Abstract
Sorting of transmembrane cargo proteins to different cellular compartments is mediated by sorting signals that are recognized by coat proteins involved in vesicle biogenesis. We have identified a sorting signal in the yeast cell fusion protein Fus1p that is required for its transport from the trans-Golgi compartment to the plasma membrane. Transport of Fus1p from the trans-Golgi to the cell surface is dependent on Chs5p, a component of the multisubunit exomer complex. We show that Fus1p transport is also dependent on the exomer components Bch1p and Bud7p. Disruption of the clathrin adaptor protein complex 1 (AP-1) restores Fus1p localization to the cell surface in the absence of exomer, possibly by promoting an alternate, exomer-independent route of transport. Mutation of an IXTPK sequence in the cytosolic tail of Fus1p abolishes its physical interaction with Chs5p, results in mislocalization of Fus1p, and therefore causes a cell fusion defect. These defects are suppressed by disruption of AP-1. We suggest that IXTPK comprises a novel sorting signal that is recognized and bound by exomer leading to the capture of Fus1p into coated vesicles en route to the cell surface.
INTRODUCTION
Sorting of transmembrane cargo proteins to the various compartments within the secretory pathway is mediated by specific sorting signals recognized by selective coat protein complexes involved in transport and vesicle budding. Coat complexes as well as the sorting signals they recognize have been identified for many of the transport steps in the secretory pathway. For example, the COPII coat recognizes diacidic and hydrophobic based sorting signals to mediate transport from the endoplasmic reticulum (ER) to the Golgi complex, and the COPI coat recognizes a dilysine-based sorting signal to mediate transport from the Golgi to the ER (Lee et al., 2004). The clathrin coat, along with clathrin adaptor proteins (APs) and the GGAs recognize dileucine and tyrosine based sorting signals to mediate transport between the trans-Golgi network (TGN), endosomes and the vacuole (Bonifacino and Traub, 2003). Much less is understood about the coat complexes that mediate TGN-to-plasma membrane transport and about the sorting signals these coats recognize.
In polarized epithelial cells, examples of sorting signals that target cargo to the basolateral membrane include tyrosine- and dileucine-based motifs (Folsch, 2008) as well as the PDZ-binding ligand of syndecan-1 (Maday et al., 2008). However, a protein coat that recognizes these signals has not been identified. Targeting of cargo to the apical membrane may involve glycosylation as well as association with lipid rafts (Folsch, 2008). In yeast and mammalian systems, diacylgycerol levels, which are maintained in part by the phosphatidylinositol-transfer protein Sec14p and the peripheral Golgi protein Nir2, are critical for protein transport to the plasma membrane (Corda et al., 2002; Litvak et al., 2005). Phosphatidylinositol-4-phosphate is also involved in TGN-to-plasma membrane transport in yeast (Hama et al., 1999; Walch-Solimena and Novick, 1999; Audhya et al., 2000). Recently, the protein complex exomer was identified as a coat complex involved in TGN-to-plasma membrane transport in yeast (Sanchatjate and Schekman, 2006; Trautwein et al., 2006; Wang et al., 2006), although a sorting signal recognized by exomer was not defined.
Exomer is a protein complex important for the transport of select membrane proteins from the TGN to the plasma membrane in yeast (Santos and Snyder, 1997, 2003). It is composed of five subunits: Chs5p and a family of four related proteins—Chs5p-Arf1p binding proteins (ChAPs) (Sanchatjate and Schekman, 2006; Trautwein et al., 2006; Wang et al., 2006). The four ChAPs include two pairs of closely related paralogues: Chs6p and Bch2p; Bch1p and Bud7p (Sanchatjate and Schekman, 2006; Trautwein et al., 2006; Wang et al., 2006). All five exomer components are cytosolic proteins that colocalize with late Golgi markers and associate with each other both in vivo and in vitro (Santos and Snyder, 1997, 2000; Sanchatjate and Schekman, 2006; Trautwein et al., 2006; Wang et al., 2006). Chs5p may play a structural role in organizing exomer and the ChAPs may confer cargo specificity (Sanchatjate and Schekman, 2006). When baculovirus-purified exomer is combined with Escherichia coli-purified Arf1p-guanosine 5′-O-(3-thio)triphosphate, exomer formed an electron-dense protein coat on synthetic liposomes reminiscent of the COPII protein coat. However, in the case of exomer, coated buds or coated vesicles were never observed as they were with COPII (Matsuoka et al., 1998; Wang et al., 2006). Therefore, exomer alone is not a complete protein coat sufficient to deform membranes and induce bud formation.
The exomer component Chs5p is known to be important for the plasma membrane localization of two transmembrane cargo molecules: Chs3p, which is required at the cell surface for synthesis of the yeast cell wall component chitin; and Fus1p, which is required at the cell surface for cell fusion during the yeast mating response (Trueheart et al., 1987; Trueheart and Fink, 1989; Santos and Snyder, 1997, 2003; Nolan et al., 2006). In chs5Δ mutants, neither Chs3p nor Fus1p localizes to the plasma membrane and instead both are found only in intracellular patches that seem to correspond to a TGN or endosomal compartment (Santos and Snyder, 1997, 2003). In addition to Chs5p, Chs3p also requires Chs6p or Bch1p and Bud7p for its plasma membrane localization (Ziman et al., 1998; Sanchatjate and Schekman, 2006; Trautwein et al., 2006).
We hypothesized that one or more of the ChAPs would play a role in Fus1p localization. We have identified Bch1p and Bud7p as the ChAPs required for localization of Fus1p to the plasma membrane. In addition, we found that the exomer-dependent localization of Fus1p to the plasma membrane relies on a novel IXTPK sorting signal found within the cytosolic tail of Fus1p and that exomer physically interacts with Fus1p in a sorting signal dependent manner.
MATERIALS AND METHODS
Yeast Strains, Media, and Growth Conditions
The yeast strains used in this study are listed in Table 1. Yeast strains were grown in standard rich media (YPD) or synthetic dextrose media (SD) supplemented with appropriate amino acids. All strains were cultured at 30°C. Strains were constructed either by integration of disruption cassettes replacing the entire coding region (Brachmann et al., 1998; Longtine et al., 1998), by integration of insertions at the COOH-terminal codon (Longtine et al., 1998) or by tetrad dissection of sporulated diploid strains. Allelic replacements or insertions were confirmed by polymerase chain reaction (PCR). chs6Δ::HIS3, chs6Δ::TRP1, bud7Δ::KANMX4, and fus1Δ::KANMX4 disruption cassettes were generated by PCR amplification from total genomic DNA isolated from TSY49, TSY52, EUROSCARF YOR299w::KANMX4, and EUROSCARF YCL027w::KANMX4, respectively.
Table 1.
