Abstract
Colorectal cancers with microsatellite instability (MSI) have clinical, pathological, genetic, and epigenetic features distinct from microsatellite stable (MSS) CRC. Examination of EGFR mRNA and protein expression levels in a panel of colon cancer cell lines identified strong expression of EGFR in multiple cell lines with microsatellite instability. While no relationship between EGFR overexpression and the length of a CA dinucleotide repeat in intron 1 was observed, a variant A13/A14 repeat sequence within the 3’-UTR of the EGFR gene was identified which was mutated by either mono or dinucleotide adenosine deletions in 64% of MSI cell lines, and 69% of MSI colon tumors. Utilizing a Tet-Off system we demonstrate that this mutation increases EGFR mRNA stability in colon cancer cells, providing a mechanistic basis for EGFR over-expression in MSI colon cancer cell lines. To determine whether this mutation is a driver or a bystander event in MSI colon cancer, we examined the effect of pharmacological and molecular inhibition of EGFR in EGFR 3’UTR mutant MSI cell lines. Cell lines with an EGFR 3’UTR mutation and which were wild type for downstream signaling mediators in the Ras/BRAF and PIK3CA/PTEN pathways were sensitive to EGFR inhibition, while those harboring mutations in these signaling mediators were not. Furthermore, in cell lines wild type for downstream signaling mediators, those with EGFR 3’UTR mutations were more sensitive to EGFR inhibition than EGFR 3’UTR WT cells, suggesting this mutation provides a growth advantage to this subset of MSI colon tumors.
Keywords: EGFR, MSI, colon cancer, mutation, 3’UTR
Introduction
The important role played by EGFR in the progression of cancer is illustrated by the clinical efficacy of multiple small molecule and biological agents that target and inhibit this receptor. In colon cancer, two antibody-based therapies, cetuximab and panitumumab, have recently been approved for the treatment of metastatic disease (1).
EGFR signaling stimulates cell proliferation, angiogenesis and metastatic spread, and inhibits apoptosis of colon cancer cells (2–4). Activation of EGFR is initiated by binding of one of several ligands (EGF, TGF, amphiregulin) which leads to EGFR phosphorylation and oligodimerization. EGFR phosphorylation in turn activates the Ras/Raf/mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K)-Akt, and STAT pathways (3, 5).
EGFR overexpression has been reported to occur in 30–85% of human colon tumors (3, 5–8), and some, though not all studies, have linked EGFR expression levels with clinical outcome (7, 9). Mechanisms by which EGFR overexpression occurs in colon cancers have been shown to be mediated by polysomy of chromosome 7, or on rare occasions (<1%) by EGFR gene amplification (10). The length of a polymorphic CA repeat element present in intron 1 has also been shown to regulate EGFR levels (11).
In the present study we observed frequent overexpression of EGFR in colon cancer cell lines with microsatellite instability. Colon cancer can be broadly classified as microsatellite stable (MSS) or as having microsatellite instability (MSI) (12). While deregulation of β-catenin-TCF signaling is the primary event driving both of these forms of CRC, several mutational, cytogenetic and epigenetic differences exist between MSS and MSI tumors. These include the propensity for MSS tumors to be aneuploid whereas MSI tumors are largely diploid, and a higher frequency for methylation-mediated tumor suppressor inactivation in MSI tumors (12–14). Differences in histopathological presentation and prognosis between these two groups have been clearly defined (15–17), while differences in response to 5FU-based adjuvant chemotherapy have been suggested (18). MSI colon cancers can be further separated into familial (HNPCC) or sporadic MSI. Patients with familial MSI colon cancer inherit mutations in one of several DNA mismatch repair genes, though mutations in MLH1 and MSH2 are most common (12, 17). Tumors arise in these patients typically in the 4th decade of life following loss of heterozygosity (12). In comparison, sporadic MSI is driven largely by epigenetic silencing of the MLH1 locus with significantly later tumor onset (19). Repetitive elements are particularly prone to mutations, and truncating mutations due to mutations in repeat elements within the coding sequences of TGFβRII (20) and Bax (21) are frequently observed in MSI colon tumors
In the present study we identify a novel deletion mutation in a polyA(13) repeat element within the 3’UTR of EGFR in a high percentage of MSI colon cancers, which is linked to EGFR overexpression. Functional studies demonstrate that this mutation enhances EGFR mRNA stability. Importantly, we demonstrate that this mutation provides a growth advantage in MSI colon cancer cell lines that are devoid of activating mutations in downstream signaling mediators.
Materials and Methods
Cell lines and cell culture
The source of all cell lines has previously been described (22). The hypomorphic Dicer knockout cell line (Dld-1-Dicer −/−) and parental line were generously provided by the Vogelstein/Kinzler laboratory (23). All cells were maintained in modified minimal media (Invitrogen), supplemented with 10% fetal bovine serum (Invitrogen), 1% HEPES, 1% nonessential aminoacids, and 1% antibiotics, and were grown in monolayer at 37°C. For actinomycin D assays, cells were treated with 5µg/ml actinomycin D (Sigma). For determining sensitivity to cetuximab, cells were treated with cetuximab (20 µg/ml) for 24 hours. Effects on cell growth were determined in vitro by PI staining and FACS analysis as previously described (24).
Isolation of RNA and RT-PCR
The methods of RNA extraction, reverse transcription and real-time PCR were as previously described (24). Primers used were as follows: EGFR, F: ATGCTCTACAACCCCACCAC and R: GCCCTTCGCACTTCTTACAC; Dicer, F: AAATTGGCGAACTGGATGAC, R: GGAATTGCTTTTGGGTAGCA; p21, F: ATGTGTCCTGGTTCCCGTTTC, R: CATTGTGGGAGGAGCTGTGA. Actin primers: L:CACCTTCACCGTTCCAGTTT, R: GATGAGATTGGCATGGCTTT.
Western Blot
Westen blot analysis was performed as previously described (24). Anti-EGFR (2232) was obtained from Cell Signaling and anti-Dicer obtained from Abcam (Ab14601).
Determination of 3’UTR polyA and CA repeat length by direct sequencing
Genomic DNA extracted from colon cancer cell lines and from fresh frozen resected primary colorectal tumors were PCR amplified with PCR supermix (Invitrogen). For amplification of the polyA repeat within the EGFR 3’UTR the following primers were used ; F: TACAGAAACGCATCCAGCAA, and R: ACTTGTGGCTTGTGCTCCTT. Primers used for amplification of the EGFR 1st intron CA repeat were, F: GGCTCACAGCAAACTTCTCC and R: GCACACTTGGCACATTGA. In each case the Forward primer was used in subsequent direct sequencing reactions.
