Abstract
On entry into mitosis, many transcription factors dissociate from chromatin, resulting in global transcriptional shutdown. During mitosis, some genes are marked to ensure the inheritance of their expression in the next generation of cells. The nature of mitotic gene marking, however, has been obscure. Brd4 is a double bromodomain protein that localizes to chromosomes during mitosis and is implicated in holding mitotic memory. In interphase, Brd4 interacts with P-TEFb and functions as a global transcriptional coactivator. We found that throughout mitosis, Brd4 remained bound to the transcription start sites of many M/G1 genes that are programmed to be expressed at the end of, or immediately after mitosis. In contrast, Brd4 did not bind to genes that are expressed at later phases of cell cycle. Brd4 binding to M/G1 genes increased at telophase, the end phase of mitosis, coinciding with increased acetylation of histone H3 and H4 in these genes. Increased Brd4 binding was accompanied by the recruitment of P-TEFb and de novo M/G1 gene transcription, the events impaired in Brd4 knockdown cells. In sum, Brd4 marks M/G1 genes for transcriptional memory during mitosis, and upon exiting mitosis, this mark acts as a signal for initiating their prompt transcription in daughter cells.
INTRODUCTION
The states of gene expression, either active or silenced, are inherited through generations of somatic cells, providing a basis for stable cellular functions (Ringrose and Paro, 2004; Egli et al., 2008; Ng and Gurdon, 2008). With respect to inheritance of gene expression, mitosis poses a mystery, because most transcription factors dissociate from chromosomes during that time (Delcuve et al., 2008; Egli et al., 2008). The massive dissociation of transcription factors accompanies global cessation of transcription, which likely erases gene expression patterns established before mitosis (Martinez-Balbas et al., 1995; Gottesfeld and Forbes, 1997; Delcuve et al., 2008). Transcription factors that dissociate from mitotic chromosomes include RNA polymerase II, Oct1,2, Sp1,3, Pax 3, E2F1, Brg1, Brm, TFIIB, and TFIID among others. Mitotic nuclei are thus thought to represent a transcriptionally uncommitted state. Consistent with this idea, mitotic nuclei serve as better donors of genome transfer compared with interphase nuclei (Egli et al., 2008). The transcriptionally inert state is reversed at the end of mitosis, when RNA polymerase II (Pol II) and other transcription factors are sequentially reloaded onto chromosomes, leading to the initiation of transcription at telophase (Prasanth et al., 2003). Although the prior modes of transcription established in parental cells would be erased upon entry into mitosis, some “memory” remains during mitosis, allowing daughter cells to reproduce an inherited pattern of gene expression after mitosis. Relevant to this memory, some core histones retain their acetylation mark during mitosis, although histone acetylation is generally reduced during mitosis (Kruhlak et al., 2001; Nishiyama et al., 2006). Particularly, regions at or near the transcription start sites (TSS) are shown to be enriched with acetylated histone H3 and H4 as well as H3 with trimethylated lysine K4 (H3K4me3) during mitosis (Kouskouti and Talianidis, 2005; Valls et al., 2005). Moreover, some transcription factors remain associated with mitotic chromosomes to mark specific genes in the genome (Delcuve et al., 2008; Egli et al., 2008). For example, a lineage specific transcription factor, Runx2 binds to mitotic chromosomes through specific DNA sequences to ensure the expression of lineage specific genes in the progeny cells (Young et al., 2007a,b). The zinc finger protein, CTCF which acts as an insulator during interphase, also remains on mitotic chromosomes to take part in the inheritance of long-range chromatin interactions at the Igf2/H19 locus (Burke et al., 2005). Similarly, transcription factors such as HSF2 and FoxI1 remain bound on mitotic chromosomes to mark target genes (Xing et al., 2005; Yan et al., 2006). In addition, a fraction of the TATA-binding protein (TBP) remains bound to certain areas of chromatin (Christova and Oelgeschlager, 2002). A recent article found that the chromosome-bound TBP marks promoter regions of many genes by inhibiting condensin-mediated chromatin compaction (Xing et al., 2008). Partial retention of polycomb proteins on mitotic chromosomes has also been documented, which is involved in heritable gene silencing (Saurin et al., 1998).
The double bromodomain protein Brd4 possesses certain characteristics that are compatible with a role in keeping transcriptional memory during mitosis. Brd4 localizes to the noncentromeric regions of mitotic chromosomes in many vertebrate cells. These cells range from cells of zebrafish embryos to mammalian embryonic stem cells, primary and established fibroblasts, virus-transformed cells to macrophages (Dey et al., 2000, 2003; You et al., 2004; Nishiyama et al., 2006; Toyama et al., 2008). Brd4 binds to acetylated tails of histones H3 and H4 with the preference for acetylated lysine (K) 9/14 of H3 and diacetylated K5/K12 of H4 (Dey et al., 2000, 2003; Nishiyama et al., 2008). Moreover, Brd4 plays a key role in Pol II–dependent transcription during interphase (Jang et al., 2005; Yang et al., 2005; Wu and Chiang, 2007). This activity of Brd4 is mediated primarily by its ability to interact with P-TEFb, a kinase that phosphorylates Serine 2 (S2) of the C-terminal domain (CTD) of Pol II, thus promoting transcriptional elongation. Through this activity, Brd4 is shown to act as a transcriptional coactivator of many cellular genes (Jang et al., 2005; Yang et al., 2005; Bisgrove et al., 2007; Mochizuki et al., 2008). For example, we previously showed that Brd4 is recruited to the promoter of many G1 genes and stimulates G1 gene transcription by corecruiting P-TEFb (Mochizuki et al., 2008). In an independent study, transcription of several G1 genes was also reported to be dependent on Brd4 (Yang et al., 2008). Recently, Brd4 was shown to be recruited to many genes in macrophages activated by bacterial lipopolysaccharides, prompting the recruitment of P-TEFb, and subsequent transcription elongation of these genes (Hargreaves et al., 2009). Brd4 was also shown to interact with NF-κB, stimulating transcription linked to inflammation (Huang et al., 2008).
In this study, we asked 1) whether Brd4 marks specific genes during mitosis to retain their transcriptional memory and 2) whether Brd4 helps retrieve the memory in daughter cells by stimulating transcription after mitosis. By chromatin immunoprecipitation (ChIP) analysis, we found that Brd4 remains bound to the TSS of many genes throughout mitosis. Brd4-bound genes mostly belonged to the M/G1 genes that are destined to be expressed at the end of and immediately after mitosis. Genes expressed at later stages of cell cycle were not marked by Brd4. Correlating with Brd4-dependent gene marking, many M/G1 genes were associated with acetylated H3 and H4 in their promoters. Brd4 binding sharply increased at telophase, the end stage of mitosis, correlating again with the increased H3/H4 acetylation. The increased Brd4 binding triggered corecruitment of P-TEFb and the onset of M/G1gene transcription, and this activity was greatly curtailed in Brd4 knockdown cells. Together, Brd4 preferentially marks the promoters of late and early postmitotic genes and directs restart of their transcription in newly divided cells.
MATERIALS AND METHODS
Cell Culture, Synchronization, and Brd4 Short Hairpin RNA Vectors
NIH3T3 cells were cultured in DMEM (Mediatech, Herndon, VA) supplemented with 10% donor bovine serum (Atlanta Biologicals. Norcross, GA) and 100 U/ml penicillin-streptomycin (Invitrogen, Carlsbad, CA). Mitotic cells were collected as described with some modifications (Whitfield et al., 2002; Nishiyama et al., 2006). Cells were first treated with 2.5 mM thymidine (Sigma-Aldrich, St. Louis, MO) for 18 h, washed, and then incubated with 50 ng/ml nocodazole (Sigma Aldrich) for 8 h, and mitotic cells were harvested by gentle shaking. Cells were replated in fresh media and allowed to proceed for indicated periods. Retroviral vectors for control and Brd4 shRNA and viral transduction procedures were described (Mochizuki et al., 2008).