S. cerevisiae strains used in this study
| Name | Genotype | Source |
|---|---|---|
| PJ69-4A | MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4D gal80D LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al. (1996) |
| YPH499 | MATa ade2-101oc his3-Δ 200 leu2-Δ1 lys2-801am trp1-Δ 63 ura3-52 | Sikorski and Hieter (1989) |
| YPH500 | MATα ade2-101oc his3-Δ 200 leu2-Δ1 lys2-801am trp1-Δ 63 ura3-52 | Sikorski and Hieter (1989) |
| YAS9 | MATa YPH499 bar1Δ::KANMX6 | J. Thorner, University of California–Berkeley |
| YRB38 | MATa YAS9 FUS1-13Myc::TRP1 | This study |
| YRB41 | MATa YAS9 bch1Δ::HIS3 | This study |
| YRB46 | MATa YPH499 bar1Δ::HIS3 bud7Δ::KANMX4 | This study |
| YRB48 | MATa YAS9 chs6Δ::HIS3 | This study |
| YRB56 | MATa YRB41 bch2Δ::LEU2 | This study |
| YRB57 | MATa YRB41 chs6Δ::TRP1 | This study |
| YRB86 | MATa YRB46 bch1Δ::LEU2 | This study |
| YRB90 | MATa YAS9 chs5Δ::HIS3 | This study |
| YRB100 | MATa YRB41 FUS1-13Myc::TRP1 | This study |
| YRB103 | MATa YRB86 FUS1-13Myc::TRP1 | This study |
| YRB106 | MATa YRB90 FUS1-13Myc::TRP | This study |
| YRB132 | MATa YPH499 bar1Δ::HIS3 bud7Δ::KANMX4 bch1Δ::LEU2 apl2Δ::TRP1 | This study |
| YRB151 | MATa YPH499 bar1Δ::HIS3 bud7Δ::KANMX4 bch1Δ::LEU2 apl2Δ::URA3 FUS1-13Myc::TRP1 | This study |
| YRB168 | MATa YPH499 bar1Δ::HIS3 bch2 Δ::URA3 bud7 Δ::KANMX4 chs6 Δ::LEU2 FUS1-13Myc::TRP1 | This study |
| YRB171 | MATa YPH499 bar1Δ::HIS3 bch2Δ::NATMX bud7Δ::KANMX4 chs6Δ::LEU2 | This study |
| YRB180 | MATa YPH499 bar1Δ::HIS3 bch1Δ::LEU2 bch2Δ::URA3 bud7Δ::KANMX4 chs6Δ::HIS3 FUS1-13Myc::TRP1 | This study |
| YRB191 | MATa YPH499 bar1Δ::HIS3 fus1Δ::KANMX4 | This study |
| YRB192 | MATa YPH499 fus1Δ::KANMX4 | This study |
| YRB193 | MATα YPH500 fus1Δ::KANMX4 | This study |
| YRB202 | MATa YPH499 chs5Δ::LEU2 | This study |
| YRB207 | MATa YRB90 apl2Δ::TRP2 | This study |
| YRB208 | MATa YRB106 apl2Δ::URA3 | This study |
| YRB209 | MATa YPH499 FUS1-13Myc::TRP1 | This study |
| YRB210 | MATa YPH499 bud7Δ::KANMX4 FUS1-13Myc::TRP1 | This study |
| YRB211 | MATa YPH499 bch1Δ::LEU2 FUS1-13Myc::TRP1 | This study |
| YRB212 | MATa YPH499 bch1Δ::LEU2 bud7Δ::KANMX4 | This study |
| YRB213 | MATa YRB202 apl2Δ::URA3 | This study |
| YRB214 | MATa YRB212 apl2Δ::URA3 | This study |
| YRB215 | MATa YRB191 apl2Δ::TRP2 | This study |
| YRB216 | MATa YRB192 apl2Δ::TRP2 | This study |
| YRB217 | MATa YRB191 chs5Δ::TRP2 | This study |
| YRB218 | MATa YRB191 SEC7-RFP::TRP | This study |
| YSS27 | MATa leu2 ura3-52 trp1 prb1-1122 pep4-3 pre1-451 CHS5-Stag-TEV-ZZ::KanMX | Sanchatjate and Schekman (2006) |
Plasmid Construction
Plasmids used in this study are listed in Table 2. pRB74 was generated by subcloning a NotI-SpeI fragment containing the entire coding region of FUS1 plus 300 bp of the FUS1 5′ untranslated region (UTR) into pRS416-GFP. pRS416-green fluorescent protein (GFP) was generated by subcloning GFP into the SpeI and EcoRI sites of pRS416 (Brachmann et al., 1998). pRB228-pRB231 and pRB238-pRB254 were generated by performing QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA) on pRB74. pRB210, pRB214, pRB212, pRB213, pRB211, and pRB215 were generated by subcloning a NotI-SpeI fragment from pRB198, pRB205, pRB200, pRB204, pRB199, and pRB206, respectively, into pRS416-GFP. pRB198, pRB205, pRB200, pRB204, pRB199, and pRB206 were generated by first subcloning a NotI-BamHI fragment containing the region coding for 300 bp of the FUS1 3′UTR plus Fus1p (1-150), Fus1p (1-200), Fus1p (1-250), Fus1p (1-300), Fus1p (1-350), and Fus1p (1-400), respectively, into pRS416, which created pRB193, pRB201, pRB196, pRB194, pRB195, and pRB202, respectively. A BamHI-XhoI fragment containing the region coding for Fus1p (201-512), Fus1p (251-512), Fus1p (301-512), Fus1p (351-512), Fus1p (401-512), and Fus1p (451-512) was subcloned into pRB193, pRB201, pRB196, pRB194, pRB195, and pRB202, respectively, which created pRB198, pRB205, pRB200, pRB204, pRB199, and pRB206, respectively. pRB135 was generated by subcloning a NotI-SpeI fragment containing the region coding for Fus1p (1-400) plus 300bp of the FUS1 5′UTR into pRS416-GFP. pER9 was generated by subcloning an EcoRI-SalI fragment containing the region coding for Chs5p (1-264) into pGBDU-C1 (James et al., 1996). pRB28 was generated by subcloning a BamHI-SalI fragment containing the region coding for Fus1p (97-513) into pGAD-C1 (James et al., 1996). To generate pRB226, pRB270-pRB275, and pRB232-pRB237, we subcloned an 842bp DraIII-NheI fragment from the corresponding FUS1-GFP plasmid into pRB28. pRB188 was generated by subcloning a BamHI-SpeI fragment containing the region coding for Fus1p (1-512) into pMET25-3xHA (Noergaard, personal communication). pRB269 was generated by subcloning a BamHI-SpeI fragment containing the region coding for Fus1p (1-512 176ALAAA180) into pMET25-3xHA. pER7 was generated by subcloning a SacI fragment containing 300 bp of the FUS1 5′UTR, a SacI-SacII fragment containing the region coding for Kex2p (1-675) and a SacII-SpeI fragment containing the region coding for Fus1p (71-96) into pRS416-GFP. pER5 was generated by subcloning a SacI fragment containing 300 bp of the FUS1 5′UTR, a SacI-SacII fragment containing the region coding for Kex2p (1-675) and a SacII-SpeI fragment containing the region coding for Fus1p (71-512) into pRS416-GFP. pER12 was generated by subcloning a SpeI fragment containing the region coding for Kex2p (700-814) into pER7. pRB255 was generated by subcloning a SacI-SacII fragment containing the entire coding region of MID2 into a plasmid containing 300 bp of the FUS1 5′UTR cloned into the SacI site of pRS416-GFP. pRB256 was generated by subcloning a SacI-SacII fragment containing the region coding for Mid2p (1-251) into a plasmid containing 300 bp of the FUS1 5′UTR cloned into the SacI site of pRS416-GFP. pRB257 was generated by subcloning a SacII fragment the region coding for Fus1 (97-512) into pRB256. pRB277 was generated by subcloning a SacI-SacII fragment containing the entire coding region for Kex2p, a SacII-SpeI fragment containing the entire coding region for monomeric red fluorescent protein (mRFP) (vector pRSETB; Invitrogen, Carlsbad, CA), and a SacI fragment containing 300bp of the FUS1 5′UTR into pRS415 (Brachmann et al., 1998). pER21 was generated by subcloning a SacI-SacII fragment containing the entire coding region for mRFP, a SacII-SpeI fragment containing the entire coding region for Tlg1p, and a SacI fragment containing 300 bp of the FUS1 5′UTR into pRS415. pER22 was generated by subcloning a SacI-SacII fragment containing the entire coding region for mRFP, a SacII-SpeI fragment containing the entire coding region for Tlg1p, and a SacI fragment containing 760bp of the ADH1 5′UTR into pRS415.
Table 2.
Plasmids used in this study
| Plasmid name | Expressinga | Source |
|---|---|---|
| pDN291 | Soluble cytosolic GFP | Ng and Walter (1996) |
| pGAD-C1 | GAL4 activation domain (AD) | James et al. (1996) |
| pGBDU-C1 | GAL4 DNA binding domain (BD) | James et al. (1996) |
| pER9 | GAL4BD-CHS5 (1-264) | This study |
| pRB28 | GAL4AD-FUS1 (97-512) | This study |
| pRB226 | GAL4AD-FUS1 (97-512Δ151-200) | This study |
| pRB270 | GAL4AD-FUS1 (97-512Δ151-163) | This study |
| pRB271 | GAL4AD-FUS1 (97-512Δ164-176) | This study |
| pRB272 | GAL4AD-FUS1 (97-512Δ177-189) | This study |
| pRB273 | GAL4AD-FUS1 (97-512Δ190-200) | This study |
| pRB274 | GAL4AD-FUS1 (97-512Δ164-174) | This study |
| pRB275 | GAL4AD-FUS1 (97-512Δ184-189) | This study |
| pRB232 | GAL4AD-FUS1 (97-512I176A) | This study |
| pRB233 | GAL4AD-FUS1 (97-512 L177A) | This study |
| pRB234 | GAL4AD-FUS1 (97-512T178A) | This study |
| pRB235 | GAL4AD-FUS1 (97-512P179A) | This study |
| pRB236 | GAL4AD-FUS1 (97-512K180A) | This study |
| pRB237 | GAL4AD-FUS1 (97-512 176-ALAAA-180) | This study |
| pRB74 | FUS1-GFP | This study |
| pRB210 | FUS1Δ151-200-GFP | This study |
| pRB214 | FUS1Δ201-250-GFP | This study |
| pRB212 | FUS1Δ251-300-GFP | This study |
| pRB213 | FUS1Δ301-350-GFP | This study |
| pRB211 | FUS1Δ351-400-GFP | This study |
| pRB215 | FUS1Δ401-450-GFP | This study |
| pRB135 | FUS1Δ400-512-GFP | This study |
| pRB228 | FUS1Δ151-163-GFP | This study |
| pRB229 | FUS1Δ164-176-GFP | This study |
| pRB230 | FUS1Δ177-189-GFP | This study |
| pRB231 | FUS1Δ190-200-GFP | This study |
| pRB238 | FUS1Δ164-174-GFP | This study |
| pRB239 | FUS1 Δ184-189-GFP | This study |
| pRB240 | FUS1H175A-GFP | This study |
| pRB241 | FUS1I176A-GFP | This study |
| pRB242 | FUS1L177A-GFP | This study |
| pRB243 | FUS1T178A-GFP | This study |
| pRB244 | FUS1P179A-GFP | This study |
| pRB245 | FUS1K180A-GFP | This study |
| pRB246 | FUS1K181A-GFP | This study |
| pRB247 | FUS1T182A-GFP | This study |
| pRB248 | FUS1V183A-GFP | This study |
| pRB249 | FUS1K184A-GFP | This study |
| pRB250 | FUS1N185A-GFP | This study |
| pRB251 | FUS1P186A-GFP | This study |
| pRB252 | FUS1Y187A-GFP | This study |
| pRB253 | FUS1W189A-GFP | This study |
| pRB254 | FUS1176-ALAAA-180-GFP | This study |
| pRB188 | pMET25-FUS1-3xHA | This study |
| pRB269 | pMET25-FUS1176-ALAAA-180-3xHA | This study |
| pER12 | KEX21-675 FUS171-96 KEX2700-814-GFP | This study |
| pER7 | KEX21-675 FUS171-96-GFP | This study |
| pER5 | KEX21-675 FUS171-512-GFP | This study |
| pRB255 | MID2-GFP | This study |
| pRB256 | MID21-251-GFP | This study |
| pRB257 | MID21-251 FUS197-512-GFP | This study |
| pRB277 | pFUS1-KEX2-RFP | This study |
| pER21 | pFUS1-RFP-TLG1 | This study |
| pER22 | pADH1-RFP-TLG1 | This study |
aAll FUS1-GFP, KEX2-GFP, and MID2-GFP constructs are expressed on the centromeric plasmid pRS416-GFP under the control of the FUS1 promoter.