Determination of EGFR 3’UTR polyA and intron 1 CA repeat length by Fragment Analysis
The same forward primers used in the direct sequencing of the 3’UTR polyA repeat and 1st intron CA repeat were used with the exception that the FAM fluorescence dye (Applied Biosystems) was conjugated to the forward primer for the 3’UTR polyA repeat analysis, and the VIC fluorescence dye conjugated to the forward primer for the CA repeat analysis. Reverse primers were also the same with the exception of addition of the 7mer FARG (Fragment Analysis Research Group) consensus sequence to the 5’ end. PCR amplification was performed using Amplitaq Gold (Applied Biosystems). Fragment analysis was performed at the DNA Sequencing Facility at Albert Einstein College of Medicine.
Construction of Tetracyline regulated Luciferase-EGFR 3’UTR reporter construct
The entire 1720 bp 3’-UTR of EGFR was PCR amplified with the following primers; F: CCACGGAGGATAGTATGAGC, R: AGAGTGGAAATGAATATAGTTTTATT using Amplitaq Gold (Applied Biosystems). The purified PCR fragment was subcloned downstream of the luciferase gene in pGEM-Luc (Promega) linearized by StuI (New England Biolab). Due to PCR introduced error, clones containing A12 and A13 repeats were also identified at this time and likewise cloned downstream of the luciferase gene in pGEM-Luc. The Luciferase-EGFR 3’-UTR fragment containing either a A11, A12, or A13 (wild type) repeat was excised with BamHI and SalI, and cloned into BamHI and SalI digested pTRE-tight-BI-AcGFP (Clonetech). All constructs were sequence verified.
Transient transfection of siRNA and TET-OFF expression vectors
A siRNA pool of 4 siRNAs targeting the EGFR mRNA was obtained from (Dharmacon). Sequences of the sense strand were as follows: GAAGGAAACUGAAUUCAAAUU; GGAAAUAUGUACUACGAAAUU; CCACAAAGCAGUGAAUUUAUU; GUAACAAGCUCACGCAGUUUU. LIM1215 and HCT116 cell lines were transfected with EGFR siRNA (100 nM) using the Dharmafect-4 transfection reagent (Dharmacon).
To determine the effect of mono and dinucleotide deletion mutations within the polyA repeat within the EGFR 3’UTR, the pTRE-luciferase-EGFR 3’UTR A13 (WT), pTRE-luciferase-EGFR 3’UTR A12, or pTRE-luciferase-EGFR 3’UTR A11 vector was transiently transfected into HCT116 cells in combination with the Tet-off vector using the Profection transfection reagent (Promega). TK-Renilla was included as a control for transfection efficiency. Twelve hours post transfection, doxycycline was added to a final concentration of 25 ng/ml, and cells harvested 24 hours later using Passive Lysis Buffer (Promega). Luciferase assays performed using the dual luciferase assay reagents (Promega) and firefly luciferase expressed relative TK-Renilla.
Xenograft assays
Colon cancer cells were grown as xenografts in SCID mice as previously described (25). For determination of sensitivity to cetuximab and panitumumab, tumors were injected IP with PBS, cetuximab or panitumumab (1mg per mouse per day), for 14 days, beginning on the day of tumor cell injection. Upon sacrifice, tumors were extracted and volume calculated from measurements of the smallest (s) and longest (l) diameter based on the following formula: Volume = [(s2× l)× π] / 6, and measurement of tumor weight. For siRNA mediated EGFR knockdown in vivo, colon cancer cells were transfected with siRNA targeting EGFR or control siRNA (NT1) (Dharmacon) and injected into SCID mice 24 hours post-transfection. Mice were sacrificed 1 week post injection and tumor volume quantified as above.
Immunohistochemistry
EGFR expression was determined in formalin fixed paraffin-embedded tissue of colon cancer cell lines grown as xenografts in SCID mice using a monoclonal anti-human EGFR antibody from Dako (Real Carinteria, USA).
Results
EGFR is overexpressed in MSI colon cancer cell lines
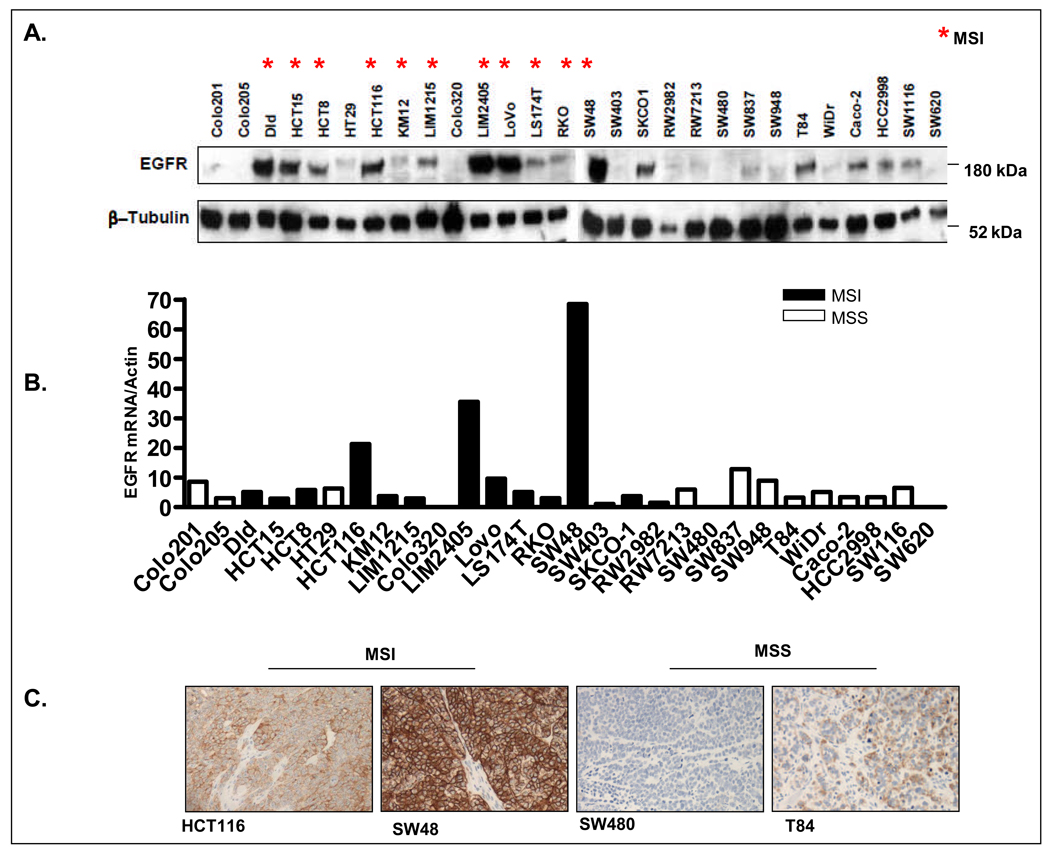
EGFR expression in a panel of 28 colon cancer cell lines was determined by western blot analysis. Remarkably, as shown in Figure 1, we observed strong EGFR expression in a number of MSI colon cancer cell lines. To determine whether this high level of EGFR protein expression correlated with increased EGFR mRNA expression, EGFR mRNA expression was determined by quantitative RT-PCR (Figure 1B). Significant correlation between EGFR mRNA and protein expression was observed across the cell line panel (r=0.65, p=0.002 Pearson’s correlation coefficient). Immunohistochemical staining for EGFR in cell lines grown as xenografts also confirmed strong EGFR expression in several MSI colon cancer cell lines (Figure 1C).
