; gray, neutral; red, negative), and the antigenic peptide is shown in space filling form (color code: carbon, green; oxygen, red; nitrogen, blue; hydrogen, white; sulfur, yellow). Several DR14 residues that have particular interactions with the antigenic peptide (α55Glu, α58Gly, α62Asn, α65Val, α72Ile, α73Met, β57Ala, β60His, β61Trp, β67Leu, β70Arg, β71Arg, β74Glu, β78Tyr) are shown in stick form with the same color-code as the antigenic peptide with the exception of carbon (orange). P1Ile is in pocket 1, p6His+ in pocket 6, and p9Ile in pocket 9 (first and third pointing into the plane of the paper, and the second only partly so) as expected of a peptide bound to DR14 with high affinity. Figure drawn with the aid of WebLabViewer version 3.5 and DSViewer Pro version 6.0, of Accelrys.
B) TCR view of pocket 4 of the complex of the peptide RIRLSNHNVINA with the HLA-DR14 allele. The arrangement of the various αβ residues in this pocket promotes the anchoring of both small and large aliphatic, but also polar and some charged amino acids at p4. The combination of β74Glu/β70Arg/β28Asp/β71Arg allows Asp and His binding (but not Arg/Lys) in this pocket. Likewise, the presence of β13Ser, β26Phe and α9Gln leaves sufficient room for anchoring of bulky aliphatic (but not aromatic) residues in this pocket. The figure has been rotated by 20° with respect to the y-axis (left-hand side into the plane of the paper, right-hand side above the plane of the paper) so that the interactions between various residues could be fully exposed. Color and depiction conventions identical to those in Figure 4A. Figure drawn with the aid of WebLabViewer version 3.5 and DSViewer Pro version 6.0, of Accelrys.
C) Side view of pocket 6 of the complex of the peptide RIRLSNHNVINA with the HLA-DR14 allele. This is seen from the Cα of β57Ala at the level of the β-sheet floor. The arrangement of the various αβ residues in this pocket, and the ability of β9Glu to be flexible, promotes the anchoring of a wide variety of residues (from positively charged Arg, His (shown here), to polar Ser, Gln and small and large aliphatics), but no acidic ones or Tyr. The presence of β11Ser provides sufficient space for the larger of the allowed residues, while the presence of four acidic residues (α11Glu, α66Asp, β9Glu, β28Asp) provides a suitable environment for the binding of positively charged residues in this pocket, excluding negatively charged ones. For orientation purposes we also show part of the α1 helix (residues 62 to 69), the β–sheet floor from where residues originate, and the peptide backbone to which p6Asp belongs. In all probability p6His would be charged even at the extracellular pH of 7.4 (energy minimisation performed at pH 5.4, the pH of the binding study) by induction, due to its proximity to α66Asp. Color and depiction conventions identical to those in Figure 4A. In addition, α-helix is in red, β-sheet is in turquoise and all other secondary structure forms (random coil, polyproline type II helix of the antigenic peptide are in gray). Figure drawn with the aid of WebLabViewer version 3.5 and DSViewer Pro version 6.0, of Accelrys.
D) TCR view of pocket 9 of the complex of the peptide RIRLSNHNVINA with the HLA-DR14 allele. The arrangement of the various αβ residues in this pocket promotes the anchoring of either large aliphatic/aromatic amino acids (but hardly tryptophan) at p9. The presence of β30Tyr limits the space available, while the combination α72Ile/α73Met/β37Phe/β57Ala/α76Arg favors this preference, excluding acidic and basic amino acids from this pocket. Contrast this to the situation in the H2-Ek allele (α72Val/α73Met/β9Glu/β30Tyr/β37Asn/α57Asp/α76Arg), where Arg/Lys are exclusively favored at this pocket. There, the b9Glu/b37Asn combination attracts the positively charged residues, while the β57Asp—α76Arg salt bridge neutralizes the electrostatic repulsion of the latter residue for basic p9 residues as they approach the mouth of the pocket (16). Color and depiction conventions identical to those in Figure 4A. Figure drawn with the aid of WebLabViewer version 3.5 and DSViewer Pro version 6.0, of Accelrys.