Fluorescence Microscopy
To visualize Fus1p-GFP, Kex2p-Fus1p-GFP, and Mid2p-Fus1p-GFP, MATa bar1Δ cells were grown in SD medium without uracil with twice the standard concentration of adenine to OD600 ∼0.3–0.4. The culture was treated with 50 ng/ml α-factor pheromone peptide (Sigma-Aldrich, St. Louis, MO) for 90 min, fixed on ice for at least 30 min with 3.7% formaldehyde (Thermo Fisher Scientific, Waltham, MA), and washed twice with water before placing on a slide. Cells were imaged at room temperature on a fluorescence microscope (IX71; Olympus, Melville, NY) equipped with a 100× 1.4 numerical aperture objective and a charge-coupled device camera (Orca II; Hamamatsu, Bridgewater, NJ). Images were acquired with MetaMorph software (Molecular Devices, Sunnyvale, CA) and processed with Photoshop (Adobe Systems, Mountain View, CA). Percentage of Fus1p-GFP shmoo tip localization was calculated as the number of cells with GFP at the shmoo tip divided by the total number of shmoos visualized.
For colocalization experiments, FUS1Δ151-200-GFP (pRB210) or FUS1176-ALAAA-180-GFP (pRB254) was transformed into YRB218 for colocalization with Sec7-RFP or cotransformed into YRB191 with pFUS1-KEX2-RFP (pRB277), pFUS1-RFP-TLG1 (pER21), or pADH1-RFP-TLG1 (pER22). Percentage of Fus1p-GFP punctae that colocalized with the TGN/endosomal red fluorescent protein (RFP) markers was calculated by selecting clear, distinct Fus1p-GFP punctae and then determining the percentage of those punctae that colocalized with Sec7p-RFP, Kex2p-RFP, or RFP-Tlg1p. There was slightly higher colocalization of Fus1p-GFP with RFP-Tlg1p driven by the ADH1 promoter compared with the FUS1 promoter.
Protease Protection Assays
MATa bar1Δ FUS1–13Myc::TRP1 cells were grown in YPD to OD600 ∼0.3–0.4. The culture was treated with 50 ng/ml α-factor pheromone peptide for 90 min and then treated on ice with NaN3 for 10 min. Nine OD600 cell equivalents were centrifuged at 2000 × g for 3 min. Cell pellets were resuspended in 600 μl of 100 mM Tris, pH 9.4, and 10 mM dithiothreitol and incubated at room temperature for 10 min. Cells were centrifuged at 2000 × g for 2 min, washed once with 1 ml of buffer B (1 M sorbitol, 40 mM HEPES, pH 7.5, 160 mM KoAC, and 5 mM MgCl2) and resuspended in 300 μl of buffer B. Cells were split into three equal aliquots and incubated at room temperature for 10 min: one aliquot was an untreated control, one aliquot was treated with 0.5 mg/ml proteinase K (Roche Diagnostics, Indianapolis, IN), and one aliquot was treated with 0.5 mg/ml proteinase K in the presence of 0.5% Triton X-100. Samples were precipitated with 17% trichloroacetic acid for 30 min on ice, rinsed with acetone, resuspended in 100 μl of sample buffer (8 M urea, 4% SDS, 125 mM Tris, pH 6.8, 20% glycerol, and 5% β-mercaptoethanol), agitated with glass beads for 7 min and solubilized at 55°C for 10 min. Aliquots (10 μl) of a 1:10 dilution of each sample were resolved by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA), and immunoblotted with anti-Myc antibodies (diluted 1:20,000; Cell Signaling Technology, Danvers, MA).
To quantify the amount of Fus1p-Myc accessible to proteinase K, we calculated the percentage of full-length Fus1p-Myc relative to the total Fus1p-Myc (the two prominent species added together) in each lane. We then divided the percentage of the full-length Fus1p-Myc species in the protease treated lane by the percentage in the untreated lane and subtracted this from 100%. Proteins were quantified using Photoshop.
Quantitative Cell Fusion Assays
Cell fusion assays were performed essentially as described previously (Heiman and Walter, 2000), with MATα fus1Δ cells expressing soluble cytosolic GFP (pDN291; Ng and Walter, 1996). Cells were incubated on YPD for 135 min and fixed on ice in 4% paraformaldehyde for at least 1 h before being washed twice in 1× phosphate-buffered saline and visualized by fluorescence microscopy. For cell fusion assays quantified in Table 4, we mated MATα fus1Δ cells with MATa fus1Δ cells expressing the various FUS1-GFP constructs.
Table 4.
Cell fusion and Fus1p-GFP shmoo tip localization for Fus1p-GFP mutants
| Plasmid name | Fus1p-GFPa | % cell fusionb | % Fus1p-GFP shmootip localizationc | ||
|---|---|---|---|---|---|
| fus1Δ | fus1Δ apl2Δ | fus1Δ | fus1Δ apl2Δ | ||
| None | None | 8 ± 3 | 2 ± 2 | NA | NA |
| pRB74 | Wild type (1-512) | 100 | 100 | 77 ± 11 | 81 ± 12 |
| 50-112 amino acid deletions | |||||
| pRB210 | Δ151-200 | 27 ± 8 | 97 ± 16 | 14 ± 5 | 76 ± 6 |
| pRB214 | Δ201-250 | 60 ± 8 | 82 ± 20 | 48 ± 5 | ND |
| pRB212 | Δ251-300 | 27 ± 11 | 42 ± 12 | 51 ± 4 | ND |
| pRB213 | Δ301-350 | 95 ± 10 | ND | 77 ± 3 | ND |
| pRB211 | Δ351-400 | 99 ± 21 | ND | 71 ± 20 | ND |
| pRB215 | Δ401-450 | 63 ± 19 | 50 ± 2 | 58 ± 11 | ND |
| pRB135 | Δ401-512 | 31 ± 12 | 34 ± 12 | 70 ± 8 | ND |
| Small deletions within amino acids 151-200 | |||||
| pRB228 | Δ151-163 | 95 ± 25 | ND | ND | ND |
| pRB229 | Δ164-176 | 68 ± 17 | ND | ND | ND |
| pRB230 | Δ177-189 | 41 ± 9 | 78 ± 2 | 26 ± 2 | 81 ± 3 |
| pRB231 | Δ190-200 | 89 ± 3 | ND | ND | ND |
| pRB238 | Δ164-174 | 89 ± 18 | ND | ND | ND |
| pRB239 | Δ184-189 | 51 ± 29 | 97 ± 16 | 31 ± 3 | 82 ± 11 |
| Point mutants within amino acids 175-189 | |||||
| pRB240 | H175A | 80 ± 16 | ND | ND | ND |
| pRB241 | I176A | 34 ± 5 | 77 ± 18 | 31 ± 7 | 72 ± 18 |
| pRB242 | L177A | 83 ± 7 | ND | ND | ND |
| pRB243 | T178A | 54 ± 11 | 82 ± 14 | 49 ± 7 | 73 ± 15 |
| pRB244 | P179A | 41 ± 9 | 86 ± 15 | 36 ± 4 | 83 ± 14 |
| pRB245 | K180A | 37 ± 13 | 89 ± 10 | 51 ± 12 | 78 ± 7 |
| pRB246 | K181A | 108 ± 14 | ND | ND | ND |
| pRB247 | T182A | 103 ± 7 | ND | ND | ND |
| pRB248 | V183A | 110 ± 2 | ND | ND | ND |
| pRB249 | K184A | 70 ± 19 | ND | ND | ND |
| pRB250 | N185A | 98 ± 6 | ND | ND | ND |
| pRB251 | P186A | 85 ± 26 | ND | ND | ND |
| pRB252 | Y187A | 79 ± 8 | ND | ND | ND |
| pRB253 | W189A | 86 ± 14 | ND | ND | ND |
| pRB254 | ALAAA | 27 ± 13 | 82 ± 21 | 27 ± 5 | 80 ± 10 |
NA, not applicable; ND, not determined.
aAll plasmids were expressed in fus1Δ cells to score cell fusion and fus1Δbar1Δ cells to score shmoo tip localization.
bPercentage of mating pairs that expressed a soluble cytosolic GFP marker in both halves of the mating pair. The cell fusion for cells expressing wild-type FUS1-GFP was normalized to 100% within each experiment.
cPercentage of shmooing cells with GFP localized to the shmoo tip after 90 min treatment with α-factor. Numbers are the average of at least three independent exeriments ± the 95% confidence interval where 50–100 cells were scored for each experiment.