Figure 1.
EGFR overexpression in MSI colon cancer cell lines. A. Measurement of basal EGFR protein expression by western blot in a panel of 28 colon cancer cell lines. B. Determinatiom of EGFR mRNA level by quantitative RT-PCR in a panel of CRC cell lines. C. EGFR protein expression as determined by immunohistochemistry in 4 colon cancer cell lines grown as xenografts in SCID mice.
Overexpression of EGFR in MSI cell lines is not due to EGFR amplification
EGFR overexpression driven by EGFR gene amplification, though rare, has been reported in colon cancer (10). While MSI tumors tend to be genomically stable, we wished to eliminate the possibility that the observed EGFR overexpression in MSI cell lines was due to EGFR gene amplification. We therefore examined EGFR gene copy number by FISH (Fluorescence In-situ Hybridization) analysis in 3 EGFR overexpressing MSI lines. As shown in Supplementary Figure 1, no EGFR amplification was observed in the cell lines examined. Two of the 3 MSI cell lines demonstrated polysomy of chromosome 7, as demonstrated by the presence of >2 EGFR and chromosome 7 signals per nucleus. EGFR polysomy was also observed in 3/3 MSS cell lines examined, and was therefore not linked to the EGFR overexpression in MSI lines (Supplementary Figure 1).
EGFR expression does not correlate with length of the polymorphic dinucleotide (CA) repeat within intron 1 of the EGFR gene
Repetitive sequences are particularly prone to mutation in MSI tumors. Previous reports have demonstrated that EGFR transcriptional efficiency is dependent upon the length of a dinucleotide CA repeat present within intron 1 of the EGFR gene, with the efficiency of transcription proportionally reduced with increasing repeat length (11). Other studies, however, have failed to demonstrate this link (26, 27). To determine whether EGFR expression was linked to the length of the CA repeat in the colon cancer cell line panel, the length of the repeat was determined by fragment analysis. As the length of the CA repeat was heterozygous in the majority of the cell lines (Supplementary Figure 2A), the sum of the repeat length of the two alleles was computed. As shown in Supplementary Figure 2B, the two allele repeat length ranged from 36 to 52 across the cell line panel, with a repeat length of 40 observed in the majority of cell lines. However, no significant correlation between EGFR intron 1 CA repeat length and EGFR mRNA expression was observed (r = 0.29 P=0.16).
The 3’UTR of EGFR contains a variant A13/A14 repeat sequence that is frequently mutated in EGFR overexpressing MSI colon cancer cell lines
To determine the basis of strong EGFR expression in MSI CRC lines, the EGFR gene was examined for the presence of mono or dinucleotide repeat sequences. While no repeat sequences were detected within the coding sequence, a 13 Adenine repeat sequence within the 3’-UTR was identified (Supplementary Figure 3A). Prior to determining whether this sequence was mutated in MSI colon cancer cell lines, it was important to determine whether the length of this repeat element varied among individuals in normal tissue. To determine this, we examined the length of the EGFR 3’UTR polyA repeat in 27 normal colon tissue samples by fragment analysis. Eleven samples (41%) contained an A14 repeat and 16 samples (59%) contained an A13 repeat at this locus. None of the normal samples showed variants at this repeat element that were fewer than 13A’s in length.
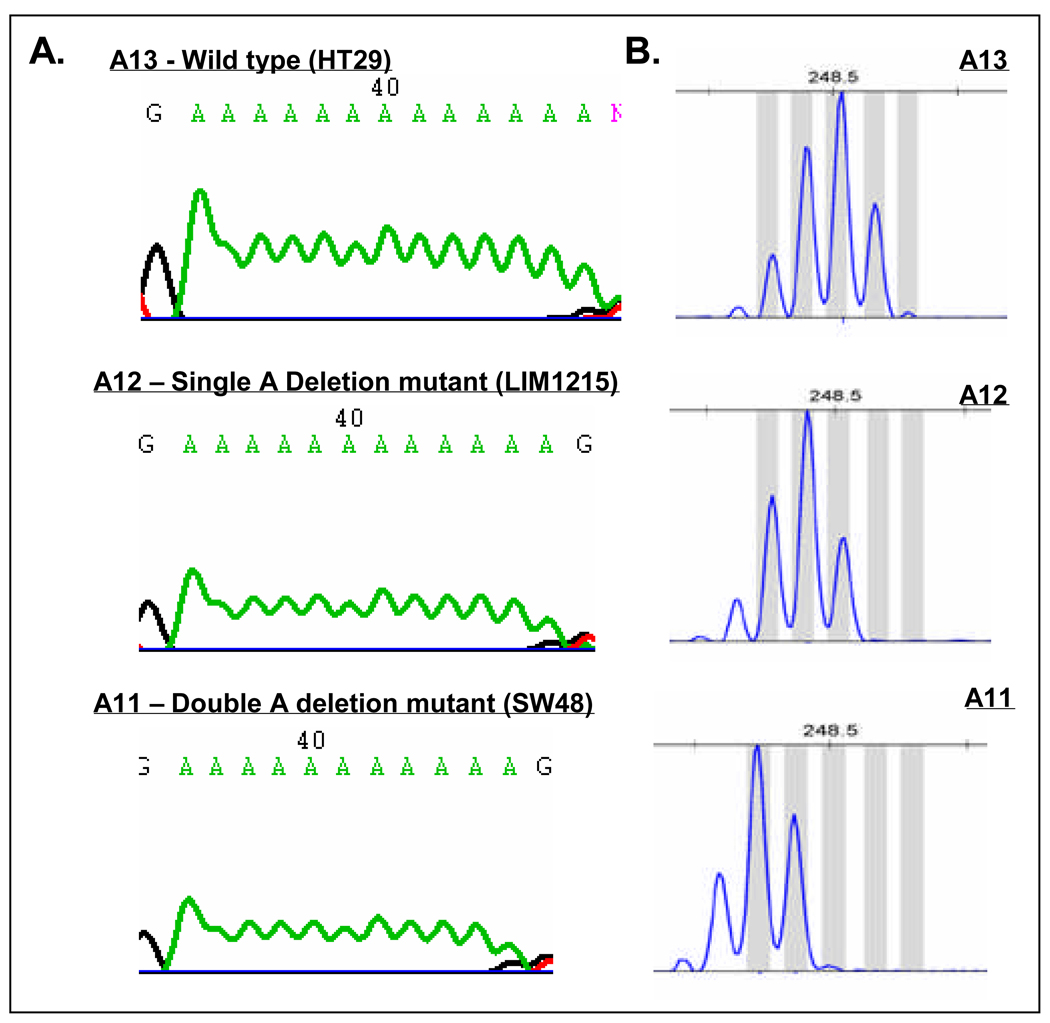
To determine whether mutations within this repeat sequence were linked to the increased EGFR expression in MSI CRC, the length of the repeat was determined in the cell line panel by fragment analysis. As shown in Supplementary Table 1, 7 out of the 11 (64%) MSI CRC cell lines analyzed demonstrated the presence of a 1 or 2 base deletion mutation within this element, resulting in an A12 or A11 repeat. The presence of 1 or 2 base deletion mutations within the A13/A14 repeat was further confirmed by PCR amplification followed by direct sequencing in the 7 mutant MSI cell lines (Figure 2). None of the MSS cell lines tested (0/17) harbored a deletion mutation in the polyA tract (Figure 2, Supplementary Table 1).