Motif Analysis of Peptides Containing DR1401 epitopes
Using the method of tetramer guided epitope mapping, a total of 11 peptides were identified that contain DR1401 restricted epitopes. These are summarized in Table II. For nine of these peptides, nonameric core epitopes consistent with the DR1401 binding motif were identified (highlighted in boldface). Two of these peptides (Flu A-PR NP 185-204 and Flu B 257-276) contain two distinct registers that could be expected to bind DR1401. For these the second motif is underlined. Flu B 105-124 is problematic because its only nonameric epitope (underlined) contains tyrosine at position 4, which would be expected to bind poorly. This peptide also contains all but position 9 of the Flu B study epitope (in boldface). Thus, it is possible that the peptide binds in this register with pocket 9 empty. Interestingly, Flu B 313-332 does not contain a particularly good motif for DR1401. However, if alanine could be accommodated at position 1, as has been reported for DR1301 (17), AKAIGNCPI (underlined) could bind. Similarly, we evaluated published peptide sequences containing putative DR1401 epitopes and found several that contain a motif consistent with our findings. These are summarized in Table III. Again, the predicted minimal DR1401 epitope is highlighted in boldface for each peptide.
Table II.
Motif Analysis for Novel DR1401 Epitopes
| Protein | Residues | Epitope |
|---|---|---|
| Flu A-PR NP | 185-204 | GVGTMVMELVRMIKRGINDR |
| 241-260 | QVRESRNPGNAEFEDLTFLA | |
| 401-420 | ASAGQISIQPTFSVQRNLPF | |
| Flu B HA | 105-124 | RQLPNLLRGYERIRLSNHNV |
| 113-132 | GYERIRLSNHNVINAEKAPG | |
| 257-276 | KTGTITYQRGILLPQKVWCA | |
| 265-284 | RGILLPQKVWCASGRSKVIK | |
| 313-332 | EHAKAIGNCPIWVKTPLKLA | |
| Flu A-PA HA | 33-52 | QIEVTNATELVQSSSTGRIC |
| Flu A-NC HA | 121-140 | QLSSVSSFERFEIFPKESSW |
| Flu A-WY HA | 265-284 | GNLIAPRGYFKIRSGKSSIM |
Table III.
Motif Analysis for Published DR1401 Epitopes
| Protein | Residues | Epitope |
|---|---|---|
| VP16 (18) | 477-487 | LMFINGSLTVR |
| P53 (8) | 193-204 | LIRVEGNLRVE |
| Actin (19) | 206-216 | REIVRDIKEKL |
| Histone H4 (Mouse) (19) | 31-45 | TKPAIRRLARRGGVK |
| Mycobacterium leprae (20) | 1-16 | VAKVKIKPLEDKILVQ |
| Mycobacterium leprae (20) | 41-56 | GTVVAVGPGRWDEDGA |
| Mycobacterium tuberculosis (20) | 65-80 | KRIPLDVAEGDTVIYS |
| Mycobacterium tuberculosis (20) | 73-88 | KYGGTEIKYNGEEYLI |
| Mycobacterium tuberculosis (20) | 81-100 | YNGEEYLILSARDLAVVSK |
| Synaptonemal complex protein 1 (21) | 635-649 | QLNVYEIKVNKLELE |
| Herpesvirus 4 B95-8 (22) | 529-543 | PQCRLTPLSRLPFGM |
| Plasmodium falciparum (23) | 239-249 | LKIRANELDVL |
| Plasmodium falciparum (23) | 249-263 | LKKLVFGYRKPLDNI |
| Plasmodium falciparum (23) | 338-350 | IDTLKKNENIKEL |
Discussion
It has been demonstrated that interactions of peptide side chains with pockets 1, 4, 6, 7, and 9 determine most of the peptide binding specificity of class II MHC proteins (24). For DR1401 in particular, initial experiments with arginine substituted peptides (Figure 2) indicated that pockets 1, 4, 6, and 9 play the most significant roles in peptide binding. As such, understanding the amino acid preferences for each of these pockets defines a distinct set of peptides that can be presented by a given class II allele. The observed peptide competition results, taken together with the modeling results allow the definition of a distinct set of peptides that can be presented by DR1401. For the ensuing discussion, each pocket will be considered in turn to construct an overall understanding of the peptide binding motif. Of the class II alleles with known binding preferences, DR1401 is most similar (based on sequence homology) to DR1104 and DR1301. Therefore, the similarities and differences between these alleles will also be discussed.