Immunofluorescence Staining, Salt Extraction Analysis, and Immunoblotting
Mitotic cells were spun on to coated slides (Shandon, Pittsburgh, PA) and fixed in 2% paraformaldehyde for 10 min. Cells were permeabilized in 1% Triton X-100 for IgM (Prasanth et al., 2003) or methanol for IgG antibody (Dey et al., 2003). After blocking with 5% BSA, cells were incubated in monoclonal antibodies for RNA polymerase II (8WG16, H14, or H5, diluted at 1:50, Covance, Madison, WI), polyclonal antibody for cyclin T1 or Cdk9 (diluted at 1:200, Santa Cruz Biotechnology, Santa Cruz, CA), lamin B (1:200) and antibody for Brd4 (1:400; Mochizuki et al., 2008) for 1 h at room temperature followed by incubation with secondary antibody conjugated to Alexa 594, Alexa 564, or Alexa 488 (1:200, Invitrogen) for 1 h. Immunofluorescent images were obtained using a 63× oil immersion lens on a Leica confocal microscope (TCS-SP2; Deerfield, IL). For differential salt extraction, mitotic cells were resuspended in polyamine buffer (15 mM Tris-HCl, pH 7.5, 0.2 mM spermine, 0.5 mM spermidine, 2 mM EDTA, 40 mM KCl, and 0.1% digitonin), homogenized, and centrifuged (Dey et al., 2003). The pellets were extracted with 50, 150, and 300 mM KCl diluted in polyamine buffer. Ten micrograms of extracts was resolved on 4–12% SDS PAGE (Invitrogen) and immunoblotted as in Mochizuki et al. (2008). Immunoblot images were quantified by the VisionWorksLS software (UVP, San Gabriel, CA).
Fluorescence Recovery after Photobleaching Assay
For fluorescence recovery after photobleaching (FRAP) assays, green fluorescent protein (GFP), and full-length Brd4 fragments were subcloned into the HpaI site of the retroviral vector MSCVpuro (Clontech, Palo Alto, CA). Full-length Brd2 was inserted into the EcoRI site at the 3′ end of the GFP-pMSCVpuro vector. Cells were transduced with the vectors, plated on chambered cover glass (Nunc, Rochester, NY), cultured in 2.5 mM thymidine for 18 h, and then transferred to fresh media for 5 h to obtain mitotic cells. GFP signals in live cells were detected using a Zeiss 510 confocal laser-scanning microscope (Thornwood, NY). Photobleaching was performed under a 100× oil immersion lens with a 488-nm argon laser at a maximum laser power with a 285-ms pulse within a small circle, set at 25 pixels (Sprague and McNally, 2005; Nishiyama et al., 2008). Before the bleach pulse, 30 prebleach images were acquired. Recovery was monitored for 14 s (s) with 49-ms intervals. Prebleach and recovery images were collected at 0.5% laser power. Each FRAP curve was generated by averaging data from 7 to 15 individual cells.
Incorporation of Bromouridine or [3H]Uridine and Measurement of Nascent Transcripts
Mitotic cells were incubated in bromouridine (BrU; 50 μM, Sigma-Aldrich) or [3H]uridine (1 μCi/ml, Amersham, Piscataway, NJ) for 15 min before harvest. BrU signals were detected by immunostaining with mAb for bromodeoxyuridine (BrU) (1:200, Sigma-Aldrich) using the procedures described above. RNA isolation and measurement of radioactivity were performed as described (Dey et al., 2000). Nascent mRNA was detected by quantitative RT-PCR (qRT-PCR), using two primer sets, one corresponding to the intron–exon boundary and the other to an exon or an adjacent intron. The primers were designed according to the Primer Express 3.0 software (Applied Biosystems, Foster City, CA), and are listed in the Supplementary Information (Supplementary Table S1).
ChIP Assay
ChIP assay was performed essentially as described (Dahl and Collas, 2007; Mochizuki et al., 2008). Briefly, 106 mitotic cells were cross-linked with 1% formaldehyde for 10 min at room temperature and sonicated for 10 s twice in Misonix Sonicator 3000 (Farmingdale, NY). One microgram of affinity-purified antibody prepared from polyclonal rabbit anti-Brd4 sera, antibody against di-acetyl H3, tetra-acetyl H4 (Stratagene, La Jolla, CA) or H3K4me3 (Abcam, Cambridge, MA), antibody against Cdk9 or Pol II (8WG16) or normal rabbit IgG were incubated with 10 μl of Dynabeads protein G (Invitrogen) for 2 h. Antibodies against phosphorylated Pol II (H5 and H14) were incubated with Dynabeads conjugated to rat anti-mouse IgM (Invitrogen). Antibody-conjugated beads were incubated with chromatin preparations corresponding to 0.1–0.2 × 106 cells overnight at 4°C. Immunoprecipitated chromatin was de-cross-linked and DNA was purified. Input DNA for individual samples was prepared from 1% of respective chromatin before precipitation. Purified DNA was subjected to qPCR using appropriate primers. The percentage input was calculated as follows: 2^ (CT input − CT IP sample)/Ct input × 100. Samples with control IgG were tested for each primer set, all of which gave values within 0.2% of the input, and SD among samples was <0.01%. The information on the primers used for ChIP assay is available on request.
RESULTS
Retention of Brd4 on Mitotic Chromosomes
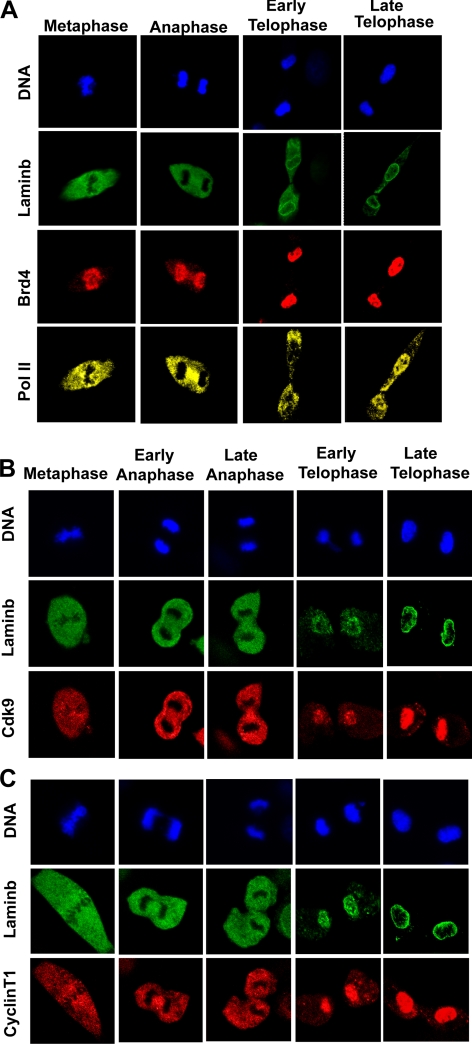
Many general and sequence specific transcription factors dissociate from chromosomes during mitosis (Gottesfeld and Forbes, 1997; Delcuve et al., 2008; Egli et al., 2008). However, Brd4 is shown to remain on chromosomes during mitosis in many cell types (Dey et al., 2000, 2003; You et al., 2004; Nishiyama et al., 2006; Toyama et al., 2008). Immunostaining experiments with NIH 3T3 cells in Figure 1A confirmed that Brd4 localizes entirely to condensed chromosomes throughout mitosis from metaphase and anaphase to telophase, in agreement with earlier reports. It should be noted here that Brd4 was reported to be only partially loaded on mitotic chromosomes in HeLa cells (Yang et al., 2008). The partial Brd4–chromosome interaction may reflect the transformed nature of HeLa cells, in which MAP kinases are aberrantly activated, inhibiting Brd4–chromosome interactions (Nishiyama et al., 2006; Chuang et al., 2008; Ghosh, S., Ghosh, A., Dey, McNally, Mueller, and Ozato, unpublished observations), whereas Pol II localized outside the chromosomes in metaphase and anaphase, but reassociated with chromosomes in late telophase, when lamin B was deposited along the nuclear periphery (Figure 1A). Because Brd4 interacts with P-TEFb during interphase (Jang et al., 2005; Yang et al., 2005; Mochizuki et al., 2008), we tested whether P-TEFb also associates with mitotic chromosomes. As shown in Figure 1, B and C, respectively, Cdk9 and cyclin T1, the core components of P-TEFb were both excluded from chromosomes until late anaphase and reassociated with chromosomes only at telophase. These data indicate that Brd4 binds to mitotic chromosomes without P-TEFb.
