Abstract
A major regulatory function has been evidenced here for HSF1, the key transcription factor of the heat-shock response, in a large-scale remodeling of the cell epigenome. Indeed, upon heat shock, HSF1, in addition to its well-known transactivating activities, mediates a genome-wide and massive histone deacetylation. Investigating the underlying mechanisms, we show that HSF1 specifically associates with and uses HDAC1 and HDAC2 to trigger this heat-shock–dependent histone deacetylation. This work therefore identifies HSF1 as a master regulator of global chromatin acetylation and reveals a cross-talk between HSF1 and histone deacetylases in the general control of genome organization in response to heat shock.
INTRODUCTION
Exposure of cells to environmental stress conditions results in the inducible expression of a family of proteins termed heat-shock proteins (HSPs) whose function is to protect the cells from stress-induced damages (reviewed in Lindquist¤ 1986; Christians et al., 2002). In parallel, heat shock also induces a global down-regulation of non-heat-shocked genes (Spradling et al., 1975; Jamrich et al., 1977; Findly and Pederson, 1981; Higashikubo and Roti Roti, 1993). Heat-shock transcription factor 1 (HSF1) is responsible for the stress-induced activation of heat-shock genes and chromatin remodeling events associated with HSF1-mediated gene activation has been thoroughly investigated in the literature (reviewed in Wu, 1995; Sorger, 1991; Morimoto et al., 1992, 1996). In contrast, mechanisms underlying gene repression have been investigated to much lesser extent.
In eukaryotic cells, the packaging of DNA into chromatin provides a dynamic structure for controlling transcription. The fundamental unit of chromatin structure, the nucleosome, is formed by a 147-base pair DNA fragment wrapped around an octamer of the four types of histones: H2A, H2B, H3 and H4, with N-terminal tails protruding from the nucleosome. Posttranslational modifications of histone tails are known to be associated with differences in gene expression (reviewed in Jenuwein and Allis, 2001; Mellor, 2006). Among other modifications, acetylation is considered as a key player in the epigenetic control of gene expression and is associated to transcriptionally active regions (reviewed in Cheung et al., 2000). The balance between opposite enzymatic activities, histone acetyl transferases (HATs), and histone deacetylases (HDACs), directly controls the level of histone acetylation and potent inhibitors of histone deacetylases, such as trichostatin A (TSA), induce a global hyperacetylation of the chromatin (Yoshida et al., 1990). Different classes of HDACs have been identified so far, with different structures and activities (reviewed in Yang and Seto, 2008). Class I and class II HDACs are classical HDACs whose activity is inhibited by TSA, whereas class III is a family of NAD+-dependent proteins the activity of which is not affected by TSA. Only one member of class IV HDACs, HDAC11, has been identified so far. Three HDACs of class I, HDAC1, 2, and 3, have been shown to play a role in chromatin remodeling associated with gene repression. HDAC1 and 2 are present in SIN3 and CoREST, and nucleosome remodeling and histone deacetylase (NURD) complexes, whereas HDAC3 belongs to the nuclear corepressor (NcoR) complex (reviewed in Cunliffe, 2008). These complexes are thought to be recruited to chromatin by sequence-specific transcriptional repressors and then to act through the deacetylation of histone tails.
The heat-shock response constitutes a potent and unique model to understand the link between gene activity and chromatin remodeling. The idea that heat shock could induce profound changes in gene expression by directly acting on the equilibrium between HAT and HDAC activities in the cell nucleus prompted us to look at the possible impact of heat shock on histone acetylation and to further examine the role of HSF1 in the control of chromatin acetylation.
MATERIALS AND METHODS
Cell Culture, Heat Shock, and Drug Treatments
The immortalized cell line derived from kidney cells of the African green monkey with a version of the SV40 genome (Cos), the cell line derived from cervical cancer cells (HeLa), and the HSF1−/− mouse embryonic fibroblast (MEF) cell lines (Dr. Ivor Benjamin, University of Utah) were grown in DMEM, 1 g/l glucose, and 4.5 g/l glucose, respectively, supplemented with 2 mM l-glutamine, 10% fetal bovine serum, and 1% penistreptomycin. Heat-shock treatment was performed in a water bath at 43°C. When needed, 330 nM of histone deacetylase inhibitor TSA (Sigma-Aldrich, St. Louis, MO) was added 1 h before heat shock.
Knockdown of Class I HDACs and Transient Transfections
Transient transfections of cells with small interfering RNA (siRNA) were performed using oligofectamine (Invitrogen, Carlsbad, CA). siRNAs targeting HDAC1, 2, and 3 were obtained from MWG Biotech (Ebersberg, Germany) and prepared according to the manufacturer's instructions: 5′-CAGCGACUGUUUGAGAACCTT-3′ for siRNA against HDAC1, 5′-UCCGUAAUGUUGCUCGAUGTT-3′ for siRNA against HDAC2, 5′GAUGCUGAACCAUGCACCUTT-3′ for siRNA against HDAC3, and 5′-CGUACGCGGAAUACUUCGATT-3′ for control nonspecific siRNA. Two cycles of transfections were performed. Cells were heat-shocked 72 h after the first transfection. Transient transfections with plasmids expressing FLAG-HDAC1, 2, or 3 (a generous gift from Dr. E. Seto, H. Lee Moffitt Cancer Center and Research Institute, San Diego, CA) were performed using Lipofectamine (Invitrogen).
Protein Extracts and Western Blot Analysis
The preparation of cytosolic and of soluble and nonsoluble nuclear fractions were obtained using the BioVision nuclear/cytosol fractionation kit (catalogue no. K266-25; Mountainview, CA). For total cell extracts used for histone code analysis by Western blot, cells were resuspended in 8 M urea. The suspension was then sonicated, and the supernatant was analyzed after centrifugation at 13,000 rpm for 10 min. Ten micrograms of whole protein extracts was run on acrylamide gels, which were subsequently stained with Coomassie blue for a visual equilibration of all extracts. For the detection of HSF1 and histone modifications, protein extracts were separated on 8 and 15% acrylamide gels, respectively. The following primary antibodies were used: rabbit polyclonal anti-HSF1 antibodies (StressGen, San Diego, CA), anti-acetyl lysin (Millipore, Bedford, MA; 06-933), HDAC1 (Millipore, 06-720), HDAC2 (Abcam, Cambridge, MA; 16032), HDAC3 (Abcam; 7030), FLAG (Sigma; F3165), H3acK9 (Abcam; 12179), H3acK14 (52946), H3acK18, and K23 (United States Biological, Swampscott, MA; H-511013 E and F, respectively), pan acetylH4 (Millipore; 06-866), H4acK16 (Abcam; 1762). H2Aac, H2Bac, H4acK5, H4acK8, H4acK12 were provided from Prof. B. M. Turner (University of Birmingham, United Kingdom). Western blot were quantified using NIH ImageJ software (http://rsb.info.nih.gov/ij/).
