Abstract
Background
One important preprocessing step in the analysis of microarray data is background subtraction. In high-density oligonucleotide arrays this is recognized as a crucial step for the global performance of the data analysis from raw intensities to expression values.
Results
We propose here an algorithm for background estimation based on a model in which the cost function is quadratic in a set of fitting parameters such that minimization can be performed through linear algebra. The model incorporates two effects: 1) Correlated intensities between neighboring features in the chip and 2) sequence-dependent affinities for non-specific hybridization fitted by an extended nearest-neighbor model.
Conclusion
The algorithm has been tested on 360 GeneChips from publicly available data of recent expression experiments. The algorithm is fast and accurate. Strong correlations between the fitted values for different experiments as well as between the free-energy parameters and their counterparts in aqueous solution indicate that the model captures a significant part of the underlying physical chemistry.
Background
The analysis of microarray data has attracted continuous interest over the past years in the Bioinformatics community (see e.g. [1]). The problem consists in obtaining the gene expression level from the experimental measurements, which are the emitted fluorescence intensities from different sites in the array. On general grounds one expects that the experimental signal can be decomposed into two contributions:
| (1) |
where ISP(c) is the specific signal due to the hybridization of the surface-bound probe sequence with a complementary target sequence. This quantity depends on the concentration c of the complementary strand in solution (target). The non-specific term Ibg has different origins. It arises due to spurious effects such as incomplete hybridization where probe sequences bind to only partially complementary targets or due to other optical effects.
Models based upon the physical chemistry of hybridization (see e.g. [2]) predict a linear increase of the specific signal until saturation is approached. In case of highly expressed genes the specific part of the signal ISP(c) dominates the total signal intensity I and hence one can safely make the approximation I ≈ ISP(c). For lowly expressed genes, as well as for sequences with a low binding affinity, the specific and the non-specific contribution to the total intensity can be of comparable magnitude. In this case an accurate estimate of Ibg is crucial to draw reliable conclusions concerning the expression level; estimates based on the intensity distribution over the whole chip suggest that this is the case for roughly a quarter or half of the probes [3]. Once the background is calculated the gene expression level is then computed from background subtracted data I - Ibg.
In this paper we present an algorithm for the calculation of the background level for Affymetrix expression arrays, also known as GeneChips. In these arrays the probe sequences come in pairs: for each perfect match (PM) probe, which is exactly complementary to the transcript sequence in solution, there is a second probe with a single non-complementary nucleotide with respect to the specific target. The latter is called mismatch (MM) probe.
Several algorithms for background analysis of Affymetrix chips are available. Some of these use the MM intensities as corrections for non-specific hybridization, while others rely on PM intensities only. For instance, the Affymetrix MAS 5.0 software (Microarray Analysis Suite version 5.0) uses the difference (IPM - IMM) as estimator of the specific signal; an adjusted MM intensity (ideal MM) is used in the case the MM intensity exceeds the PM signal [4]. The Robust Multiarray Algorithm (RMA) [5] uses a different type of subtraction scheme which does not involve the MM intensities. The more recent version of this algorithm GCRMA performs background subtraction using information on the probe sequence composition through the calculation of binding affinities [6]. The position-dependent nearest-neighbor model (PDNN) [7] fits the background intensity using weight factors which depend on the position along the probe. The free energy parameters then enter in a nonlinear function. In the VSN algorithm [8] a generalized log transform is used to background correct the data. A study dedicated to the performance of different algorithms showed that the type of background subtraction used has a large effect on the global performance of the algorithms [9]. It is therefore not surprising that the background issue has attracted a lot of interest by the scientific community.
In this paper we present an algorithm for background estimation which combines information from the sequence composition and physical neighbors on the chip. This algorithm relies on previous work by the authors [10]. While the previous algorithm performed well with respect to the accuracy of the background estimation, the computational effort (per probe) involved was a severe limiting factor concerning its practical usability. The main cause of this significant computational effort was the iterative minimization of a cost function with nonlinear terms. The algorithm presented in this work involves a different cost function which is quadratic in the parameters. Its minimization can be performed via standard matrix computations of linear algebra. The algorithm is fast and accurate and is therefore suited for large scale analysis.
This paper is organized as follows. In Methods we discuss the optimization step from singular value decomposition and we provide the details of the selected cost function. In Results a test of the algorithm on about 360 Genechips from recent (2006 onwards) experiments from the Gene Expression Omnibus (GEO) is presented. Finally, the advantages of this scheme and its overall performance as background subtraction method is highlighted.
Methods
Approach
The general assumption is that the (natural) logarithm of the background intensity can be approximated by a function linear in some fitting parameters ωα. Once these parameters are set to their optimal values  α an estimate of the background intensity for the i-th (i = 1, 2, ..., Ndim) PM probe can be obtained as
α an estimate of the background intensity for the i-th (i = 1, 2, ..., Ndim) PM probe can be obtained as
| (2) |
where Nf is the number of fitting parameters. Ωiα is a sequence- and position-dependent element of the Ndim × Nf -dimensional matrix Ω, which will be defined below.
The optimized values for the model parameters  α are obtained by minimizing the difference (of the logarithms) of the observed and estimated intensities
α are obtained by minimizing the difference (of the logarithms) of the observed and estimated intensities
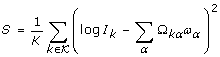 |
(3) |
i.e. solving a linear least square problem. The sum extends over the training set  which is a subset (with K elements) of the intensities of all annotated features. The choice of the elements of
which is a subset (with K elements) of the intensities of all annotated features. The choice of the elements of  will be discussed later (see Data Set - Parameter Optimization). The minimum is found by imposing vanishing partial derivatives of S w.r.t. ωα. This yields the following Nf linear equations
will be discussed later (see Data Set - Parameter Optimization). The minimum is found by imposing vanishing partial derivatives of S w.r.t. ωα. This yields the following Nf linear equations
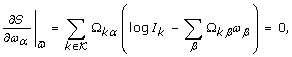 |
(4) |
which can be rewritten as
| (5) |
Here, we have introduced the matrix
| (6) |
and the vector Γα
| (7) |
(Note that M = ΩTΩ is symmetric and dim(M) = Nf × Nf).
The solution of Eq. (5) is given by the vector
| (8) |
of the optimal parameter values. If the matrix M is singular, M-1 has to be replaced by its pseudoinverse M+which can be obtained by means of Singular value decomposition (SVD). In this work a standard SVD algorithm based on Golub and Reinsch is used (see e.g. [11,12]).
Due to the symmetry of M only half of the off-diagonal elements need to be generated, hence reducing the computational effort. For the chips tested in this paper with dimensions up to 1164 × 1164 features, the computational time on a standard PC (×86_64 Intel Core 2 Duo with 3 GHz, 3 GB RAM) required to estimate the background intensities is 8 to 10 seconds for the larger chips, and faster for the smaller ones.
