Abstract
Reporter gene transactivation by human p53 is inhibited in budding yeast lacking the TRR1 gene encoding thioredoxin reductase. To investigate the role of thioredoxin in controlling p53 activity, the level of reporter gene transactivation by p53 was determined in yeast lacking the TRX1 and TRX2 genes encoding cytosolic thioredoxin. Surprisingly, p53 activity was unimpaired in yeast lacking thioredoxin. Subsequent analyses showed that thioredoxin deletion suppressed the inhibitory effect of thioredoxin reductase deletion, suggesting that accumulation of oxidized thioredoxin in mutant yeast was necessary for p53 inhibition. Purified human thioredoxin and p53 interacted in vitro (Kd = 0.9 µM thioredoxin). To test the idea that dithio-disulfide exchange reactions between p53 and thioredoxin were responsible for p53 inhibition in mutant yeast, each p53 cysteine was changed to serine and the effect of the substitution on p53 activity in TRR1 and Δtrr1 yeast was determined. Substitutions at Zn-coordinating cysteines C176, C238 or C242 resulted in p53 inactivation. Unexpectedly, substitution at cysteine C275 also inactivated p53, which was the first evidence for a non-zinc-coordinating cysteine being essential for p53 function. Cysteine substitutions at six positions (C124, C135, C141, C182, C229 and C277) neither inactivated p53 nor relieved the requirement for thioredoxin reductase. Furthermore, no tested combination of these six cysteine substitutions relieved thioredoxin reductase dependence. The results suggested that p53 dependence on thioredoxin reductase either was indirect, perhaps mediated by an upstream activator of p53, or was due to oxidation of one or more of the four essential cysteines.
Several lines of evidence suggest that the activity of the p53 tumor suppressor protein is subject to redox modulation. Electrophoretic mobility shift assays (EMSAs) have shown that at least one redox-sensitive cysteine residue influences DNA binding affinity, as treatment of p53 in vitro with cysteine alkylating agents diamide and N-ethylmaleimide abrogates binding to specific DNA target sequences (1–5), while the reductant dithiothreitol (DTT) greatly enhances binding (1–4). The presence of redox active proteins thioredoxin or Ref1 also stimulates p53 binding to target DNA in vitro (1, 6), and overexpression of thioredoxin or Ref1 in transfected mammalian cells stimulates p53-dependent reporter gene expression (1). The ability of human p53 to transactivate reporter gene expression is inhibited in fission yeast carrying a point mutation in the trr1 gene encoding thioredoxin reductase (7) and in budding yeast lacking the TRR1 gene (8). Thioredoxin reductase deletion has no effect on p53 protein levels (7, 8) or nuclear localization (7), suggesting that the deletion affects p53 specific activity. Thioredoxin reductase is not required for efficient transactivation of reporter gene expression by a fusion protein consisting of the p53 activation domain and LexA DNA binding domain (9), therefore excluding downstream effectors of p53 activity as the source of thioredoxin reductase dependence.
Thioredoxin reductase is a disulfide oxidoreductase that specifically catalyzes the transfer of electrons from NADPH to the active site cysteines of thioredoxin. Thioredoxin functions as the electron donor in several biological reduction reactions, including those catalyzed by ribonucleotide reductase, methionine sulfoxide reductase, 3'-phosphoadenosine-5'-phosphosulfate (PAPS) reductase, and thioredoxin-dependent peroxidase. Thioredoxin has also been implicated in controlling the DNA binding activity of NFκB, AP1, E2A and p53 in vitro (1, 6, 10–12).
Thioredoxin reductase deletion in budding yeast results in significant oxidation of both thioredoxin and glutathione (13). However, inhibition of p53 activity in thioredoxin reductase null yeast is not due to oxidative stress imposed by the higher GSSG:GSH ratio, as p53 activity remains inhibited when the normal GSSG:GSH ratio is restored by ectopic overexpression of the glutathione reductase gene. Furthermore, p53 activity is not affected by deletion of the glutathione reductase gene, a condition that markedly increases the GSSG:GSH ratio. Thus, p53 is specifically dependent on the thioredoxin system, and is not sensitive to the general redox state of the cell as defined by the GSSG:GSH ratio.
Saccharomyces cerevisiae, an organism in which all genes have been identified, and in which genes can easily be disrupted or ectopically expressed, offers a system in which the role of thioredoxin in controlling transcription factor activity can be analyzed. We investigated whether the inhibition of p53 activity observed in yeast lacking thioredoxin reductase was also observed in yeast lacking cytosolic thioredoxin. We found that deletion of the yeast cytosolic thioredoxin genes did not inhibit p53 activity, and in fact suppressed the inhibitory effect of deleting the thioredoxin reductase gene. The genetic suppression results, coupled with the biochemical observation that purified thioredoxin and p53 physically interact in vitro, supported a model in which p53 is in redox equilibrium with thioredoxin. According to the model, oxidized thioredoxin accumulates in the absence of thioredoxin reductase and promotes the oxidative inactivation of p53. To test this model, a comprehensive set of single Cys-to-Ser replacement alleles of human p53 was constructed, reasoning that replacement of redox-sensitive cysteines with non-oxidizable serines should produce an oxidation-resistant, thioredoxin reductase-independent p53 protein. Six replacements (C124S, C135S, C141S, C182S, C229S and C277S) retained at least partial activity, but in none of these cases did cysteine replacement result in an oxidation-resistant p53 protein. Furthermore, no tested combination of these six cysteine substitutions relieved thioredoxin reductase dependence. The other four single Cys-to-Ser replacements (C176S, C238S, C242S and C275S) resulted in an inactive p53 protein that thus could not be assayed for thioredoxin reductase-dependence. The results suggested that p53 dependence on thioredoxin reductase either was indirect, perhaps mediated by an upstream activator of p53, or was due to oxidation of one or more of the four essential cysteines.
EXPERIMENTAL PROCEDURES
Yeast strains
Saccharomyces cerevisiae strains used in the study are listed in Supplemental Table 1. The strain background was W303-1a (14). Standard yeast genetic techniques were used for tetrad dissections and random spore preparations (15). Yeast were grown with vigorous shaking at 25°C. The medium was either yeast extract peptone dextrose (YEPD) or yeast nitrate broth (YNB) containing 2% glucose and required supplements (15). For all assays, yeast were harvested at 107 cells/ml as determined by A600.
Plasmids
The ADE2-selectable ARS-CEN plasmids pASZ11-TRX1 and pASZ11-TRX2 (abbreviated pTRX1 and pTRX2, respectively, in Fig. 3A) were constructed by subcloning a 3.2-kb BamHI-SalI fragment of pLI833, containing the TRX1 gene and its native promoter, or a 2.6-kb BamHI-SalI fragment of pLI800, containing the TRX2 gene and its native promoter, into the polylinker region of pASZ11 (16). The pLI833 and pLI800 plasmids were obtained from Eric Muller (U. of Washington) (17). The pASZ11GPD expression vector was made by cloning a SacI/KpnI restriction enzyme fragment containing the GPD1 promoter, polylinker region and CYC1 terminator sequence from pRS414GPD (18) into SphI/KpnI-cleaved pASZ11. The SacI and SphI sites were destroyed by a T4 polymerase blunting reaction during the subcloning procedure. The pASZ11GPD-TRX1 and pASZ11GPD-TRX2 plasmids (abbreviated pGPD-TRX1 and pGPD-TRX2, respectively, in Fig. 3B) were constructed in two steps. First — using W303-1a yeast DNA as template — PCR products corresponding to the TRX1 or TRX2 coding regions were generated and cloned into the EcoRV site of pBluescript-KS to generate pBS-TRX1 and pBS-TRX2, respectively. For TRX1, the forward primer was 5'-GCATTAGTGTAATAGAAGACT-3' and the reverse primer was 5'-ATATCGGTCATTGGGTGAGTT-3'. For TRX2, the forward primer was 5'-TACACGAGAGTCTACGATATC-3' and the reverse primer was 5'-CATGATGTACTTTACGTAGCG-3'. Second, the thioredoxin open reading frames of pBS-TRX1 and pBS-TRX2 were subcloned as SpeI/XhoI restriction enzyme fragments into the pASZ11GPD expression vector. The plasmid pASZ11GPD-hTRX (abbreviated pGPD-hTRX in Fig. 4) was made in two steps. First, a PCR product corresponding to the hTRX coding region, but with an NdeI site introduced at the AUG start codon, was cloned into the EcoRV site of pBluescript KS to yield pBS-hTRX (orientation was such that the hTRX start codon was proximal to the pBluescript SacI site). The PCR reaction was done using the plasmid hTrx-pBlue II KS+ (Not1), obtained from Garth Powis (U. of Arizona), as template, the oligonucleotide 5'-CCATATGGTGAAGCAGATCGAGAGC-3' as forward primer (NdeI site underlined), and the oligonucleotide 5'-GAAAACATGATTAGACTAATTC-3' as reverse primer. Second, the thioredoxin open reading frame of pBS-hTRX was subcloned as a SpeI/XhoI fragment into the pASZ11GPD expression vector. The bacterial expression plasmid pET28a-hTRX was constructed by subcloning the thioredoxin open reading frame of pBS-hTRX as a NdeI/XhoI fragment into NdeI/XhoI-cleaved pET28a (Stratagene, La Jolla, CA).
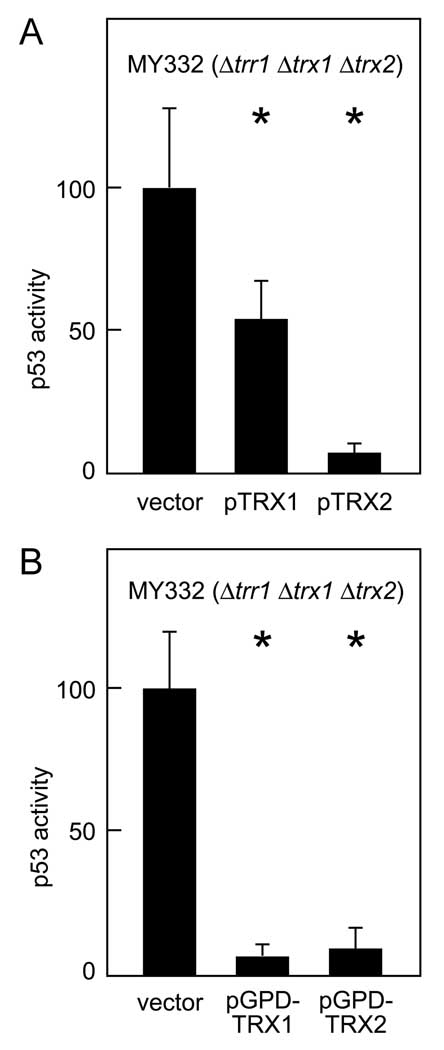
Figure 3.