Immunofluorescence and Microscopy
Immunofluorescence was performed on formaldehyde-fixed cells as described previously (Jolly et al., 2002). DNA was counterstained with 250 ng/ml DAPI. Secondary antibodies conjugated to either FITC or rhodamine were used. To allow a comparative analysis between cells samples submitted to different experimental conditions, images were acquired with the same exposure time with an apotome microscope using the 63×, 1.25 NA oil immersion objective and an intermediate magnification of 1.25×.
Immunoprecipitation
Cells (106) were resuspended in lysis buffer containing 50 mM HEPES, pH 7.4, 60 mM KCl, 15 mM NaCl, 0.65 mM spermidine, 0.34 M saccharose, 2 mM EDTA, 0.5 mM EGTA, 0.5 mM DTT, 0.05%Triton, protease inhibitors (Roche, Indianapolis, IN), and phosphatase inhibitors (Sigma-Aldrich). Samples were incubated on ice for 15 min and then sonicated (250 J). Immunoprecipitation of FLAG-conjugated proteins were performed using 20 μl of FLAG-Sepharose beads (Sigma-Aldrich) with 200 μg of protein extracts. If needed, elution of immunoprecipitates was done using FLAG peptide (Sigma-Aldrich). For endogenous immunoprecipitation of HDAC1 and 2, cells were lysed in LSDB (500 mM NaCl containing 0.02% NP40, 1 mM DTT, 2.5 mM CaCl2, 2 mM EDTA, 5 mM NaF, 25 mM ß-glycerophosphate, 100 μM orthovanadate, and protease and phosphatase inhibitors). Preclearing was performed using 500 μg of protein extracts incubated with insulin-precoated Sepharose A beads. HDAC1 or HDAC2 antibodies (5 μg; Tebu-Bio, Santa Cruz Biotechnology, Santa Cruz, CA; H51 and H54, respectively) and protein A-Sepharose beads were used for immunoprecipitation.
Enzymatic Assays
HDAC assays were performed with a BioVision fluorometric HDAC activity assay kit according to the manufacturer instructions (BioVision). The fluorescence was analyzed using a fluorescence plate reader (excitation, 380 nm; Em, 440 nm). Based on the reference graph, the absolute amount of deacetylated lysine generated in the samples was determined, and HDAC activity was expressed as arbitrary units. HAT assays were performed according to the manufacturer's instruction (BioVision).
Quantification of HDAC Activity from HSF1 Immunoprecipitates by Mass Spectrometry Analysis
Cos cells were plated in 10-cm dishes and transfected with 5 μg Myc-HSF1 with or without 5 μg Flag-HDAC1 and HDAC2 (2.5 μg + 2.5 μg). Twenty-four hours after transfection, half of the cells were heat-shocked and lysed in LSDB500 (500 mM KCl, 50 mM HEPES, pH 7.9, 3 mM MgCl2, and 20% glycerol) containing 0.2% NP40, 1 mM DTT, 2.5 mM CaCl2, 2 mM EDTA, protease inhibitor cocktail (complete mini-EDTA free, Roche), and phosphatase inhibitor cocktail (phosSTOP, Roche). After centrifugation, LSDB0 (the same as lysis buffer without KCl) was added to have 250 mM KCl, and the lysate was incubated for 1 h on ice with anti-myc antibody (Abcam; ab9106). Protein A-Sepharose beads (Amersham, Piscataway, NJ) were then added and incubated at 4°C on a rotoshake for 1 h. After three washes with LSDB250 (same as the lysis buffer with 250 mM KCl), beads were washed twice with deacetylation reaction buffer (20 mM Tris, pH 8, and 150 mM NaCl). Beads were then incubated with 5 pmol/μl acetylated H4 peptide (Ac-SGRGKGGKGLGKGGAK(Ac)-MCA) in reaction buffer at 37°C overnight. Supernatant was then kept, and peptide was purified with Agilent Cleanup C18 pipette tips (Wilmington, DE) according to the manufacturer's instructions. Deacetylation was evaluated using MALDI-TOF (Applied Biosystems, 4800 MALDI-TOF/TOF analyzer) mass spectrometry. Briefly, 1 μl of purified peptide was spotted onto the MALDI target, dried, and 1 μl of 20% CHCa (α-cyano-4-hydroxy-cinnamic acid, Sigma-Aldrich; ref C2020) in 50% acetonitrile, 0.1% trifluoroacetic acid was added. Beads were eluted with 1× Laemmli sample buffer and analyzed by Western blotting.
RESULTS
Heat Shock Induces a Deacetylation of Core Histones
To assess the impact of a thermal stress on chromatin structure, we used DAPI staining of DNA to first analyze in situ the overall chromatin changes induced by heat exposure. In heat-shocked cells, regions of highly condensed chromatin were clearly visible, whereas a more homogeneous fluorescence pattern was observed in the nuclei of control cells (Figure 1A). Because changes in gene expression and chromatin structure are correlated with posttranslational modifications of histone tails, in particular acetylation, we next analyzed histone acetylation in control and heat-shocked cells by immunofluorescence with an antiacetylated lysine antibody, which mainly recognizes acetylated histones. We observed a fine punctate staining homogeneously distributed throughout the nucleoplasm both in control and in heat-shocked cells. As expected, we also observed the presence, in heat-shocked cells, of acetylation foci (Figure 1A) corresponding to nuclear structures known to be enriched in HSF1 (Cotto et al., 1997; Jolly et al., 1997, 2004) and termed nuclear stress bodies (nSBs; reviewed in Biamonti, 2004). Interestingly, heat shock also induced a clear decrease in the overall intensity of the fluorescence staining, suggesting a deacetylation of nuclear proteins, likely to correspond to histones (Figure 1A).