Figure 2.
Mono and dinucleotide deletion mutations within polyA repeat region in the EGFR 3’UTR. A. Representative chromatograms from wild type and deletion mutant cell lines. B. Validation of direct sequencing of EGFR3’UTR by fragment analysis. The most dominant peak represents the true fragment length. The first peak to the right of the dominant peak represents an adenine addition. Other peaks likely represent polymerase slippage.
To determine the link between mutations within the 3’UTR of EGFR and EGFR expression, we rank ordered the cell lines by EGFR mRNA expression (Supplementary Table 2). This analysis demonstrated that 4 of the 5 cell lines that most highly expressed EGFR mRNA were MSI lines harboring a deletion mutation. In contrast, none of the 5 cell lines with the lowest EGFR mRNA expression were EGFR mutant. A t-test revealed that cell lines harboring a deletion mutation in the EGFR 3’UTR had significantly higher EGFR mRNA (P=0.032) and protein (P=0.023) expression compared to EGFR 3’UTR wild-type cell lines.
Deletion mutations in the poly A13 repeat sequence of the EGFR 3’UTR occur at a high frequency in MSI colon tumors in vivo
To determine whether the mutation identified in the 3’UTR of EGFR in colon cancer cell lines was also evident in freshly resected MSI colon tumors, we extended this analysis to DNA extracted from a cohort of 16 MSI tumors. Consistent with the cell line data, 11 out of 16 (68.7%) MSI tumors harbored a deletion mutation (Supplementary Table 1). Of the 11 mutations identified, 5 were mononucleotide (A12) deletions (29.4%), 2 were dinucleotide (A11) deletions (11.7%) and 4 were heterozygous (A12/A11) deletions (23.5%). None of the MSS tumors tested (0/15) harbored a deletion mutation in the polyA tract (Supplementary Table 1).
Inactivation of Dicer in colon cancer cells has no effect on EGFR mRNA expression
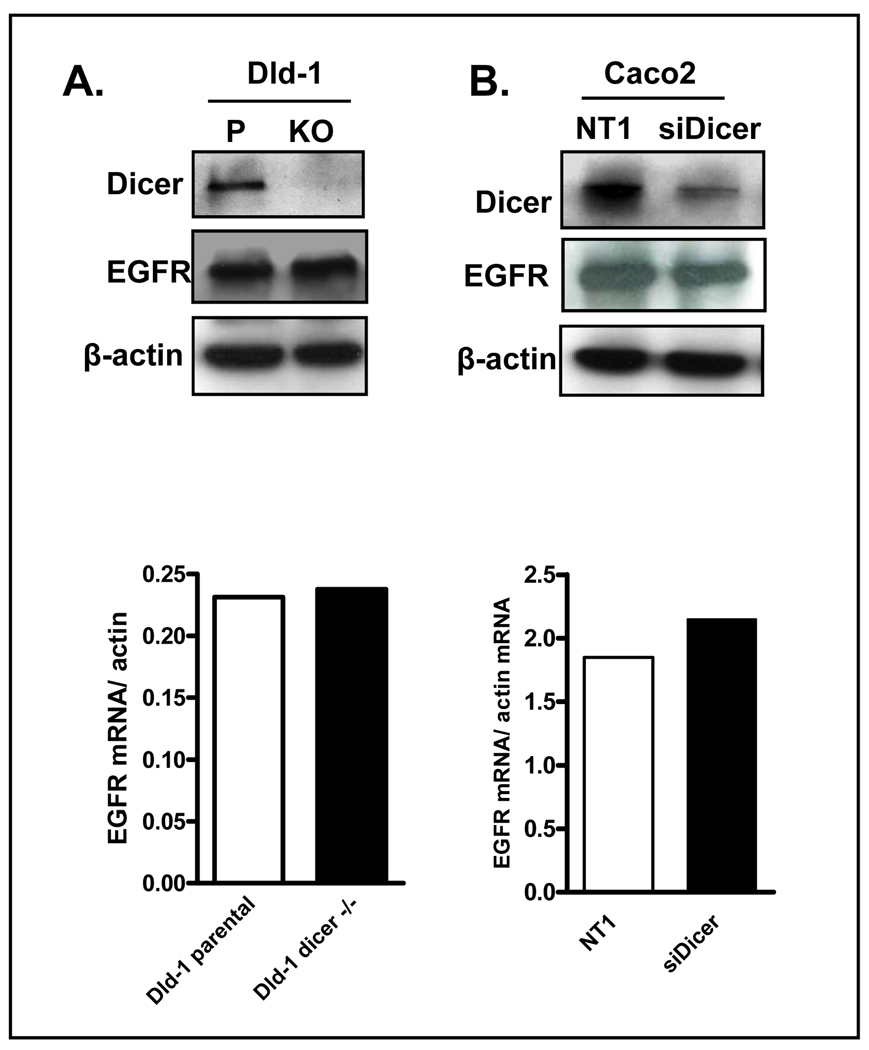
As the 3’-UTR of mRNA transcripts is a major target for microRNA-mediated transcriptional and translational regulation, we next tested the hypothesis that mutation within the 3’UTR of EGFR may lead to decreased binding of a microRNA, thus leading to increased EGFR expression. To test this hypothesis we utilized the Dld1 Dicer−/− cell line in which the helicase domain of the Dicer gene, a key processor of pre-microRNA into mature microRNA, has been deleted by homologous recombination (23). The Dld cell line was selected as it was WT for the repeat element within the EGFR 3’UTR. However, as shown Figure 3A, no difference in EGFR mRNA or protein expression was observed between parental and Dicer−/− Dld1 cells, indicating Dicer processed miRNAs do not play a role in regulating EGFR expression. To further confirm this finding, we examined the effect of transient siRNA-mediated downregulation of Dicer in the MSS Caco-2 colon cancer cell line. As shown in Figure 3B, despite efficient downregulation of Dicer protein, no change in EGFR protein or mRNA expression was observed.