Yeast Two-Hybrid
The two-hybrid strain PJ69-4A was cotransformed with pGAD-C1 (empty vector or pGAD-FUS1) and pGBDU-C1 (empty vector or pGBDU-CHS5) (James et al., 1996). Cells were grown to saturation in SD lacking leucine and uracil and 4 μl of 1:5 serial dilutions were spotted on SD lacking leucine and uracil and SD lacking leucine, uracil, and histidine.
Chs5p-TAP Purification
Tandem affinity purification of Chs5p, including cross-linking with 5 mM dithiobis(succunimidylpropionate) (DSP), was performed essentially as described previously (Sanchatjate and Schekman, 2006). The Chs5-TAP strain was transformed with wild-type or mutant pMET25-FUS1–3xHA and grown in SD lacking uracil and methionine.
RESULTS
BCH1 and BUD7 Are Redundantly Required for Fus1p Plasma Membrane Localization
Fus1p is an O-glycosylated type I membrane protein that localizes to the plasma membrane at the tip of shmoos, or mating arrested cells, during the yeast mating response (Trueheart and Fink, 1989). Fus1p is required for cells of opposite mating type to fuse (Trueheart et al., 1987; Nolan et al., 2006). In addition to its cell surface localization, Fus1p is found in intracellular punctae thought to represent a late Golgi or post-Golgi compartment (Santos and Snyder, 2003; Figure 1Aa). The cell surface localization of Fus1p is dependent on the exomer complex component Chs5p. In chs5Δ cells, Fus1p-GFP is not found at the cell surface and is only seen intracellularly (Santos and Snyder, 2003; Figure 1Ab). We hypothesized that in addition to Chs5p, Fus1p shmoo tip localization would also require one of the four ChAPs that are part of the exomer complex. To test this hypothesis, we examined Fus1p-GFP localization in ChAP single mutant cells treated with α-factor for 90 min, conditions that induce expression and localization of Fus1p to the shmoo tip. We found that each of the four ChAP single mutants exhibited Fus1p-GFP tip localization similar to wild-type cells (Table 3 and Figure 1Ad). Even triple mutant cells where BCH2, CHS6, and BUD7 were deleted showed Fus1p-GFP shmoo tip localization indistinguishable from wild-type cells (Table 3 and Figure 1Ac). However, when BCH1 was deleted in combination with any of the other three ChAPs, Fus1p-GFP was at least partially mislocalized (Table 3). In bch1Δ bud7Δ double mutants, the mislocalization defect was as severe as in a chs5Δ mutant; very few cells showed Fus1-GFP tip localization but instead demonstrated a punctate intracellular localization pattern similar to chs5Δ mutants (Table 3 and Figure 1A, b and e). This mislocalization defect was not due to a decrease in Chs5p expression; expression of Chs5p was similar in all ChAP mutant combinations (Supplemental Figure 1).
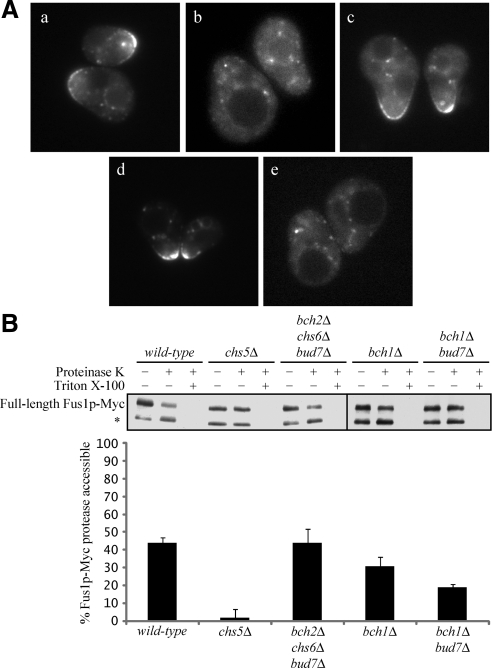
Figure 1.
BCH1 and BUD7 are redundantly required for Fus1p plasma membrane localization. (A) Fus1p-GFP localization in (a) wild-type, (b) chs5Δ, (c) bch2Δ chs6Δ bud7Δ, (d) bch1Δ, and (e) bch1Δ bud7Δ cells. All cells were also mutant for BAR1. Cells were treated with α-factor for 90 min and then visualized by fluorescence microscopy. (B) Fus1p-Myc protease accessibility in exomer mutants. Asterisk (*) indicates endogenously cleaved Fus1p-Myc and exogenously cleaved Fus1p-Myc (see text for details). Quantifications are the average of five or more independent experiments. Error bars represent the 95% confidence interval.
Table 3.
Fus1p-GFP shmoo tip localization in exomer mutants
| Genotypea | % Fus1p-GFP shmoo tip localizationb |
|---|---|
| WT | 81 ± 6 |
| chs5Δ | 24 ± 6 |
| bch2Δ | 73 ± 4 |
| chs6Δ | 74 ± 6 |
| bch1Δ | 66 ± 8 |
| bud7Δ | 67 ± 11 |
| bch2Δ chs6Δ bud7Δ | 68 ± 15 |
| bch2Δ bch1Δ | 48 ± 15 |
| chs6Δ bch1Δ | 40 ± 8 |
| bch1Δ bud7Δ | 26 ± 8 |
| chs5Δ apl2Δ | 82 ± 14 |
| bch1Δ bud7Δ apl2Δ | 72 ± 4 |
aAll strains were also bar1Δ and were carrying a wild-type Fus1p-GFP plasmid (pRB74).
bPercentage of shmooing cells with GFP localized to the shmoo tip after a 90 min treatment with α-factor. Numbers are the average of at least three independent exeriments ± the 95% confidence interval, where 50–100 cells were scored for each experiment.
To confirm these results biochemically, we conducted protease protection experiments to determine the amount of Fus1p at the cell surface in wild-type, chs5Δ, and ChAP mutant cells. For these experiments, we chromosomally tagged Fus1p at its C terminus with a 13xMyc tag. Fus1p-Myc was functional as assessed by a cell fusion assay (data not shown). Fus1p-Myc cells were treated with α-factor for 90 min and then treated with proteinase K in the presence or absence of 0.5% Triton X-100. The samples were analyzed by SDS-PAGE followed by anti-Myc immunoblotting to determine the amount of Fus1p-Myc accessible to protease, i.e., the amount of Fus1p-Myc localized to the plasma membrane. In the absence of proteinase K, two predominant Fus1p-Myc species were detected. The higher molecular weight species is the full-length form of Fus1p-Myc that is O-glycosylated at its N terminus. The lower molecular weight species is probably an endogenously cleaved form of Fus1p-Myc that is generated in the Golgi (Proszynski et al., 2004). In the presence of proteinase K, the higher molecular weight species probably represented intracellular, full-length, O-glycosylated Fus1p-Myc that was inaccessible to protease. The lower molecular weight species corresponded to the endogenously cleaved Golgi species and the plasma membrane localized species that was accessible to and degraded by proteinase K (Figure 1B).
In wild-type cells treated with proteinase K, almost 45% of Fus1p-Myc was at the cell surface and accessible to protease (Figure 1B). These data indicate that although the majority of wild-type cells have cell surface expression of Fus1p-GFP by microscopy, a large portion of the total Fus1p is localized intracellularly. In contrast to wild-type cells, <5% of Fus1p-Myc was accessible to protease in chs5Δ mutant cells, consistent with the Fus1p-GFP microscopy data (Table 3 and Figure 1B). bch2Δ chs6Δ bud7Δ triple mutants had wild-type levels of protease accessible Fus1p-Myc, and bch1Δ single mutants showed a slight decrease in protease accessibility (Figure 1B). However, deleting BCH1 and BUD7 together had a much more dramatic effect on protease accessibility; 19% of Fus1p-Myc was accessible in bch1Δ bud7Δ double mutants, less than half of the wild-type level (Figure 1B). Surprisingly, this accessibility was still well above what we measured in chs5Δ cells. These data indicate that although the percentage of cells with Fus1p localized to the shmoo tip (as measured by fluorescence microscopy) was similar in chs5Δ and bch1Δ bud7Δ cells, the proportion of Fus1p molecules that is localized to the shmoo tip (as measured by protease protection) was much less in chs5Δ cells compared with bch1Δ bud7Δ cells. Even the ChAP quadruple mutant (bch2Δ chs6Δ bch1Δ bud7Δ) showed 13 ± 7% protease accessibility, suggesting that Chs5p can partially function without the four ChAPs in the transport of Fus1p.