Figure 1.
Heat shock induces a deacetylation of core histones. (A) Heat shock induces a decrease in nuclear acetylation level. Acetylated proteins were detected by immunofluorescence with an anti-acetylated lysine antibody in HeLa cells exposed (+) or not (−) to a 1-h heat shock at 43°C. Nuclei were counterstained with DAPI. Acetylated nuclear foci corresponding to the nuclear stress bodies are present in heat-shocked cells. Bar, 5 μm. (B) Heat shock induces a reversible deacetylation of H4. Whole cell extracts from HeLa exposed to different conditions of heat shock were analyzed by Western blot using an anti-acetylated H4 antibody: 37°C (−), heat shock 1 h at 43°C (0), 3 h (3), or 5 h (5) of recovery after the 1-h heat shock. H2B was used as loading control. (C) Heat shock does not affect histone level and solubility. Cytosolic (C), nuclear soluble (NS), and nuclear insoluble (NI) protein extracts from cells exposed (+) or not (−) to a 1-h heat shock at 43°C or recovery were run on 15% acrylamide and stained with Coomassie blue, and the same fractions were analyzed by Western blot with an anti-H2B antibody.
To confirm this observation, we next performed Western blot analyses with a specific antibody against acetylated H4. As shown in Figure 1B, a significant deacetylation of H4 was observed after a 1-h heat shock. Moreover, the deacetylation was progressively reversed when cells were allowed to recover at 37°C but was not yet fully back to normal after 5 h of recovery. To exclude the possibility that histone deacetylation could be the mere consequence of a heat-induced degradation of histones tails, the overall amount of core histones and migration was analyzed by Coomassie staining and Western blot with an anti H2B antibody, in the cytosolic (C) fraction and in both soluble (NS) and insoluble (NI) nuclear fractions of unstressed and heat-shocked cells. As shown in Figure 1C, similar amounts of histones, with no change in size, were detected in the two nuclear fractions from control and heat-shocked cells. Altogether these observations show that heat-shock results in a massive chromatin remodeling which involves a rapid and reversible deacetylation of core histones.
Identification of the Deacetylation Sites on Core Histones
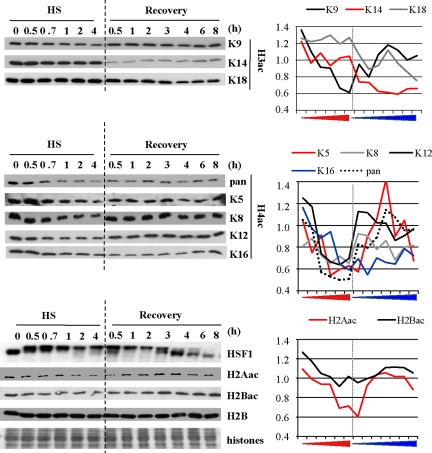
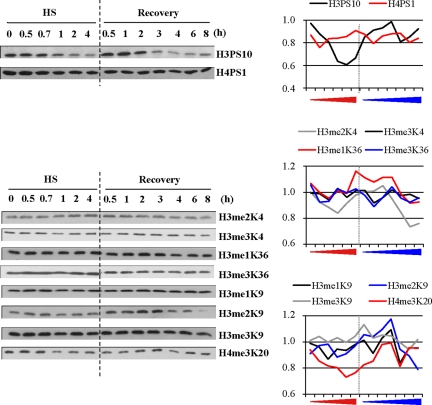
We next tried to determine which histones were targeted by deacetylation and when possible, which specific lysine residues were deacetylated. To address this question, we performed Western blot analyses with a panel of antibodies against specific acetylated histones on extracts from HeLa cells exposed to different lengths of heat-shock exposure (30 min to 4 h) and to different lengths of recovery (30 min to 8 h) conditions after a 1-h heat shock. Equal loading was controlled either by Coomassie staining or Western blot analysis against H2B (Figure 2). The activation of the heat-shock response was controlled by Western blot against HSF1, whose electrophoretic mobility slows down when the factor is active. HSF1 activation began after 30 min of heat shock and was maintained throughout the entire heat-shock treatment. The persistence of HSF1 activation is in agreement with recent data by Westerheide et al. (2009), who found that the DNA-binding capacity of HSF1 is only attenuated after 6 h of continuous heat shock at 43°C in Hela cells, at a time where the percentage of apoptotic cells was found to be <10% (data not shown). After a 1-h heat shock, HSF1 inactivation occurred between 3 and 4 h of recovery. We next analyzed the same protein extracts with a panel of antibodies directed against histone H2A acetylated on K5, H2B acetylated on K12/K15, and H3 acetylated on K9, K14, or K18 and H4 acetylated on K5, K8, K12, or K16. As shown in Figure 2, a deacetylation of all histones was observed, albeit with different intensities and kinetics. For instance, deacetylation of H2BK12/K15, H3K9, H4K5, H4K8, H4K12, and H4K16 was observed after 0.5–1 h of heat shock. This suggests that these modifications represent early epigenetic marks of the heat-shock response. In contrast, deacetylation of H3K14 and H3K18 was only detected at the onset of the recovery period and was still visible after 8 h of recovery, therefore representing a late epigenetic mark. Reacetylation of H3K14 was observed later in the recovery period, after 18 h of recovery (data not shown). Likewise, reacetylation of H2A, H3K9, H4K5, H4K8, and H4K12 was observed in early recovery, whereas reacetylation of H2BK12/K15 and H4K16 appeared as a later event, occurring after 0.5 and 6 h of recovery, respectively, concomitantly, in the case of H4K16, with HSF1 inactivation. Besides changes in histone acetylation, we also found that phosphorylation and histone methylation are both affected by heat shock (Figure 3). However, in contrast to what was observed in the case of acetylation, heat-induced loss of phosphorylation and methylation was only observed for H3PS10 and H3K20me3, whereas increase of H3me2K9 labeling was observed at the onset of the recovery period. Taken together, these results show that heat shock induces a modification of the epigenome, affecting specific epigenetic marks in a time-dependent manner, thus highlighting the complexity of the remodeling events occurring in response to stress.
Figure 2.