Figure 1.
Retention of Brd4 on mitotic chromosomes: reloading of Pol II and P-TEFb on chromosomes after telophase. Synchronized NIH3T3 cells were allowed to proceed through the indicated mitotic stages, fixed with paraformaldehyde, permeabilized, and immunostained for (A) Brd4 and Pol II, (B) Cdk9, and (C) Cyclin T1. Cells were coimmunostained for lamin B and counterstained for DNA with Hoechst 33342.
Brd4–Chromatin Interactions Increase at Telophase
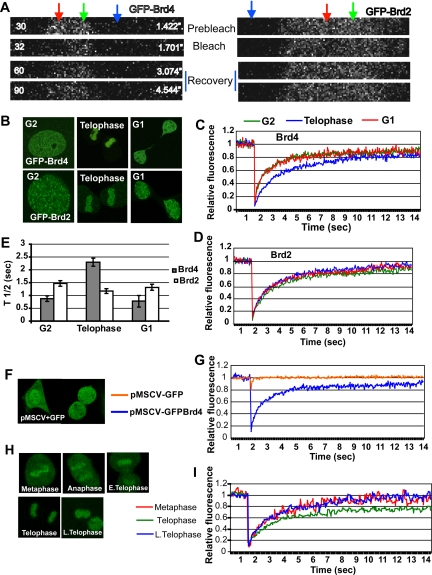
FRAP assays provide an excellent means by which to assess interactions of a protein with chromatin in living cells (Phair et al., 2004; Sprague and McNally, 2005). Our earlier FRAP analysis showed Brd4 is mobile during interphase, and its mobility reflects interaction of Brd4 with acetylated histones (Dey et al., 2003; Nishiyama et al., 2008). Histones become generally hypoacetylated during mitosis, although some acetylated residues persist on H3 and H4 in some genes (Kruhlak et al., 2001; Kouskouti and Talianidis, 2005; Valls et al., 2005). We performed FRAP assays for mitotic NIH3T3 cells expressing Brd4 fused to the enhanced GFP, GFP-Brd4. We have previously shown that ectopic GFP-Brd4 localizes to chromatin and functions in a manner comparable to the endogenous Brd4 (Dey et al., 2003; Nishiyama et al., 2008). Fluorescent images of live cells in Figure 2B verified that GFP-Brd4 resided in the nucleus during G2, then localized on chromosomes during mitosis (telophase in Figure 2B), and redistributed to the nuclei in G1 after cell division. In Figure 2A a small region of a mitotic chromosome with GFP-Brd4 or GFP-Brd2 (telophase in both cases) was briefly bleached (marked by red), and fluorescent recovery in that region was detected after bleach. Green and blue arrows indicate an unbleached region and a region of background fluorescence, respectively. Based on images such as these, fluorescence recovery was quantified for cells at G2, M (at telophase), and G1 using 15 independent samples at each stage. The recovery of GFP-Brd4 was rapid in G1 and G2 cells (red and green, respectively, in Figure 2C), as it reached a plateau within 7 s, showing a 50% recovery time (t1/2) of ∼1 s, similar to the previously noted recovery profile (Dey et al., 2003; Nishiyama et al., 2008). In contrast, mitotic cells showed distinctly slower recovery kinetics (blue in Figure 2C). The delayed recovery was substantiated by an increase in the t1/2 value (∼2.3 s), indicating that Brd4–chromatin interactions become tighter during telophase. A delay in FRAP recovery was unexpected, because overall histone acetylation is reduced during mitosis, whereas chromatin interaction of double bromodomains of Brd4 occurs via acetylated (Kruhlak et al., 2001; Dey et al., 2003). To test if the slowed Brd4 recovery was due to general changes in chromatin configuration/architecture, we performed FRAP analysis for another double bromodomain protein Brd2 that also associates with chromosomes during mitosis (Figure 2, A and B; Kanno et al., 2004). As presented in Figure 2D, GFP-Brd2 showed essentially identical recovery patterns at G2, M, and G1. Accordingly, t1/2 values for the three stages were very similar to each other (Figure 2E). In Figure 2, F and G, recovery of free GFP was tested as another control. As expected, free-GFP did not localize to chromosomes during mitosis and showed extremely rapid and complete recovery within a second, which obscured bleaching effects (Sprague and McNally, 2005). These results indicate that the slowed Brd4 recovery reflects a change in the property of Brd4–chromatin interactions, rather than changes in chromatin architecture itself. To further investigate the altered mobility of Brd4 during mitosis, we performed FRAP assays from metaphase, anaphase, and telophase (Figure 2, H and I) and found that the delayed recovery was restricted to telophase, because Brd4-GFP in metaphase and anaphase showed recovery comparable to that in G2 and G1 cells. These results suggest that at telophase Brd4 acquires an increased binding affinity for chromatin, at the time when other nuclear factors begin to reassociate with chromatin and transcription restarts.
Figure 2.
Real-time mobility of GFP-Brd4 during mitosis. (A) Live cell images of telophase chromatin in NIH3T3 cells expressing GFP-Brd4 (left) or GFP Brd2 (right) at prebleach (image 30), bleach (image 32), and recovery (image 60 and 90) from typical FRAP experiments. The red, green, and blue arrows indicate the photobleached area, the area without photobleach, or the area taken as background, respectively (25 pixels each). (B) Fluorescent images were acquired from live cells expressing GFP-Brd4 or GFP-Brd2 at indicated stages of cell cycle. (C) Cells expressing GFP-Brd4 were photobleached at the indicated stages of the cell cycle and analyzed for recovery. Fluorescence recovery was quantified at indicated times (sec) from 15 separate cells. (D) FRAP analysis was performed with cells expressing GFP-Brd2 at the indicated stages of the cell cycle. Recovery was quantified from the measurement of 15 independent cells. (E) The averages of half recovery times (t1/2) recorded in C and D are plotted. (F) Fluorescent images of cells expressing free GFP (pMSCV-GFP). (G) Fluorescence recovery of free-GFP was quantitatively compared with that of GFP-Brd4 from analyses of 10 independent cells each. (H) Images of cells at different stages of mitosis with GFP-Brd4. (I) Recovery was quantified from the analyses six independent cells at indicated stages of mitosis.