This makes our algorithm an order of magnitude faster than our previous version [10], 3 to 5 times faster than GC-RMA, PDNN and MAS5, and about twice as slow as RMA and DFCM (Bioconductor packages were used for the testing). Note that for our algorithm, the time estimate includes both reading in the CEL-file and the background calculation, as it is done in one step.
This computation involves the generation of the matrix M and vector Γ (from Eqs. (6) and (7)), the SVD of M to solve Eq. (5) and the estimation of the background intensity for all PM probes through Eq. (2). Differently from other approaches in which the cost function is minimized by means of Monte Carlo methods [13] or other dynamical algorithms [10], the solution of SVD provides the exact minimum of the cost function Eq. (3). Hence, there is no risk in getting stuck in local minima different from the global one.
Data Set - Parameter Optimization
As mentioned above, probes in Affymetrix form PM/MM pairs. Consider now a target sequence at a concentration c in solution. The analysis of Affymetrix spike-in data (see e.g. [14]) shows that not only the PM signal increases with increased target concentration c but also the MM intensity. This is an indication that a single MM nucleotide only partially prevents probe-target hybridization. Therefore the intensity of MM probes can also be decomposed in a non-specific and specific part as in Eq. (1). Supported by Affymetrix spike-in data analysis, our assumption is that the non-specific part of the hybridization is about equal for PM and MM probes: 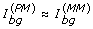 . The specific part of the signal is different in those two cases; equilibrium thermodynamics suggests a constant ratio
. The specific part of the signal is different in those two cases; equilibrium thermodynamics suggests a constant ratio 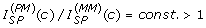 , independent of the target concentration, as observed in experiments [3].
, independent of the target concentration, as observed in experiments [3].
These insights are useful for the selection of probes for the optimization set 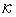 in Eq. (3):
in Eq. (3): 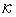 includes all MM probes whose intensities are below a certain threshold I0 and whose corresponding PM intensities also fulfill
includes all MM probes whose intensities are below a certain threshold I0 and whose corresponding PM intensities also fulfill 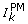 <I0 (a similar selection criterion was recently used by Chen et al. [15]). The threshold I0 is chosen on the basis of the total distribution of the intensities.
<I0 (a similar selection criterion was recently used by Chen et al. [15]). The threshold I0 is chosen on the basis of the total distribution of the intensities. 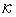 contains a significant fraction of the mismatch probes: typically 35%. Since the specific signal of MM intensities is lower than that of their corresponding PM's, they provide more reliable information on the background. The coordinates and sequences of the probes in
contains a significant fraction of the mismatch probes: typically 35%. Since the specific signal of MM intensities is lower than that of their corresponding PM's, they provide more reliable information on the background. The coordinates and sequences of the probes in 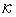 are then fitted to the intensities of these probes yielding the parameters ω. With those newly acquired parameters ω the background signal of all MM probes is estimated based upon the assumption
are then fitted to the intensities of these probes yielding the parameters ω. With those newly acquired parameters ω the background signal of all MM probes is estimated based upon the assumption 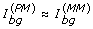 .
.
The matrix Ω
The choice of the matrix elements of Ω is dictated by input from physical chemistry as well as by the architecture of the microarray. Different schemes involving different choices for Ω with a varying number of parameters Nf were tested. Given a choice of Ω and in particular the number Nf of fitting parameters, the accuracy of the background estimation is reflected by the value of S from the minimization of Eq. (3). While the addition of fitting parameters always yields lower values of S, a too large set of fitting parameters runs the risk of "overfitting". The final choice of Ω is a compromise between a minimization of S and the use of the smallest possible set of parameters.
In the present model the number of parameters is Nf = 50. Similarly to the previous work [10] these parameters can be split into two groups: a first group describes the correlation of the background intensities with features which are physical neighbors on the chip; the second group are nearest-neighbor parameters which describe affinities for non-specific hybridization to the chip.
Physical Neighbors on the Chip
The first 18 parameters ωα describe the correlation of the background intensity with physical neighboring sequences in the array. Let (xi, yi) be the coordinate of the i-th MM sequence s(i). Then, its eight neighbors are located at (xi ± 1, yi), (xi ± 1, yi ± 1), and (xi, yi ± 1). As there is evidence that base-specific interactions (purine/pyrimidine asymmetry) might influence the hybridization process in general [16], we furthermore distinguish between probes whose central nucleotide is either A/G (purine) or C/T (pyrimidine). Then, the corresponding matrix elements in case of purines can be written as
| (9) |
with
| (11) |
and
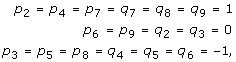 |
so that the intensities of the neighboring features explicitly enter the calculation of the background intensity of (xi, yi) as matrix elements Ωiα (2 ≤ α ≤ 9). In analogy to Eqs. (9,10) we define Ωiα with 10 ≤ α ≤ 18 corresponding to the sequences with a central pyrimidine.
Nearest-Neighbor Free Energy Parameters
The second contribution to the background model arises from the sequence composition. Let us first label the 16 dinucleotides according to the order {CC, GC, AC, TC, CG, GG, AG... TT}. We then define the next 16 matrix elements as
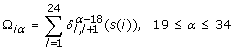 |
(12) |
where
| (13) |
according to the order given above. The sum runs over all the 24 dinucleotides along a probe sequence.
The matrix element Ωαi is equal to the number of dinucleotides of a given type in the sequence s(i). For instance, if the sequence s(i) contains 4 dinucleotides of type CC and 2 of type GC, then Ωi,19 = 4 and Ωi,20 = 2. Hybridization thermodynamics predicts log I ∝ ΔG where ΔG is the hybridization free energy.
In the nearest-neighbor model [17] the free energy is written as a sum of dinucleotide terms. Therefore, the parameters ωα (19 ≤ α ≤ 35) are the analogues of the free energy parameters of the nearest-neighbor model.
Position-Dependent Nearest-Neighbors
Finally, we consider the possibility that the hybridization strength is "modulated" along the sequence by a parabolic weight as
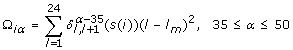 |
(14) |
where lm = 12.5, i.e. each dinucleotide is given a parabolic weight according to its position relative to the center at lm of the sequence. Thus, possible "unzipping" effects of the DNA-RNA duplex are approximately accounted for by Eq. (14).
The introduction of a position-dependence effect is in analogy with work done by other groups [7,16,18,19]. However, we do not introduce a position-dependent weight for each position along the 25-mer sequences. Instead, we limit ourselves to a parabolic modulation of the parameters along the chain, which drastically reduces the number of parameters involved in the model.