Figure 3.
Role of microRNAs in EGFR mRNA and protein expression. A. Western blot of dicer and EGFR in Dicer disrupted Dld-1 cell lines compared to their parental cell lines, demonstrating no increase in EGFR protein expression. Bottom panel, Measurement of EGFR mRNA expression by quantitative RT-PCR in parental and Dicer−/− Dld cells. B. Western blot of dicer and EGFR in dicer-silenced Caco-2 cells. Bottom panel: Measurement of EGFR mRNA expression by quantitative RT-PCR in parental and Dicer−/− Dld cells.
Mutation of the polyA tract within the EGFR 3’UTR stabilizes EGFR mRNA
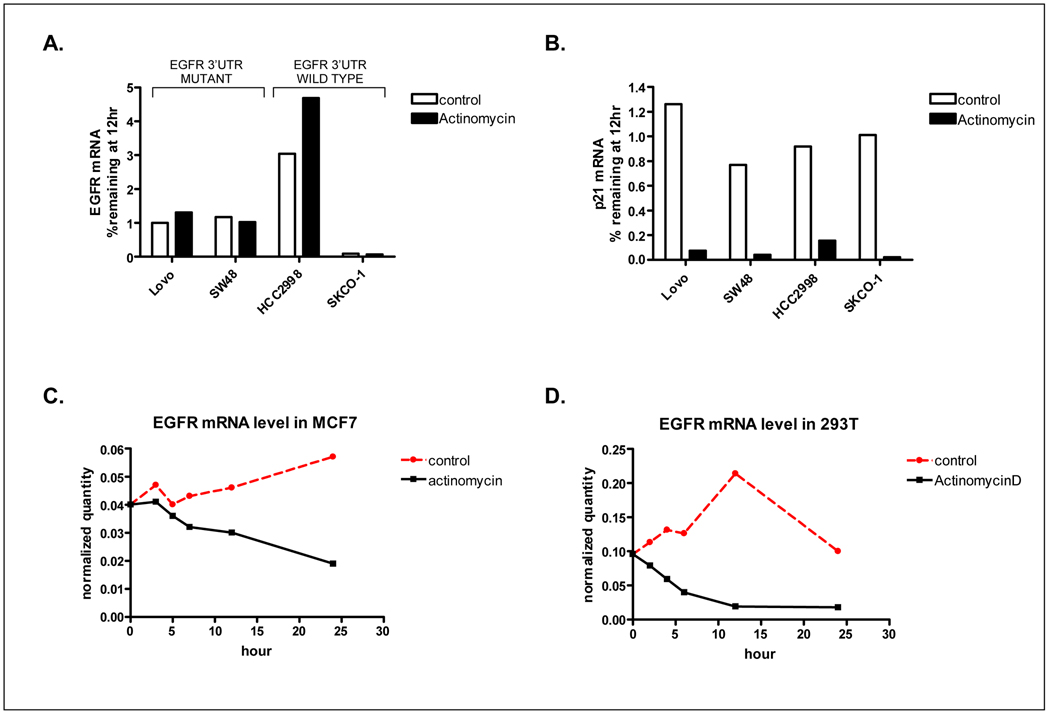
An additional role of the 3’UTR is in the regulation of mRNA stability. Initially, to determine whether mutations in the EGFR 3’UTR resulted in increased endogenous EGFR mRNA stability, we attempted to compare the rate of EGFR degradation in EGFR 3’UTR WT and mutant cell lines following treatment with the inhibitor of transcription, actinomycin D. However, as shown in Figure 4A, we observed minimal decay of EGFR mRNA following actinomycin D treatment in each of the colon cancer cell lines tested. This stability was specific to EGFR as expression of p21 which was tested in parallel was reduced by 90% in each cell line 12h following actinomycin D treatment (Figure 4B). Furthermore, this observation was unique to colon cancer cell lines, as robust decay of EGFR following actinomycin D treatment was observed in MCF7 breast cancer cells and 293T kidney epithelial cells (Figure 4C and D).
Figure 4.
(A) Determination of EGFR mRNA stability in EGFR 3’UTR WT and mutant cell lines by actinomycin D treatment. Two EGFR 3’UTR mutant cell lines (Lovo, SW48), and 2 WT lines (HCC2998, SK-CO-1) were treated with actinomycin D for 12h. mRNA was isolated and EGFR levels determined by Q-RT-PCR. (B) Parallel determination of p21 mRNA levels. (C–D) Determination of EGFR mRNA stability by actinomycin D treatment of breast (C) and kidney epithelial (D) cells.