Biochemical Evidence for an Increased Brd4-Chromatin Interaction at Telophase
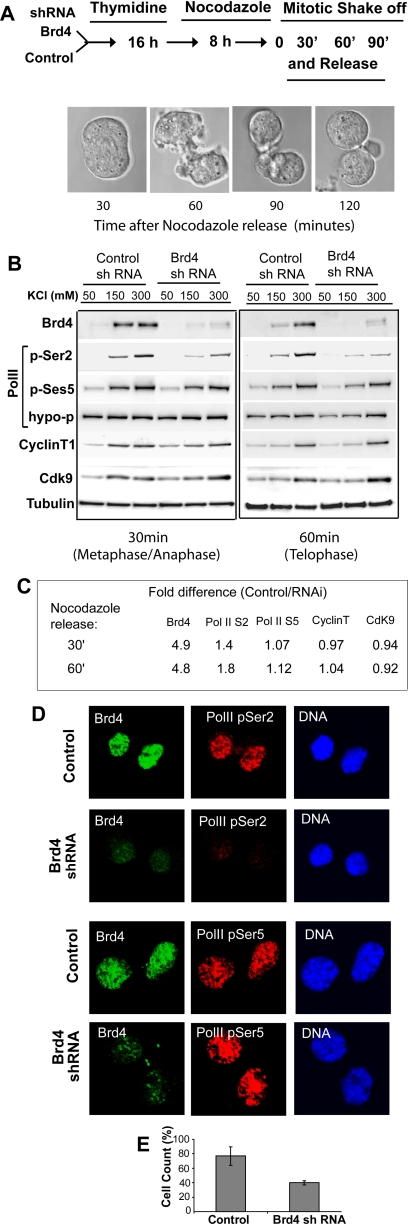
To ascertain whether telophase-selective reduction in Brd4 mobility was attributed to increased binding of Brd4 to chromatin, differential salt extraction experiments were carried out, in which the extraction profiles were compared between control and Brd4 knockdown cells. In the latter cells, Brd4 protein expression was reduced by ∼90% (Dey et al., 2003; Mochizuki et al., 2008; Nishiyama et al., 2008). Cells with control short hairpin RNA (shRNA) or with Brd4 shRNA were synchronized by thymidine block and nocodazole treatment and released into fresh media, allowing cells to proceed through mitosis (see the diagram in Figure 3A for the procedure). At 30 min after release, more than 98% of cells were in metaphase/anaphase, reaching early telophase at 60 min and late telophase at 90 min. Cells at metaphase/anaphase and early telophase were lysed and subjected to extraction with a buffer containing increasing salt concentrations (from 50 to 300 mM KCl), and extracts were tested for Brd4 by immunoblot. At metaphase/anaphase, Brd4 was extracted at 150 and 300 mM KCl in similar amounts in control cells (control in Figure 3B). On the other hand, at telophase the bulk of Brd4 was extracted at 300 mM KCl, with very small amounts of Brd4 extracted at 150 mM KCl. With Brd4 knockdown cells, little Brd4 was extracted at any salt concentrations tested, reflecting low Brd4 expression in these cells (Figure 3B, Brd4 shRNA). This was confirmed by the quantification of Brd4 in the gels shown in Figure 3C. Similar results were obtained with extracts from cells at late telophase (Supplementary Figure S1A). These results indicate that Brd4 binds to chromatin more tightly at telophase relative to metaphase/anaphase, consistent with the FRAP results in Figure 2B.
Figure 3.
Reduction of postmitotic S2-phosphorylation of Pol II CTD in Brd4 knockdown cells. (A) A diagram of the synchronization procedure. Cells with control shRNA or Brd4 shRNA were synchronized by thymidine and nocodazole, released, and allowed to proceed for the indicated times. Mitotic progression was viewed by DIC images. (B) Extracts from control (control shRNA) and Brd4 knockdown cells (Brd4 shRNA) at 30 and 60 min after release were extracted with buffers with increasing KCl concentrations. Extracts were immunoblotted with the indicated antibodies. Pol II phosphorylated at S2, or at S5 in the CTD, and hypophosphorylated Pol II were detected by H5, H14, and 8WG16 antibodies, respectively. (C) Quantification of immunoblot data. Band intensity of each protein was normalized by respective tubulin bands. Values represent the ratio of band intensity in control cells/Brd4 knockdown cells collected from all salt concentrations. Note that ratios are higher only for Brd4 and S2-phosphorylated Pol II. Similar results were seen with separate preparations of extracts. (D) Control and Brd4 knockdown cells at 90 min were coimmunostained for Brd4 (green) and Pol II with phospho-S2 CTD (top panels, red) or with phospho-S5 (bottom panels) and counterstained for DNA. (E) The percentage of phospho-S2 positive cells in control and Brd4 knockdown cells was obtained by counting 200–250 cells in three independent fields.
Salt extraction profiles were also examined for three forms of Pol II: the hypophosphorylated form, the form that phosphorylated at serine (S) 5 of the CTD, and the form that phosphorylated at S2 of the CTD. On activation, hyperphosphorylated Pol II becomes phosphorylated at S5 in the CTD, conferring an initiation competent state. This is followed by P-TEFb-dependent phosphorylation of S2 in the CTD that signifies an elongation state of Pol II (Price, 2000; Sims et al., 2004). Phosphorylation of S2 in the Pol II CTD takes place toward the end of mitosis subsequent to S5-phosphorylation (Prasanth et al., 2003). Hypophosphorylated Pol II was extracted at all salt concentrations similarly in control and Brd4 knockdown cells. Likewise, the extraction profiles of S5-phosphorylated Pol II were largely comparable in control and Brd4 knockdown cells at both times (Figure 3, B and C). However, extraction profiles of S2-phopshorylated Pol II were markedly different between control and Brd4 knockdown cells, in that the amount of S2-phosphorylated Pol II in Brd4 knockdown cells was lower than in control cells (Figure 3, B and C). Similarly, S2-phosphorylated Pol II was lower in Brd4 knockdown cells at 90 min after release (Supplementary Figure S1A, not shown). These results indicate that Pol II associates with chromatin and gains S5-phosphorylation without requiring Brd4, whereas S2-phosphorylation depends on Brd4. Because during interphase Brd4 interacts with P-TEFb, which phosphorylates S2 of Pol II CTD, salt extraction patterns were next examined for cyclin T1 and Cdk9, the core P-TEFb component. At telophase, both proteins were extracted at 300 mM KCl, a higher salt concentration than that observed at metaphase/anaphase. Importantly, the extraction patterns were comparable between control and Brd4 knockdown cells (Figure 3, B and C), indicating that P-TEFb reassociates with chromatin after mitosis without requiring Brd4, although the subsequent S2-phosphorylation is dependent on Brd4.
To further investigate the role of Brd4 in Pol II S2-phosphorylation, immunostaining was performed for cells at telophase (Figure 3D). Nuclei from control cells were intensely stained with antibodies for S2-phosphorylated Pol II, S5-phosphorylated Pol II, and Brd4 (Figure 3D). In Brd4 knockdown cells, staining of S2-phosphorylated Pol II and Brd4 was both very faint, although staining of S5-phosphorylated Pol II was comparable in control and Brd4 knockdown cells. Cell count data in Figure 3E showed that although nearly 80% of control telophase cells were S2-phosphorylation positive, <40% of Brd4 knockdown cells showed S2-phosphorylation signals. These values are likely to be an underestimate, because fluorescence intensity in signal-positive cells was much lower in Brd4 knockdown cells than in control cells. As expected, staining of hypophosphorylated Pol II, Cdk9, and cyclinT1 was likewise similar in control and Brd4 knockdown cells (Supplementary Figure S1B). These results are consistent with the above biochemical data and indicate that Pol II and P-TEFb are reloaded on chromatin after mitosis independently of Brd4, but the subsequent P-TEFb–mediated S2-phosphorylation of Pol II depends on Brd4.