Identification of the heat-induced deacetylation lysine targets. Analysis by Western blot of the evolution of different epigenetic marks during heat shock (HS) and recovery in total cell extracts from HeLa cells. Unmodified H2B was used as a loading control, together with Coomassie staining of histones. H2A and H2B acetylation profiles, H3 acetylation profile on K9, K14, and K18. H4 acetylation profile using a pan-acetyl H4 antibody or antibodies to specific acetylated residues (K5, K8, K12, and K16). All fractions were loaded on the same gel. White bar indicate the place where blank lanes were cropped. HSF1 was detected as a marker of the heat-shock response. Delay in HSF1 migration is indicative of its hyperphosphorylated activated state (only in the case of HSF1, fractions were not equilibrated before loading). Quantification of the Western blot signals are also represented with a dash line indicating the transition between the heat-shock and recovery period. Red arrow, increasing time of heat exposure; blue arrow, increasing time of recovery.
Figure 3.
Evolution of histone H3- and H4-specific methylated and phosphorylated residues in the course of the heat-shock response. Analysis by Western blot of the evolution of phosphorylated and methylated epigenetic marks during heat shock (HS) and recovery in total cell extracts from HeLa cells. As in Figure 2, unmodified H2B was used as a loading control, together with Coomassie staining of histones (shown in Figure 2). H3 and H4 phosphorylation profiles on S10 and S1. H3 and H4 methylation profiles using antibody or antibodies specific to methylated residues (me2K4, me3K4, me1K36, me3K36, me1K9, me2K9a, and me3K9 of histone H3 and me3K20 of histone H4). All fractions were loaded on the same gel. White bar indicates the place where blank lanes were cropped. HSF1 was detected as a marker of the heat-shock response. Delay in HSF1 migration is indicative of its hyperphosphorylated, activated state (only in the case of HSF1, fractions were not equilibrated before loading). Quantification of the Western blot signals are also represented, with a dash line indicating the transition between the heat-shock and recovery period. Red arrow, increasing time of heat exposure; blue arrow, increasing time of recovery.
HSF1 Controls Histone Acetylation in Heat-shocked Cells
HSF1 is the key factor involved in the heat-induced activation of both hsp gene and sat III repeated sequences (reviewed in Biamonti, 2004). Besides this role, several observations suggest that upon heat shock, HSF1 also plays a role as a transcriptional repressor on specific genes (Chen et al., 1997; Singh et al., 2002; Xie et al., 2002). Given its transactivating and repressive role, we thought that HSF1 might be a good candidate to regulate the epigenetic changes occurring during heat shock. Therefore, we prepared protein extracts after a kinetic of heat shock and recovery in control (WT) and HSF1 knockout (KO) MEFs (McMillan et al., 1998) and analyzed these extracts by Western blot with an anti-acetylated H4 antibody. As shown in Figure 4, the heat-induced deacetylation of H4 was no longer observed in KO MEF cells, whereas histones were clearly deacetylated in WT cells. We also found that the level of histone acetylation was higher in non-heat-shocked cells than in heat-shocked cells, suggesting that HSF1 exerts either direct or indirect controls on the global level of histone acetylation in normal conditions.
Figure 4.
HSF1 controls acetylation and heat-induced deacetylation of core histones. Western blot analysis of H4 acetylation level in parental (WT) and HSF1 KO mouse cells exposed to different conditions: 37°C (−), 30-min heat shock at 43°C (0.5), 1-h heat shock at 43°C (1), or 3 h of recovery following a 1-h heat shock (rec). Unmodified H2B was used as a loading control. No heat-induced deacetylation is observed in heat-shocked samples in HSF1 KO cells.
HDAC1 and 2 Are Required for Heat-induced Histone Deacetylation
We next investigated the molecular mechanisms involved in the heat-induced deacetylation. Because the heat-induced deacetylation could result from an inhibition of HATs and/or an activation of HDACs, we first tested the implication of HDAC in the deacetylation process with TSA, a potent inhibitor of class I and class II HDACs. Western blot analysis of extracts from cells treated or not with TSA before heat shock revealed that the deacetylation of core histones was no longer observed after TSA treatment (Figure 5A), thus demonstrating that the heat-induced deacetylation of core histones involved class I and/or class II HDACs.
Figure 5.
HDAC1 and HDAC2 mediate the changes in histone acetylation profile upon heat shock. (A) Impact of TSA on heat-induced deacetylation of histones. Protein extracts were obtained from heat-shocked cells or from cells allowed to recover for 3 h after a 1-h heat shock at 37°C (rec). Protein extracts were analyzed by Western blot with an antibody against pan-acetylated H4. H2B was used as a loading control. Inhibition of class I and II HDACs by TSA prevents the heat-induced deacetylation of histone H4. (B) Impact of HDAC1 and 2 knockdown on heat-induced global nuclear protein acetylation level determined by immunofluorescence with an anti-acetylated lysine antibody. Heat-shocked cells were treated with scrambled siRNA (top) or with siRNAs against HDAC1 and 2 (middle) or HDAC3 (bottom). Anti-HDAC1 and 2 or Anti-HDAC3 were used to identify the cells in which the corresponding protein expressions were lowered (right column). Knockdown of HDAC1 and 2 prevents heat-induced deacetylation. Bar, 5 μm. (C) Impact of HDAC1 and 2 knockdown on the level of heat-induced histone deacetylation. Western blot analysis was performed on cellular extracts from non-heat-shocked (−) or heat-shocked (+) HeLa cells treated with siRNA against HDAC1 and 2. H4 acetylation level was analyzed in nontransfected HeLa cells (lanes 1 and 2), in HeLa cells transfected with a scrambled siRNA (lanes 3 and 4), and in HeLa cells treated with an siRNA against HDAC1 (lanes 5 and 6), or HDAC2 (lanes 7 and 8) and in HeLa cells treated with both HDAC1 and HDAC2 (lanes 9 and 10). H4 acetylation level was analyzed using a pan acetyl-H4 antibody (H4ac). The active slow-migrating hyperphosphorylated form of HSF1 is present in both normal cells and in siRNA-treated heat-shocked cells. Unmodified H2B was used as a loading control, as well as Coomassie staining of histones (bottom panel). HDAC1 and 2 were detected with specific antibodies.