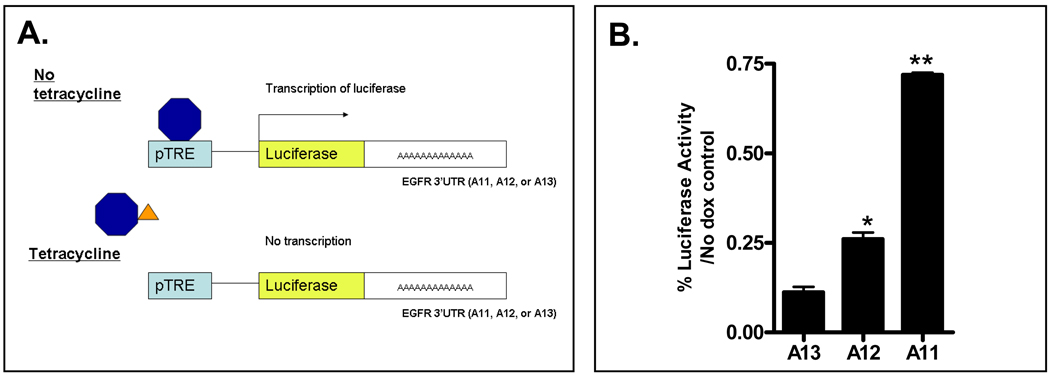
Therefore, to directly determine whether mutations within the polyA tract of the EGFR 3’UTR results in increased EGFR mRNA stability, we designed a Tet-Off system in which the entire 1720bp EGFR 3’-UTR, containing either the wild type polyA(13) tract, or A(12) or A(11) deletion mutations, was subcloned downstream of the luciferase gene. This construct was in turn subcloned into a pTRE-BI-AcGFP vector in which addition of doxycycline turns off transcription of the luciferase-EGFR 3’UTR construct (Figure 5A). As shown in Figure 5B, the decay in luciferase activity over time following doxycycline addition was significantly slower from constructs containing mutant polyA(12) or A(11) 3’UTRs compared to the WT A(13) construct, indicating truncating mutations within the A13 repeat result in a reduced rate of EGFR mRNA degradation.
Figure 5.
Single and dinucleotide deletion mutations within the polyA repeat of the EGFR 3’UTR increase mRNA stability. HCT116 cells were transiently transfected with pTRE-luciferase-EGFR 3’UTR A13 (WT), pTRE-luciferase-EGFR 3’UTR A12, or pTRE-luciferase-EGFR 3’UTR A11 in combination with the Tet-off vector, and the TK-Renilla transfection efficiency control. Twelve hours post transfection, doxycycline was added to a final concentration of 25 ng/ml. Cells were harvested at 24 hours and Luciferase activity determined. Values shown are the percentage of firefly luciferase in doxycycline treated cells relative to untreated cells. All values are corrected for TK-Renilla luciferase activity. Values shown are mean ± SEM from a representative experiment performed in triplicate (*P<0.05, **P<0.0005).
Inhibition of EGFR reduces growth of some but not all EGFR 3’UTR mutant colon cancer cell lines
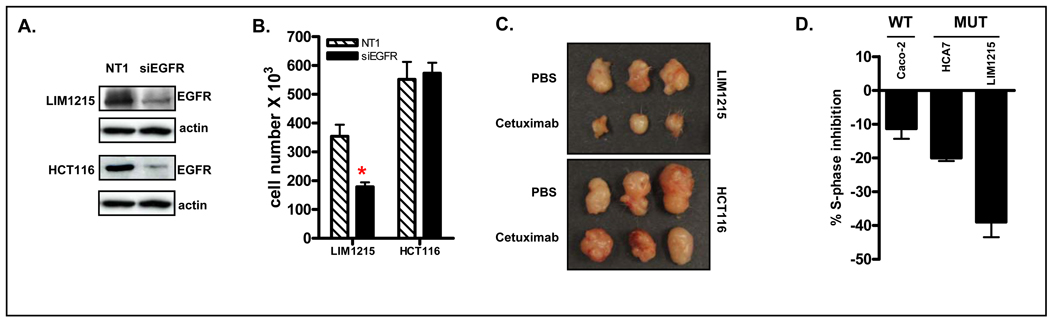
Having identified mutations within the 3’UTR of EGFR which in turn was linked to increased EGFR mRNA expression, it was important to determine whether this was a driver or bystander mutation in MSI colon cancer. To address this, EGFR mRNA was downregulated using siRNA in the LIM1215 MSI cell line, which harbors a mutation within the EGFR 3’UTR. As shown in Figure 6A, efficient siRNA-mediated downregulation of EGFR was achieved. Relative to cells transfected with a non-targeting siRNA, a 40% inhibition of cell growth was observed 48 hours after EGFR siRNA transfection (Figure 6B). Similar effects were observed in vivo where siRNA-mediated EGFR downregulation resulted in reduced growth of LIM1215 xenografts compared to cells transfected with non-targeting siRNA (Supplementary Figure 5). Furthermore, targeted inhibition of EGFR using the anti-EGFR antibodies panitumumab or cetuximab resulted in significant inhibition of LIM1215 cell growth in vivo and in vitro (Figure 6C, Supplementary Figure 4A). In contrast, however, siRNA-mediated EGFR downregulation or pharmacological inhibition had minimal effects on growth of a second MSI EGFR mutant cell line, HCT116 (Figure 6, Supplementary Figure 4A). Notably, HCT116 cells harbor activating mutations in Ras and PIK3CA, two critical signaling mediators located downstream of EGFR, and which have been shown to confer resistance to EGFR inhibition (24), whereas the LIM1215 is WT at these loci.
Figure 6.
EGFR inhibition reduces cell proliferation in some but not all colon cancer cell lines harboring an EGFR 3’UTR mutation. (A). siRNA-mediated downregulation of EGFR in HCT116 and LIM1215 colon cancer cells. (B). siRNA-mediated downregulation of EGFR inhibits growth of LIM1215 but not HCT116 cells in vitro. (C) Targeted inhibition of EGFR using cetuximab inhibits growth of LIM1215 but not HCT116 colon cancer cells in vivo. (D) Comparison of the magnitude of cetuximab-induced growth inhibition in EGFR 3’UTR WT (Caco-2) and mutant (LIM1215 and HCA7) cell lines. Shown is the percentage change of cells in S-phase following 24h treatment with cetuximab (20 µg/ml). Values shown are mean±SEM, n=4 independent experiments.
As all the MSI lines with EGFR 3’UTR mutations screened in our original panel harbored mutations in signaling mediators downstream of EGFR (24), or in the case of SW48 cells, an activating G719S mutation in the kinase domain of EGFR itself (COSMIC), we screened an additional 5 MSI cell lines to identify further lines that harbored a mutation within the EGFR 3’UTR but which were WT for the downstream signaling regulators, K-Ras, BRAF, PIK3CA and PTEN. A second cell line, HCA7, that contained a single A deletion mutation in the EGFR 3’UTR but which was wild type for K-Ras, BRAF, PIK3CA and PTEN was identified. As expected, EGFR mRNA and protein levels were elevated in this line compared to 4 EGFR 3’UTR WT lines (Supplementary Figure 5A).
Next, to directly test the functional significance of EGFR 3’UTR mutations we compared the effect of EGFR inhibition on cell growth in 2 EGFR 3’UTR mutant (LIM1215 and HCA7) and the EGFR 3’UTR wild type cell line, Caco-2, but which were all WT for Ras, BRAF, PIK3CA and PTEN. As shown in Figure 6D, cetuximab treatment resulted in greater inhibition of the percentage of cells in S-phase in EGFR 3’UTR mutant (LIM1215 and HCA7) cells compared to EGFR 3’UTR WT Caco-2 cells. Collectively therefore, these results indicate that mutations within the 3’UTR of EGFR provides a growth advantage in a subset of MSI colon cancer cell lines, specifically those which do not harbor activating mutations in downstream signaling mediators.