The binding preferences for pocket 1 are easily explainable because the b86Val residue of this allele limits pocket size (as compared to b86Gly alleles) allowing aliphatic but not bulky aromatic side chains to bind. Although phenyalanine was tolerated, it is almost certain that tryptophan would be excluded since tyrosine was excluded. At the amino acid positions that comprise pocket 1, DR1401, DR1104, and DR1301 are completely identical. In agreement with our current observations, DR1104 and DR1301 have been shown to prefer Ile, Leu, and Val at this position (17, 25).
The results for pocket 4 are more complex. The modeling results indicate that the b70Arg/71Arg/74Glu combination is prohibitive to basic residues as anchors, barely acceptable to acidic residues, but permissive to polar ones. At the base of the binding pocket, it would appear that there is sufficient space available for aliphatic residues to contact b26Phe. However, a slow association constant might be expected for this interaction. This coincides with the binding results, in which pocket 4 preferred small to moderate sized hydrophobic and neutral residues. It is interesting that this pocket also bound histidine and tolerated aspartic acid, since these residues are charged. Given the fact that this pocket preferred asparagine and tolerated glutamine and aspartic acid, it is likely that glutamic acid would be tolerated. For the residues that comprise pocket 4, DR1104 is most similar to DR1401, sharing b13Ser, b26Phe, b28Asp, b78Tyr, and b71Arg while differing at b70 (Arg and Asp respectively) and b74 (Glu and Ala respectively). DR1104 has been shown to accept Leu, Val, Met, Ala, Phe, and Tyr at this position (25). Apparently, the smaller size of b70Asp allows DR1104 to accept bulky and aromatic residues such as Tyr and Phe. The reversal of charge at this position may also be less favorable for the binding of Asn and Gln.
For pocket 6, the modeling suggests that acidic residues from the a-chain along with b9Glu/11Ser/30Tyr allow basic residues to anchor. In this arrangement polar residues have a chance to bind but not acidic or aromatic ones. In agreement with these findings, the binding studies revealed that pocket 6 preferred positively charged residues, but also allowed some hydrophobic and neutral residues (Ser, Gln, Ala, Leu, and Pro). Aspartic acid and tyrosine were excluded, suggesting that negative charges are problematic and indicating a possible size limit for this position. Thus it is likely that glutamic acid and tryptophan would also be excluded. DR1401, DR1104, and DR1301 are completely identical at the amino acid positions that comprise pocket 6, so binding preferences should be the same at this position. In agreement with our observations, DR1104 and DR1301 have been shown to prefer Arg, Lys, and His at this position (17, 25).
Pocket 9 was perhaps the most interesting pocket for this allele. This position preferred moderately sized aliphatic residues. However, tyrosine, a large polar residue, and alanine, a small hydrophobic residue, were also tolerated. Thus, it is not surprising the moderately sized polar residues serine and glutamine were also accepted. It is striking that both Asp and Arg were not allowed, given the presence of b57Ala, which has been shown in other alleles to favor acidic residues at p9. The key difference for the DR1401 allele is the presence of b37Phe in addition to the standard hydrophobic residues a72Ile and a73Met. It appears that these four residues form a barrier such that positively or negatively charged residues cannot pass easily. This agrees with observations for DRB1*1201, which has a b9Glu/37Leu/57Val combination that likewise excludes basic residues but allows hydrophobic ones (26). In contrast, I-Ek which has b9Glu/37Asn/57Asp, accepts positively charged residues at p9 indicating that no such barrier exists for that arrangement of residues. For the residues that comprise pocket 9, DR1104 is similar to DR1401 sharing b9Glu, b30Tyr, and b61Trp while differing at b37 (Phe and Tyr respectively) and b57 (Ala and Asp respectively). DR1104 has been shown to accept Ala, Gly, Ser, and Pro (25). The presence of b57Asp allows the formation of a salt bridge with a76Arg, narrowing the opening of the p9 pocket; furthermore, the presence of b37Tyr renders the binding of bulky residues (to say nothing of basic ones) very difficult thereby shifting the preference of DR1104 toward smaller residues.