Invariances
Given the definition of Ω above it can be shown that Eq. (5) permits multiple solutions. Therefore, the optimal parameters  α are not unique. Consider a set of optimal parameters
α are not unique. Consider a set of optimal parameters  α which minimizes the cost function S (Eq. (3)). Let us then add a constant λ to the 16 nearest-neighbor parameters
α which minimizes the cost function S (Eq. (3)). Let us then add a constant λ to the 16 nearest-neighbor parameters
| (15) |
From Eq. (12) we obtain
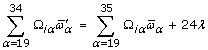 |
(16) |
because Ωiα counts the frequency of the dinucleotide α in the sequence corresponding to feature i and because there are 24 dinucleotides in a 25-mer probe sequence. Now, also consider
| (17) |
while 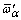 =
=  α for all other α. We conclude that the reparametrization of Eqs. (15) and (17) yields
α for all other α. We conclude that the reparametrization of Eqs. (15) and (17) yields
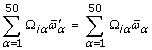 |
(18) |
since the shifting of Eq. (17) compensates the one introduced by Eq. (16). This reparametrization, valid for any real λ, leaves S invariant, and produces a zero eigenvalue of the matrix M of Eq. (6).
Similarly, one can verify that there is at least a second zero eigenvalue: a shift of the position-dependent nearest-neighbor parameters 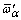 =
=  α + λ (for 35 ≤ α ≤ 50) as well as of
α + λ (for 35 ≤ α ≤ 50) as well as of 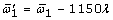 ,
, 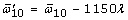 leaves S invariant. To obtain the latter equations Eq. (14) and
leaves S invariant. To obtain the latter equations Eq. (14) and 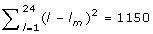 have to be applied.
have to be applied.
Having zero eigenvalues, the matrix M is therefore not invertible; the SVD thus provides the appropriate pseudo-inverse as discussed above. Accidental degeneracies or quasi-degeneracies of M could also occur, yielding eigenvalues close to zero in machine precision. These are, however, rare, and were actually never found in the calculations presented here.
Results
We analyzed a total of 366 CEL-files which are publicly available from the GEO server http://www.ncbi.nlm.nih.gov/geo. Table 1 gives an overview of the distribution of CEL-files over the twelve different organisms considered in this study. The array size for an organism might vary depending on the GSE accession number, since the most recent Affymetrix chips tend to use smaller features, thus more probes can be accommodated on the same surface area. For instance the Human HGU-133A contains 7122 features, while the Human Genome U133 Plus 2.0 Array goes to 11642 features. The last column of Table 1 lists the attained minimum value 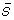 min, which ranges from 0.017 for Escheria Coli to 0.11 for Oryza Sativa.
min, which ranges from 0.017 for Escheria Coli to 0.11 for Oryza Sativa. 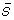 min estimates the mean squared deviation of the logarithm of the estimated background intensity from the actual background value. For instance
min estimates the mean squared deviation of the logarithm of the estimated background intensity from the actual background value. For instance 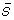 min = 0.01 corresponds to a 10% deviation, while
min = 0.01 corresponds to a 10% deviation, while 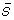 min = 0.1 corresponds to a 37% deviation. Table 2 provides a summary of the optimal parameters as calculated from the Singular Value Decomposition for the minimization of the cost function of Eq. (3). The parameters
min = 0.1 corresponds to a 37% deviation. Table 2 provides a summary of the optimal parameters as calculated from the Singular Value Decomposition for the minimization of the cost function of Eq. (3). The parameters  1 and
1 and  10 are associated to constant intensities for purines and pyrimidines. Their magnitudes are not unique due to the reparametrization as discussed in Eqs. (15) to (18). If information on neighboring probes is disregarded, the value of S typically increases by 45%; if the sequence information is not directly used, then it will increase by 52%. In the following we analyze the parameters associated to the local physical neighbors and to the nearest-neighbor free energy.
10 are associated to constant intensities for purines and pyrimidines. Their magnitudes are not unique due to the reparametrization as discussed in Eqs. (15) to (18). If information on neighboring probes is disregarded, the value of S typically increases by 45%; if the sequence information is not directly used, then it will increase by 52%. In the following we analyze the parameters associated to the local physical neighbors and to the nearest-neighbor free energy.
Table 1.
Overview over organisms and number of CEL-files analyzed
| Organism | GEO # | Chiptype (dimension) | # files |
 min min
|
|---|---|---|---|---|
| A. Thaliana | GSE4847 | ATH1-121501 (712 × 712) | 18 | 0.0259 |
| GSE7642 | ATH1-121501 (712 × 712) | 12 | 0.0544 | |
| GSE9311 | ATH1-121501 (712 × 712) | 8 | 0.0546 | |
| C. Elegans | GSE6547 | Celegans (712 × 712) | 25 | 0.0361 |
| GSE8159 | Celegans (712 × 712) | 7 | 0.0396 | |
| D. Melanogaster | GSE3990 | Drosophila_2 (732 × 732) | 6 | 0.0620 |
| GSE6558 | DrosGenome1 (640 × 640) | 24 | 0.0605 | |
| D. Rerio | GSE4859 | Zebrafish (712 × 712) | 8 | 0.0357 |
| E. Coli | GSE11779 | E_coli_2 (478 × 478) | 3 | 0.0869 |
| GSE2928 | Ecoli (544 × 544) | 12 | 0.0172 | |
| GSE6195 | E_coli_2 (478 × 478) | 4 | 0.0664 | |
| H. Sapiens | GSE10433 | HG-U133A_2 (732 × 732) | 12 | 0.0757 |
| GSE5054 | HG-U133A (712 × 712) | 20 | 0.0392 | |
| HG-U133A_2 (732 × 732) | ||||
| M. Musculus | GSE7148 | HG-U133A (712 × 712) | 14 | 0.0296 |
| GSE8514 | HG-U133_Plus_2 (1164 × 1164) | 15 | 0.0738 | |
| GSE11897 | MOE430A (712 × 712) | 11 | 0.0640 | |
| MOE430B (712 × 712) | ||||
| Mouse430_2 (1002 × 1002) | ||||
| GSE6210 | Mouse430_2 | 12 | 0.0594 | |
| GSE6297 | Mouse430_2 | 24 | 0.0325 | |
| O. Sativa | GSE15071 | Rice (1164 × 1164) | 20 | 0.1157 |
| R. Norvegicus | GSE4494 | RG_U34A (534 × 534) | 59 | 0.0488 |
| GSE7493 | Rat230_2 (834 × 834) | 9 | 0.0497 | |
| GSE8238 | Rat230_2 (834 × 834) | 4 | 0.0640 | |
| S. Aureus | GSE7944 | S_aureus (602 × 602) | 6 | 0.0746 |
| S. Cerevisiae | GSE6073 | YG_S98 (534 × 534) | 12 | 0.0283 |
| GSE8379 | YG_S98 (534 × 534) | 8 | 0.0180 | |
| X. Laevis | GSE3368 | Xenopus_laevis (712 × 712) | 20 | 0.0514 |
Table 2.