Discussion
In this study we identify and characterize a novel mutation in a polyA repeat sequence located within the 3’UTR of the critical signaling mediator, EGFR, in MSI colon cancers. Mono or dinulceotide deletion mutations were identified in approximately 68% of MSI colon cancer cell lines and primary colon cancer specimens. Importantly, mutations within the EGFR 3’UTR were associated with increased EGFR mRNA and protein expression. Notably, we also observed that this site is polymorphic, with A13 and A14 repeats observed at frequencies of 59% and 41% respectively, in normal colonic tissue.
The 3’UTR serves an important function in the post-transcriptional regulation of transcribed sequences. Mechanistically, this has been shown to be mediated by altered RNA stability regulated by binding of RNABPs, or as is becoming increasingly appreciated, through binding of microRNAs. Our findings that EGFR levels were unchanged in Dld-Dicer−/− cells or following transient Dicer knockdown in Caco-2 cells suggests microRNAs do not play a significant role in EGFR regulation in colon cancer cells. However, we note that while elimination of the helicase domain of Dicer results in the failure to process the majority of pre-microRNAs, a subset of microRNAs were found to be still processed into the mature form in these cells (23). The possibility that a microRNA within this subset may contribute to the post-transcriptional regulation of EGFR mRNA therefore cannot be eliminated.
In comparison, utilizing a Tet-Off system, we observed that mutations in the poly(A) repeat of the EGFR 3’UTR resulted in increased EGFR mRNA stability. This finding is consistent with a previous report which demonstrated that EGFR expression is post-transcriptionally regulated in breast cancer cells, with a 260 nt cis acting regulatory element containing 4 AU-rich elements within the 3’UTR of EGFR shown to play an important regulatory role (28). Remarkably, and as shown in Supplementary Figure 3, the polyA tract is located within the center of this element, flanked on the 5’ and 3’ ends by two ARE elements. Furthermore, our observation that EGFR mRNA is stabilized in response to actinomycin D treatment is consistent with previous studies demonstrating actinomycin D-mediated stabilization of transcripts containing class I and II AU-rich elements (29). One possibility therefore, is that mutations within the polyA repeat modulates the ability of AU-rich element binding proteins to interact with the EGFR 3’UTR, thus altering mRNA stability.
The present finding that mutations in the 3’UTR of a gene can result in increased mRNA expression is consistent with a recent report by Ruggiero et al who demonstrated that a single A deletion within a polyA(8) element within the 3’UTR of the CEACAM gene was linked to increased CEACAM expression (30). Furthermore, the possibility that mutations within the 3’UTR of genes may contribute in altered gene expression in MSI colon cancers on a global scale is suggested by the finding of diPietro et al who observed that a significant percentage of genes upregulated in MSI relative to MSS colon cancer contained simple repeat sequences within their 3’-UTR (31).
Given the high mutation frequency observed in MSI colon tumors, particularly in repeat elements of increasing length (32), it is important to determine whether specific mutations are driver or bystander mutations. In this regard, we were able to demonstrate that siRNA-mediated knockdown of EGFR results in growth inhibition in some but not all EGFR mutant MSI lines. Notably, the EGFR 3’UTR mutant LIM1215 and HCA7 cell lines, in which EGFR silencing induced growth inhibition, are devoid of mutations in the Ras/BRAF/MAPK and PI3K/PTEN/AKT pathways, which mediate downstream EGFR signaling. In contrast, HCT116 cells which harbor mutations in both K-Ras and PIK3CA, were not sensitive to EGFR downregulation or pharmacological inhibition of the EGFR. Furthermore, when response to EGFR inhibitors was examined specifically in cells lines with no known mutations in downstream signaling mediators, we observed greater growth inhibition in cell lines with mutant EGFR 3’UTR. Collectively these findings indicate that EGFR overexpression driven by EGFR 3’UTR mutations provides a growth advantage specifically in colon cancer cells devoid of constitutively activating downstream mutations.
Mutations in the EGFR 3’UTR and subsequent EGFR overexpression may also provide a growth advantage through other mechanisms. First, activating mutations in the EGFR kinase domain, though rare, occur in a small percentage of colon tumors (33, 34). Consistent with this finding, among the cell lines examined in the present study, SW48 cells, which we showed to have a mutation in the EGFR 3’UTR and express high levels of EGFR, also harbor an activating G719S mutation in the EGFR kinase domain (COSMIC database, Sanger Institute), as observed in lung tumors (35). Mutations in the polyA repeat region of the EGFR 3’UTR which increase EGFR expression may therefore serve to further enhance EGFR-dependent cell growth in a subset of colon tumors harboring activating mutations in the EGFR kinase domain.
Second, a novel role for EGFR in the activation of SGLT1, independent of its kinase activity, was recently demonstrated (36). Notably, in this study the authors found that the KM12 cell line, which we found to harbor a mutation in the EGFR 3’UTR, underwent apoptosis in response to siRNA-mediated EGFR downregulation when cultured in low glucose media. This finding raises the possibility that mutations in the EGFR 3’UTR and subsequent overexpression may contribute to cell survival in a larger subset of MSI tumors depending upon the growth conditions of the tumor.
In conclusion, we have identified a novel mutation in the 3’UTR of EGFR in a high percentage of MSI colon tumors. This mutation, through enhanced EGFR mRNA stability, results in EGFR overexpression. This in turn provides a growth advantage to a subset of MSI colon cancer cells.