Brd4 Directs Late-Telophase and Early Postmitotic Transcription
Data in the preceding section showed that Brd4 binds to mitotic chromosomes and promotes phosphorylation of S2 in the Pol II CTD. These data are compatible with the idea that Brd4 marks chromatin during mitosis and plays a role in restarting transcription in newly divided cells. To assess the role of Brd4 in postmitotic, de novo RNA synthesis, we tested BrU incorporation in control and knockdown cells. Immunostaining data in Figure 4A showed that although control cells robustly took up BrU, Brd4 knockdown cells showed little BrU incorporation. More than 85% of 200 cells counted were strongly BrU-positive in control cells, whereas <45% of cells displayed BrU signals in Brd4 knockdown cells. Furthermore, signals within BrU-positive cells were much weaker in Brd4 knockdown cells than control cells. In Figure 4B, postmitotic RNA synthesis was quantitatively estimated by measuring [3H]uridine incorporation. In control cells, the [3H]uridine uptake was low at metaphase/early anaphase (30 min), but increased thereafter when cells moved through telophase (60 and 90 min) and early G1 (120 min). [3H]uridine incorporation was consistently lower in Brd4 knockdown cells than control cells at all time points tested (Figure 4B). These data indicate that Brd4 has a role in restarting transcription after mitosis.
Figure 4.
Reduction of postmitotic de novo transcription in Brd4 knockdown cells. (A) Cells synchronized as in Figure 3A were allowed to reach telophase, pulsed with BrU for 15 min and immunostained for Brd4 (green) and BrU (red), counterstained for DNA. (B) Control and Brd4 knockdown cells synchronized as above were pulsed with [3H]uridine for 15 min and amounts of incorporated 3H were quantified. The values represent the average of three assays ± SD. (C) Control and Brd4 knockdown cells synchronized as above were tested for de novo transcription of indicated genes by qRT-PCR. Transcript levels were normalized by 18S RNA. Values represent the average of three determinations ± SD. Results of additional genes are shown in Supplementary Figure S2. The primer information is in Supplementary Table S1.
We next sought to identify individual genes that depend on Brd4 for postmitotic transcription. A previous study of synchronized HeLa cells reported that among ∼1100 genes expressed in a cell cycle–regulated manner, ∼17% are specifically expressed at the end of mitosis through early G1 (Whitfield et al., 2002). These genes are classified as “M/G1 genes,” many of which are important for establishing basic cellular functions in newly divided cells, such as those involved in forming structural components, chromatin metabolism, and cell cycle control. We tested expression of 23 M/G1 genes in control and Brd4 knockdown cells. qRT-PCR was performed using primers that detected nascent transcripts, excluding mature RNAs that may have been carried over from prior stages. Data in Figure 4C show transcript levels of nine M/G1 genes among 23 measured at prometaphase, metaphase/anaphase, telophase, and late telophase/early G1 (0, 30, 60, and 90 min after release). As expected, their expression was low in prometaphase and metaphase/anaphase, but sharply increased at telophase in control cells. In Brd4 knockdown cells, however, transcript levels of eight M/G1 genes (Ran, Kif5b, Tgf1, Ywhah, Gnb1, Rad21, Ctnnd1, and Topo1) were substantially lower than those of control cells. One exception was Parathymosin, which was expressed at similar levels in control and Brd4 knockdown cells. Of remaining 14 M/G1 genes tested, 11 showed a clear reduction in transcript in Brd4 knockdown cells (Supplementary Figure S2A). Together, 19 of 23 M/G1 genes examined in this study showed reduced expression in Brd4 knockdown cells, indicating that Brd4 has a large impact on restarting transcription of late M and early G1genes. As expected, G1/S genes (Mcm2 and E2f1), S genes (Hdac3 and Rad51), a gene not regulated by cell cycle (Rcc1), and a silent gene (β-globin) showed negligible transcript expression throughout the mitotic and postmitotic stages, in both control and Brd4 knockdown cells.
Brd4 Marks TSS of M/G1 Genes During Mitosis
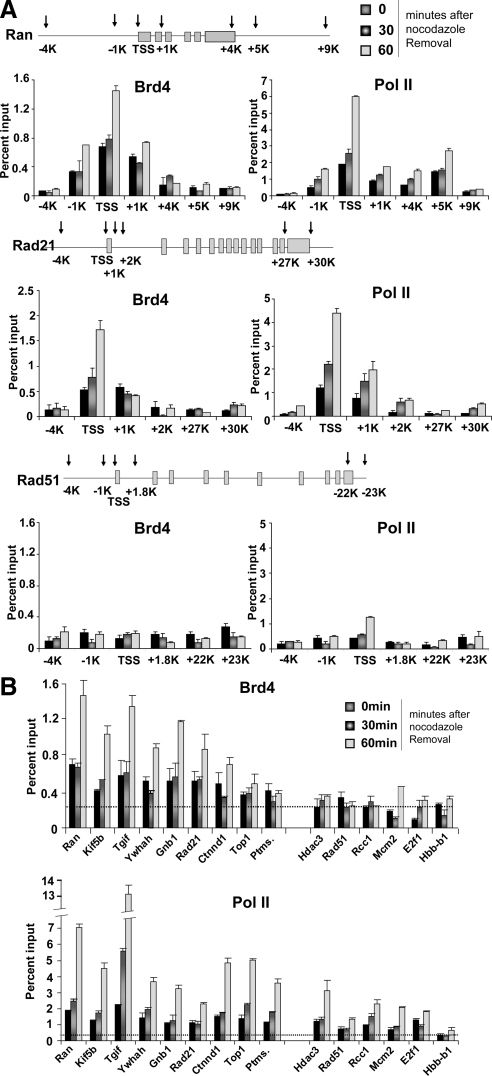
To determine whether Brd4 specifically binds to M/G1 genes and marks them during mitosis for postmitotic transcription, ChIP analysis was performed with Brd4 antibody at different stages of mitosis: prometaphase, metaphase/anaphase, and telophase (0, 30, and 60 min after release, respectively). We found that Brd4 remained bound at or near the TSS of the majority of M/G1 genes throughout mitosis. Figure 5A (left panel) shows examples of ChIP results obtained for Rad21 and Ran, typical M/G1 genes. Brd4 binding to the TSS regions was evident already at prometaphase and metaphase/anaphase, and the binding to the TSS markedly increased at telophase. However, Brd4 binding on other regions within these genes (i.e., upstream promoter regions and the coding regions) was much lower. Rad51, an S phase gene, however, showed essentially background levels of Brd4 binding over the entire gene including the TSS throughout mitosis. Figure 5B shows binding of Brd4 to the TSS of all nine M/G1 tested in Figure 4C. Eight M/G1 genes showed significantly above-background Brd4 binding at prometaphase through anaphase followed by a sharp increase at telophase. These genes were expressed at telophase in a Brd4-dependent manner (Figure 4C). Four additional M/G1 genes (Cbx3, Cdkn3, Cdk7, and Vcam1; Supplementary Figure S2A) also showed significant Brd4 binding throughout mitosis (Supplementary Figure S2B). However, little Brd4 binding was seen on Ptms (Parathymosin), an M/G1 gene, whose expression was not dependent on Brd4. Importantly, Brd4 did not bind to genes expressed at later times after mitosis, i.e., Mcm2, E2f1 (expressed at G1/S), Hdac3, and Rad51(expressed at S), nor did it bind to the silent Hbb-b1 (β-globin) gene throughout mitosis. These results indicate that Brd4 selectively marks M/G1 genes during the course of mitosis and its binding increases at telophase.