Restoration of p53 inhibition in Δtrr1 Δtrx1 Δtrx2 yeast by ectopic expression of thioredoxin. For each transformation group, six independent transformants were assayed for β-galactosidase activity. Bars represent activity levels (mean ± S.D.), normalized to the level in MY332 yeast (Δtrr1 Δtrx1 Δtrx2) expressing native p53. Asterisk (*) indicates genotypes where p53 activity in wild-type and mutant yeast differed significantly (p < 0.05, by t-test). A. Restoration of p53 inhibition in Δtrr1 Δtrx1 Δtrx2 yeast transformed with plasmids containing the native TRX1 or TRX2 gene. Yeast strain MY332 (Δtrr1 Δtrx1 Δtrx2) was transformed with a plasmid containing the TRX1 gene (pTRX1), a plasmid containing the TRX2 gene (pTRX2), or an empty pASZ11 vector (vector). Transformants also received the pRS314PGK-p53 effector and pRS316-p53RE-LacZ reporter plasmids. B. Restoration of p53 inhibition in Δtrr1 Δtrx1 Δtrx2 yeast transformed with plasmids expressing the TRX1 or TRX2 coding regions from the constitutive GPD1 promoter. Yeast strain MY332 (Δtrr1 Δtrx1 Δtrx2) was transformed with a plasmid expressing TRX1 from the GPD1 promoter (pGPD-TRX1), a plasmid expressing TRX2 from the GPD1 promoter (pGPD-TRX2), or an empty pASZ11GPD expression vector (vector). Transformants also received the pRS314PGK-p53 effector plasmid and pRS316-p53RE-LacZ reporter plasmid.
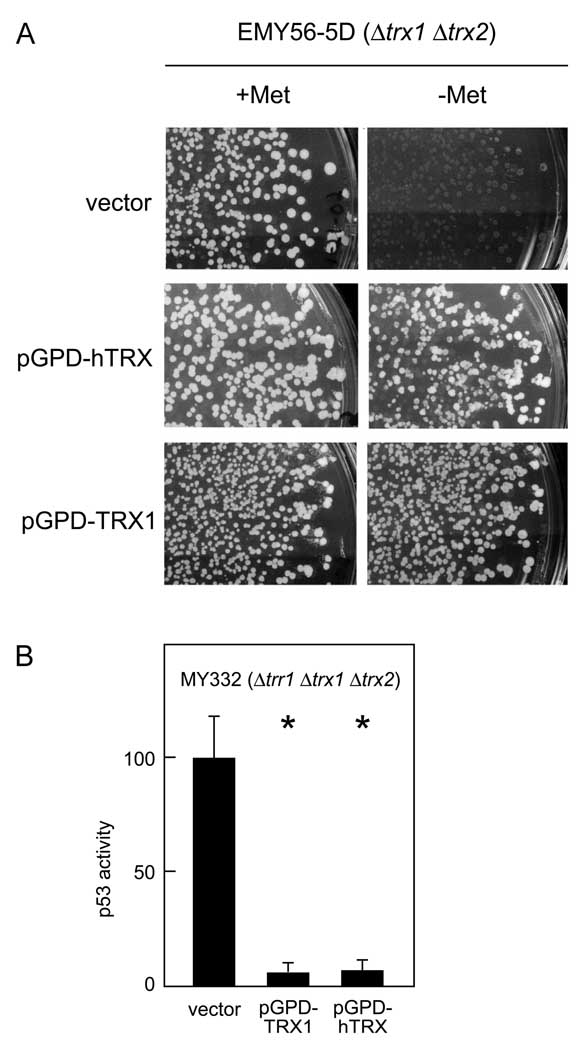
Figure 4.
Complementation of thioredoxin null phenotype by human thioredoxin. A. Effectiveness of human thioredoxin in complementing the methionine auxotrophy of yeast lacking thioredoxin. Strain EMY56-5D (Δtrx1 Δtrx2) was transformed with a plasmid expressing human thioredoxin from the yeast GPD1 promoter (pGPD-hTRX), a plasmid expressing yeast Trx1 from the GPD1 promoter (pGPD-TRX1), or an empty pASZ11GPD expression vector (vector). Primary transformant cultures were replicaplated to medium containing or lacking methionine, and colony re-growth was scored after a 3-day incubation at 25°C. An identical area of each replica plate is shown. B. Effectiveness of human thioredoxin in restoring p53 inhibition in Δtrr1 Δtrx1 Δtrx2 yeast. Strain MY322 (Δtrr1 Δtrx1 Δtrx2) was transformed with a plasmid expressing human thioredoxin from the yeast GPD1 promoter (pGPD-hTRX), a plasmid expressing yeast Trx1 from the GPD1 promoter (pGPD-TRX1), or the empty pASZ11GPD expression vector (vector). All transformants additionally received the pRS314PGK-p53 expression and pRS316-p53RE-LacZ reporter plasmids. For each transformation group, six independent transformants were assayed for β-galactosidase activity. Bars represent activity levels (mean ± S.D.), normalized to the level in MY332 yeast (Δtrr1 Δtrx1 Δtrx2) expressing native p53. Asterisk (*) indicates genotypes where p53 activity in wild-type and mutant yeast differed significantly (p < 0.05, by t-test).
The TRP1-selectable ARS-CEN plasmid expressing human p53 from the yeast PGK1 promoter (pRS314PGK-p53, formerly called pRS314-PGK-SN) and the URA3-selectable ARS-CEN plasmid carrying a p53-responsive LacZ reporter gene (pRS316-p53RE-LacZ, formerly called pRS316-PG-β-gal) have been described previously (19). The integrative reporter plasmid pRS305-p53RE-Z was derived from pRS316-p53RE-LacZ and pRS305. For each p53 mutation, three PCR reactions were done. Primers used are listed in Supplemental Table 2. Using pRS314PGK-p53 (19) as template, the vector forward primer and mutagenic reverse primer were used to generate an upstream PCR subfragment, and the corresponding mutagenic forward primer and vector reverse primer were used to generate a downstream PCR subfragment. A fifty-fold dilution of gel-purified upstream and downstream PCR subfragments were mixed with vector forward and reverse primers and used in a third PCR reaction to generate a full-length fragment containing the mutation. For wild-type p53 control, full-length p53 was amplified from pRS314PGK-p53 using vector upstream and downstream primers. Gel-purified full-length fragments were blunt end ligated into the EcoRV site of pBlueScript KS+ (Stratagene, La Jolla, CA). Ligation mixtures were used to transform DH5α Δlac bacteria, which were plated on LB-IPTG/XGAL plates. White transformant colonies were screened for proper orientation of the p53 alleles in pBluescript KS+, and p53 alleles were subcloned using vector-derived EcoR1 and Sal1 sites into the yeast expression plasmid p414GPD (18). Expression plasmids were purified using Plasmid Midi-Kits (Qiagen, Hilden, Germany). Each allele was sequenced to confirm its sequence and proper insertion into the expression vector. For alleles containing multiple mutations, either natural restriction sites in the p53 coding region were used to make recombinants or additional mutations were serially introduced by PCR site-directed mutagenesis as described above. For mammalian cell studies, mutated p53 alleles were subcloned as EcoR1/Sal1 fragments into the mammalian expression plasmid pTL1 (20), which expresses inserts from the constitutive SV40 promoter.
β-galactosidase assay
β-galactosidase activity was determined by the rapid-freeze/sarkosyl method (21), as described previously (8). Briefly, cells were grown to 107 cells per ml as measured by A600. Aliquots (200 µl) were removed from each culture, frozen at 80°C, and thawed in a 30°C water bath after addition of 400 µl Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM Mg2SO4, 0.2% sarkosyl, 50 mM β–mercaptoethanol, pH 7.0). After 45 min, samples received 150 µl Z-buffer containing 4 mg/ml ortho-nitrophenyl-β-D-galactopyranoside (ONPG) and were incubated an additional 20 min at 37°C. Reactions were stopped by addition of 400 µl 1.5 M Na2CO3 and the extent of ONPG cleavage by β–galactosidase was measured by A420. β–galactosidase activity was normalized by dividing the β–galactosidase activity for each individual sample (A420 value) by the cell density (A600 value). To facilitate comparison between alleles and strains, the mean and standard deviation for each group of normalized β–galactosidase activities was divided by the mean normalized β–galactosidase activity exhibited by wild-type (MY320) cells transformed with wild-type p53 allele (p414GPD–p53).
Interaction of purified human p53 and thioredoxin
Purified baculovirus-expressed human p53 was obtained from Byron Hann (UCSF). Histidine-tagged human thioredoxin was isolated from BL21(DE3) E. coli transformed with the pET28a-hTRX plasmid. Mid-log cultures (50 ml) maintained in LB medium containing 50 µg/ml kanamycin were induced by addition of 1 mM IPTG. After a 3 hr induction at 37°C, bacteria were harvested by centrifugation and lysed by incubation for 20 min at room temperature in 8 ml TALON extraction buffer (50 mM sodium phosphate, pH 7, 300 mM NaCl) containing 0.75 mg/ml lysozyme, followed by three 10 sec sonications on ice at maximal power. Lysate was clarified by centrifugation at 12,000 × g for 20 min and applied to a 0.5 ml slurry of pre-equilibrated TALON metal affinity resin (Clontech, Mountain View, CA). After tumbling in a 15 ml Falcon tube for 20 min at 30°C, the slurry was transferred to a column and washed twice with 5 ml TALON extraction buffer and once with 5 ml TALON extraction buffer supplemented with 5 mM imidazole. Bound protein was eluted using 5 ml TALON extraction buffer supplemented with 150 mM imidizole. Eluate was collected in 500 µl fractions, and fractions containing pure histidine-tagged thioredoxin were identified by SDS-PAGE and pooled. Protein concentration was determined by A280, using a calculated extinction coefficient of 13,700 (1 OD = 856 µg/ml).