We next sought to identify more specifically the nuclear HDACs involved in the deacetylation of histones upon heat shock. To address this question, we used previously reported siRNA to down-regulate the expression of HDAC1, 2, or 3 in HeLa cells. For negative control, cells were either untransfected or transfected with a scrambled siRNA.
To determine the impact of HDAC1 and 2 knockdown on the heat-induced deacetylation of histones, we performed immunofluorescence with the antiacetylated lysine antibody to compare the global acetylation level in control and heat-shocked cells (Figure 5B). As expected, in unstressed conditions, cells knocked down for HDAC1 and 2 displayed a higher level of histone acetylation when compared with control cells. After heat shock, cells transfected with a scrambled siRNA and cells knocked down for HDAC3 both displayed a deacetylation of the whole nucleus and acetylated foci known to correspond to transcription sites that form at the 9q12 locus (Jolly et al., 2004; Rizzi et al., 2004), also known as nSBs (reviewed in Biamonti, 2004). In contrast, cells knocked down for HDAC1 and 2 displayed no decrease of fluorescent staining in the whole nucleus, suggesting that these cells were no longer able to deacetylate histones after heat shock and that HDAC1 and HDAC2 were involved in the deacetylation process. In addition, acetylated foci were no more detected in these cells, most likely masked by the higher level of histone acetylation. To confirm the potential role of HDAC1 and 2 in the heat-induced deacetylation of core histones, protein extracts from cells transfected with the different siRNAs and exposed or not to a heat shock were analyzed by Western blot with a pan acetyl H4 antibody, because H4 was identified as a major specific target of histone deacetylation in heat-shocked cells. In agreement with the in situ observations, cells knocked down for HDAC1 and 2 displayed deacetylation of histone H4 weaker than that seen in control cells transfected with scrambled siRNA. A stronger effect of histone deacetylation was observed in cells transfected with the combination of siRNA against HDAC1 and 2 (Figure 5C). Indeed, although efficiently knocked down by their respective siRNA, the existence of compensatory mechanisms leading to an increased level of HDAC1 upon HDAC2 knockdown and vice versa (Senese et al., 2007) probably explain the better efficiency of experiments combining siRNA against HDAC1 and 2 on the inhibition of histone deacetylation. HSF1 activation in heat-shocked HDAC1 and 2 siRNA-treated cells was not affected, as revealed by the presence of the hyperphosphorylated slow migrating form of HSF1 in the corresponding fractions.
Altogether our findings show that the heat-induced deacetylation of core histones is mediated by HDAC1 and/or HDAC2. We thus identify HDAC1 and HDAC2 as new actors of the heat-shock response that mediate important epigenetic remodeling during stress.
HDAC1 and HDAC2 Activities Are Regulated by Heat Shock
We then tried to identify the mechanisms underlying HDAC1 and 2 activation upon stress. We first analyzed by immunofluorescence the intracellular distribution of HDAC1 and 2 before and after heat shock in HeLa cells. HDAC1 and HDAC2 displayed a diffuse nuclear localization, both at 37°C and in heat-shocked cells. Likewise, HDAC3, which was used as a negative control, displayed a diffuse cytoplasmic and nuclear labeling.
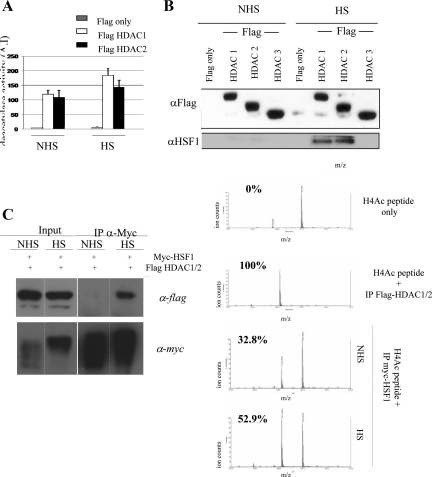
Because neither subcellular redistribution nor quantitative increase of HDAC1 and 2 (Figure 5C) was observed in heat-shocked cells, we then made the assumption that heat shock could specifically enhance the enzymatic activity of HDAC1 and/or 2. To test this hypothesis, we immunoprecipitated overexpressed FLAG-tagged HDAC1 and 2 from non-heat-shocked and heat-shocked COS cells. The control experiments were done with cells transfected with FLAG empty vector. Immunoprecipitated complexes were eluted by competition using a FLAG peptide, and a HDAC fluorescent enzymatic assay was then performed on the immunoprecipitates. Results are shown in Figure 6A, which show that heat shock significantly increases HDAC1 and 2 activities. Loading of equal amounts of each HDAC for the enzymatic assay was evaluated by Western blot using an anti-FLAG antibody (Figure 6B). These results suggested that increase in the heat-induced activity of HDAC1- and 2-containing complexes may account for the global deacetylation of core histones.
Figure 6.
HSF1 binding to HDAC 1 and HDAC2 in heat-shocked cells correlates with increased HDAC activities. (A) Increased HDAC1 and 2 activities after heat shock. Immunoprecipitation of transiently transfected Flag-tagged HDAC1 and 2 or Flag tag alone from non-heat-shocked (NHS) or heat-shocked (HS) HeLa cells were performed with anti-Flag antibodies. The same amounts of immunoprecipitates were used in a deacetylase assay to quantify the HDAC activity in each sample. Increased HDAC1 and 2 activities were both observed in heat-shocked cells. Each sample was analyzed in duplicates in two independent experiments; error bars, SEM. (B) Endogenous HSF1 coimmunoprecipitates with Flag-HDAC1 and HDAC2 but not with HDAC3 in heat-shocked cells. Flag immunoprecipitations were performed as in A on cellular extracts from unstressed and heat-shocked cells transiently transfected with Flag-HDAC1, -HDAC2, or -HDAC3. Western blot analysis of the immunoprecipitates were performed with anti-Flag (top panel) and anti-HSF1 (bottom panel) antibodies. (C) Increased HDAC activity in HSF1 immunoprecipitates after heat shock. Cos cells were transiently cotransfected with Myc-tagged HSF1 and a mixture of flag-tagged HDAC1/2. Immunoprecipitations from non-heat-shocked (NHS) or heat-shocked (HS) cells were performed with anti-Myc or anti-Flag antibodies as indicated. The level of HSF1 and HDAC1/2 in NHS and HS cells and the level of HSF1 and of HDAC1/2 in the corresponding immunoprecipitates are analyzed by Western blot using the anti-Flag and anti-Myc antibodies. Each immunoprecipitate was incubated ON with an acetylated H4 synthetic peptide. The percentage of deacetylated and acetylated peptides in the reaction was determined by MALDI-TOF mass spectrometry. The peak at 1655 Da (left peak) corresponds to the acetylated H4 peptide and the peak at 1613 Da (right peak) to the deacetylated H4 peptide. The percentages of deacetylated H4 peptides is indicated on each spectrum.