Optimized parameter values as obtained from the minimization of Eq. (3).
| A. Thaliana | C. Elegans | D. Melanogaster | D. Rerio | |||||
|---|---|---|---|---|---|---|---|---|
| GEO no | GSE4847 | GSE7642 | GSE9311 | GSE6547 | GSE8159 | GSE3990 | GSE6558 | GSE4859 |
 1 1
|
0.178 | 0.196 | 0.261 | 0.379 | 0.520 | 0.270 | 0.421 | 0.338 |
 2 2
|
0.050 | 0.056 | 0.042 | 0.047 | 0.050 | 0.022 | 0.025 | 0.041 |
 3 3
|
0.051 | 0.056 | 0.041 | 0.034 | 0.042 | 0.024 | 0.024 | 0.050 |
 4 4
|
-0.013 | -0.012 | -0.013 | -0.012 | -0.016 | -0.005 | -0.011 | -0.012 |
 5 5
|
-0.013 | -0.011 | -0.012 | -0.010 | -0.012 | -0.003 | -0.010 | -0.008 |
 6 6
|
0.186 | 0.198 | 0.224 | 0.168 | 0.271 | 0.228 | 0.195 | 0.140 |
 7 7
|
0.004 | 0.005 | 0.003 | 0.003 | 0.005 | 0.001 | 0.005 | 0.006 |
 8 8
|
0.002 | 0.009 | 0.003 | 0.002 | 0.005 | 0.001 | 0.006 | 0.002 |
 9 9
|
0.013 | 0.027 | 0.010 | 0.008 | 0.008 | 0.009 | 0.012 | 0.014 |
 10 10
|
-0.174 | -0.192 | -0.258 | -0.375 | -0.517 | -0.267 | -0.418 | -0.334 |
 11 11
|
0.063 | 0.060 | 0.059 | 0.058 | 0.062 | 0.042 | 0.057 | 0.068 |
 12 12
|
0.069 | 0.064 | 0.064 | 0.052 | 0.059 | 0.046 | 0.061 | 0.075 |
 13 13
|
-0.017 | -0.015 | -0.016 | -0.011 | -0.012 | -0.012 | -0.013 | -0.016 |
 14 14
|
-0.018 | -0.016 | -0.019 | -0.010 | -0.009 | -0.011 | -0.014 | -0.012 |
 15 15
|
0.258 | 0.299 | 0.328 | 0.301 | 0.459 | 0.336 | 0.316 | 0.246 |
 16 16
|
0.004 | 0.006 | 0.005 | 0.005 | 0.009 | 0.002 | 0.008 | 0.007 |
 17 17
|
0.002 | 0.007 | 0.003 | 0.003 | 0.006 | 0.001 | 0.007 | 0.004 |
 18 18
|
0.013 | 0.025 | 0.012 | 0.012 | 0.015 | 0.009 | 0.016 | 0.016 |
 19 19
|
0.068 | 0.138 | 0.143 | 0.149 | 0.155 | 0.159 | 0.217 | 0.140 |
 20 20
|
0.052 | 0.191 | 0.169 | 0.090 | 0.157 | 0.124 | 0.152 | 0.140 |
 21 21
|
-0.021 | -0.038 | 0.006 | -0.033 | 0.030 | -0.060 | -0.033 | -0.069 |
 22 22
|
-0.021 | 0.051 | 0.056 | -0.025 | 0.072 | -0.084 | 0.003 | -0.068 |
 23 23
|
-0.032 | -0.162 | -0.147 | -0.038 | -0.121 | -0.060 | -0.094 | -0.104 |
 24 24
|
0.041 | 0.075 | 0.072 | 0.024 | 0.015 | 0.030 | 0.048 | 0.035 |
 25 25
|
-0.063 | -0.221 | -0.162 | -0.122 | -0.140 | -0.172 | -0.200 | -0.190 |
 26 26
|
-0.060 | -0.130 | -0.119 | -0.118 | -0.109 | -0.173 | -0.154 | -0.181 |
 27 27
|
0.000 | -0.008 | -0.047 | 0.035 | -0.041 | 0.052 | 0.035 | 0.055 |
 28 28
|
0.036 | 0.148 | 0.076 | 0.062 | 0.063 | 0.105 | 0.096 | 0.128 |
 29 29
|
-0.026 | -0.058 | -0.069 | -0.046 | -0.044 | -0.065 | -0.085 | -0.052 |
 30 30
|
-0.050 | -0.032 | -0.093 | -0.065 | -0.052 | -0.106 | -0.102 | -0.078 |
 31 31
|
0.057 | 0.037 | 0.056 | 0.082 | 0.012 | 0.133 | 0.103 | 0.130 |
 32 32
|
0.093 | 0.180 | 0.173 | 0.125 | 0.112 | 0.211 | 0.181 | 0.221 |
 33 33
|
0.011 | -0.074 | -0.021 | -0.008 | -0.026 | 0.015 | -0.036 | 0.001 |
 34 34
|
-0.009 | -0.017 | -0.016 | -0.023 | -0.014 | -0.035 | -0.047 | -0.022 |
| Corr. Coeff | 0.775 | 0.686 | 0.738 | 0.814 | 0.700 | 0.754 | 0.826 | 0.705 |
 35 35
|
0.020 | 0.016 | 0.016 | 0.019 | 0.013 | 0.016 | 0.012 | 0.018 |
 36 36
|
0.020 | 0.010 | 0.012 | 0.021 | 0.011 | 0.016 | 0.016 | 0.017 |
 37 37
|
0.025 | 0.025 | 0.022 | 0.029 | 0.019 | 0.028 | 0.027 | 0.031 |
 38 38
|
0.024 | 0.019 | 0.019 | 0.028 | 0.016 | 0.028 | 0.023 | 0.031 |
 39 39
|
0.026 | 0.036 | 0.034 | 0.030 | 0.030 | 0.027 | 0.031 | 0.033 |
 40 40
|
0.021 | 0.021 | 0.020 | 0.026 | 0.021 | 0.022 | 0.023 | 0.026 |
 41 41
|
0.028 | 0.039 | 0.035 | 0.036 | 0.031 | 0.035 | 0.038 | 0.039 |
 42 42
|
0.027 | 0.032 | 0.031 | 0.035 | 0.028 | 0.033 | 0.033 | 0.038 |
 43 43
|
0.023 | 0.024 | 0.026 | 0.024 | 0.023 | 0.019 | 0.021 | 0.021 |
 44 44
|
0.020 | 0.013 | 0.017 | 0.022 | 0.015 | 0.015 | 0.017 | 0.016 |
 45 45
|
0.024 | 0.028 | 0.028 | 0.030 | 0.024 | 0.027 | 0.029 | 0.029 |
 46 46
|
0.025 | 0.025 | 0.029 | 0.030 | 0.024 | 0.028 | 0.029 | 0.030 |
 47 47
|
0.020 | 0.022 | 0.020 | 0.022 | 0.021 | 0.015 | 0.019 | 0.016 |
 48 48
|
0.017 | 0.012 | 0.012 | 0.019 | 0.013 | 0.010 | 0.014 | 0.011 |
 49 49
|
0.023 | 0.029 | 0.026 | 0.028 | 0.023 | 0.023 | 0.028 | 0.026 |
 50 50
|
0.023 | 0.023 | 0.023 | 0.028 | 0.021 | 0.024 | 0.026 | 0.026 |
| Corr coeff | -0.679 | -0.618 | -0.679 | -0.759 18 | -0.631 | -0.683 | -0.783 | -0.658 |
Magnitudes and signs of the parameters are approximately constant across different experiments. The correlation coefficients are the Pearson correlations between the nearest-neighbor parameters and the free energy parameters measured in hybridization experiments in aqueous solution [20].