For binding assays, 0.8 pmol p53 and 80 to 8,000 pmol histidine-tagged thioredoxin were mixed in 1.5-ml tubes containing 100 µl 0.1 M HEPES, pH 7, 1 mM EDTA buffer. After a 30 min incubation at 37°C, 1 ml TALON extraction buffer and 80 µl 50% TALON resin slurry (pre-equilibrated with TALON extraction buffer) was added, and the mixture was tumbled for 20 min at 30°C. The resin was collected by centrifugation (45 sec at 11,000 × g), and rinsed three times by tumbling for 10 min at 30°C in 1 ml TALON extraction buffer and collecting by centrifugation between rinses. Protein was eluted from the rinsed resin by adding 20 µl TALON elution buffer (50 mM sodium phosphate, pH 7, 300 mM NaCl, 150 mM imidizole, 150 mM EDTA) and incubating samples for 5 min at 25°C. After centrifugation for 60 sec at 11,000 × g, 10 µl of the supernatant was boiled with 3.5 µl SDS-PAGE sample buffer (250 mM Tris, pH 6.8, 4% SDS, 50% glycerol, 280 mM β-mercaptoethanol, 1 mM EDTA, 0.05% bromphenol blue) and applied to a 0.75 mm × 8 cm × 10 cm SDS polyacrylamide gel (9% acrylamide, 0.25% bis-acrylamide). After electrophoresis (1 hr at 35 mA constant current), proteins were transferred (1 hr at 60 mA constant current) to Hybond ECL membrane (Amersham Biosciences, Arlington Heights, IL) using a Trans-blot SD semi-dry transfer cell (BioRad, Hercules, CA) and Towbin transfer buffer (192 mM glycine, 25 mM Tris-OH,, pH 8.6, 20% MeOH, 0.1% SDS). After transfer, blots were incubated 1 hr in TBST (15 mM Tris, pH 7.6, 140 mM NaCl, 0.1% Tween) containing 5% non-fat dried milk as blocking agent. Blots were then incubated 1 hr in TBST containing a 1:10,000 dilution of DO-1 mouse anti-human p53 1° antibody (Santa Cruz Biotechnology, Santa Cruz, CA), washed three times for 10 min in TBST, incubated 1 hr in TBST containing a 1:10,000 dilution of HRP-conjugated goat anti-mouse IgG 2° antibody (Santa Cruz Biotechnology), and washed three times for 10 min in TBST. Antibody binding was detected using ECL chemiluminescence reagents (Amersham Biosciences), and quantitated using a Molecular Dynamics densitometer and ImageQuant software (Amersham Biosciences). Binding data were analyzed using MicroMath Scientist software (Scientific Instrument Services, Ringoes, NJ).
For determination of thioredoxin concentration in MCF-7 cells, 80% confluent cultures were lysed in urea lysis buffer (8M urea, 100 mM Tris, pH 8.2, 1 mM EDTA) supplemented with 3.5 mM DTT and incubated at 37°C for 15 min. Free thiols were alkylated by addition of iodoacetic acid to 10 mM and incubation for 15 min at 37°C. Lysate containing 5 × 105 cell equivalents of protein was resolved by SDS-PAGE, along with 50, 20 and 5 pmol purified histidine-tagged thioredoxin, as described above. Immunoblots were then incubated 1 hr in TBST containing a 1:10,000 dilution of goat anti-human thioredoxin 1° antibody (American Diagnostica, Greenwich, CT), washed three times for 10 min in TBST, incubated 1 hr in TBST containing a 1:10,000 dilution of HRP-conjugated rabbit anti-goat IgG 2° antibody (Accurate, Westbury, NY), and washed three times for 10 min in TBST. Antibody binding was detected using ECL chemiluminescence reagents (Amersham Biosciences), and quantitated using a Molecular Dynamics densitometer and ImageQuant software (Amersham Biosciences). Intracellular thioredoxin concentration was calculated from the immunoblot signal, the number of cell equivalents loaded, an average cell diameter of 13 microns and an estimated cell volume of 1.3 picoliters.
SDS-PAGE and immunoblotting of yeast lysates
Yeast were grown to 107 cells per ml as measured by A600 in YNB. Yeast were collected from 50 ml aliquots of cell cultures by centrifugation, resuspended in SDS-PAGE loading buffer, and transferred to screw-cap microfuge tubes filled with acid-washed 0.6 mm glass beads (Sigma, St. Louis, MO). Cells were disrupted using four 30 sec pulses of a reciprocating bead beater (BioSpec, Bartlesville, OK) set to 5000 rpm, with incubation on ice for 1 min between pulses. Microscopic monitoring showed cell lysis routinely exceeded 95% by this method. Protein concentration was determined by RC-DC assay (Bio-Rad, Hercules, CA). Precast 10% acrylamide SDS-PAGE Tris-Cl gels (Bio-Rad) were used. After transfer to nitrocellulose membranes (Bio-Rad) using a Mini TransBlot instrument (Bio-Rad) at 200 mA overnight (approximately 2.5 amp-hours), total transferred protein was detected by SYPRO Ruby staining and a Molecular Imager FX Pro Plus scanner (Bio-Rad). Membranes were probed for p53 protein using DO-1 antibody as described above.
Isolation of yeast nuclei
Yeast nuclei were isolated by the lyticase spheroplasting method (15). Briefly, yeast were collected by centrifugation, rinsed in wash buffer (50 mM potassium phosphate, pH 7.6, 10 mM MgCl2, 30 mM DTT, 0.5 mM PMSF and 1 M sorbitol), and resuspended in lysis buffer (25 mM potassium phosphate, 25 mM sodium succinate, pH 7.6, 10 mM MgCl2, 30 mM DTT, 0.5 mM PMSF and 1 M sorbitol). Lyticase (Sigma) was added to a final concentration of 0.1 mg/ml and, after a 30 min incubation at 37°C, spheroplasts were collected by centrifugation at 1500 × g, resuspended in lysis buffer without lyticase and disrupted by 10 strokes with a Dounce homogenizer. Nuclei were collected by centrifugation at 1100 × g, resuspended in gradient buffer (50% Percoll, 40 mM PIPES, pH 6.8, 10 mM MgC l2, 0.5 mM PMSF and 0.5% Triton X-100) and centrifuged at 21,000 rpm for 45 min using a Beckman Type 30 rotor. Banded nuclei were removed from tubes, collected at 1100 × g, and washed once in gradient buffer without Percoll. Nuclei were lysed in urea lysis buffer (8M urea, 100 mM Tris, pH 8.2, 1 mM EDTA) supplemented with 3.5 mM DTT and incubated at 37°C for 15 min. Free thiols were alkylated by addition of iodoacetic acid to 10 mM and incubation for 15 min at 37°C. Protein concentration was determined by RC-DC assay, and 10 µg of each nuclear protein sample was resolved by SDS-PAGE for immunoblot analysis. Membranes were probed for p53 protein using DO-1 antibody as described above. Membranes were probed for histone H2A protein using a 1:5000 dilution of rabbit anti-histone H2A 1° antibody (Active Motif, Carlsbad, CA) and a 1:10000 dilution of HRP-conjugated goat anti-rabbit 2°antibody (Bio-Rad, Hercules, CA).
Human cell transfections
Human lung carcinoma line H1299 (22), which is homozygous for a p53 null mutation, was used. Cells were cultured in 50% Dulbecco’s modified Eagle’s medium and 50% Hamm’s F12 medium (F/D), containing 10% bovine serum (HyClone, Logan, UT) and 1% penicillin-streptomycin. For transfection, 5 × 105 cells were dispensed to 1.5-cm wells of 12-well plates and allowed to adhere for 24 hours. Cells were washed free of serum and antibiotic with F/D, and co-transfected using lipofectamine (Stratagene) with 0.5 µg of transfection control plasmid pCMV-β-gal (Invitrogen, Carlsbad, CA), 0.5 µg of p53-responsive luciferase reporter gene plasmid PG-luc (p53-luc, Stratagene), and 0.5 µg of the designated pTL1-p53 expression construct. Culture medium was adjusted to 10% serum at 24 h after transfection. Cells were harvested at 48 h after transfection. Luciferase and β–galactosidase activities were determined using the Luciferase Assay System (Promega, Madison, WI) and ONPG, respectively, using Promega protocols. A liquid scintillation counter, with coincidence correction disabled, was used to quantify luminescence. The luciferase activity of each sample was normalized to the β–galactosidase activity of the sample, and the mean and standard deviation for each group of normalized luciferase activities was divided by the mean normalized luciferase activity exhibited by cells transfected with wild-type p53. Identical conditions were used for analyzing p53 transactivation of alternate luciferase reporter genes (gift from Dr. Byron Hann, UCSF), excepting substitution of 0.5 µg MDM2-luc, 14-3-3-luc, cyclin G-luc or PIG3-luc in place of PG-luc.
Statistical treatments
One-tailed Student’s t-test for unpaired samples was used throughout.
RESULTS
Regulation of p53 activity is dependent on thioredoxin
We previously showed that deletion of the yeast TRR1 gene encoding thioredoxin reductase inhibits the ability of human p53 to transactivate a p53-responsive reporter gene in a yeast model system (8). The defining function of thioredoxin reductase is to catalyze the reduction of the active site disulfide of thioredoxin to the dithiol form, which in turn donates electrons to disulfide-bonded cysteines on target proteins. Deletion of the yeast TRR1 gene shifts the redox state of thioredoxin from primarily reduced (34% in disulfide form) to primarily oxidized (70% in disulfide form), as shown by protein electrophoretic mobility shift assay (13). It was not clear whether inhibition of p53 reporter gene transactivation in Δtrr1 yeast was due to abnormally low levels of reduced thioredoxin or abnormally high levels of oxidized thioredoxin. Reduced thioredoxin may be involved in maintaining p53, or a protein controlling p53, in a reduced and active state. Alternatively, oxidized thioredoxin may be involved in actively converting p53, or a protein controlling p53, to an oxidized and inactive state. Finally, thioredoxin reductase may control p53 activity by a mechanism that does not involve thioredoxin.