To confirm that heat shock leads to enhanced HDAC activity associated with HSF1, a deacetylation assay was performed on a synthetic acetylated peptide of H4 with myc-HSF1 immunoprecipitates of cells overexpressing Flag-HDAC1 and 2 as well as MALDI-TOF mass spectrometry analysis. The mass spectra in Figure 6C show that the percentage of deacetylated peptide significantly increases when the assay is performed with myc-HSF1 immunoprecipitates from stressed cells (52.9 vs. 32.8% for unstressed cells).
Given the role played by HSF1 in the control of global histone deacetylation, we next tested the possibility that increased HDAC1 and 2 activities could involve their transient association with HSF1.
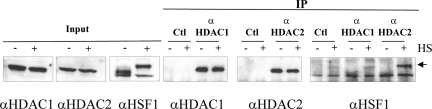
Using the same FLAG-HDAC constructs, we immunoprecipitated HDAC1, 2, or 3 from cells exposed or not to a heat shock and then looked for the presence of HSF1 in the immunoprecipitated complexes. We found that endogenous HSF1 is associated with HDAC1 and 2 mainly in heat-shocked cells and not with HDAC3 or with the Flag alone (Figure 6B). The interaction between HSF1 and HDAC1 and 2 in heat-shocked cells was also confirmed by endogeneous immunoprecipitation with anti-HDAC1 and anti-HDAC2 antibodies (Figure 7).
Figure 7.
Endogenous HSF1 immunoprecipitates with endogenous HDAC1 and 2. Immunoprecipitations anti HDAC1 and HDAC2 were performed with anti HDAC1 and 2. Equal amounts of HDAC1 and 2 and HSF1 in non-heat-shocked and heat-shocked extracts was controlled by Western blot in total cell extracts (Input). Immunoprecipitations were performed on cellular extracts from unstressed (−) and heat-shocked (+) cells. Equal level of HDAC1 and 2 in the immunoprecipitates was also controlled by Western blot. The presence of HSF1 in the immunoprecipitates from heat-shocked cells was determined by Western blot with anti-HSF1 (the arrow indicates the signal corresponding to HSF1). Immunoprecipitations were also performed with nonspecific rabbit IgG used as controls of specificity (Ctl).
Altogether our results reveal a new role for HSF1 as a regulator of genome-wide epigenetic changes, most likely, at least in heat-shocked cells, through its association with HDAC1 and 2.
DISCUSSION
Histone Acetylation and Large-Scale Genome Organization
The work presented here and few known examples reported in the literature now allow pointing to a genome-wide modulation of histone acetylation as a key event in the genome response to various types of stimuli. Indeed, in the literature only two cases have already been reported describing a major modulation of histone acetylation in physiological settings, but the underlying mechanisms have remained poorly understood. The first example concerns spermatogenesis, where a genome-wide histone acetylation is known to precede their removal and replacement by testis-specific basic proteins (reviewed in Boussouar et al., 2008). The second example concerns experimentally denervated skeletal muscle cells, where it was shown that innervation also controls a global chromatin deacetylation. In this latter case, further investigations showed a key function for MITR, an HDAC9 splice variant lacking its deacetylase domain (Méjat et al., 2005). Cell response to heat shock provides a third example. Indeed, more than 20 years ago the existence of a global deacetylation of core histone in heat-shocked cells had already been described in Drosophila melanogaster (Arrigo, 1983; Desrosiers and Tanguay, 1986), but the functional significance and underlying mechanisms have remained obscure. Here, investigating the molecular basis of this phenomenon, we show that a major heat-shock transcription factor is capable of mediating a global deacetylation of all core histones, demonstrating for the first time the implication of a transcription factor in the control of genome-wide global deacetylation. We further demonstrate the underlying mechanisms of heat-induced chromatin deacetylation and highlight the implication of HSF1, most likely through its transient association with transcriptional repressors HDAC1 and 2. Unexpectedly, we also found that HSF1 controls the level of chromatin acetylation in non-heat-shocked cells and that cells deficient in HSF1 display an hyperacetylated chromatin. These observations considerably enlarge the role of HSF1, leading to the new concept that HSF1 may represent a general orchestrator of chromatin acetylation. Our own observation therefore indicates that stress-induced deacetylation of core histones is a common feature of higher eukaryotes.
Stress and Modification of the Histone Code
We find that heat shock, through HSF1 activation, induces a series of epigenetic modifications, providing evidence for the existence of a stress-related histone code. Several studies have shown that other types of stress (hypoxia, arsenite) are associated with posttranslational changes of histones including a deacetylation of H3K9 and a methylation of the same residue (Chen et al., 2006). In our study, a particular interest was brought to the analysis of the acetylation status of some of the lysine residues histones H3 and H4: H3K9, K14 and K18 and H4K5, K8, K12, and K16. One interesting observation is that heat-shock targets all lysine residues. Another interesting result of this analysis is the observation that the kinetics of deacetylation varies depending on the residue considered. Indeed we show that H3K14 and H3K18 are not deacetylated during the course of heat shock itself, but are rather deacetylated in the early minutes of the recovery period, at a time where, in contrast, the reacetylation of most other lysines occurs. Besides changes in histone acetylation, we also found that phosphorylation and histone methylation are both affected by heat shock. H3S10P is known to be enriched on mitotic chromosomes. Decrease of H3S10P, a mitotic chromosome marker, in the course of the heat-shock response has been reported in the literature as a consequence of a blockade of the cell cycle at the G2/M transition (reviewed in Dyson et al., 2005). Transient increase of H3S10P was observed at the onset of the recovery period, which most likely corresponds to reentry of the cells into mitosis. Interestingly, a similar profile was also observed for H3K9me2. In contrast, Western blot analysis did not reveal any clear quantitative changes of other epigenetic residues. These observations highlight the existence of a tightly orchestrated control of each lysine residue in the course of the heat-shock response. The significance of this sophisticated histone code remains to be clarified. Evidence suggests that the epigenetic modifications that occur in response to different types of stress lead to specific signatures of the epigenome. For example, although both associated with an activation of HSF1, hypoxia is accompanied by a global deacetylation of H3K9 (Chen et al., 2006) and to an increase of H3K14 acetylation (Johnson et al., 2008), whereas oxidative stress induces an increase of histone H4 acetylation (Rahman et al., 2002).