Parameters of physical neighbors in the Chip
The parameters  2 to
2 to  9 and
9 and  11 to
11 to  18 describe the coupling of the background intensities to the physically neighboring features on the chip. As already mentioned, our estimate of the PM background is based on the non-specific intensity of the MM sequence. An Affymetrix chip is designed such that MM and PM are found in rows at equal y-coordinates. In addition, given a PM at (x, y), the corresponding MM feature is at (x, y + 1).
18 describe the coupling of the background intensities to the physically neighboring features on the chip. As already mentioned, our estimate of the PM background is based on the non-specific intensity of the MM sequence. An Affymetrix chip is designed such that MM and PM are found in rows at equal y-coordinates. In addition, given a PM at (x, y), the corresponding MM feature is at (x, y + 1).
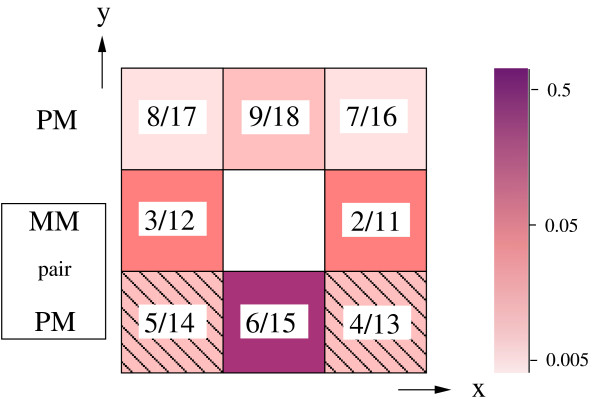
Figure 1 schematically represents the influence of the neighboring intensities on the background value. The strength of the correlation of the eight neighboring spots (i.e. the magnitude and sign of the corresponding  α) with the central MM feature is indicated by the color. The numbers in the figure identify the associated parameters
α) with the central MM feature is indicated by the color. The numbers in the figure identify the associated parameters  α. For example
α. For example  6 and
6 and  15 are associated to the intensity at (x, y - 1) with respect to the reference MM intensity with coordinates (x, y). There does not seem to be any evidence that the middle-nucleotide classification in purines and pyrimidines reveals any insight concerning the background. Instead, our results show that the absolute values of two "corresponding" (purine-pyrimidine pair)
15 are associated to the intensity at (x, y - 1) with respect to the reference MM intensity with coordinates (x, y). There does not seem to be any evidence that the middle-nucleotide classification in purines and pyrimidines reveals any insight concerning the background. Instead, our results show that the absolute values of two "corresponding" (purine-pyrimidine pair)  α 's are generally of the same order of magnitude. Across all species, we find typical average outputs of
α 's are generally of the same order of magnitude. Across all species, we find typical average outputs of
Figure 1.
Correlations with neighboring features. Schematic representation of the neighboring parameters on the array. The inset numbers indicate the corresponding parameter(s)  α, e.g. 3/12 represents
α, e.g. 3/12 represents  3,
3,  12 respectively.
12 respectively.  2 through
2 through  9 are related to purines,
9 are related to purines,  11 to
11 to  18 to pyrimidines. The bar to the right gives the intensity scale of the parameters. The dashed pattern indicates negative values. Box on the left indicates the rows of corresponding PM and MM pairs. The central feature (position (x, y)) is a MM; its corresponding PM is located just below it (x, y - 1), its intensity has the strongest correlation with the central MM feature. The features in (x ± 1, y - 1)
18 to pyrimidines. The bar to the right gives the intensity scale of the parameters. The dashed pattern indicates negative values. Box on the left indicates the rows of corresponding PM and MM pairs. The central feature (position (x, y)) is a MM; its corresponding PM is located just below it (x, y - 1), its intensity has the strongest correlation with the central MM feature. The features in (x ± 1, y - 1)
Since  6,
6,  15 respectively, reflects the correlation of the MM signal with its corresponding PM, as expected its magnitude is greatest among all parameter values. Next, the MM intensity shows strong correlations with its direct nearest neighbors positioned at (x ± 1, y), i.e.
15 respectively, reflects the correlation of the MM signal with its corresponding PM, as expected its magnitude is greatest among all parameter values. Next, the MM intensity shows strong correlations with its direct nearest neighbors positioned at (x ± 1, y), i.e.  2/11 and
2/11 and  3/12. Hence, the stronger the MM-neighboring intensities, the stronger their influence on the background signal of MM. However, in order to somehow compensate for strong MM-neighboring signals which might be caused by the presence of complementary target sequences, parameters
3/12. Hence, the stronger the MM-neighboring intensities, the stronger their influence on the background signal of MM. However, in order to somehow compensate for strong MM-neighboring signals which might be caused by the presence of complementary target sequences, parameters  4/13 and
4/13 and  5/14 have negative sign. The remaining three parameters, i.e. the top three neighbors limit their influence to a basically negligible minimum which appears to indicate the weak sequence correlation in y-direction as previously found. The results indicate that the influence of neighboring intensities on the background noise is significant. In fact, our analysis shows that ≈ 30% of log
5/14 have negative sign. The remaining three parameters, i.e. the top three neighbors limit their influence to a basically negligible minimum which appears to indicate the weak sequence correlation in y-direction as previously found. The results indicate that the influence of neighboring intensities on the background noise is significant. In fact, our analysis shows that ≈ 30% of log  are constituted by neighboring probes in terms of absolute intensities (see Table 3). It appears that for a few probes, the latter might even play a crucial role. It is unlikely that the correlations summarized in Fig 1 could be explained only as simple optical effects, as light from bright probes spilling into their neighborhood. Indeed, optical effects should produce an isotropic correlation pattern in the x and y directions which is not seen in our analysis. Another cause for this correlation might be that neighboring probes share common sequences, as is the case in Affymetrix chips [10].
are constituted by neighboring probes in terms of absolute intensities (see Table 3). It appears that for a few probes, the latter might even play a crucial role. It is unlikely that the correlations summarized in Fig 1 could be explained only as simple optical effects, as light from bright probes spilling into their neighborhood. Indeed, optical effects should produce an isotropic correlation pattern in the x and y directions which is not seen in our analysis. Another cause for this correlation might be that neighboring probes share common sequences, as is the case in Affymetrix chips [10].
Table 3.