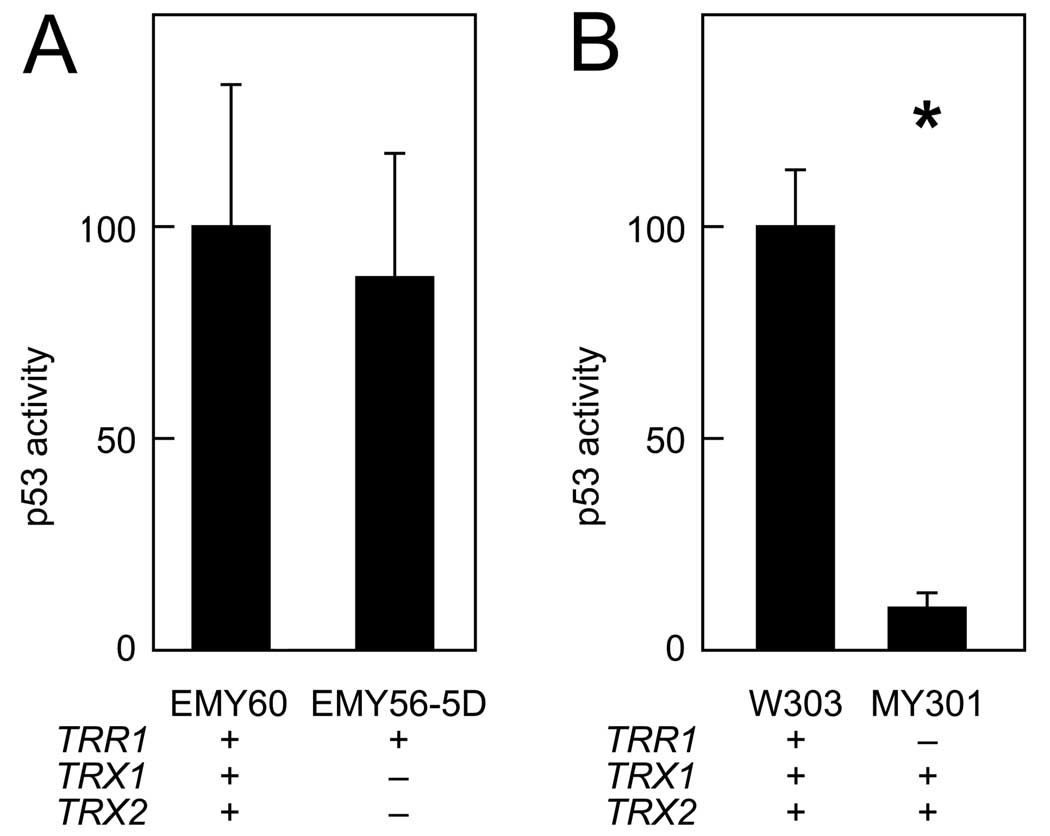
To investigate the first possibility, we determined whether deleting TRX1 and TRX2, the two yeast genes encoding cytosolic thioredoxin, had the same inhibitory effect on p53 activity as deleting the TRR1 gene‥ As shown in Fig. 1A, the Δtrx1 Δtrx2 double mutation resulted in no significant reduction of p53 reporter gene transactivation. Consistent with previous observations (8), the Δtrr1 mutation resulted in a 10-fold reduction in p53-dependent reporter gene transactivation (Fig. 1B). The failure of thioredoxin deletion to mimic the effect of thioredoxin reductase deletion suggested that p53 inhibition in Δtrr1 cells was not due to abnormally low levels of reduced thioredoxin, and was therefore due either to abnormally high levels of oxidized thioredoxin or to disregulation of a pathway independent of thioredoxin.
Figure 1.
Effect of deleting thioredoxin on p53-dependent reporter gene expression. Strains with the indicated genotypes were co-transformed with the pRS314PGK-p53 effector and pRS316-p53RE-LacZ reporter plasmids, and β-galactosidase activity in six independent transformants was assayed. Bars represent activity levels (mean ± S.D.), normalized to the level in wild-type yeast expressing native p53. Asterisk (*) indicates genotypes where p53 activity in wild-type and mutant yeast differed significantly (p < 0.05, by t-test). A. Reporter gene activity in strain EMY56-5D (Δtrx1 Δtrx2) and congenic wild-type strain EMY60. B. Reporter gene activity in strain MY301 (Δtrr1) and congenic wild-type strain W303-1a.
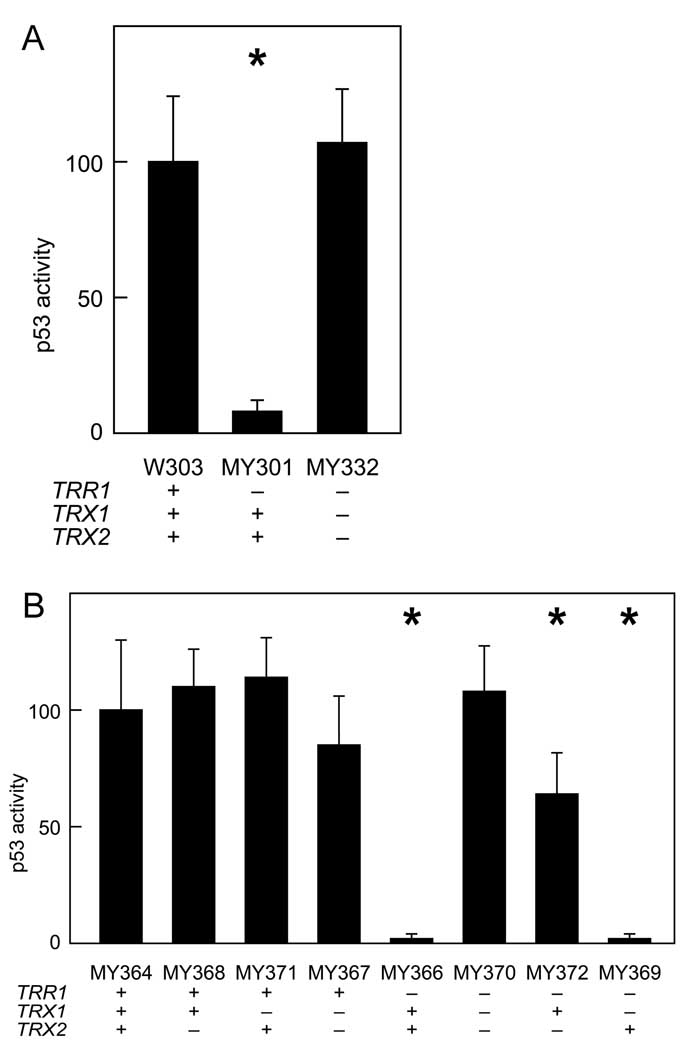
If accumulation of oxidized thioredoxin was necessary for p53 inhibition in Δtrr1 cells, then deletion of the TRX1 and TRX2 genes should suppress the inhibitory effect of deleting the TRR1 gene. To test this prediction, the ability of p53 to transactivate reporter gene expression was determined in Δtrr1 Δtrx1 Δtrx2 cells lacking both thioredoxin reductase and thioredoxin. As shown in Fig. 2A, p53 transactivation of reporter gene expression in Δtrr1 Δtrx1 Δtrx2 triple mutants was as efficient as in wild-type cells. The observation that thioredoxin gene deletion suppressed the inhibitory effect of thioredoxin reductase gene deletion indicated that p53 inhibition in Δtrr1 yeast required thioredoxin and was likely due to the abnormally high levels of oxidized thioredoxin present in these cells. To test whether deletion of both thioredoxin genes was necessary for suppression of the inhibitory effect of the Δtrr1 mutation, a diploid that was heterozygous for the Δtrr1, Δtrx1 and Δtrx2 null mutations was sporulated, and segregants inheriting various combinations of the disrupted alleles were isolated by tetrad dissection. As shown in Fig. 2B, reporter gene activity was inhibited in Δtrr1 Δtrx1 segregants — which retained an intact TRX2 gene — to the same ten-fold extent as in Δtrr1 cells. In contrast, reporter gene activity was inhibited less than twofold in Δtrr1 Δtrx2 segregants — which retained an intact TRX1 gene. The results indicated that the TRX2 gene was a more potent inhibitor of p53 activity than the TRX1 gene in Δtrr1 cells.
Figure 2.
Effect of thioredoxin deletion on p53 activity in thioredoxin reductase null yeast. For each transformation group, six independent transformants were assayed for β-galactosidase activity. Bars represent activity levels (mean ± S.D.), normalized to the level in wild-type yeast expressing native p53. Asterisk (*) indicates genotypes where p53 activity in wild-type and mutant yeast differed significantly (p < 0.05, by t-test). A. Effect of deleting thioredoxin on p53-dependent reporter gene expression in thioredoxin reductase null cells. Strains with the indicated genotypes were co-transformed with the pRS314PGK-p53 effector and pRS316-p53RE-LacZ reporter plasmids. B. Levels of p53-dependent reporter gene expression in yeast inheriting different combinations of the Δtrr1, Δtrx1 and Δtrx2 null alleles. A diploid that was heterozygous for null mutations in the TRR1, TRX1 and TRX2 genes was sporulated, and tetrad segregants representing the eight possible haplotypes were isolated and co-transformed with the pRS314PGK-p53 effector and pRS316-p53RE-LacZ reporter plasmids.
The results in Fig. 2B were obtained with segregants isolated from a diploid that was heterozygous for the Δtrr1, Δtrx1 and Δtrx2 mutations. To confirm that the observed differences in reporter gene expression were due to the presence or absence of the Δtrr1, Δtrx1 and Δtrx2 alleles, and not to cryptic mutations or compensatory epigenetic changes that were selected during strain derivation, the wild-type TRX1 or TRX2 genes were ectopically expressed from plasmids pTRX1 or pTRX2 in a Δtrr1 Δtrx1 Δtrx2 triple mutant. As shown in Fig. 3A, ectopic expression of TRX1 gave 50% inhibition of p53 activity, and ectopic expression of TRX2 gave 90% inhibition. Thus, the results obtained when thioredoxin genes were restored on a plasmid (Fig. 3A) closely matched those obtained when thioredoxin genes were restored by mating and tetrad dissection (Fig. 2B). In both cases, re-introduction of TRX2 fully reinstated p53 inhibition, and re-introduction of TRX1 partially reinstated p53 inhibition.
Weaker inhibition by TRX1 could be due to an intrinsic difference in the substrate specificity of the Trx1 and Trx2 proteins. Alternatively, it could be due to differences in the level of expression of the TRX1 and TRX2 genes. Indeed, whole genome expression assays, as well as northern blot analyses, have shown that TRX2, but not TRX1, is induced several fold in Δtrr1 cells (23). Immunoblots suggest that levels of Trx2 protein, but not Trx1 protein, are elevated in Δtrr1 yeast (data not shown). To distinguish whether differential inhibition of p53 activity by TRX1 and TRX2 was due to different levels of gene expression or to intrinsic differences in the substrate selectivity of the encoded proteins, the TRX1 and TRX2 coding regions were placed under the control of the constitutively active GPD1 promoter. As shown in Fig. 3B, when TRX1 and TRX2 were expressed from this heterologous promoter, both thioredoxins were equally effective in inhibiting p53 activity in Δtrr1 Δtrx1 Δtrx2 yeast. The results thus suggested that there was no intrinsic difference between the Trx1 and Trx2 polypeptides in the degree to which they affected p53 activity, and that the earlier noted stronger inhibition by the native TRX2 gene was likely due to higher levels of TRX2 expression in Δtrr1 cells.