In addition to the differences listed above, DR1401 differs from DR1104 and DR1302 at b47 (Tyr and Phe respectively) and b60 (Tyr and His respectively). In particular, the presence of b60His could influence pocket 9. Although it was not included in the binding studies for this allele, the modeling results suggest that pocket 7 may prefer acidic and polar residues because of the presence of b28Asp/71Arg. The complete findings for the binding pockets of DR1401 are summarized in Table IV. Together, these results define a population of peptides that can be bound and presented by DR1401 that is distinct, even from similar alleles such as DR1104 and DR1302. These findings should allow a better understanding of DR1401 restricted T cell epitopes and responses.
Table IV.
DR1401 Binding Motif
| Position | ||||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| M | M | H | M | |||||
| L | S | R | I | |||||
| I | L | S | Y | |||||
| V | A | Q | A | |||||
| F | N | A | S | |||||
| H | L | Q | ||||||
| D | P | |||||||
| Q | ||||||||
Conclusions
Using a combination of tetramer guided epitope mapping, cell free peptide competition assay, and molecular modeling, this study characterized the peptide-binding properties of HLA-DRB1*1401 (DR1401). The peptide binding motif was inferred from the binding affinities of 33 peptides derived from the Flu B HA 116-128 sequence and confirmed by molecular modeling studies. The observed motif had distinct features, including the exclusion of large anchor residues at pocket 1, a preference for polar and aliphatic residues at pocket 4, positively charged or polar residues at pocket 6, and the exclusion of all charged residues at pocket 9. The observed DR1401 binding motif was in agreement with 10 of 11 novel 20mer peptides identified by tetramer guided epitope mapping and with several published peptides reported as putative DR1401 epitopes. These findings represent the first definition of a DR1401 peptide binding motif.
Figure 3.
Amino acid preferences for pocket 1 (upper left panel), pocket 4 (upper right panel), pocket 6 (lower left panel), and pocket 9 (lower right panel) of the DR1401 motif. Black bars represent values within 2.5-fold of the reference peptide (RBA ≥ 0.4, “preferred”), hatched bars represent values within 10-fold of the reference peptide (RBA ≥ 0.1, “tolerated”), and open bars represent values less than 10-fold of the reference peptide (RBA ≤ 0.1, “excluded”).
Acknowledgments
This work was supported in part by NIH contract HHSN266200400028C
Abbreviations
- DR1401
DRA1/B1*1401
- Flu A-PR NP
Influenza A/Puerto Rico/8/34 Nucleoprotein
- Flu B HA
Influenza B/Hong Kong/330/2001 Hemagglutinin
- Flu A-PA HA
Influenza A/Panama/2007/99 Hemagglutinin
- Flu A-NC HA
Influenza A/New Caledonia/20/99 Hemagglutinin
- Flu A-WY HA
Influenza A/Wyoming/3/03 Hemagglutinin
- PBMC
Peripheral blood mononuclear cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bondinas GP, Moustakas AK, Papadopoulosm GK. The spectrum of HLA-DQ and HLA-DR alleles, 2006: a listing correlating sequence and structure with function. Immunogenetics. 2007;59:539–553. doi: 10.1007/s00251-007-0224-8. [DOI] [PubMed] [Google Scholar]
- 2.Geluk A, van Meijgaarden KE, Southwood S, Oseroff C, Drijfhout JW, de Vries RR, Ottenhoff TH, Sette A. HLA-DR3 molecules can bind peptides carrying two alternative specific submotifs. J Immunol. 1994;152:5742–5748. [PubMed] [Google Scholar]