Because Fus1p must be present at the cell surface to promote cell fusion, we predicted that bch1Δ bud7Δ double mutants would have a cell fusion defect similar to fus1Δ mutants. To test this hypothesis, we conducted filter mating assays to measure the ability of bch1Δ bud7Δ cells to function in cell fusion. A severe defect in cell fusion is seen only when both mating types are mutant for FUS1 (Trueheart et al., 1987); therefore, we combined MATα fus1Δ mutants carrying a soluble cytosolic GFP marker (Ng and Walter, 1996) with MATa bch1Δ bud7Δ mutants. After concentrating the cells on filter paper and mating on YPD, we fixed the cells and used fluorescence microscopy to score mating pairs as fused if the cytosolic GFP marker was observed in both halves of the mating pair or unfused if the cytosolic GFP marker was restricted to one-half of the mating pair. As predicted, bch1Δ bud7Δ double mutants showed a cell fusion defect similar to fus1Δ mutants (28 ± 11 and 16 ± 7% relative to wild-type, respectively), whereas bch1Δ and bud7Δ single mutants showed similar cell fusion to wild-type (98 ± 26 and 87 ± 13% relative to wild-type, respectively). One caveat however, is that bch1Δ bud7Δ double mutants are also defective in localizing Chs3p to the cell surface (Sanchatjate and Schekman, 2006; Trautwein et al., 2006) and CHS3 is also required for cell fusion (Trilla et al., 1999). Therefore, the cell fusion defect seen in bch1Δ bud7Δ double mutants could be due to the inability of these cells to properly localize Fus1p, Chs3p, or both. However, based on our microscopy data and our protease protection experiments, we conclude that BCH1 and BUD7, the two most highly similar ChAPs, are redundantly required for the localization of Fus1p to the plasma membrane.
AP-1 Is Important for the Intracellular Localization of Fus1p
In addition to its localization at the cell surface, Fus1p localizes to intracellular punctae (Santos and Snyder, 1997; Figure 1Aa) reminiscent of the intracellular compartments where another exomer cargo, Chs3p, resides (Chuang and Schekman, 1996; Santos and Snyder, 1997). The intracellular stores of Chs3p seem to be maintained by recycling between the TGN and early endosome. This recycling event is dependent on the clathrin adaptor AP-1, although the direction in which AP-1 transports cargo is not clear. Disruption of the AP-1 complex results in relocalization of Chs3p to the cell surface in the absence of exomer (Valdivia et al., 2002). Our lab has suggested that in the absence of AP-1 and exomer, Chs3p accesses an alternate pathway to the plasma membrane (Valdivia et al., 2002).
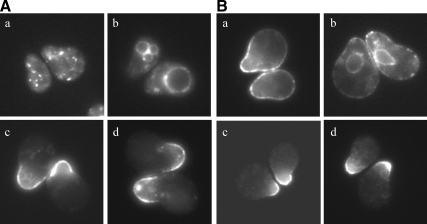
We considered the possibility that Fus1p and Chs3p might share an alternate path to the cell surface in an exomer AP-1 double mutant. Fus1p-GFP localization was examined in chs5Δ and bch1Δ bud7Δ cells also deleted for APL2, the large subunit of the AP-1 complex. These combined mutations restored Fus1p tip localization to wild-type levels (Table 3 and Figure 2A, c and e). To further validate these findings, we conducted protease protection assays and found that the Fus1p-Myc protease inaccessibility phenotype seen in chs5Δ and bch1Δ bud7Δ mutants was suppressed by a mutation in APL2 (Figure 2B). The cell fusion defect seen in chs5Δ and bch1Δ bud7Δ mutants was also suppressed by a mutation in APL2. chs5Δ apl2Δ mating pairs fuse 65 ± 5% of the time compared with 9 ± 5% for chs5Δ cells, relative to wild type. bch1Δ bud7Δ apl2Δ mating pairs fuse 62 ± 23% of the time compared with 22 ± 13% for bch1Δ bud7Δ cells, relative to wild type. These results suggest that the intracellular localization of Fus1p is maintained by recycling between the TGN and early endosome in an AP-1–dependent manner. When this recycling is not maintained, Fus1p accesses the cell surface by an exomer-independent mechanism.
Figure 2.
AP-1 is important for the intracellular retention of Fus1p in chs5Δ and bch1Δ bud7Δ mutants. (A) Fus1p-GFP localization in (a) wild-type, (b) chs5Δ, (c) chs5Δ apl2Δ, (d) bch1Δ bud7Δ, and (e) bch1Δ bud7Δ apl2Δ cells. All cells were also mutant for BAR1. Cells were treated with α-factor for 90 min and then visualized by fluorescence microscopy. (B) Fus1p-Myc protease accessibility in exomer AP-1 mutants. Asterisk (*) indicates endogenously cleaved Fus1p-Myc and exogenously cleaved Fus1p-Myc (see text for details). Quantifications are the average of three or more independent experiments. Error bars represent the 95% confidence interval.
Fus1p 151-200 Is Necessary for the Exomer-dependent Plasma Membrane Localization of Fus1p
We know that the exomer components Chs5p (Santos and Snyder, 2003), Bch1p and Bud7p are important for localizing Fus1p to the plasma membrane. We also know that the exomer component Chs5p physically interacts in a yeast two-hybrid assay with the cytosolic C-terminal domain of Fus1p (Nelson et al., 2004). Therefore, we sought to identify an exomer-dependent Golgi to plasma membrane sorting signal in the C-terminal cytosolic domain of Fus1p. We reasoned that deletion of a plasma membrane sorting signal in Fus1p would delay transport of Fus1p to the plasma membrane and would therefore interfere with cell fusion. Fus1p is a 512 amino acid protein; residues 1-71 comprise its predicted extracellular N terminus, residues 72-96 comprise its predicted transmembrane domain, and residues 97-512 comprise its predicted cytosolic C-terminus (Trueheart et al., 1987). To identify the plasma membrane sorting signal, we made eight 50-112 amino acid deletions along the cytosolic domain of Fus1p (from amino acids 100-512) and tested the ability of these deletion constructs to complement the cell fusion defect caused by disruption of FUS1 (Figure 3A).
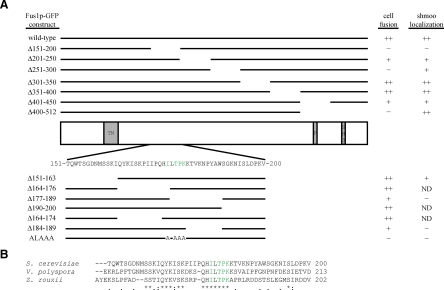
Figure 3.
Fus1p-GFP constructs. Green residues indicate critical residues for Fus1p-GFP shmoo tip localization. (A) Cartoon of Fus1p with transmembrane domain (TM), proline-rich domain (P), and Src homology 3 domain (SH3). Fus1p-GFP construct is indicated on the left. Top, the 50-112-amino acid deletion constructs are depicted. Bottom, residues 151-200 are shown and the small deletion constructs within this region and the 176ILTPK180 to 176ALAAA180 construct are depicted. Ability to rescue fus1Δ cell fusion defect (cell fusion) and GFP localization to the shmoo tip are indicated on the right. ++, +, and − indicate 67–100, 34–66, and 0–33%, respectively. (B) Protein sequence alignment of Fus1p-like proteins from S. cerevisiae, Vanderwaltozyma polyspora, and Zygosaccharomyces rouxii. Identical residues are indicated by an asterisk (*), very similar residues by a colon (:), and somewhat similar residues by a period (.).
To test the Fus1p deletion constructs for their ability to function in cell fusion, we conducted filter mating assays. For these assays, we mated MATα fus1Δ cells carrying a cytosolic GFP marker with MATa fus1Δ cells carrying no plasmid, a wild-type FUS1-GFP plasmid or one of eight FUS1-GFP deletion plasmids. All FUS1 constructs were maintained on a low copy plasmid under the control of the FUS1 promoter. The wild-type FUS1-GFP construct was completely functional and all FUS1-GFP constructs were expressed at similar levels as detected by immunoblot (Supplemental Figure 2). Cells expressing FUS1Δ100–150-GFP did not form mating pairs well and therefore were not analyzed further.
Table 4 shows the percentage of fus1Δ mating pairs that fused when the different FUS1-GFP constructs were expressed. We normalized the cell fusion percentage of fus1Δ cells expressing wild-type FUS1-GFP to 100%. FUS1Δ301-350-GFP and FUS1Δ351-400-GFP constructs complemented the fus1Δ cell fusion defect as well as wild-type. FUS1Δ201-250-GFP and FUS1Δ401-450-GFP constructs partially complemented the fus1Δ cell fusion defect. FUS1Δ151-200-GFP, FUS1Δ251-300-GFP and FUS1Δ401-512-GFP constructs failed to complement the fus1Δ cell fusion defect (Table 4 and Figure 3).
We considered the possibility that defective cell fusion resulted from a loss of Fus1p function or a failure to localize Fus1p to the cell surface. To distinguish these possibilities, we visualized cells carrying the various deletion constructs by fluorescence microscopy. We predicted that cells expressing a deletion construct lacking the Fus1p exomer sorting signal would show a mislocalization defect similar to chs5Δ and bch1Δ bud7Δ mutants, whereas loss of a functional domain may not affect the normal cell surface distribution of mutant protein or may result in the ER retention of a misfolded protein.