As a means of testing the relevance of results obtained in yeast to higher systems, we investigated whether human thioredoxin was functional when expressed in yeast and whether it could substitute for yeast thioredoxin in inhibiting p53 activity in thioredoxin reductase null cells. Yeast lacking thioredoxin are unable to reduce sulfate via PAPS reductase and are thus auxotrophic for methionine. To confirm the functionality of human thioredoxin in yeast, a Δtrx1 Δtrx2 strain was transformed with a plasmid expressing a human thioredoxin cDNA from the yeast GPD1 promoter, and transformants were checked for growth in the absence of methionine. As shown in Fig. 4A, Δtrx1 Δtrx2 yeast transformed with a plasmid expressing either human thioredoxin (pGPD-hTrx) or yeast thioredoxin (pGPD-TRX1) were able to grow in the absence of methionine, but yeast transformed with an empty pASZ11GPD expression vector were not. Thus, human thioredoxin was able to serve as a functional homolog of yeast thioredoxin in regard to restoration of PAPS reductase function.
To determine whether human thioredoxin functions as an inhibitor of p53 activity in the absence of thioredoxin reductase, a Δtrr1 Δtrx1 Δtrx2 yeast strain was transformed with the human thioredoxin expression plasmid, and transformants were assayed for p53-dependent reporter gene transactivation. As shown in Fig. 4B, transformation with the human thioredoxin plasmid pGPD-hTRX gave the same ten-fold inhibition of p53 activity in Δtrr1 Δtrx1 Δtrx2 cells as transformation with the yeast thioredoxin plasmid pGPD-TRX1. The observation that human p53 activity was sensitive to the redox state of human thioredoxin in the genetically tractable yeast system supported the idea that p53 activity could be sensitive to the redox state of thioredoxin in human cells.
Purified thioredoxin and p53 interact at physiological thioredoxin concentrations
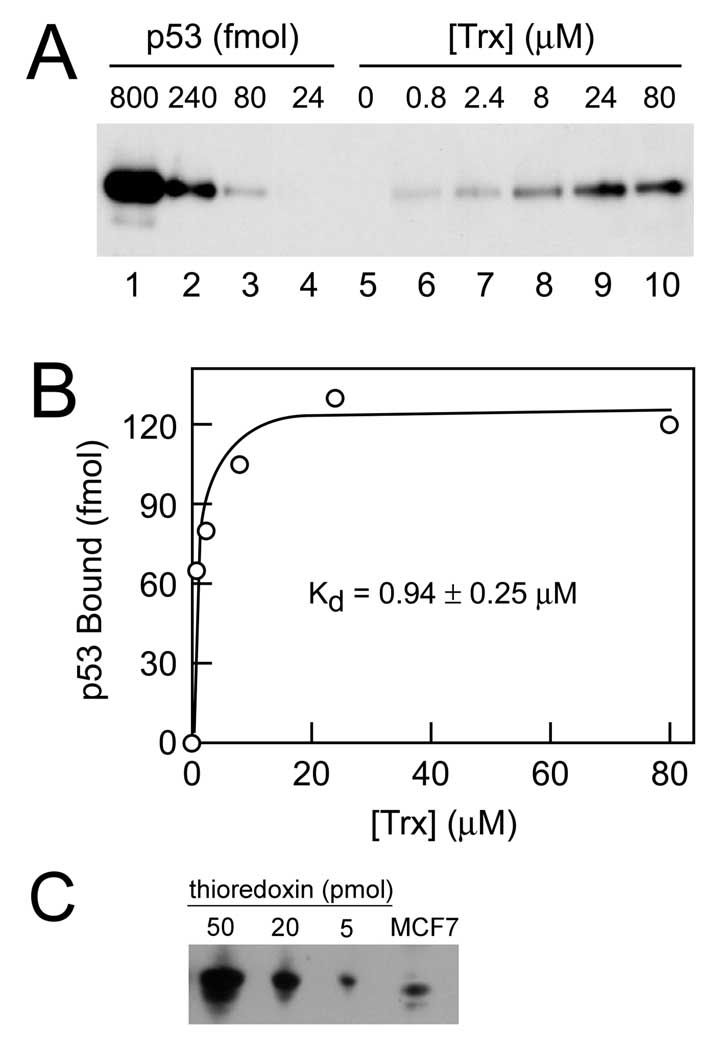
Inhibition of p53 activity in Δtrr1 yeast could be due to either a direct or indirect effect of oxidized thioredoxin on p53 activity. Co-immunoprecipitation experiments have thus far failed to detect a physical interaction between thioredoxin and p53 in vivo (data not shown). As an alternative approach to demonstrate a direct interaction between thioredoxin and p53, increasing amounts of purified histidine-tagged human thioredoxin (80–8000 pmol; final concentration of 0.8–80 µM) were incubated with purified recombinant human p53 (0.8 pmol; final concentration of 8 nM), and binding of p53 to thioredoxin was measured by cobalt resin precipitation and immunoblotting. Typical results of such a pull-down assay are shown in Fig. 5A. Dilutions of the input p53 added to the binding reactions (lanes 1–4) allowed construction of a standard curve relating densitometric signal intensity to p53 level. No p53 was detected in the cobalt resin precipitate when thioredoxin was omitted from the binding reaction (lane 5). When increasing amounts of thioredoxin were present during the binding reaction, increasing amounts of p53 were detected in the cobalt resin precipitate (lanes 6–10). Quantitation of the amount of p53 bound at each thioredoxin concentration is shown in Fig. 5B. Binding data yielded a Kd of 0.94 µM thioredoxin and a Bmax of 0.12 picomoles p53, which corresponded to 15% of the p53 polypeptide added to the binding reaction. Immunoblot analyses on human MCF-7 lysates indicated that the cellular levels of thioredoxin were in the 10 µM range (Fig. 5C). Thus, the interaction between purified p53 and thioredoxin shown in Fig. 5A and B was achieved at physiological thioredoxin concentrations.
Figure 5.
Physical interaction of purified human p53 and thioredoxin. A. Indicated concentrations of histidine-tagged human thioredoxin (lanes 4–10) were incubated with 0.8 pmol of baculovirus-produced human p53, and the amount of p53 bound by thioredoxin was determined by precipitation with TALON resin and immunoblotting. Dilutions of the input p53 added to the binding reactions were analyzed in parallel lanes (lanes 1–4). B. Densitometry was used to determine the amount of p53 bound at each thioredoxin concentration. Analysis of the binding data using MicroMath curve-fitting software indicated 15% of the p53 in the preparation was competent to bind thioredoxin and that the Kd ± S.D. for binding was 0.94 ± 0.25 µM. C. Quantitative immunoblot analysis of thioredoxin concentration in MCF-7 cells. The intensity of thioredoxin signal in 5 × 105 cell equivalents of MCF-7 cell lysate (lane 4) was compared to a standard curve generated using indicated amounts purified histidine-tagged thioredoxin (lanes 1–3). An intracellular thioredoxin concentration of 15 µM was calculated based a cell diameter of 13.5 µm and an estimated cell volume of 1.3 picoliters.
In addition to in vitro binding assays, we used a yeast one-hybrid interaction assay to test whether thioredoxin interacted with p53 in vivo. Strong transactivation of a Lex operator-containing reporter gene was observed in yeast transformed with plasmids encoding human p53 and a LexA-Trx1 fusion protein, but not in yeast transformed with plasmids encoding human p53 and unfused LexA, or a plasmid encoding a LexA-Trx1 fusion protein and an empty vector carrying no p53 allele (Table 1). The yeast data, combined with the in vitro binding data obtained at physiological thioredoxin concentrations, suggested that thioredoxin and p53 interacted in vivo.
Table 1.
Interaction of thioredoxin and p53 in yeast one-hybrid assay
| Transformation | Plasmids a | β-gal activity b |
|---|---|---|
| nmol ONP | ||
| min 107 cells | ||
| 1 | pRS415-p53 (p53) pBTM116-Trx (LexA-Trx) |
6.56 ± 0.42 |
| 2 | pRS415-p53 (p53) pBTM116 (LexA) |
0.72 ± 0.14 |
| 3 | pRS415 (none) pBTM116-Trx (LexA-Trx) |
0.65 ± 0.22 |
| 4 | pBTM116-p53 (LexA-p53) | 64.0 ± 4.16 |
Yeast were transformed with a LexA-operator/β-galactosidase reporter gene plasmid (pAS18–35) and the indicated effector plasmids. The protein expressed from the effector plasmid is shown in parentheses.
Three independent transformants from each transformation were assayed for β-galactosidase activity (mean ± SD).
Effects of cysteine replacement on p53 activity in yeast
One model consistent with the data thus far obtained was that p53 contains oxidation-prone cysteines that form intramolecular or intermolecular disulfide bonds that result in p53 inactivation. Under the reducing environment prevailing in non-stressed wild-type cells, disulfide formation is antagonized and active p53 predominates. Conversely, under the oxidizing environment prevailing in oxidatively stressed cells or cells lacking thioredoxin reductase, disulfide bond formation is favored and inactive p53 predominates. If this model is correct, changing the redox-sensitive cysteines to structurally similar but non-oxidizable serines should make the protein resistant to oxidative inactivation.