Interplay between HSF1 and HDACs in the Control of the Epigenome, in Heat-shocked Cells
Our study identifies HDAC1 and 2 as new important actors of the heat-shock response regulating the heat-induced epigenetic changes. The emerging role of HDACs as essential actors of the stress responses is not restricted to class I HDACs. Indeed, we and others have recently identified HDAC6 and SIRT1 as critical regulators of HSF1 activity (Boyault et al., 2007; Westerheide et al., 2009). Recently, an interaction between HSF1 and HDAC1/2 was reported in the repression of estrogen-dependent transcription by heregulin (Khaleque et al., 2007). Moreover the protein CoREST, which belongs to HDAC1 and 2 repressor complexes, has recently been implicated in the control of hsp70 expression in non-heat-shocked and heat-shocked cells (Gómez et al., 2008). Our study explores a totally new aspect of the heat-shock response and identifies HDAC1 and 2 as important general regulators of genome wide organization in heat-shocked cells, regulating the heat-induced epigenetic changes. Our data shed light on a new activity of HSF1. HSF1 specifically associates with and uses HDAC1 and 2 to trigger heat-shock–dependent histone deacetylation. Our contribution considerably enlarges the role of HSF1 in the general control of chromatin acetylation and establishes a new functional cross-talk between HSF1 and general chromatin acetylation.
Heat Shock, HDACs, and Cancer Therapy
Finally the new interplay between HSF1 and HDACs that we highlight may be useful to better understand the emerging role of HSF1 in tumorigenesis (Dai et al., 2007). The deacetylation of H4K16 that we observe during heat shock is also thought to represent a common hallmark of human cancer cells (Fraga et al., 2005). Hypoacetylation in cancer cells may reveal the implication of HSF1 in carcinogenesis (Dai et al., 2007). A number of HDAC inhibitors are used in clinical trials for anticancer therapy. Today, we believe that histone deacetylation, similar to the induction of HSPs, is an essential event in the cellular defense to stress. Our work thus establishes a new link between two targets that are the subject of numerous studies in oncology: HSPs and HDACs. Strategies combining specific HDACs inhibitors with thermotherapy could pothole the effect of these inhibitors on tumor cells.
ACKNOWLEDGMENTS
We are grateful to Drs. S. Gazzeri, C. Souchier, and C. Caron (Inserm U823, Grenoble, France) and to Drs. B. Turner (Birmingham, UK) and E. Seto (H. Lee Moffitt Cancer Center and Research Institute) for help in data analysis and for providing antibodies and plasmids. The medical proteomic platform of the Center for Innovation in Biology (CIB) receives financial support from the Lyon-Auvergne-Rhône-Alpes Canceropole (CLARA), University Joseph Fourier-Grenoble 1, Grenoble University Hospital, INSERM, and the French Research Ministry. This work was supported by grants from INSERM and Université J. Fourier, by ARC (Research Grant 3449 to C.V) by grants from the InCa (Epistress project to C.V. and S.K.), by CLARA (EPIPRO and EPIMED projects to C.V. and S.K.), by grants from University Toulouse III (BQR to E.C.), and by grants from AIRC (Associazione Italiana Ricerca sul Cancro to S.C.). S.F. was supported by the French ministry of research and ARC.
Abbreviations used:
- HAT
histone acetyl transferase
- HDAC
histone deacetylase
- HSF1
heat-shock factor 1
- HSP
heat-shock protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-04-0295) on September 30, 2009.
REFERENCES
- Arrigo P. Acetylation and methylation patterns of core histones are modified after heat or arsenite treatment of Drosophila tissue culture cells. Nucleic Acids Res. 1983;11:1389–1404. doi: 10.1093/nar/11.5.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G. Nuclear stress bodies: a heterochromatin affair? Nat. Rev. Mol. Cell Biol. 2004;5:493–498. doi: 10.1038/nrm1405. [DOI] [PubMed] [Google Scholar]
- Boussouar, Rousseaux F., Khochbin S., S. A new insight into male genome reprogramming by histone variants and histone code. Cell Cycle. 2008;7:3499–3502. doi: 10.4161/cc.7.22.6975. [DOI] [PubMed] [Google Scholar]
- Boyault C., Zhang Y., Fritah S., Gilquin B., Kwon S. H., Garrido C., Yao T. P., Vourc'h C., Matthias P., Khochbin S. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007;21:2172–2181. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Xie Y., Stevenson M. A., Auron P. E., Calderwood S. K. Heat shock factor 1 represses Ras-induced transcriptional activation of the c-fos gene. J. Biol. Chem. 1997;272:26803–26806. doi: 10.1074/jbc.272.43.26803. [DOI] [PubMed] [Google Scholar]
- Chen H., Yan Y., Davidson T. L., Shinkai Y., Costa M. Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer Res. 2006;66:9009–9016. doi: 10.1158/0008-5472.CAN-06-0101. [DOI] [PubMed] [Google Scholar]
- Cheung W. L., Briggs S. D., Allis C. D. Acetylation and chromosomal functions. Curr. Opin. Cell Biol. 2000;12:326–333. doi: 10.1016/s0955-0674(00)00096-x. [DOI] [PubMed] [Google Scholar]
- Christians E. S., Yan L. J., Benjamin I. J. Heat shock factor 1 and heat shock proteins: critical partners in protection against acute cell injury. Crit. Care Med. 2002;30:S43–50. [PubMed] [Google Scholar]
- Cotto J., Fox S., Morimoto R. HSF1 granules: a novel stress-induced nuclear compartment of human cells. J. Cell Sci. 1997;110:2925–2934. doi: 10.1242/jcs.110.23.2925. [DOI] [PubMed] [Google Scholar]
- Cunliffe V. T. Eloquent silence: developmental functions of Class I histone deacetylases. Curr. Opin. Genet. Dev. 2008;18:404–410. doi: 10.1016/j.gde.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C., Whitesell L., Rogers A. B., Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R., Tanguay R. M. Further characterization of the posttranslational modifications of core histones in response to heat and arsenite stress in Drosophila. Biochem. Cell Biol. 1986;64:750–757. doi: 10.1139/o86-102. [DOI] [PubMed] [Google Scholar]