In Table 4, the percentage of cells that have Fus1p-GFP localized to the shmoo tip is indicated. Of the five deletion constructs that did not fully complement the fus1Δ cell fusion defect, only Fus1pΔ151-200-GFP was as severely mislocalized as wild-type Fus1p-GFP in chs5Δ and bch1Δ bud7Δ mutants (Tables 3 and 4). Cells expressing Fus1pΔ151-200-GFP showed a nearly complete lack of GFP localization at the tip and had a very clear intracellular punctate localization pattern (Figure 4Ab). Fus1pΔ201-250-GFP partially localized to the vacuolar membrane (Figure 4Ac), Fus1pΔ251-300-GFP either localized to the plasma membrane or was undetectable (Figure 4Ad) and Fus1pΔ401-450-GFP and Fus1pΔ401-512-GFP localized predominantly to the shmoo tip (Figure 4Ae). These data suggest that cells expressing FUS1Δ201-250-GFP, FUS1Δ251-300-GFP, FUS1Δ401-450-GFP, and FUS1Δ401-512-GFP may be defective in some functional aspect of cell fusion whereas cells expressing FUS1Δ151-200-GFP are most likely defective in Golgi-to-cell surface traffic.
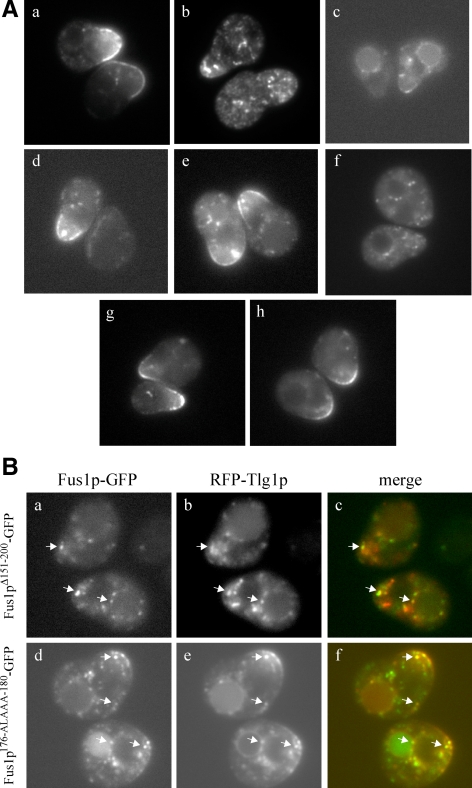
Figure 4.
Fus1p 176IXTPK180 is necessary for the plasma membrane localization of Fus1p. (A) Localization of (a) wild-type Fus1p-GFP, (b) Fus1pΔ151-200-GFP, (c) Fus1pΔ201-250-GFP, (d) Fus1pΔ251-300-GFP, (e) Fus1pΔ401-512-GFP, and (f) Fus1p176-ALAAA-180-GFP in wild-type cells and (g) Fus1pΔ151-200-GFP and (h) Fus1p176-ALAAA-180-GFP in apl2Δ cells. (B) Colocalization of (a–c) Fus1pΔ151-200-GFP and (d–f) Fus1p176-ALAAA-180-GFP with pFUS1-RFP-Tlg1p. Arrows indicate examples of colocalized punctae. All cells in A and B were also mutant for endogenous FUS1 and BAR1. Cells were treated with α-factor for 90 min and then visualized by fluorescence microscopy.
To further distinguish a functional or folding defect from a transport defect, we expressed FUS1Δ151-200-GFP in cells deficient in AP-1. Because deleting an exomer-dependent plasma membrane sorting signal on Fus1p should have the same effect on Fus1p transport as deleting exomer itself, we reasoned that deleting APL2 would allow Fus1pΔ151-200-GFP to reach the cell surface. Indeed, in apl2Δ cells, Fus1pΔ151-200- GFP localized to the shmoo tip as well as wild-type Fus1p-GFP (Table 4 and Figure 4Ag). We next conducted filter mating assays using apl2Δ cells expressing FUS1Δ151-200-GFP. FUS1Δ151-200-GFP fully complemented the fus1Δ cell fusion defect when APL2 was deleted, whereas the other FUS1-GFP deletion constructs showed little or no change in the absence of APL2 (Table 4). These data indicate that FUS1Δ151-200-GFP fails to complement fus1Δ due to a failure of the mutant protein to localize to the plasma membrane but not because of a defect in the biological function of Fus1p. Our data showing that deletion of residues 151-200 caused a severe cell fusion defect, a mislocalization defect as severe as deletion of exomer, and that both of these phenotypes were suppressed to wild-type levels by disruption of AP-1, suggest that this region alone contains the residues essential for exomer-dependent transport to the cell surface.
Fus1p 176IXTPK180 Is Necessary for the Exomer-dependent Plasma Membrane Localization of Fus1p
To refine the sorting signal region, we made four smaller deletions within amino acids 151-200 in the context of Fus1p-GFP and tested the ability of these deletion constructs to complement the fus1Δ cell fusion defect (Figure 3A). FUS1Δ151-163-GFP and FUS1Δ190-200-GFP fully complemented fus1Δ (Table 4). FUS1Δ164-176-GFP partially complemented fus1Δ whereas FUS1Δ164-174-GFP complemented almost as well as wild type (Table 4). FUS1Δ177-189-GFP complemented 41% relative to wild type (Table 4). Thus, we focused on the region of Fus1p from residues 175-189.
FUS1Δ184-189-GFP showed reduced complementation in fus1Δ, but of the alanine substitutions of residues 175-189, only four—I176A, T178A, P179A, and K180A—showed reduced complementation in fus1Δ (Table 4). Surprisingly, FUS1I176A-GFP was more defective than FUS1Δ164-176-GFP, a construct that deletes I176. However, we noticed that in deleting residues 164–176, we juxtaposed I163 to L177, creating the wild-type IL sequence. Although individual alanine substitutions of residues I176, T178, P179, and K180 displayed partial effects, a combined mutant, 176ILTPK180 to 176ALAAA180, was as severely defective in fusion as FUS1Δ151-200-GFP (Table 4).
We found that the shmoo tip localization of the mutant forms of Fus1p correlated with the cell fusion defect (Table 4). For example, Fus1p176-ALAAA-180-GFP, which had the most severe cell fusion defect, was as severely mislocalized as wild-type Fus1p-GFP in chs5Δ or bch1Δ bud7Δ mutants (Tables 3 and 4 and Figure 4Af). Furthermore, the cell fusion and mislocalization defects were suppressed by a mutation in APL2 (Table 4 and Figure 4Ah).
Although the size, number and intensity of Fus1p-GFP intracellular punctae vary from cell to cell in cells expressing wild-type and mutant FUS1-GFP (probably due to expression from a CEN plasmid), punctae seen in cells expressing FUS1176-ALAAA-180-GFP or FUS1Δ151-200-GFP are indistinguishable from each other and resemble TGN/endosomal punctae. We coexpressed these mutant FUS1-GFP constructs with each of three late Golgi/endosomal markers—Sec7p-RFP, Kex2p-RFP, or RFP-Tlg1p—and determined that Fus1p176-ALAAA-180-GFP and Fus1pΔ151-200-GFP had similar colocalization patterns with all three markers. Fus1pΔ151-200-GFP punctae showed 15% colocalization with Sec7p-RFP, 27% colocalization with Kex2p-RFP, and 32 or 52% colocalization with RFP-Tlg1, depending on the promoter used (Figure 4B, a–c). Fus1p176-ALAAA-180-GFP punctae showed 19% colocalization with Sec7p-RFP, 30% colocalization with Kex2p-RFP and 40 or 46% colocalization with RFP-Tlg1, depending on the promoter used (Figure 4B, d–f). Therefore, Fus1pΔ151-200-GFP and Fus1p 176-ALAAA-180-GFP are indistinguishable in their inability to rescue cell fusion, their mislocalization, and the colocalization of their intracellular punctae with TGN/endosomal markers.
An alignment between Saccharomyces cerevisiae Fus1p and Fus1p from other yeasts show that IXTPK is conserved whereas residues 184-189 are not (Figure 3B). Because residues 184-189 are not individually important for cell fusion (Table 4) and are not conserved among other Fus1p species, whereas the IXTPK signal is important for cell fusion and Fus1p tip localization and is conserved, we suggest that residues 184-189 may serve a structural role for the Fus1p 176IXTPK180 sorting signal.