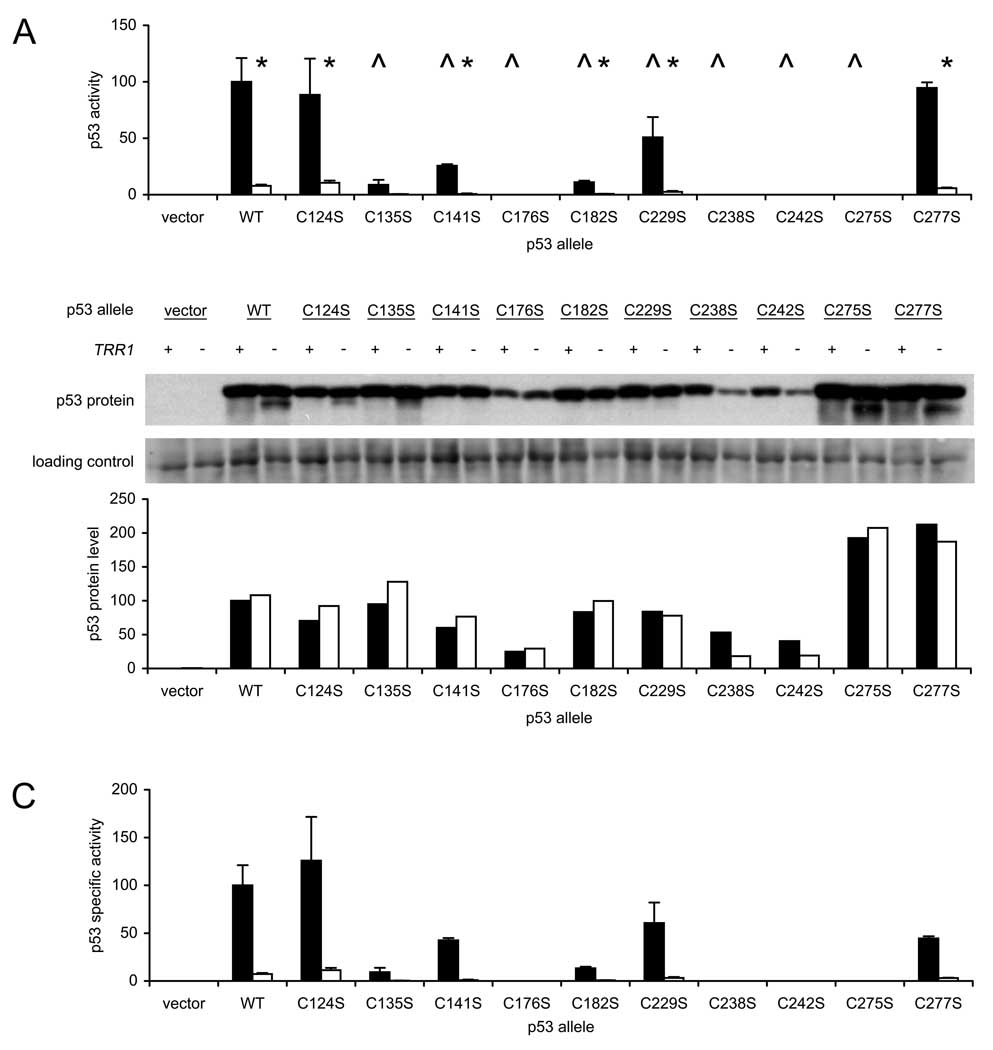
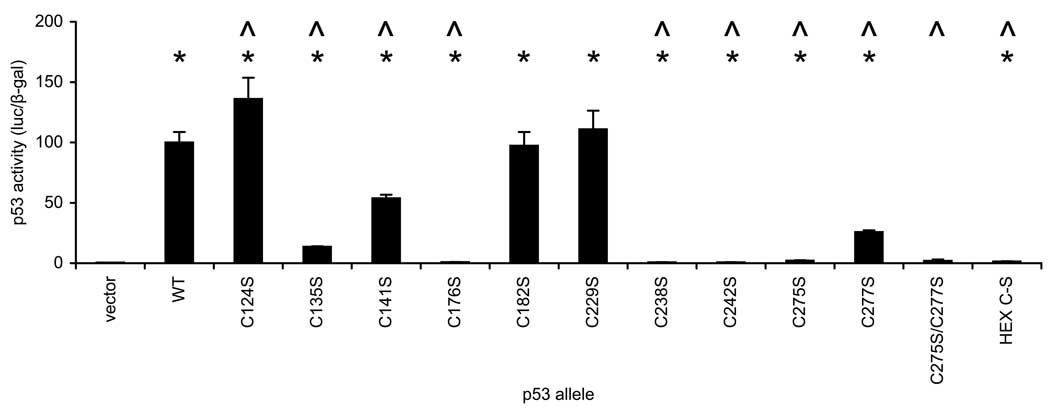
To identify oxidation-prone p53 cysteines, a series of mutant alleles was constructed in which each Cys codon was individually changed to a Ser codon by site-directed mutagenesis. Each allele was expressed in wild-type (MY320) and Δtrr1 (MY321) yeast that contained an integrated p53-dependent LacZ reporter gene (pRS305-p53RE-Z), and the ability of each p53 allele to stimulate reporter gene expression was determined (Fig. 6A). Excepting the C124S and C277S replacements, all mutated p53 alleles exhibited significantly lower activity than the native gene. Four p53 alleles (C176S, C238S, C242S and C275S) were completely inactive in transactivating the reporter gene. Three of the inactive alleles corresponded to replacements of zinc-coordinating residues Cys 176, 238 and 242, the murine homologs of which were known previously to be essential for p53 activity (3). Unexpectedly, replacement of Cys 275 with Ser also completely abrogated reporter gene transactivation. The absence of activity for the C176S, C238S, C242S and C275S alleles precluded assessment of thioredoxin reductase dependence in these cases. Six p53 alleles (C124S, C135S, C141S, C182S, C229S and C277S) efficiently stimulated reporter gene activity in wild-type yeast but not Δtrr1 yeast. The ratio of p53 activity in wild-type versus Δtrr1 yeast for these functional p53 alleles was similar to that observed for native p53. Thus, no single Cys-to-Ser replacement of these six residues relieved the dependence of p53 on thioredoxin reductase.
Figure 6.
Activity and thioredoxin reductase-dependence of p53 alleles carrying single Cys-to-Ser mutations. A. Wild-type (TRR1, dark bars) and thioredoxin-null (Δtrr1, light bars) yeast carrying an integrated p53-dependent LacZ reporter gene were transformed with single-copy plasmids expressing the indicated p53 allele, and three independent transformants were assayed for β-galactosidase activity. Bars represent activity levels (mean ± S.D.), normalized to the level in wild-type yeast expressing native p53. Carat (^) indicates genotypes where activity of native and mutated p53 alleles differed significantly when expressed in wild-type yeast (p < 0.05, by t-test). Asterisk (*) indicates genotypes where p53 activity in wild-type and mutant yeast differed significantly (p < 0.05, by t-test). B. Immunoblot analysis of p53 protein levels in yeast transformed with mutated p53 alleles and specific activity of each protein in transactivating reporter gene expression. Equal amounts of lysate protein (20 µg) from representative transformants was resolved by SDS-PAGE, and p53 protein was detected by immunoblotting using DO-1 antibody (upper panel). An 80-kDa protein, detected by SYPRO Ruby staining prior to immunostaining, was used as a loading control (center panel). The relative amount of p53 protein in each sample was calculated by dividing the p53 band intensity by the 80-kDa protein band intensity, and dividing the resulting ratio by the ratio observed in wild-type yeast expressing native p53. TRR1, dark bars; Δtrr1, light bars (lower panel). C. The specific activity of p53 in stimulating reporter gene expression was calculated by dividing the β-galactosidase levels shown in Figure 6A by the p53 protein levels determined in Figure 6B. Bars represent mean ± S.D. for three transformants.
To confirm that reduced reporter expression in Δtrr1 yeast transformed with mutated p53 alleles was due to a reduction in p53 specific activity, rather than p53 protein levels, immunoblot assays were used to measure p53 protein levels (Fig. 6B, upper panel). To correct for variation in loading between samples, a prominent 80-kD protein in each lane was quantified by SYPRO Ruby blot staining (Fig. 10B, lower panel) and used to normalize p53 protein levels (Fig. 6C). Mutation of the Zn-coordinating cysteines (C176, C238 and C242) resulted in lower protein levels, suggesting that protein stability was affected. However, the other seven mutated alleles expressed protein at levels roughly equivalent to that of the wild-type p53 allele. Importantly, the Δtrr1 deletion mutation had no consistent inhibitory effect on p53 protein levels. Thus, decreased reporter gene activity in thioredoxin reductase null cells was due to reduced p53 specific activity rather than reduced p53 protein levels.
Figure 10.
Analysis of alternative p53 alleles carrying codon 275 mutations. A. Activity and thioredoxin reductase-dependence of p53 allele carrying Cys-to-Ala rather than Cys-to-Ser mutation at residue 275. Wild-type (TRR1, dark bars) and mutant (Δtrr1, light bars) yeast carrying an integrated p53-dependent reporter gene were transformed with indicated p53 alleles, and β-galactosidase activity in three independent transformants was determined. Bars represent activity levels (mean ± S.D.), normalized to the level in wild-type yeast expressing native p53. B. Activity and thioredoxin reductase-dependence of p53 allele carrying a potentially compensatory C277S mutation in addition to a C275S mutation. Wild-type (TRR1, dark bars) and mutant (Δtrr1, light bars) yeast carrying an integrated p53-dependent reporter gene were transformed with indicated p53 alleles, and β-galactosidase activity in three independent transformants was determined. Bars represent activity levels (mean ± S.D.), normalized to the level in wild-type yeast expressing native p53.
Effect of combinatorial Cys-to-Ser mutations on p53 activity
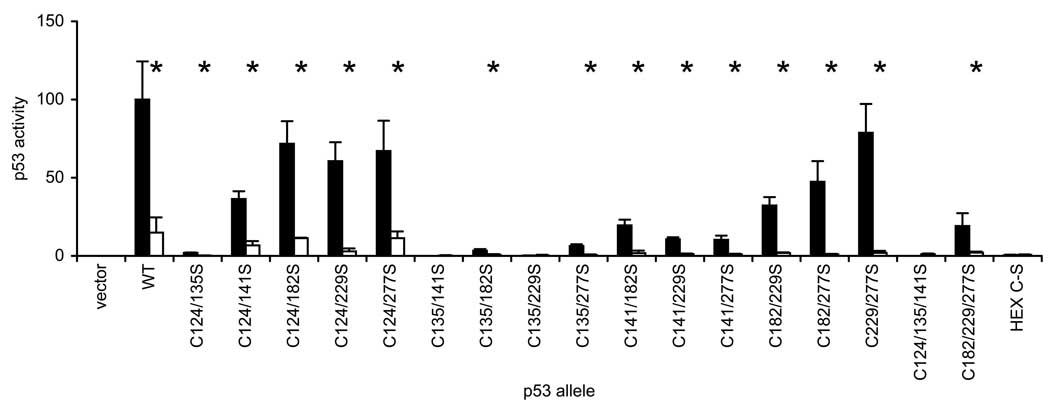
One explanation for continued thioredoxin reductase dependence of p53 alleles carrying single Cys-to-Ser mutations was that more than a single cysteine or pair of cysteines was prone to oxidation. To test this idea, a series of p53 alleles was constructed comprising all pair-wise combinations of Cys-to-Ser mutations, excepting the essential residues C176, C238, C242 and C275. In addition, alleles containing three replacements (C124/135/141S and C182/229/277S) or six replacements (C124/135/141/182/229/277S, labeled HEX C–S) were also constructed. All alleles were tested for reporter gene transactivation in MY320 and MY321 yeast (Fig. 7). All alleles that retained p53 activity (C124/135S, C124/141S, C124/182S, C124/229S, C124/277S, C135/182S, C135/C277S, C141/182S, C141/229S, C141/277S, C182/229S, C182/277S, C229/277S, and C182/229/277S) remained thioredoxin reductase dependent. No pair-wise combination of C124S, C141S, C182S, C229S and C277S mutations was sufficient to relieve dependence on thioredoxin reductase. In addition, the pair-wise combinations involving the C135S mutation and either the C182S or C277S mutations were active and regulated. We therefore concluded that putative oxidations involving these combinations of cysteines were not the basis for thioredoxin reductase dependence. However, several combinations (C135/141S, C135/229S, C124/135/141S and C124/135/141/182/229/277S) resulted in severe impairment of reporter gene activation, which precluded assessment of thioredoxin reductase dependence. A common feature of the inactive combinatorial alleles was the C135S mutation, which by itself gave a 20-fold decrease in activity (Fig. 6), and in combination with C141S or C229S exhibited near-complete elimination of activity (Fig. 7). We therefore could not exclude the possibility that putative pair-wise oxidations involving Cys 135 and either Cys 141 or Cys 229 were the basis for thioredoxin reductase dependence based on this data.