- Dyson M. H., Thomson S., Mahadevan L. C. Heat shock, histone H3 phosphorylation and the cell cycle. Cell Cycle. 2005;4:13–17. doi: 10.4161/cc.4.1.1362. [DOI] [PubMed] [Google Scholar]
- Findly R. C., Pederson T. Regulated transcription of the genes for actin and heat-shock proteins in cultured Drosophila cells. J. Cell Biol. 1981;88:323–328. doi: 10.1083/jcb.88.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga M. F., et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Gómez A. V., Galleguillos D., Maass J. C., Battaglioli E., Kukuljan M., Andrés M. E. CoREST represses the heat shock response mediated by HSF1. Mol Cell. 2008;31:222–231. doi: 10.1016/j.molcel.2008.06.015. [DOI] [PubMed] [Google Scholar]
- Higashikubo R., Roti Roti J. L. Alterations in nuclear protein mass and macromolecular synthesis following heat shock. Radiat. Res. 1993;134:193–201. [PubMed] [Google Scholar]
- Jamrich M., Haars R., Wulf E., Bautz F. A. Correlation of RNA polymerase B and transcriptional activity in the chromosomes of Drosophila melanogaster. Chromosoma. 1977;64:319–326. doi: 10.1007/BF00294939. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C. D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Johnson A. B., Denko N., Barton M. C. Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutat. Res. 2008;640:174–179. doi: 10.1016/j.mrfmmm.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C., Morimoto R., Robert-Nicoud M., Vourc'h C. HSF1 transcription factor concentrates in nuclear foci during heat shock: relationship with transcription sites. J. Cell Sci. 1997;110:2935–2941. doi: 10.1242/jcs.110.23.2935. [DOI] [PubMed] [Google Scholar]
- Jolly C., Konecny L., Grady D. L., Kutskova Y. A., Cotto J. J., Morimoto R. I., Vourc'h C. In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress. J. Cell Biol. 2002;156:775–781. doi: 10.1083/jcb.200109018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C., Metz A., Govin J., Vigneron M., Turner B. M., Khochbin S., Vourc'h C. Stress-induced transcription of satellite III repeats. J. Cell Biol. 2004;164:25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleque M. A., Bharti A., Gong J., Gray P. J., Sachdev V., Ciocca D. R., Stati A., Fanelli M., Calderwood S. K. Heat shock factor 1 represses estrogen-dependent transcription through association with MTA1. Oncogene. 2007;27:1886–1893. doi: 10.1038/sj.onc.1210834. [DOI] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu. Rev. Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- McMillan D. R, Xiao X., Shao L., Graves K., Benjamin I. J. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J. Biol. Chem. 1998;273:7523–7528. doi: 10.1074/jbc.273.13.7523. [DOI] [PubMed] [Google Scholar]
- Méjat A., Ramond F., Bassel-Duby R., Khochbin S.S., Olson E. N., Schaeffer L. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat. Neurosci. 2005;8:313–321. doi: 10.1038/nn1408. [DOI] [PubMed] [Google Scholar]
- Mellor J. Dynamic nucleosomes and gene transcription. Trends Genet. 2006;22:320–329. doi: 10.1016/j.tig.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Morimoto R. I., Sarge K. D., Abravaya K. Transcriptional regulation of heat shock genes. A paradigm for inducible genomic responses. J. Biol. Chem. 1992;267:21987–21990. [PubMed] [Google Scholar]
- Morimoto R. I., Kroeger P. E., Cotto J. J. The transcriptional regulation of heat shock genes: a plethora of heat shock factors and regulatory conditions. EXS. 1996;77:139–163. doi: 10.1007/978-3-0348-9088-5_10. [DOI] [PubMed] [Google Scholar]
- Rahman I., Gilmour P. S., Jimenez L. A., MacLee W. Oxidative stress and TNF-α induce histone acetylation and NF-kB/AP-1 activation in alveolar epithelial cells: potential mechanism in gene transcription in lung inflammation. Mol. Cell Biochem. 234/ 2002;235:239–248. [PubMed] [Google Scholar]
- Rizzi N., Denegri M., Chiodi I., Corioni M., Valgardsdottir R., Cobianchi F., Riva S., Biamonti G. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol. Biol. Cell. 2004;15:543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senese S., et al. Role for histone deacetylase 1 in human tumor cell proliferation. Mol. Cell. Biol. 2007;27:4784–4795. doi: 10.1128/MCB.00494-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I. S., He J. R., Calderwood S., Hasday J. D. A high affinity HSF-1 binding site in the 5′-untranslated region of the murine tumor necrosis factor-alpha gene is a transcriptional repressor. J. Biol. Chem. 2002;277:4981–4988. doi: 10.1074/jbc.M108154200. [DOI] [PubMed] [Google Scholar]
- Sorger S. K. Heat shock factor and the heat shock response. Cell. 1991;65:363–366. doi: 10.1016/0092-8674(91)90452-5. [DOI] [PubMed] [Google Scholar]
- Spradling A., Penman S., Pardue M. L. Analysis of Drosophila mRNA by in situ hybridization: sequences transcribed in normal and heat shocked cultured cells. Cell. 1975;4:395–404. doi: 10.1016/0092-8674(75)90160-9. [DOI] [PubMed] [Google Scholar]
- Westerheide S. D., Anckar J., Stevens S. M., Jr., Sistonen L., Morimoto R. I. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Heat shock transcription factors: structure and regulation. Annu. Rev. Cell Dev. Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- Xie Y., Chen C., Stevenson M. A., Auron P. E., Calderwood S. K. Heat shock factor 1 represses transcription of the IL-1beta gene through physical interaction with the nuclear factor of interleukin 6. J. Biol. Chem. 2002;277:11802–11810. doi: 10.1074/jbc.M109296200. [DOI] [PubMed] [Google Scholar]
- Yang X. J., Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Kijima M., Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]