Influence of neighboring spot on background intensity in % of eight (randomly chosen) CEL-files of different organisms.
| X. Laevis | 74500.CEL 35% | 76190.CEL 37% |
|---|---|---|
| C. Celegans | 201989.CEL 43% | 201994.CEL 52% |
| H. Sapiens | 263931.CEL 41% | 263930.CEL 39% |
| S. Cerevisiae | 207569.CEL 29% | 207570.CEL 29% |
Nearest-Neighbor Parameters
The parameters  α (19 ≤ α ≤ 34) are the analogues of the nearest-neighbor free energy parameters. The nearest-neighbor model is commonly used to study the thermodynamics of hybridization of nucleic acids in solution (see e.g. [17]). In this model it is assumed that the stability and thus the hybridization free energy ΔG of a dinucleotide depends on the orientation and identity of the neighboring base pairs. For RNA/DNA duplexes there are 16 hybridization free energy parameters which were measured in aqueous solution by Sugimoto et al. [20].
α (19 ≤ α ≤ 34) are the analogues of the nearest-neighbor free energy parameters. The nearest-neighbor model is commonly used to study the thermodynamics of hybridization of nucleic acids in solution (see e.g. [17]). In this model it is assumed that the stability and thus the hybridization free energy ΔG of a dinucleotide depends on the orientation and identity of the neighboring base pairs. For RNA/DNA duplexes there are 16 hybridization free energy parameters which were measured in aqueous solution by Sugimoto et al. [20].
Recent experiments [21] focusing on specific hybridization show a good degree of correlation between the hybridization free energies in solution and those directly determined from microarray data. Concerning background data, we also expect a certain degree of correlation between the parameters  α (19 ≤ α ≤ 34) and their corresponding Sugimoto free energy parameters.
α (19 ≤ α ≤ 34) and their corresponding Sugimoto free energy parameters.
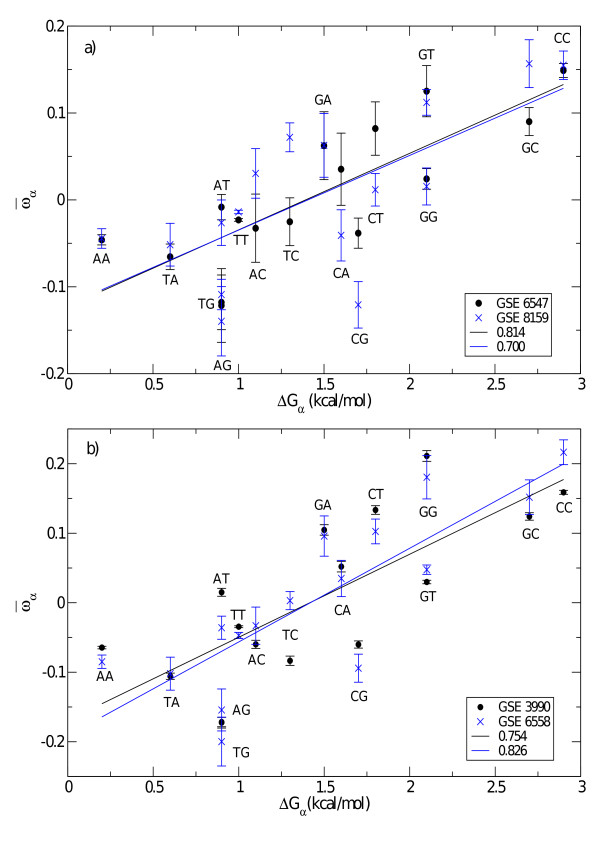
In order to test the relationship between the experimentally determined so-called Sugimoto parameters and our results, we calculate the correlation coefficient between these two sets. The results are reported in Table 2. In general, the correlation coefficients vary between 0.53 and 0.83 with a median value of 0.71. Figure 2 shows two typical results for C. Elegans and D. Melanogaster. Both plots indicate that the relationship between  α (19 ≤ α ≤ 34) and the nearest-neighbor free energy parameters of [20] is approximately linear.
α (19 ≤ α ≤ 34) and the nearest-neighbor free energy parameters of [20] is approximately linear.
Figure 2.
Fitted vs. solution hybridization free energies. Pair Parameters  α (19 ≤ α ≤ 34), as obtained from the minimization of Eq. (4) as function of ΔGα, the nearest-neighbor stacking free energy obtained from DNA/RNA hybridization in solution [20]. Plots refer to two GSE training data sets of a) C. Elegans and b) Drosophila Melanogaster. Each point on the y-axis is the averaged value over all CEL-files representing its GSE-set. The error bars are the standard deviation. Notation of DNA pairs are from 5' to 3' end. The straight lines are linear fits; correlation coefficients of each GSE set in legend.
α (19 ≤ α ≤ 34), as obtained from the minimization of Eq. (4) as function of ΔGα, the nearest-neighbor stacking free energy obtained from DNA/RNA hybridization in solution [20]. Plots refer to two GSE training data sets of a) C. Elegans and b) Drosophila Melanogaster. Each point on the y-axis is the averaged value over all CEL-files representing its GSE-set. The error bars are the standard deviation. Notation of DNA pairs are from 5' to 3' end. The straight lines are linear fits; correlation coefficients of each GSE set in legend.
Position-dependent nearest neighbor parameters
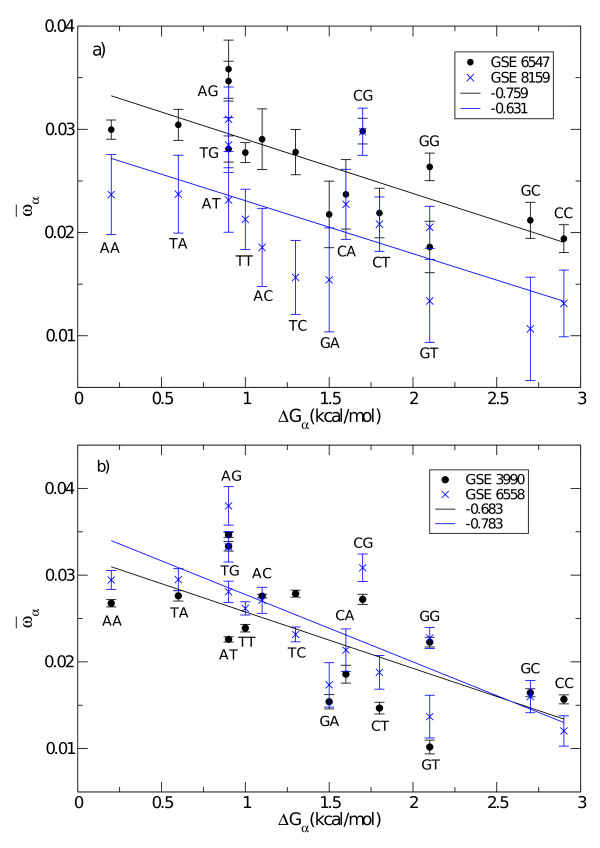
Figure 3 shows a plot of position-dependent nearest-neighbor parameters  35 to
35 to  50 as obtained from the minimization of the cost function of Eq. (3) for four different sets of experiments. The data are plotted as function of the corresponding nearest neighbor free energy parameters of [20]. We note that in all experiments shown there is a negative correlation between the two data sets. The parameters
50 as obtained from the minimization of the cost function of Eq. (3) for four different sets of experiments. The data are plotted as function of the corresponding nearest neighbor free energy parameters of [20]. We note that in all experiments shown there is a negative correlation between the two data sets. The parameters  35 to
35 to  50 reflect the difference in effective free energy between the ends and the middle of the probe sequence. Since weakly binding probes suffer more from end-effects (unzipping, etc.),
50 reflect the difference in effective free energy between the ends and the middle of the probe sequence. Since weakly binding probes suffer more from end-effects (unzipping, etc.),  35 to
35 to  50 and their corresponding nearest-neighbor free energy parameters are negatively correlated. Hence, the negative correlation in Fig. 3 and those observed in all other cases (see correlation coefficients reported in Table 2) indicates that the dominant contribution to the background intensity comes from the middle nucleotides. This conclusion complies with other types of analysis which use position-dependent free energy parameters (see e.g. [7,16]).