Figure 7.
Activity and thioredoxin reductase-dependence of p53 alleles with combinatorial Cys-to-Ser mutations. Wild-type (TRR1, dark bars) and thioredoxin-null (Δtrr1, light bars) yeast carrying an integrated p53-dependent LacZ reporter gene were transformed with single-copy plasmids expressing indicated p53 alleles, and three independent transformants were assayed for β-galactosidase activity. Bars represent activity levels (mean ± S.D.), normalized to the level in wild-type yeast expressing native p53. Asterisk (*) indicates genotypes where p53 activity in wild-type and mutant yeast differed significantly (p < 0.05, by t-test).
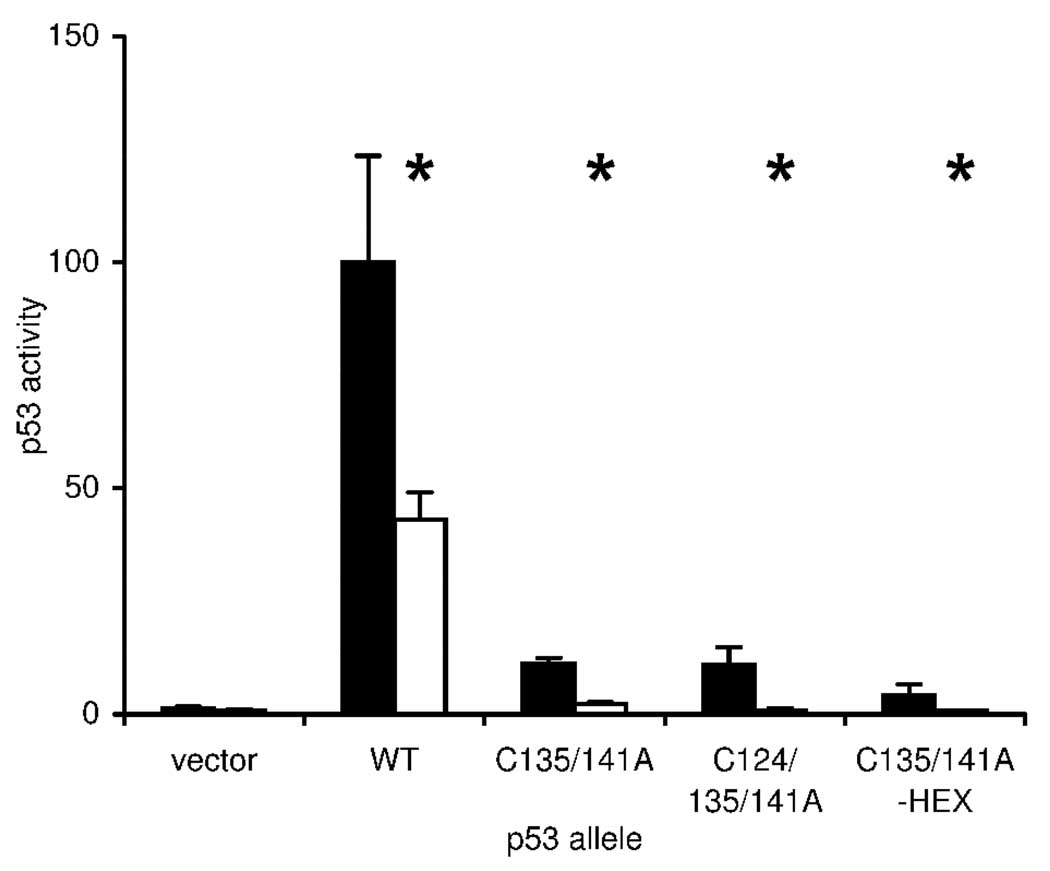
As C124, C135 and C141 reside in a buried helix-loop-helix motif (24), it was possible that serine replacement at these positions was deleterious to p53 activity due to destabilization of native structure. If so, then alanine replacement would be more compatible with proper packing and might preserve activity. We therefore constructed C135/141A, C124/135/141A and C135/141A-C124/182/229/277S (henceforth C135/141A-HEX) alleles and assessed reporter activity in MY320 and MY321 yeast (Fig. 8). In all cases, alanine replacement produced an allele with higher activity than the corresponding cysteine replacement. Importantly, all alanine replacement alleles remained dependent on thioredoxin reductase, with C135/141A and C124/135/141A exhibiting greater than ten-fold regulation of reporter gene expression and C135/141A-HEX maintaining over six-fold regulation. Thus, it was unlikely that p53 cysteines 124, 135, 141, 182, 229 and 277 were prone to oxidation and were the basis for p53 dependence on thioredoxin reductase in yeast.
Figure 8.
Activity and thioredoxin reductase-dependence of p53 alleles carrying combinatorial Cys-to-Ala mutations. Wild-type (TRR1, dark bars) and thioredoxin-null (Δtrr1, light bars) yeast carrying an integrated p53-dependent LacZ reporter gene were transformed with single-copy plasmids expressing indicated p53 alleles, and three independent transformants were assayed for β-galactosidase activity. Bars represent activity levels (mean ± S.D.), normalized to the level in wild-type yeast expressing native p53. Asterisk (*) indicates genotypes where p53 activity in wild-type and mutant yeast differed significantly (p < 0.05, by t-test).
Nuclear localization of p53 in yeast
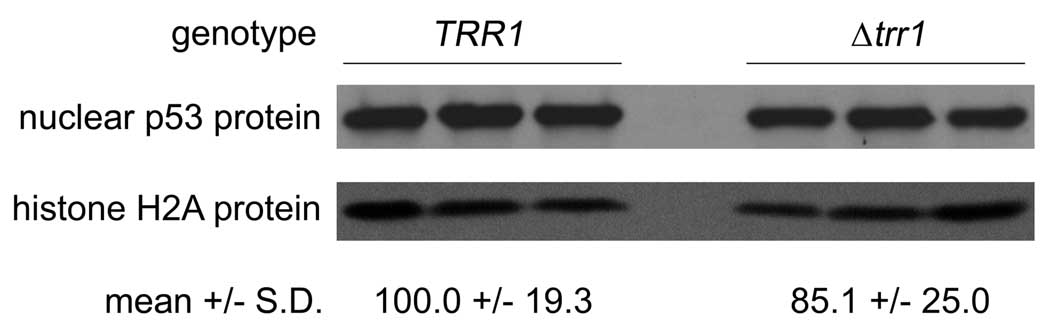
Thioredoxin reductase gene deletion produces a wide variety of gene expression changes (23), including reduction of messages for several nuclear import proteins (karyopherins, importins). Therefore, differential import of p53 into the nucleus in TRR1 and Δtrr1 yeast was investigated as a possible explanation for the observed differences in p53 activity. To test this idea, spheroplasts of wild-type and Δtrr1 yeast transformed with native p53 were prepared, and isolated nuclei were assayed for p53 protein by immunoblotting (Fig. 9). Levels of histone H2A protein were also determined, and served as a nuclear protein loading control. A 15% decrease in normalized p53 protein levels was observed in mutant nuclei, but the decrease was not significant (p = 0.23). Thus, p53 mislocalization, while possibly a contributing factor, was not responsible for the majority of the 1400% reduction in p53 activity in mutant yeast.
Figure 9.
Nuclear levels of p53 in wild-type and Δtrr1 yeast transformants. Equal amounts of nuclear protein (20 µg) from triplicate samples of wild-type (TRR1) and mutant (Δtrr1) yeast transformed with wild-type p53 allele were resolved by SDS-PAGE, and p53 and histone H2A were detected by immunoblotting. Numbers below each set of lanes show densitometric intensity of p53 bands normalized to histone H2A bands (mean ± S.D.), with wild-type results arbitrarily set at 100 and Δtrr1 ratio normalized accordingly.