- 3.Sette A, Sidney J, Oseroff C, del Guercio MF, Southwood S, Arrhenius T, Powell MF, Colón SM, Gaeta FC, Grey HM. HLA DR4w4-binding motifs illustrate the biochemical basis of degeneracy and specificity in peptide-DR interactions. J Immunol. 1993;151:3163–3170. [PubMed] [Google Scholar]
- 4.Wucherpfennig KW, Sette A, Southwood S, Oseroff C, Matsui M, Strominger JL, Hafler DA. Structural requirements for binding of an immunodominant myelin basic protein peptide to DR2 isotypes and for its recognition by human T cell clones. J Exp Med. 1994;179:279–290. doi: 10.1084/jem.179.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya T, Daniels M, Heckerman D, Foley B, Frahm N, Kadie C, Carlson J, Yusim K, McMahon B, Gaschen B, Mallal S, Mullins JI, Nickle DC, Herbeck J, Rousseau C, Learn GH, Miura T, Brander C, Walker B, Korber B. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–1586. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- 6.Redondo MJ, Kawasaki E, Mulgrew CL, Noble JA, Erlich HA, Freed BM, Lie BA, Thorsby E, Eisenbarth GS, Undlien DE, Ronningen KS. DR- and DQ-associated protection from type 1A diabetes: comparison of DRB1*1401 and DQA1*0102-DQB1*0602*. J Clin Endocrinol Metab. 2000;85:3793–3797. doi: 10.1210/jcem.85.10.6920. [DOI] [PubMed] [Google Scholar]
- 7.Jee SH, Tsai TF, Tsai WL, Liaw SH, Chang CH, Hu CY. HLA-DRB1*0701 and DRB1*1401 are associated with genetic susceptibility to psoriasis vulgaris in a Taiwanese population. Br J Dermatol. 1998;139:978–983. doi: 10.1046/j.1365-2133.1998.02552.x. [DOI] [PubMed] [Google Scholar]
- 8.Fujita H, Senju S, Yokomizo H, Saya H, Ogawa M, Matsushita S, Nishimura Y. Evidence that HLA class II-restricted human CD4+ T cells specific to p53 self peptides respond to p53 proteins of both wild and mutant forms. Eur J Immunol. 1998;28:305–316. doi: 10.1002/(SICI)1521-4141(199801)28:01<305::AID-IMMU305>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 9.Novak EJ, Liu AW, Nepom GT, Kwok WW. MHC class II tetramers identify peptide-specific human CD4 (+) T cells proliferating in response to influenza A antigen. J Clin Invest. 1999;104:R63–67. doi: 10.1172/JCI8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novak EJ, Liu AW, Gebe JA, Falk BA, Nepom GT, Koelle DM, Kwok WW. Tetramer-guided epitope mapping: rapid identification and characterization of immunodominant CD4+ T cell epitopes from complex antigens. J Immunol. 2001;166:6665–6670. doi: 10.4049/jimmunol.166.11.6665. [DOI] [PubMed] [Google Scholar]
- 11.Ettinger RA, Kwok WW. A peptide binding motif for HLA-DQA1*0102/DQB1*0602, the class II MHC molecule associated with dominant protection in insulin-dependent diabetes mellitus. J Immunol. 1998;160:2365–2373. [PubMed] [Google Scholar]
- 12.Reichstetter S, Papadopoulos GK, Moustakas AK, Swanson E, Liu AW, Beheray S, Ettinger RA, Nepom GT, Kwok WW. Mutational analysis of critical residues determining antigen presentation and activation of HLA-DQ0602 restricted T-cell clones. Hum Immunol. 2002;63:185–193. doi: 10.1016/s0198-8859(01)00377-9. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh P, Amaya M, Mellins E, Wiley DC. The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3. Nature. 1995;378:457–462. doi: 10.1038/378457a0. [DOI] [PubMed] [Google Scholar]
- 14.Ettinger RA, Papadopoulos GK, Moustakas AK, Nepom GT, Kwok WW. Allelic variation in key peptide-binding pockets discriminates between closely related diabetes-protective and diabetes-susceptible HLA-DQB1*06 alleles. J Immunol. 2006;176:1988–1998. doi: 10.4049/jimmunol.176.3.1988. [DOI] [PubMed] [Google Scholar]