50 and their corresponding nearest-neighbor free energy parameters are negatively correlated. Hence, the negative correlation in Fig. 3 and those observed in all other cases (see correlation coefficients reported in Table 2) indicates that the dominant contribution to the background intensity comes from the middle nucleotides. This conclusion complies with other types of analysis which use position-dependent free energy parameters (see e.g. [7,16]).
Figure 3.
Fitted vs. solution hybridization (position dependent) free energies. Same as in Fig. 2 for parabolic pair parameters, i.e. ΔGα vs.  α (35 ≤ α ≤ 50) for a) C. Elegans and b) D. Melanogaster.
α (35 ≤ α ≤ 50) for a) C. Elegans and b) D. Melanogaster.
Comparing estimated vs. measured background
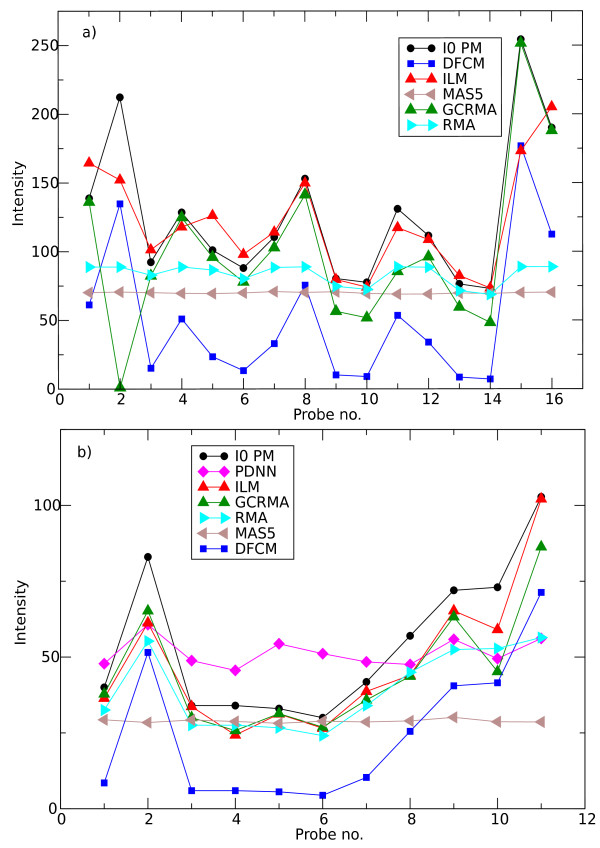
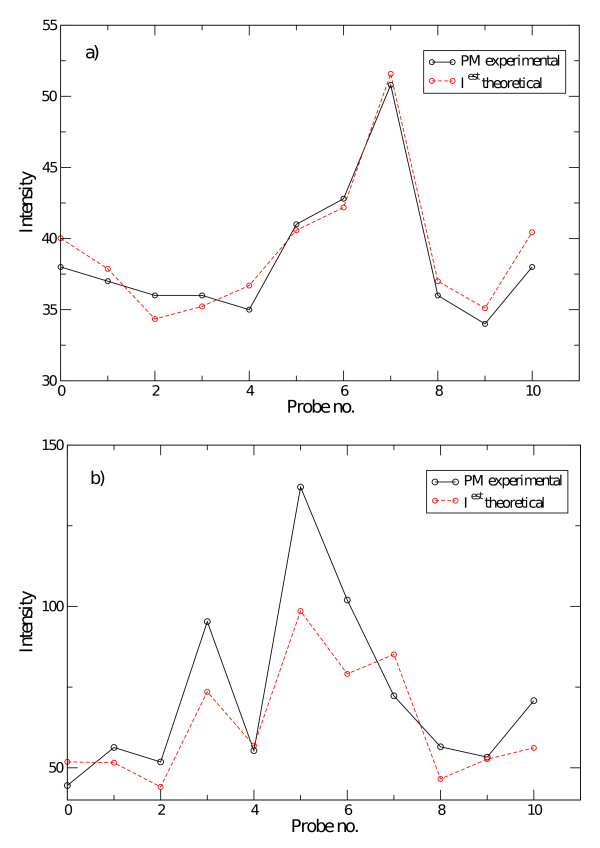
In this section we compare the estimated background signal (as given in Eq. (2)) with the intensities of given probe sets corresponding to non-expressed genes in the samples analyzed. We start with publicly available data taken from spike-in experiments on HGU95A chips http://www.affymetrix.com where genes have been spiked-in at known concentrations, ranging from 0 to 1024 pM (picoMolar). The data at 0 pM correspond to the absence of transcript in solution. Figure 4a compares the measured and predicted background for probeset 37777_at. Except for probes 2 and 15 for which the measured signal is higher than the predicted background, there is a nice agreement between our prediction and the experimental background intensity: the standard deviation of the absolute difference between the intensity of the PM and Iest is 28 in Affymetrix intensity units. Also shown are the background estimations as obtained with other algorithms (the data for PDNN is missing in Fig. 4a due to the unavailability of one of the supporting files from Bioconductor packages). Figure 4b presents the same information for probeset 209606_at. Here, the standard deviation is 7 in Affymetrix intensity units.
Figure 4.
Computed vs. true background (spike-in experiments). Experimental results (I0P M) and theoretical predictions of the algorithms discussed here for a) probeset 37777_at (HGU95A, 1521a99 hpp_av06.CEL) and b) 209606_at (HGU133A,12_13_02_U133A_Mer_Latin_Square_Expt10_R1.CEL) spiked-in at concentration c = 0, i.e. absent from the hybridizing solution. All comparison were performed using freely available packages from the Bioconductor project.