Supplementary Material
Acknowledgements
This work was supported by National Institute of Health grants, CA100823 and CA123316 (JMM) and Spanish Fondo de Investigaciones Sanitarias (Grant Number: FIS 05/1394, CP05/00256) and Fundación de Investigación Médica Mutua Madrileña (DA). We thank David Reynolds from the AECOM DNA sequencing facility for advice and assistance with fragment analysis. We thank Dr. Kathyryn Tanaka for assistance with EGFR immunohistochemistry
REFERENCES
- 1.Hegde SR, Sun W, Lynch JP. Systemic and targeted therapy for advanced colon cancer. Expert review of gastroenterology & hepatology. 2008;2:135–149. doi: 10.1586/17474124.2.1.135. [DOI] [PubMed] [Google Scholar]
- 2.Mendelsohn J, Baselga J. The EGF receptor family as targets for cancer therapy. Oncogene. 2000;19:6550–6565. doi: 10.1038/sj.onc.1204082. [DOI] [PubMed] [Google Scholar]
- 3.Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- 4.Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Critical reviews in oncology/hematology. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 5.Quesnelle KM, Boehm AL, Grandis JR. STAT-mediated EGFR signaling in cancer. J Cell Biochem. 2007;102:311–319. doi: 10.1002/jcb.21475. [DOI] [PubMed] [Google Scholar]
- 6.McKay JA, Murray LJ, Curran S, et al. Evaluation of the epidermal growth factor receptor (EGFR) in colorectal tumours and lymph node metastases. Eur J Cancer. 2002;38:2258–2264. doi: 10.1016/s0959-8049(02)00234-4. [DOI] [PubMed] [Google Scholar]
- 7.Galizia G, Lieto E, Ferraraccio F, et al. Prognostic significance of epidermal growth factor receptor expression in colon cancer patients undergoing curative surgery. Ann Surg Oncol. 2006;13:823–835. doi: 10.1245/ASO.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 8.Shia J, Klimstra DS, Li AR, et al. Epidermal growth factor receptor expression and gene amplification in colorectal carcinoma: an immunohistochemical and chromogenic in situ hybridization study. Mod Pathol. 2005;18:1350–1356. doi: 10.1038/modpathol.3800417. [DOI] [PubMed] [Google Scholar]
- 9.Galizia G, Lieto E, Orditura M, et al. Epidermal growth factor receptor (EGFR) expression is associated with a worse prognosis in gastric cancer patients undergoing curative surgery. World J Surg. 2007;31:1458–1468. doi: 10.1007/s00268-007-9016-4. [DOI] [PubMed] [Google Scholar]
- 10.Shia J, Klimstra DS, Li AR, et al. Epidermal growth factor receptor expression and gene amplification in colorectal carcinoma: an immunohistochemical and chromogenic in situ hybridization study. Mod Pathol. 2005;18:1350–1356. doi: 10.1038/modpathol.3800417. [DOI] [PubMed] [Google Scholar]
- 11.Gebhardt F, Zanker KS, Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J Biol Chem. 1999;274:13176–13180. doi: 10.1074/jbc.274.19.13176. [DOI] [PubMed] [Google Scholar]
- 12.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 13.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goel A, Nagasaka T, Arnold CN, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 15.Kim H, Jen J, Vogelstein B, Hamilton SR. Clinical and pathological characteristics of sporadic colorectal carcinomas with DNA replication errors in microsatellite sequences. Am J Pathol. 1994;145:148–156. [PMC free article] [PubMed] [Google Scholar]
- 16.Samowitz WS, Curtin K, Ma KN, et al. Microsatellite instability in sporadic colon cancer is associated with an improved prognosis at the population level. Cancer Epidemiol Biomarkers Prev. 2001;10:917–923. [PubMed] [Google Scholar]
- 17.Markowitz S. DNA repair defects inactivate tumor suppressor genes and induce hereditary and sporadic colon cancers. J Clin Oncol. 2000;18:75S–80S. [PubMed] [Google Scholar]
- 18.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cunningham JM, Christensen ER, Tester DJ, et al. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- 20.Markowitz S, Wang J, Myeroff L, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 21.Rampino N, Yamamoto H, Ionov Y, et al. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 22.Mariadason JM, Arango D, Shi Q, et al. Gene expression profiling-based prediction of response of colon carcinoma cells to 5-fluorouracil and camptothecin. Cancer Res. 2003;63:8791–8812. [PubMed] [Google Scholar]
- 23.Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jhawer M, Goel S, Wilson AJ, et al. PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab. Cancer Res. 2008;68:1953–1961. doi: 10.1158/0008-5472.CAN-07-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo FR, Yang Z, Dong H, et al. Correlation of pharmacokinetics with the antitumor activity of Cetuximab in nude mice bearing the GEO human colon carcinoma xenograft. Cancer Chemother Pharmacol. 2005;56:455–464. doi: 10.1007/s00280-005-1022-3. [DOI] [PubMed] [Google Scholar]
- 26.McKay JA, Murray LJ, Curran S, et al. Evaluation of the epidermal growth factor receptor (EGFR) in colorectal tumours and lymph node metastases. Eur J Cancer. 2002;38:2258–2264. doi: 10.1016/s0959-8049(02)00234-4. [DOI] [PubMed] [Google Scholar]
- 27.Buisine MP, Wacrenier A, Mariette C, et al. Frequent mutations of the CA simple sequence repeat in intron 1 of EGFR in mismatch repair-deficient colorectal cancers. World J Gastroenterol. 2008;14:1053–1059. doi: 10.3748/wjg.14.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balmer LA, Beveridge DJ, Jazayeri JA, Thomson AM, Walker CE, Leedman PJ, et al. Identification of a novel AU-Rich element in the 3' untranslated region of epidermal growth factor receptor mRNA that is the target for regulated RNA-binding proteins. Mol Cell Biol. 2001;21:2070–2084. doi: 10.1128/MCB.21.6.2070-2084.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruggiero T, Olivero M, Follenzi A, Naldini L, Calogero R, Di Renzo MF. Deletion in a (T)8 microsatellite abrogates expression regulation by 3'-UTR. Nucleic Acids Res. 2003;31:6561–6569. doi: 10.1093/nar/gkg858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.di Pietro M, Sabates Bellver J, Menigatti M, et al. Defective DNA mismatch repair determines a characteristic transcriptional profile in proximal colon cancers. Gastroenterology. 2005;129:1047–1059. doi: 10.1053/j.gastro.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 32.Sammalkorpi H, Alhopuro P, Lehtonen R, et al. Background mutation frequency in microsatellite-unstable colorectal cancer. Cancer Res. 2007;67:5691–5698. doi: 10.1158/0008-5472.CAN-06-4314. [DOI] [PubMed] [Google Scholar]
- 33.Nagahara H, Mimori K, Ohta M, et al. Somatic mutations of epidermal growth factor receptor in colorectal carcinoma. Clin Cancer Res. 2005;11:1368–1371. doi: 10.1158/1078-0432.CCR-04-1894. [DOI] [PubMed] [Google Scholar]
- 34.Barber TD, Vogelstein B, Kinzler KW, Velculescu VE. Somatic mutations of EGFR in colorectal cancers and glioblastomas. The New England journal of medicine. 2004;351:2883. doi: 10.1056/NEJM200412303512724. [DOI] [PubMed] [Google Scholar]
- 35.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 36.Weihua Z, Tsan R, Huang WC, et al. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385–393. doi: 10.1016/j.ccr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.