Fus1p 176IXTPK180 Is Required for the Physical Interaction between Fus1p and Exomer
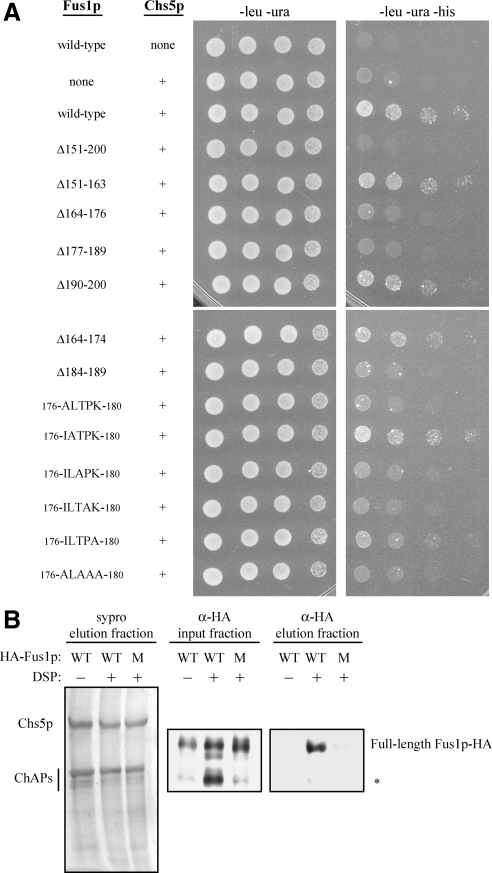
We have shown that Chs5p, Bch1p, Bud7p, and Fus1p residues 176IXTPK180 are important for the plasma membrane localization of Fus1p. Others have shown that the cytosolic domain of Fus1p physically interacts with the exomer component Chs5p in a yeast two-hybrid assay (Nelson et al., 2004). Based on these observations, we predicted that Fus1p 176IXTPK180 would be important for a physical interaction with Chs5p. To test this hypothesis, we examined the ability of IXTPK mutant versions of the Fus1p cytosolic domain (97–512) to bind to Chs5p in a yeast two-hybrid assay. We found that mutation of any of the residues critical for Fus1p-GFP plasma membrane localization (T178A, P179A, K180A, and 176ILTPK180 to 176ALAAA180) abolished binding to Chs5p, whereas mutation of L177 or deletion of other residues within amino acids 151-200, which do not affect Fus1p-GFP plasma membrane localization, showed wild-type levels of binding to Chs5p (Figure 5A).
Figure 5.
Fus1p 176IXTPK180 is necessary for the physical interaction between Fus1p and Chs5p. (A) Yeast two-hybrid. Wild-type Fus1p (97-512) or a version of Fus1p (97-512) containing a small deletion or a mutation(s) in the 176ILTPK180 region was fused to the C terminus of the GAL4 activation domain (LEU). Chs5p (1-264) was fused to the C terminus of the GAL4 binding domain (URA). −leu −ura plates measure growth, whereas −leu −ura −his plates measure binding of the GAL4 activation domain to the GAL4 binding domain. (B) In vivo cross-linking and coimmunoprecipitation of Chs5p-TAP and Fus1p. Chromosomally TAP-tagged Chs5p was purified from cells expressing either wild-type (WT) or a 176ILTPK180 to 176ALAAA180 mutant (M) version of pMET25-FUS1-HA in the presence (+) or absence (−) of 5 mM DSP cross-linker. Purified Chs5p, which copurifies with all four ChAPs, was visualized in the final elution fraction with sypro staining. Fus1p-HA was visualized in input and final elution fractions by immunoblot analysis using an anti-HA antibody (diluted 1:1000; HA.11, Covance Research Products, Princeton, NJ). Fus1p-HA WT without DSP and Fus1p-HA mutant with DSP were purified 15 and 29% as well as Fus1p-HA WT with DSP, respectively, when factoring Fus1p-HA in the input fraction and purified Chs5p in the final elution fraction. Asterisk (*) indicates endogenously cleaved Fus1p-HA species (see text for details).
We used in vivo cross-linking of Chs5p to Fus1p to assess the role of the IXTPK signal. We immunoprecipitated a tandem affinity purification (TAP)-tagged Chs5p from yeast (Sanchatjate and Schekman, 2006) expressing either wild type or the 176ALAAA180 mutant version of Fus1p-hemagglutinin (HA) under control of the MET25 promoter. In the absence of the chemical cross-linker DSP, the four ChAPs copurified with Chs5p (Sanchatjate and Schekman, 2006, Figure 5B), whereas Fus1p-HA did not (Figure 5B). In the presence of 5 mM DSP, wild type but not mutant Fus1p-HA copurified with Chs5p. Although full-length wild-type Fus1p-HA copurified with Chs5p in the presence of cross-linker, the endogenously cleaved form of wild-type Fus1p-HA that is generated in the Golgi (Proszynski et al., 2004) did not copurify. Proszynski et al. (2004) showed that an O-glycosylated 33 amino acid region of Fus1p that is missing in this cleaved form of Fus1p is sufficient for transport of an Invertase-Fus1p chimera to the cell surface. Perhaps this glycosylated region is required for Fus1p to bind Chs5p in vivo. We conclude that IXTPK is an important sorting signal that is recognized by exomer and that both the signal and the exomer complex are required for Fus1p traffic late in the secretory pathway.
The Exomer-dependent IXTPK Sorting Signal Functions in a Restricted Context
We showed that Fus1p 176IXTPK180 is necessary for the interaction between Fus1p and exomer and for its transport to the cell surface, but is it sufficient? Some signals, such as an ER export signal, can be transplanted to mobilize a neutral reporter protein (Ma et al., 2001). To test whether IXTPK is sufficient for plasma membrane localization, we made chimeric proteins containing the lumenal N terminus of Kex2p, a resident TGN protein (Redding et al., 1991), and the transmembrane and cytosolic C terminus of Fus1p. This Kex2p-Fus1p chimera contained GFP at its C terminus and its expression was driven by the FUS1 promoter. The Kex2p-Fus1p-GFP fusion protein trafficked to the cell surface whereas a truncated fusion lacking the C terminus of Kex2p or Fus1p was mostly retained intracellularly (Figure 6A, a–c). However, the plasma membrane localization of this Kex2p-Fus1p-GFP chimera was not dependent on the exomer component CHS5 (Figure 6A, c and d).
Figure 6.
The exomer-dependent IXTPK sorting signal functions in a restricted context. (A) GFP localization of (a) full-length Kex2p-GFP (percentage of cells with plasma membrane GFP: 2 ± 2), (b) C-terminally deleted Kex2p-GFP (24 ± 5), and (c) Kex2p-Fus1p-GFP (89 ± 7) in wild-type cells and (d) Kex2p-Fus1p-GFP (76 ± 13) in chs5Δ cells. All constructs contained the Fus1p transmembrane domain and were under control of the FUS1 promoter. (B) GFP localization of (a) full-length Mid2p-GFP (88 ± 9), (b) C-terminally deleted Mid2p-GFP (ND), and (c) Mid2p-Fus1p-GFP (58 ± 0) in wild-type cells and (d) Mid2p-Fus1p-GFP (60 ± 10) in chs5Δ cells. All constructs contained the Mid2p transmembrane domain and were under control of the FUS1 promoter. All cells in A and B were also mutant for endogenous FUS1 and BAR1. Cells were treated with α-factor for 90 min and then visualized by fluorescence microscopy. Numbers in parentheses are the percentage of shmooing cells with GFP localized to the plasma membrane ± the 95% confidence interval. Numbers are the average of at least two independent experiments where 50–200 cells were scored for each experiment. Plasma membrane localization was not determined for the C-terminally deleted Mid2p-GFP due to its ER localization; cortical ER localization was difficult to distinguish from plasma membrane localization.
We next made chimeras linking the lumenal N terminus and transmembrane domain of Mid2p, a type I plasma membrane protein (Philip and Levin, 2001), to the cytosolic domain of Fus1p. A truncated Mid2p-GFP fusion lacking its C terminus was ER localized (Figure 6Bb). Mid2p-Fus1p-GFP was localized mostly to the shmoo tip, but again, this cell surface localization was not dependent on exomer (Figure 6B, c and d). Because our Kex2p-Fus1p-GFP and Mid2p-Fus1p-GFP chimeras reached the cell surface despite lacking the 33 residue stretch that Proszynski et al. (2004) showed to be sufficient for transport of an Invertase-Fus1p chimera to the cell surface, we did not include this region for further sufficiency experiments. Although the C-terminal cytosolic domain of Fus1p is sufficient for plasma membrane localization of the Kex2p-Fus1p-GFP and Mid2p-Fus1p-GFP chimeras, exomer is not required for transport when the C terminus of Fus1p is transplanted to an unrelated reporter protein. These data suggest that the ability of exomer and the IXTPK sorting signal to direct cargo to the cell surface is context dependent.
DISCUSSION
We have shown that Fus1p 176IXTPK180 is necessary for the localization of Fus1p to the cell surface (Table 4 and Figure 4Af). Because of the requirement for Fus1p at the cell surface for cell fusion during the yeast mating process, mutating 176ILTPK180 to 176ALAAA180 renders cells unable to fuse with their mating partner (Table 4). When the intracellular retention of Fus1p 176ALAAA180 is inhibited by disrupting the AP-1 complex, Fus1p 176ALAAA180 is redirected to the cell surface and cell fusion is restored (Table 4 and Figure 4Ah). Fus1p 176IXTPK180 is not only necessary for the plasma membrane localization of Fus1p but also for the physical interaction between Fus1p and the exomer component Chs5p (Figure 5). These data suggest that Fus1p 176IXTPK180 comprises a plasma membrane sorting signal that is recognized and bound by exomer. Once exomer binds to this IXTPK signal, Fus1p can be captured into exomer-coated vesicles and transported to the cell surface.