Next, we go beyond the spike-in experiments. To select non-expressed genes we considered probe sets with very low expression values as obtained from the RMA algorithm. In Figure 5 the PM intensities as well as the calculated background signal of two probesets from A. Thaliana and C. Elegans experiments are shown. The absolute intensities of both probesets are very low. As a consequence, we can safely assume these genes are not expressed and hence any measured signal can be attributed to the background. Figures 4 and 5 both show that the present model captures the essentials of and correctly predicts background intensities. Both Figures are representative for the CEL-files analyzed in this work.
Figure 5.
Computed background vs. low expressed genes data. Experimental results (solid black line) and theoretical prediction (dashed red line) of probeset a) 256610_at (A. Thaliana, GSE4847, GSM109107.CEL) and b) 175270_at (C. Elegans, GSE8159, GSM201995.CEL).
Discussion
We have presented a background subtraction scheme for Affymetrix GeneExpression arrays which is both, accurate and usable on a standard ×86_64 Intel Core 2 PC. The algorithm centers around a cost function which is quadratic in its fitting parameters. This allows for a rapid minimization, through linear algebra, in particular through singular value decomposition. The accuracy of the present algorithm is very similar to that of a background algorithm previously presented by the authors [10]. The latter had been tested on Affymetrix spike-in data and its performance was compared to background schemes such as MAS5, RMA and GCRMA. Regarding spike-in data, the analysis had shown that the proposed algorithm is definitely more accurate than background computations done with MAS5 and RMA, but also improves on GCRMA [10].
The proposed algorithm has two categories of fitting parameters. The first category exploits correlations between features which are neighbors on the chip. The second category is based on the strong similarity between probe-target hybridization and duplex stability in solution, and involves stacking free energies in analogy to those in the nearest-neighbor model. Existing algorithms are either of the first [4] or the second [6,7,9] category, but not both.
The background subtraction scheme has been tested on 360 GeneChips from publicly available data of recent expression experiments. Since the fitted values for the same parameters in different experiments do not show much variation, the algorithm is robust and can be easily transferred to other experiments. Due to its speed and accuracy the present method is suited for large scale computations. An R-package integrating the background analysis scheme with the computation of expression values from background subtracted data will be made freely available to the community (a preliminary version of this package can be found in http://itf.fys.kuleuven.ac.be/~enrico/ilm.html). The performance of this approach is discussed in [22].
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
EC and GB planned and supervised the research. KMK wrote the code for data analysis and analyzed the data. KMK and EC wrote the paper.
Contributor Information
K Myriam Kroll, Email: myriam.kroll@fys.kuleuven.be.
Gerard T Barkema, Email: G.T.Barkema@phys.uu.nl.
Enrico Carlon, Email: enrico.carlon@fys.kuleuven.be.
Acknowledgements
We acknowledge financial support from FWO (Research Foundation - Flanders) grant n. G.0311.08. Stimulating discussions with N. Naouar are gratefully acknowledged.
References
- Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Statistics for Biology and Health, Springer; 2003. [Google Scholar]
- Held GA, Grinstein G, Tu Y. Modeling of DNA microarray data by using physical properties of hybridization. Proc Natl Acad Sci. 2003;100:7575–7580. doi: 10.1073/pnas.0832500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrantini A, Allemeersch J, Van Hummelen P, Carlon E. Thermodynamic scaling behavior in genechips. BMC Bioinformatics. 2009;10:3. doi: 10.1186/1471-2105-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New statistical algorithms for monitoring gene expression on genechip probe arrays. Tech rep, Affymetrix; 2001. [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostat. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry R, Gentleman R, Martinez-Murillo F, Spencer F. A Model-Based Background Adjustment for Oligonucleotide Expression Arrays. Journal of the American Statistical Association. 2004;99(468):909. doi: 10.1198/016214504000000683. [DOI] [Google Scholar]
- Zhang L, Miles MF, Aldape KD. A model of molecular interactions on short oligonucleotide microarrays. Nature Biotech. 2003;21:818. doi: 10.1038/nbt836. [DOI] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sütmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Wu Z, Jaffee HA. Comparison of Affymetrix GeneChip expression measures. Bioinformatics. 2006;22(7):789. doi: 10.1093/bioinformatics/btk046. [DOI] [PubMed] [Google Scholar]
- Kroll KM, Barkema GT, Carlon E. Modeling background intensity in DNA microarrays. Phys Rev E. 2008;77:061915. doi: 10.1103/PhysRevE.77.061915. [DOI] [PubMed] [Google Scholar]
- Golub GH, Reinsch C. Singular value decomposition and least squares solutions. Numer Math. 1970;14:403–402. doi: 10.1007/BF02163027. [DOI] [Google Scholar]
- Golub GH, Van Loan CF. Matrix computations. The Johns Hopkins University Press, London; 1996. [Google Scholar]
- Ono N, Suzuki S, Furusawa C, Agata T, Kashiwagi A, Shimizu H, Yomo T. An improved physico-chemical model of hybridization on high-density oligonucleotide microarrays. Bioinformatics. 2008;24(10):1278–1285. doi: 10.1093/bioinformatics/btn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden CJ, Pittelkow Y, Wilson SR. Adsorption models of hybridization and post-hybridization behaviour on oligonucleotide microarrays. J Phys: Cond Matt. 2006;18(23):5545. doi: 10.1088/0953-8984/18/23/024. [DOI] [Google Scholar]
- Chen Z, McGee M, Liu Q, Kong M, Deng Y, Scheuermann R. A distribution-free convolution model for background correction of oligonucleotide microarray data. BMC Genomics. 2009;10(Suppl 1):S19. doi: 10.1186/1471-2164-10-S1-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder H, Preibisch S. Specific and nonspecific hybridization of oligonucleotide probes on microarrays. Biophys J. 2005;89:337. doi: 10.1529/biophysj.104.055343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield VA, Crothers DM, Tinoco I. , Jr. Nucleic Acids Structures, Properties and Functions. University Science Books, Mill Valley; 2000. [Google Scholar]
- Naef F, Magnasco MO. Solving the riddle of the bright mismatches: Labeling and effective binding in oligonucleotide arrays. Phys Rev E. 2003;68:011906. doi: 10.1103/PhysRevE.68.011906. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wu C, Carta R, Zhao H. Free energy of DNA duplex formation on short oligonucleotide microarrays. Nucleic Acids Res. 2007;35(3):e18. doi: 10.1093/nar/gkl1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic Parameters To Predict Stability of RNA/DNA Hybrid Duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- Hooyberghs J, Van Hummelen P, Carlon E. The effects of mismatches on hybridization in DNA microarrays: determination of nearest neighbor parameters. Nucleic Acids Res. 2009;37(7):e53. doi: 10.1093/nar/gkp109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulders GC, Barkema GT, Carlon E. Inverse Langmuir method for oligonucleotide microarray analysis. BMC Bioinformatics. 2009;10:64. doi: 10.1186/1471-2105-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]