Figure 5.
Brd4 marks the TSS regions of M/G1 genes throughout mitosis. (A) ChIP analysis was performed with affinity-purified anti-Brd4 antibody (left) or antibody against hypophosphorylated Pol II (8WG16, right) at indicated sites in the Ran, Rad21, and Rad51 genes for cells at indicated times after release. Values in A and B (and all ChIP data below) represent the average of three determinations ± SD. (B) ChIP analysis was performed with the antibodies against Brd4 or hypophosphorylated Pol II as in A for the TSS regions of indicated M/G1 genes. Dotted lines represent averaged values for normal IgG binding (SD, <0.01%).
We next assessed binding of Pol II to these genes. As shown in Figure 5A (right panels), Pol II also showed TSS-preferred binding to Ran and Rad 21, which increased at telophase. Unlike Brd4, however, Rad51, an S phase gene, showed a clear, above baseline Pol II binding at the TSS. Consistent with this result, Pol II binding was found at or near the TSS of not only all M/G1 genes, but genes expressed at G1/S and S phase, although Pol II did not bind to Hbb-b1, silent in NIH3T3 cells (Figure 5B). Furthermore, binding of Pol II did not seem to depend on Brd4, because Pol II showed appreciable binding to the Ptms gene as well (see below). These data indicate that Pol II binds at the TSS of many genes, regardless of their expression at telophase or later times. In line with these Pol II results, genomewide studies have shown that Pol II is constitutively bound to the promoters of many expressed and nonexpressed genes in interphase (Guenther et al., 2007; Core et al., 2008).
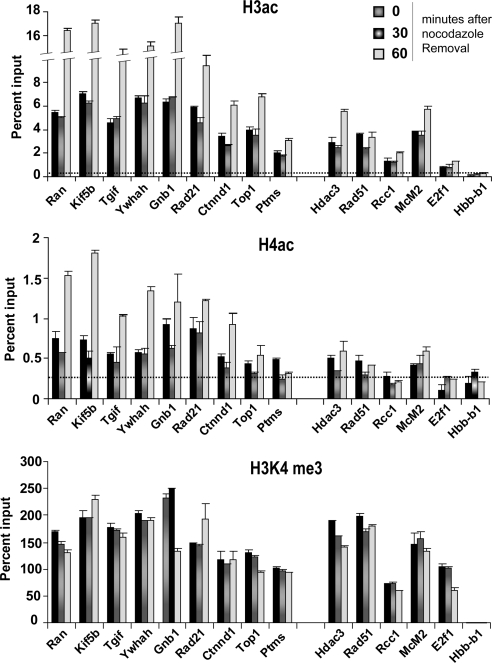
Elevated Acetylation of Histone H3 and H4 in M/G1 Genes
Many transcriptionally active genes carry acetylated histone H3 and H4 in their promoters during interphase (Guenther et al., 2007; Wang et al., 2008). Similarly, promoters of active genes are marked with H3 trimethylated at lysine 4 (H3K4me3) in interphase. Although histones are generally hypoacetylated during mitosis, enrichment of acetylated histones is reported for some genes during mitosis (Kruhlak et al., 2001; Kouskouti and Talianidis, 2005; Valls et al., 2005; Nishiyama et al., 2006). Because Brd4 preferentially binds to acetylated H3 and H4 in interphase, we performed ChIP assays for the above M/G1 genes on mitotic chromatin for acetylated H3 and H4 and H3K4me3. Figure 6 (top and middle panels), and Supplementary Figure S2B summarizes the status of H3/H4 acetylation at the TSS of genes tested in Figure 5 and Supplementary S2B. All 12 Brd4-dependent M/G1 genes showed elevated acetylation for H3 and H4 throughout mitosis, and their acetylation levels further rose at telophase. The elevated H3/H4 acetylation was confined largely to the TSS and +1-kb coding region, as shown in Supplementary Figure S3. However, the Ptms gene, which did not require Brd4 for expression, showed low histone acetylation. Similarly, histone acetylation was at a low-to-negligible level for G1/S and S genes as well as Hbb-b1 (Figure 6, top and middle panels). These results indicate that many M/G1 genes carry acetylation-rich histone H3 and H4 during mitosis and that acetylation levels markedly increase toward the end of mitosis. In contrast, the H3K4me3 mark was observed on not only M/G1 genes, but on G1/S and S genes equally prominently throughout mitosis, showing little change at telophase (Figure 6, bottom panel, and Supplementary Figure S2B). Together, the profiles for histone acetylation matched well with that of Brd4 binding, suggesting that Brd4 marking of M/G1 genes is accounted for by increased histone H3 and H4 acetylation. On the other hand, the H3K4me3 profile matched more closely with that of Pol II binding, consistent with the idea that Pol II and H3K4me3 identify many nonsilent genes, irrespective of active transcription.
Figure 6.
Histone acetylation marks correlate with Brd4 gene marking on mitotic cells. ChIP analysis was performed with antibody against di-acetyl H3 (K9, 14), tetra-acetyl H4 (K5, 8, 12, and 16), and H3K4me3 on the TSS of M/G1 genes. Dotted lines represent averaged values for normal IgG binding (SD, <0.01).
To ascertain whether Brd4 marking of M/G1 genes is a common event during mitosis, we examined another cell type, a macrophage line, RAW264.7. This cell line is widely studied for its immune functions (Hargreaves et al., 2009). ChIP analysis was performed to test Brd4 binding to M/G1 genes on mitotic RAW264.7 cells synchronized at prometaphase and telophase were tested for binding of Brd4. Brd4 bound to all four M/G1 genes, but not an S phase gene, similar to the results with NIH3T3 cells above (Supplementary Figure S4). Furthermore, these M/G1 genes, but not the S phase gene showed elevated acetylation in histones H3 and H4, again in line with the data with NIH3T3 cells (Supplementary Figure S4).
Brd4 Directs Telophase Recruitment of P-TEFb
To further verify M/G1-selective Brd4 marking, ChIP analysis was performed for Brd4 knockdown cells. In Supplementary Figure S5, Brd4 binding was negligible in four Brd4-dependent M/G1 genes not only at prometaphase but at telophase, whereas these genes showed abundant Brd4 binding in control cells. These data support the notion that mitotic Brd4 gene marking is directly linked to the onset of their transcription at telophase.