- 15.Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, Sahin U, Braxenthaler M, Gallazzi F, Protti MP, Sinigaglia F, Hammer J. Generation of tissue-specific and promiscuous HLA ligand database using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17:555–561. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 16.Fremont DH, Hendrickson WA, Marrack P, Kappler J. Structures of an MHC class II molecule with covalently bound single peptides. Science. 1996;272:1001–1004. doi: 10.1126/science.272.5264.1001. [DOI] [PubMed] [Google Scholar]
- 17.Davenport MP, Quinn CL, Chicz RM, Green BN, Willis AC, Lane WS, Bell JI, Hill AV. Naturally processed peptides from two disease-resistance-associated HLA-DR13 alleles show related sequence motifs and the effects of the dimorphism at position 86 of the HLA-DR beta chain. Proc Natl Acad Sci USA. 1995;92:6567–6571. doi: 10.1073/pnas.92.14.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwok WW, Gebe JA, Liu A, Agar S, Ptacek N, Hammer J, Koelle DM, Nepom GT. Rapid epitope identification from complex class-II-restricted T-cell antigens. Trends Immunol. 2001;22:583–588. doi: 10.1016/s1471-4906(01)02038-5. [DOI] [PubMed] [Google Scholar]
- 19.Harris PE, Maffei A, Colovai AI, Kinne J, Tugulea S, Suciu-Foca N. Predominant HLA-class II bound self-peptides of a hematopoietic progenitor cell line are derived from intracellular proteins. Blood. 1996;87:5104–5112. [PubMed] [Google Scholar]
- 20.Chua-Intra B, Peerapakorn S, Davey N, Jurcevic S, Busson M, Vordermeier HM, Pirayavaraporn C, Ivanyi J. T-cell recognition of mycobacterial GroES peptides in Thai leprosy patients and contacts. Infect Immun. 1998;66:4903–4909. doi: 10.1128/iai.66.10.4903-4909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neumann F, Wagner C, Preuss KD, Kubuschok B, Schormann C, Stevanovic S, Pfreundschuh M. Identification of an epitope derived from the cancer testis antigen HOM-TES-14/SCP1 and presented by dendritic cells to circulating CD4+ T cells. Blood. 2005;106:3105–3113. doi: 10.1182/blood-2005-04-1487. [DOI] [PubMed] [Google Scholar]
- 22.Taylor GS, Long HM, Haigh TA, Larsen M, Brooks J, Rickinson AB. A role for intercellular antigen transfer in the recognition of EBV-transformed B cell lines by EBV nuclear antigen-specific CD4+ T cells. J Immunol. 2006;177:3746–3756. doi: 10.4049/jimmunol.177.6.3746. [DOI] [PubMed] [Google Scholar]
- 23.Guttinger M, Romagnoli P, Vandel L, Meloen R, Takacs B, Pink JR, Sinigaglia F. HLA polymorphism and T cell recognition of a conserved region of p190, a malaria vaccine candidate. Int Immunol. 1991;3:899–906. doi: 10.1093/intimm/3.9.899. [DOI] [PubMed] [Google Scholar]
- 24.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, Wiley DC. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 25.Verreck FA, van de Poel A, Drijfhout JW, Amons R, Coligan JE, Konig F. Natural peptides isolated from Gly86/Val86-containing variants of HLA-DR1, -DR11, -DR13, and -DR52. Immunogenetics. 1996;43:392–397. doi: 10.1007/BF02199809. [DOI] [PubMed] [Google Scholar]
- 26.Falk K, Rötzschke O, Stevanović S, Jung G, Rammensee HG. Pool sequencing of natural HLA-DR, DQ, and DP ligands reveals detailed peptide motifs, constraints of processing, and general rules. Immunogenetics. 1994;39:230–242. doi: 10.1007/BF00188785. [DOI] [PubMed] [Google Scholar]