IXTPK Defines a Novel Plasma Membrane Sorting Signal in Yeast
To date, there has been no report of a sorting signal in yeast capable of directing cargo to the plasma membrane. Because the Fus1p IXTPK motif shares little or no similarity to other known traffic signals, it seems to represent a novel sorting signal important for the transport of a cargo molecule to the plasma membrane in yeast. In S. cerevisiae, there are no predicted plasma membrane proteins other than Fus1p that contain an IXTPK motif. However, if conservative substitutions are allowed, other plasma membrane proteins contain what may be an equivalent signal. Chs3p, which is also dependent on exomer for its transport to the plasma membrane, contains an LITPK sequence in its second predicted cytosolic loop. However, mutation of this sequence did not affect transport of Chs3p to the cell surface (data not shown). We know that multiple ChAPs (Chs6p or Bch1p and Bud7p) can direct Chs3p to the cell surface (Sanchatjate and Schekman, 2006, Trautwein et al., 2006) so it is possible that multiple sorting signals exist within Chs3p, some of which might be redundant. Future analysis of other IXTPK-like signals and the identification of additional exomer-dependent cargo proteins may help explain the nature of exomer-cargo interactions.
How Does Exomer Interact with Its Cargo?
The simplest interpretation of our data are that exomer directly recognizes and binds Fus1p at the IXTPK sequence. However, it is entirely possible that one or more additional proteins intervene. For example, we showed that exomer forms an electron dense coat on synthetic liposomes but is not sufficient to induce membrane curvature or vesicle budding (Wang et al., 2006). Therefore, exomer could function as a coat adaptor and cooperate with other coat or cytoskeletal components needed for vesicle budding from the TGN. Perhaps an unidentified component of the coat is directly responsible for interacting with Fus1p. Attempts to demonstrate a direct in vitro interaction between Fus1p and exomer have thus far not been successful.
Our two-hybrid interaction experiments were conducted using Chs5p as bait because Bch1p and Bud7p were highly self-activating. However, we suggest that the ChAPs provide cargo specificity. The two transmembrane cargo known to require exomer for transport, Chs3p and Fus1p, both require Chs5p but have different requirements for the ChAPs. In addition, Sanchatjate and Schekman found that Chs5p was required for the integrity of the exomer complex whereas mutation of the ChAPs required for Chs3p transport affected binding of exomer to Chs3p (Sanchatjate and Schekman, 2006). We show that Bch1p and Bud7p are redundantly required for the transport of Fus1p to the cell surface. Perhaps each ChAP or redundant pair recognizes distinct sorting motifs or posttranslational modifications on cargo proteins. Bch1p and Bud7p may each be able to recognize the Fus1p IXTPK signal or a modification on this sequence whereas Chs6p and Bch2p cannot.
The IXTPK Sorting Signal Is Context Dependent
We showed that Fus1p 176IXTPK180 is necessary for the interaction between Fus1p and exomer and for its transport to the cell surface, but when this signal is transplanted to an unrelated reporter protein, it is not sufficient for exomer-dependent transport to the cell surface (Figure 6). We suggest that the C-terminal cytosolic domain of Fus1p is sufficient for plasma membrane localization and, in the context of full-length Fus1p, the IXTPK signal is necessary. However, exomer is not required for transport when the cytosolic domain of Fus1p is transplanted to an unrelated reporter protein. Although some traffic signals can be transplanted, the nature of TGN to plasma membrane signals may not be as straightforward. For example, we know that Fus1p and Chs3p can take multiple routes to the cell surface: an exomer-dependent pathway and an alternative exomer-independent pathway that is revealed in the absence of AP-1. Perhaps, the Kex2p-Fus1p and Mid2p-Fus1p chimeric proteins are able to access yet another exomer-independent pathway to the cell surface.
Supplementary Material
ACKNOWLEDGMENTS
We thank past and present members of the Schekman laboratory for valuable discussions, especially Alenka Čopič, Silvere Pagant, Siraprapha Sanchatjate, Danny Scott, Trevor Starr, and Chao-Wen Wang. We thank Elizabeth Roth and Trevor Starr for extremely helpful technical assistance. We thank Bob Fuller and Jennifer Lippincott-Schwartz for advice about Kex2p-Fus1p chimeric proteins and Alex Engel, Eric Grote, Max Heiman, Robin Klemm, and Chris Toret for helpful discussions about Fus1p. We are grateful to the laboratories of Georjana Barnes, David Drubin, Jasper Rine, Jeremy Thorner, and Peter Walter for reagents and use of equipment. This work was supported by National Institutes of Health National Research Service Award grant F32-GM-071164-03 (to R. B.) and grant GM-26755 (to R. S.) and funds from the Howard Hughes Medical Institute (to R. S.).
Abbreviations used:
- AP
adaptor protein complex.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-04-0324) on October 7, 2009.
REFERENCES
- Audhya A., Foti M., Emr S. D. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J. S., Traub L. M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Chuang J. S., Schekman R. W. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J. Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corda D., Hidalgo Carcedo C., Bonazzi M., Luini A., Spano S. Molecular aspects of membrane fission in the secretary pathway. Cell Mol. Life Sci. 2002;59:1819–1832. doi: 10.1007/PL00012508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsch H. Regulation of membrane trafficking in polarized epithelial cells. Curr. Opin. Cell Biol. 2008;20:208–213. doi: 10.1016/j.ceb.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H., Schnieders E. A., Thorner J., Takemoto J. Y., DeWald D. B. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:34294–34300. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- Heiman M. G., Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 2000;151:719–730. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. C., Miller E. A., Goldberg J., Orci L., Schekman R. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- Litvak V., Dahan N., Ramachandran S., Sabanay H., Lev S. Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat. Cell Biol. 2005;7:225–234. doi: 10.1038/ncb1221. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Ma D., Zerangue N., Lin Y. F., Collins A., Yu M., Jan Y. N., Jan L. Y. Role of ER export signals in controlling surface potassium channel numbers. Science. 2001;291:316–319. doi: 10.1126/science.291.5502.316. [DOI] [PubMed] [Google Scholar]
- Maday S., Anderson E., Chang H. C., Shorter J., Satoh A., Sfakianos J., Folsch H., Anderson J. M., Walther Z., Mellman I. A PDZ-binding motif controls basolateral targeting of syndecan-1 along the biosynthetic pathway in polarized epithelial cells. Traffic. 2008;9:1915–1924. doi: 10.1111/j.1600-0854.2008.00805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K., Orci L., Amherdt M., Bednarek S. Y., Hamamoto S., Schekman R., Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- Nelson B., Parsons A. B., Evangelista M., Schaefer K., Kennedy K., Ritchie S., Petryshen T. L., Boone C. Fus1p interacts with components of the Hog1p mitogen-activated protein kinase and Cdc42p morphogenesis signaling pathways to control cell fusion during yeast mating. Genetics. 2004;166:67–77. doi: 10.1534/genetics.166.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D. T., Walter P. ER membrane protein complex required for nuclear fusion. J. Cell Biol. 1996;132:499–509. doi: 10.1083/jcb.132.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan S., Cowan A. E., Koppel D. E., Jin H., Grote E. FUS1 regulates the opening and expansion of fusion pores between mating yeast. Mol. Biol. Cell. 2006;17:2439–2450. doi: 10.1091/mbc.E05-11-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip B., Levin D. E. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Biol. Cell. 2001;21:271–280. doi: 10.1128/MCB.21.1.271-280.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proszynski T. J., Simons K., Bagnat M. O-glycosylation as a sorting determinant for cell surface delivery in yeast. Mol. Biol. Cell. 2004;15:1533–1543. doi: 10.1091/mbc.E03-07-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K., Holcomb C., Fuller R. S. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J. Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchatjate S., Schekman R. Chs5/6 complex: a multiprotein complex that interacts with and conveys chitin synthase III from the trans-Golgi network to the cell surface. Mol. Biol. Cell. 2006;17:4157–4166. doi: 10.1091/mbc.E06-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B., Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J. Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B., Snyder M. Sbe2p and sbe22p, two homologous Golgi proteins involved in yeast cell wall formation. Mol. Biol. Cell. 2000;11:435–452. doi: 10.1091/mbc.11.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B., Snyder M. Specific protein targeting during cell differentiation: polarized localization of Fus1p during mating depends on Chs5p in Saccharomyces cerevisiae. Eukaryot. Cell. 2003;2:821–825. doi: 10.1128/EC.2.4.821-825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautwein M., Schindler C., Gauss R., Dengjel J., Hartmann E., Spang A. Arf1p, Chs5p and the ChAPs are required for export of specialized cargo from the Golgi. EMBO J. 2006;25:943–954. doi: 10.1038/sj.emboj.7601007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trilla J. A., Duran A., Roncero C. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:1153–1163. doi: 10.1083/jcb.145.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueheart J., Boeke J. D., Fink G. R. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol. Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueheart J., Fink G. R. The yeast cell fusion protein FUS1 is O-glycosylated and spans the plasma membrane. Proc. Natl. Acad. Sci. USA. 1989;86:9916–9920. doi: 10.1073/pnas.86.24.9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia R. H., Baggott D., Chuang J. S., Schekman R. W. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev. Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- Walch-Solimena C., Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat. Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- Wang C. W., Hamamoto S., Orci L., Schekman R. Exomer: a coat complex for transport of select membrane proteins from the trans-Golgi network to the plasma membrane in yeast. J. Cell Biol. 2006;174:973–983. doi: 10.1083/jcb.200605106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M., Chuang J. S., Tsung M., Hamamoto S., Schekman R. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol. Biol. Cell. 1998;9:1565–1576. doi: 10.1091/mbc.9.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.