During interphase, Brd4 interacts with P-TEFb and recruits it to the promoter of active genes to prompt transcriptional elongation from these genes (Jang et al., 2005; Yang et al., 2005; Wu and Chiang, 2007; Mochizuki et al., 2008; Yang et al., 2008). Thus, it was of importance to determine whether Brd4 recruits P-TEFb to M/G1 genes during mitosis to start their transcription. We sought to address this question for another reason, i.e., to resolve an apparent dichotomy where Brd4 was not required for reassociation P-TEFb with chromosomes at telophase, whereas it was required for phosphorylation of S2 in the Pol II CTD (see Figure 3). ChIP analysis was performed for P-TEFb and three forms of Pol II: the hypophosphorylated, the S5 phosphorylated, and the S2 phosphorylated Pol II in control and Brd4 knockdown cells (Figure 7). In control cells, P-TEFb bound to the M/G1 genes at telophase, although was not present on these genes at earlier times, indicating that P-TEFb was recruited to the genes only after telophase. In Brd4 knockdown cells, however, P-TEFb recruitment was reduced by ∼40–60%. Similarly, binding of S2-phosphorylated Pol II was reduced in Brd4 knockdown cells by 40–50% relative to control cells. On the other hand, hypophosphorylated Pol II and S5-phosphoylated Pol II were present on these genes at comparable levels in control and Brd4 knockdown cells. As might have been expected, Rad 51, a non-M/G1 gene to which Brd4 did not bind, showed little P-TEFb recruitment, and very low S2 phosphorylation. Nevertheless, Rad51 showed significant binding of hypophosphorylated Pol II and S5 phosphorylated Pol II in both control and knockdown cells, in line with the data in Figure 5B. These results indicate that while P-TEFb returns to the telophase nucleus in a Brd4 independent manner, the subsequent recruitment to the M/G1 promoters depends on Brd4.
Figure 7.
Brd4-dependent recruitment of P-TEFb and S2 phosphorylation of Pol II CTD at telophase. Binding of Brd4, Cdk9 (P-TEFb), and three forms of Pol II to the TSS of indicated M/G1 genes was tested for control and Brd4 knockdown cells at the indicated times after release.
DISCUSSION
Our analysis revealed that Brd4 remains bound to many M/G1 genes throughout mitosis, at a time when other chromatin-binding factors are dispersed and the genome is rendered transcriptionally inert. On the basis of this and our additional observations that Brd4 directs M/G1 gene expression at telophase, we suggest that Brd4 belongs to the class of proteins that contribute to the epigenetic inheritance of gene expression through cell division (John and Workman, 1998; Sarge and Park-Sarge, 2005). A number of proteins deposit distinct marks on chromatin during mitosis to ensure expression of particular genes in the progeny cells. These proteins include the transcription factors: Runx2, CTCF, and FoxI1 (Burke et al., 2005; Yan et al., 2006; Young et al., 2007a,b). They mark chromosomes by binding to specific DNA sequences. In addition, HSP2 marks mitotic chromatin not only by binding to specific DNA, but also by regulating chromatin compaction through the interaction with a phosphatase (Xing et al., 2005). A recent report shows that TBP also marks mitotic chromatin that accompanies an interaction with a phosphatase and prevention of chromatin compaction (Xing et al., 2008). Compared with gene marking by other proteins, marking by Brd4 has two distinguishing features. First, Brd4 marking relies on the recognition of acetylated histones, H3 and H4, but not that of specific DNA sequences. Second, Brd4 selectively marks M/G1 genes to the exclusion of other genes expressed later in cell cycle.
M/G1-restricted Gene Marking by Brd4
Brd4 remained bound to many M/G1 genes throughout mitosis, with the binding sites typically limited to the TSS regions. Our results that Brd4 occupies these genes not only in mitotic NIH3T3 cells, but in the RAW264.7 macrophages further support the M/G1-selectivity of Brd4 binding. Remarkably, Brd4 marking was essentially undetectable in non-M/G1 genes, in that genes that are expressed at later stages or silent in these cells were not occupied by Brd4 during mitosis. This result was striking, given that Brd4 binds and stimulates expression of many G1/S and S genes when cells are in G1 and S phase (Mochizuki et al., 2008) and highlights the dynamic nature of Brd4 occupancy. The fluid nature of Brd4 binding is also evident as Brd4 is induced to bind to the promoter of genes stimulated by inflammatory signals (Huang et al., 2008; Hargreaves et al., 2009). Together, M/G1 selectivity of Brd4 marking, although striking, likely represents an important feature of mitotic memory (see below).
Coinciding with Brd4 marking, histone H3 and H4 showed elevated acetylation on M/G1 genes, which was largely confined at/near the TSS regions. Moreover, elevated H3 and H4 acetylation was predominantly seen on M/G1 genes, but not on other genes tested here. Further underscoring the correlation between Brd4 binding and histone acetylation, both Brd4 binding and H3/H4 acetylation greatly increased toward the end of mitosis. Our findings support the previous reports that promoters of some genes bear acetylated chromatin during mitosis (Kouskouti and Talianidis, 2005; Valls et al., 2005). Extending these reports, our data indicate that it is Brd4 that recognizes the mitotic acetyl histone marks and translates them into transcription at telophase (Dey et al., 2003; Nishiyama et al., 2008).
In light of the information gleaned from the ChIP data, earlier microscopic observations that Brd4 localizes to noncentromeric regions of mitotic chromosomes are best explained by Brd4's selective binding to M/G1 genes (Dey et al., 2000). It is important to note that not only Brd4 but other BET family proteins such as Brd2 may take part in mitotic gene marking, given that they also remain on mitotic chromosomes through acetylated histones (Kanno et al., 2004; Wu and Chiang, 2007; Shang et al., 2009).
Brd4 Gene Marking and the Role in Postmitotic Transcription
Levels of Brd4 binding to M/G1 genes markedly increased when mitosis proceeded from anaphase to telophase. This increase coincided with reduced real-time Brd4 mobility and the decreased salt solubility, both indicative of Brd4's increased affinity for chromatin at this stage of mitosis. At telophase, many other transcription factors as well as Pol II return to chromatin, and Pol II CTD becomes phosphorylated at S5 (Prasanth et al., 2003). This is followed by S2 phosphorylation of Pol II and initiation of postmitotic transcription. We noted high Pol II binding to a broad array of genes at this stage: unlike Brd4, Pol II binding was not limited to M/G1 genes, but was found on many other genes, including G1/S and S genes that are expressed at later times. Furthermore, unlike Brd4, Pol II binding did not correlate well with elevated histone acetylation, which was largely limited to M/G1 genes. Rather, Pol II binding displayed a better concordance with H3K4me3, because this mark was present on many genes active throughout interphase. Moreover, binding of hypophosphorylated Pol II and S5 phosphorylated Pol II was comparable in control and Brd4 knockdown cells, the data consistent with the salt solubility results. Together, it may be suggested that Pol II returns to chromatin and binds to a large set of genes beyond M/G1 genes, irrespective of Brd4. This view is in line with the reports that in interphase, Pol II binds to the promoter regions of numerous genes, regardless of their transcriptional states (Guenther et al., 2007; Core et al., 2008).
Although postmitotic Pol II binding to M/G1 genes did not require Brd4, recruitment of P-TEFb and Pol II CTD S2 phosphorylation of these genes was critically dependent on Brd4, as evidenced by inhibition of both events in Brd4 knockdown cells. Because Brd4 physically interacts with P-TEFb, it is likely that Brd4 stabilizes binding of P-TEFb and confers an elongation competent state upon Pol II. Considering that P-TEFb returned to chromatin at telophase without requiring Brd4, it is clear that reassociation with chromatin itself was not sufficient for P-TEFb to secure stable association with the Pol II complex, but rather, it requires Brd4 to achieve this and to phosphorylate the substrate S2 in the Pol II CTD. Although the molecular mechanism by which Brd4 recruits P-TEFb is not fully understood, it may involve regulation of P-TEFb's inhibitory complex, HEXIM1/7SK RNA (Price, 2000; Jang et al., 2005; Yang et al., 2005).
A picture that emerges from these observations is that mitotic gene marking by Brd4 is not universal, as non-M/G1 genes were not marked by Brd4, and that Brd4's memory is internally coupled with postmitotic M/G1gene transcription. What is the significance of the lack of Brd4 marking on non-M/G1 genes? How important is the coupling of Brd4 marking with postmitotic transcription? It is noted that proper execution of early postmitotic transcription is essential for determining subsequent gene expression programs in the daughter cells, as it shapes their identity and future potentials (Egli et al., 2008). Under this line of thinking, M/G1 selective marking would ensure faithful execution of early postmitotic step, but would leave subsequent gene expression open to modifications. By limiting mitotic memory to M/G1 genes, the genome would have ample room for flexible reprogramming of gene expression in newly divided cells, allowing them to readjust to a shifting environment.
In conclusion, Brd4 binds to early postmitotic genes throughout mitosis. By recruiting P-TEFb, it directs the onset of de novo M/G1 gene transcription. This transcription-coupled Brd4 marking is distinct from Pol II binding that takes place in a broader array of genes beyond immediate postmitotic genes. Further, by the restricting the range of mitotic Brd4 gene marking, it provides the daughter cells a license to reprogram subsequent gene expression patterns in response to various external events.
Supplementary Material
ACKNOWLEDGMENTS
We thank Carl Wu, David Levens, Tom Misteli, and David Clark for critical reading of the manuscript; S. Barton and D. Huang for technical assistance; and members of Ozato lab for valuable discussions.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-05-0380) on October 7, 2009.
REFERENCES
- Bisgrove D. A., Mahmoudi T., Henklein P., Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. USA. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke L. J., et al. CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. EMBO J. 2005;24:3291–3300. doi: 10.1038/sj.emboj.7600793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christova R., Oelgeschlager T. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat. Cell Biol. 2002;4:79–82. doi: 10.1038/ncb733. [DOI] [PubMed] [Google Scholar]
- Chuang J. Y., Wang Y. T., Yeh S. H., Liu Y. W., Chang W. C., Hung J. J. Phosphorylation by c-Jun NH2-terminal kinase 1 regulates the stability of transcription factor Sp1 during mitosis. Mol. Biol. Cell. 2008;19:1139–1151. doi: 10.1091/mbc.E07-09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L. J., Waterfall J. J., Lis J. T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl J. A., Collas P. Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells. 2007;25:1037–1046. doi: 10.1634/stemcells.2006-0430. [DOI] [PubMed] [Google Scholar]
- Delcuve G. P., He S., Davie J. R. Mitotic partitioning of transcription factors. J. Cell. Biochem. 2008;105:1–8. doi: 10.1002/jcb.21806. [DOI] [PubMed] [Google Scholar]
- Dey A., Chitsaz F., Abbasi A., Misteli T., Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A., Ellenberg J., Farina A., Coleman A. E., Maruyama T., Sciortino S., Lippincott-Schwartz J., Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D., Birkhoff G., Eggan K. Mediators of reprogramming: transcription factors and transitions through mitosis. Nat. Rev. Mol. Cell Biol. 2008;9:505–516. doi: 10.1038/nrm2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M., Forbes D. J. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves D. C., Horng T., Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Yang X., Zhou M. M., Ozato K., Chen L. F. Brd4 coactivates transcriptional activation of NF-κB via specific binding to acetylated RelA. Mol. Cell. Biol. 2008;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M. K., Mochizuki K., Zhou M., Jeong H.-S., Brady J. N., Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- John S., Workman J. L. Bookmarking genes for activation in condensed mitotic chromosomes. Bioessays. 1998;20:275–279. doi: 10.1002/(SICI)1521-1878(199804)20:4<275::AID-BIES1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Kanno T., Kanno Y., Siegel R. M., Jang M. K., Lenardo M. J., Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- Kouskouti A., Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M. J., Hendzel M. J., Fischle W., Bertos N. R., Hameed S., Yang X. J., Verdin E., Bazett-Jones D. P. Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J. Biol. Chem. 2001;276:38307–38319. doi: 10.1074/jbc.M100290200. [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas M. A., Dey A., Rabindran S. K., Ozato K., Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Nishiyama A., Jang M. K., Dey A., Ghosh A., Tamura T., Natsume H., Yao H., Ozato K. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J. Biol. Chem. 2008;283:9040–9048. doi: 10.1074/jbc.M707603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R. K., Gurdon J. B. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat. Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- Nishiyama A., Dey A., Miyazaki J.-I., Ozato K. Brd4 is required for recovery from antimicrotubule drug-induced mitotic arrest: preservation of acetylated chromatin. Mol. Biol. Cell. 2006;17:814–823. doi: 10.1091/mbc.E05-08-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A., Mochizuki K., Mueller F., Karpova T., McNally J. G., Ozato K. Intracellular delivery of acetyl-histone peptides inhibits native bromodomain-chromatin interactions and impairs mitotic progression. FEBS Lett. 2008;582:1501–1507. doi: 10.1016/j.febslet.2008.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R. D., Scaffidi P., Elbi C., Vecerova J., Dey A., Ozato K., Brown D. T., Hager G., Bustin M., Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth K. V., Sacco-Bubulya P. A., Prasanth S. G., Spector D. L. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol. Biol. Cell. 2003;14:1043–1057. doi: 10.1091/mbc.E02-10-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D. H. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L., Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Sarge K. D., Park-Sarge O. K. Gene bookmarking: keeping the pages open. Trends Biochem Sci. 2005;30:605–610. doi: 10.1016/j.tibs.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Saurin A. J., Shiels C., Williamson J., Satijn D. P., Otte A. P., Sheer D., Freemont P. S. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 1998;142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang E., Wang X., Wen D., Greenberg D. A., Wolgemuth D. J. Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev. Dyn. 2009;238:908–917. doi: 10.1002/dvdy.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R. J., 3rd, Belotserkovskaya R., Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Sprague B. L., McNally J. G. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Toyama R., Rebbert M. L., Dey A., Ozato K., Dawid I. B. Brd4 associates with mitotic chromosomes throughout early zebrafish embryogenesis. Dev. Dyn. 2008;237:1636–1644. doi: 10.1002/dvdy.21576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valls E., Sanchez-Molina S., Martinez-Balbas M. A. Role of histone modifications in marking and activating genes through mitosis. J. Biol. Chem. 2005;280:42592–42600. doi: 10.1074/jbc.M507407200. [DOI] [PubMed] [Google Scholar]
- Wang Z., et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield M. L., et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. Y., Chiang C. M. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- Xing H., Vanderford N. L., Sarge K. D. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat. Cell Biol. 2008;10:1318–1323. doi: 10.1038/ncb1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H., Wilkerson D. C., Mayhew C. N., Lubert E. J., Skaggs H. S., Goodson M. L., Hong Y., Park-Sarge O.-K., Sarge K. D. Mechanism of hsp70i gene bookmarking. Science. 2005;307:421–423. doi: 10.1126/science.1106478. [DOI] [PubMed] [Google Scholar]
- Yan J., Xu L., Crawford G., Wang Z., Burgess S. M. The forkhead transcription factor FoxI1 remains bound to condensed mitotic chromosomes and stably remodels chromatin structure. Mol. Cell. Biol. 2006;26:155–168. doi: 10.1128/MCB.26.1.155-168.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., He N., Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol. Cell. Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Yik J.H.N., Chen R., He N., Jang M. K., Ozato K., Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- You J., Croyle J. L., Nishimura A., Ozato K., Howley P. M. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- Young D. W., et al. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007a;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- Young D. W., et al. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc. Natl. Acad. Sci. USA. 2007b;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.