Analysis of essential cysteine residue C275
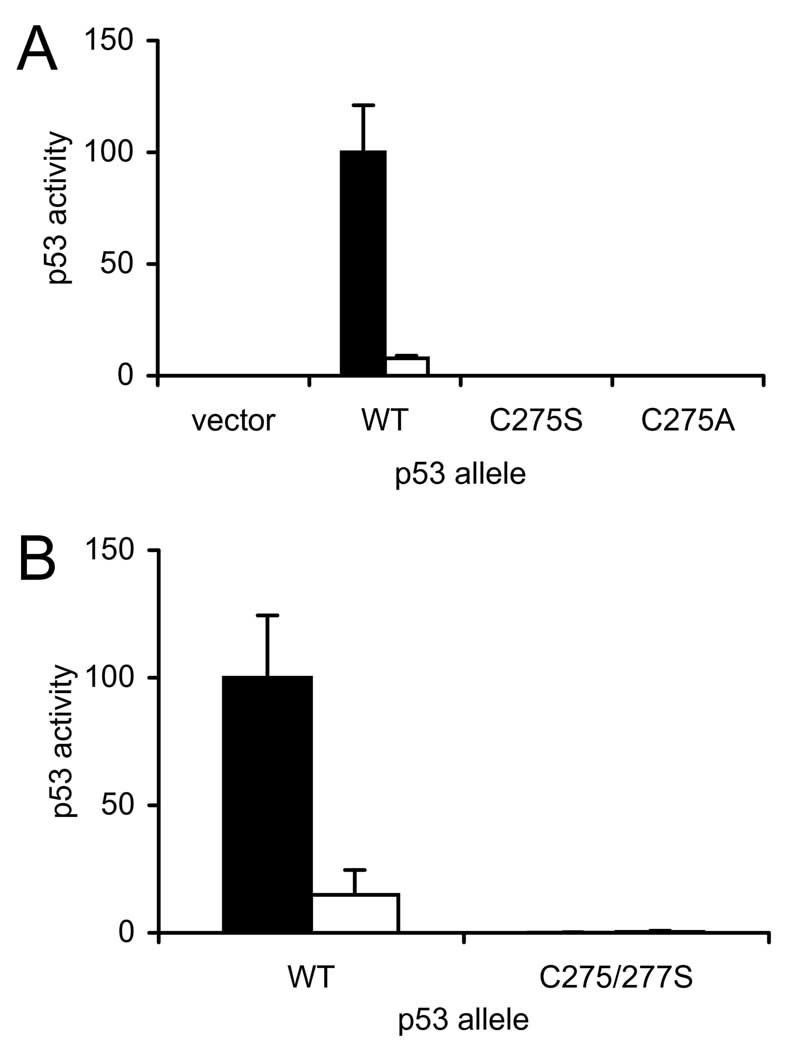
The inactivating effect of the C275S mutation was unexpected. Therefore, several confirmatory experiments were performed. Although the structure of all mutated p53 alleles had been confirmed by sequencing, it was possible that an alteration elsewhere in the plasmid expressing the C275S allele resulted in the observed phenotype. To eliminate this possibility, the C275S allele was independently reconstructed. In protein folding and crystallography studies, alanine rather than serine is often used when replacing cysteines, with the idea that a small hydrophobic methyl group is less likely to disrupt protein structure than a polar hydroxyl group. Thus, we constructed a p53 allele in which Cys 275 was replaced with Ala. As shown in Fig. 10A, the independently-reconstructed C275S allele was inactive, and the Cys-to-Ala mutation had the same inactivating effect on p53 activity as the Cys-to-Ser mutation. Finally, replacement of Cys 275 with Ser was shown to inactivate p53 reporter gene transactivation in human cells (see below). We therefore concluded that Cys 275 was essential for p53 activity.
The inactivating effect of the C275 replacement was intriguing because C275 and C277 were within disulfide bonding distance of each other and were part of a sheet-loop-helix motif that contacts DNA (24). It was possible that interactions between these vicinal residues were involved in regulating p53 activity. For example, activation of the yeast transcription factor Yap1 during oxidative stress is known to involve transient formation of an intermolecular disulfide between Yap1 residue Cys 598 and glutathione peroxidase 3 (Gpx3) residue Cys 36, which is subsequently resolved by reaction with a second Yap1 residue, Cys 303. The resulting intramolecular Cys 595-Cys 303 disulfide results in Yap1 activation (25). Similarly, p53 residue Cys 277 may tend to form an inactivating disulfide with another polypeptide or thiol compound in the cell and may need to be resolved by Cys 275. Another study showed that binding of p53 to the GADD45 gene response element is much more transient than binding to the p21 gene response element due to different bases at position 3 in the fourth p53 binding site pentamer (GADD45, 5'-TGCTG-3'; p21, 5'-TGTTG-3') (26). Changing Cys277 to Ser stabilizes the binding of p53 to the GADD45 response element, suggesting the possibility that Cys277 is prone to oxidation and that oxidation of this residue differentially affects the affinity of p53 for various p53 response elements. A third study, using cells expressing a truncated p53 peptide containing C275 and C277 but no other cysteines, demonstrated increased reactivity of the peptide toward thiol reagents when cells were treated with selenomethionine (27).
To investigate the possibility that C277 forms an inactivating disulfide resolved by C275, a C275S/277S double mutation was constructed. If Cys 275 was essential because it resolved an intermolecular disulfide involving Cys 277, mutation of Cys 277 to Ser should prevent disulfide trapping and thereby relieve the requirement for Cys 275. In contrast to this expectation, the doubly mutated C275/277S allele, like the singly mutated C275S allele, was completely inactive in stimulating reporter gene expression (Fig. 10B). The absence of transactivation by the doubly mutated p53 allele precluded making conclusions concerning the existence of redox communication between C275 and C277. However, the essential nature of the C275 residue for p53 transactivation activity must stem from factors other than disulfide trapping with C277.
Effect of cysteine replacements on p53 activity in human cells
The ability of native p53, each single Cys-to-Ser replacement allele, the C275/277S double replacement allele and the C124/135/141/182/229/277S hextuple replacement allele (HEX C-S) to transactivate reporter gene expression was assessed in co-transfected human H1299 cells that lack an endogenous p53 gene (Fig. 11). The relative levels of reporter gene transactivation exhibited by the mutated p53 alleles in H1299 cells were generally similar to that previously observed in yeast (compare Fig. 11 to Fig. 6). Exceptions include C182S and C229S, which exhibited essentially wild-type activity in the H1299 system; C277S, which exhibited significantly lower activity than native p53 in the H1299 system; and C124S, which exhibited significantly higher activity than native p53 in the H1299 system. Unexpectedly, the high sensitivity of the luciferase reporter system revealed very low (<2% of wild-type activity) but significant residual transactivation activity by the C176S, C238S, C242S and C275S alleles.
Figure 11.
Effect of p53 Cys-to-Ser mutations on reporter gene transactivation in co-transfected human cells. Human H1299 cells, which lack an endogenous p53 gene, were co-transfected with a pTL1-effector plasmid expressing the indicated p53 allele, a p53-dependent luciferase reporter gene (PG-luc), and a CMV promoter-driven LacZ transfection control plasmid (pCMV-β-gal). Luciferase activity and β-galactosidase activity in three independent transfectants was determined. Bars represent luciferase activity normalized to β-galactosidase activity activity (mean ± S.D.), relative to activity in cells expressing wild-type p53. Carat (^) indicates genotypes where activity of native and mutated p53 alleles differed significantly (p < 0.05, by t-test). Asterisk (*) indicates genotypes where activities of p53 allele and vector control differed significantly (p < 0.05, by t-test).
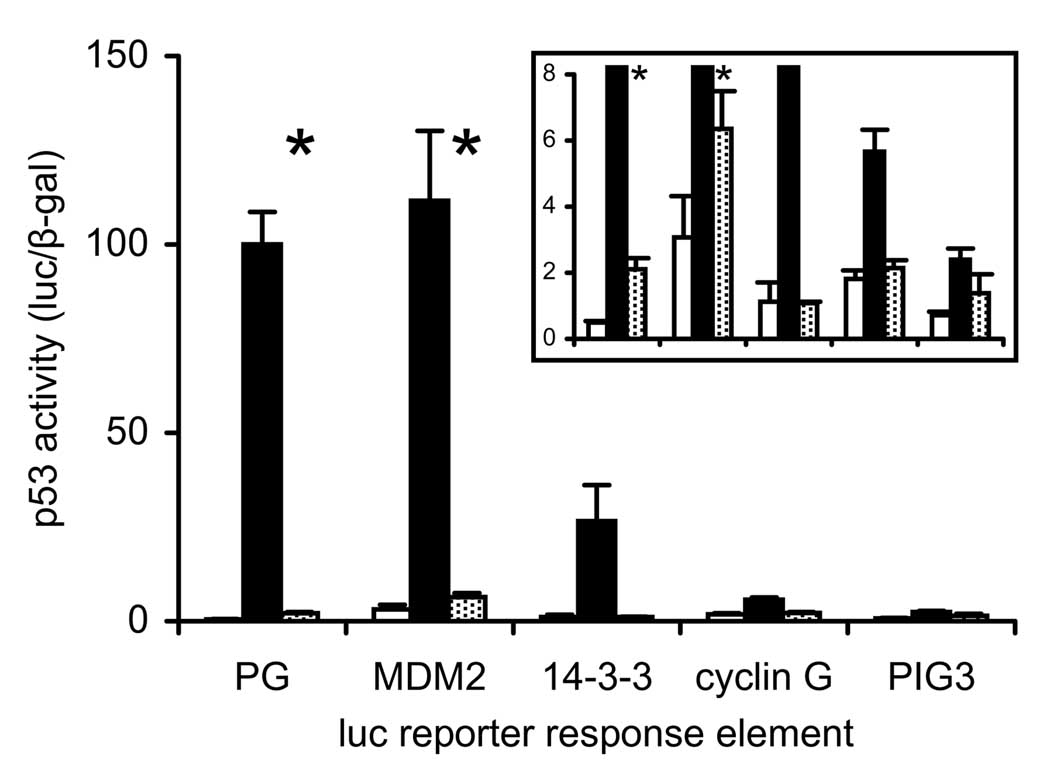
The possibility that Cys 275 was essential for discriminating between specific DNA binding sites was considered. As the reporter constructs used in all of the above analyses were based on the PG repeat binding site (19, 28, 29), it was possible that Cys 275 had higher affinity for other p53 response element sequences. This would not be unprecedented, as mutation of p53 Cys 277 was previously shown to alter the affinity with which p53 binds different p53 target sequences (26, 30). We therefore transfected H1299 cells with either the wild-type or C275S p53 allele, along with an array of p53-responsive luciferase reporter constructs containing the p53 binding sites from different p53 target genes (MDM2, 14-3-3, cyclin G and PIG3). The previously analyzed PG-luc reporter was included for comparison to earlier results. The C275S allele gave very low but significant transactivation of the PG-luc and MDM2-luc reporters over vector control (1.6% and 3.0% of wild-type p53, respectively), but gave no significant transactivation of the 14-3-3-1-luc, cyclin G-luc and PIG3-luc reporters (Fig. 12). Thus, the data suggests that mutation of C275 is likely to be strongly inactivating independent of response element sequence.
Figure 12.
Effect of wild-type and C275S p53 alleles on transactivation of reporter genes carrying p53 response elements from different p53 target genes. H1299 cells were co-transfected with the pCMV-β-gal transfection control plasmid; an effector plasmid containing no insert (light bars), wild-type p53 allele (dark bars) or C275S p53 allele (dotted bars) (vector, WT or C275S, respectively); and a luciferase reporter gene plasmid containing p53 response elements from either the human PG genomic fragment or the p53 target genes MDM2, 14-3-3, cyclin G or PIG3 (PG-luc, MDM2-luc, 14-3-3-luc, cyclin G-luc and PIG3-luc, respectively). Luciferase activity was normalized to β-galactosidase activity for three independent transfectant populations. Bars show normalized luciferase reporter gene activity (mean ± S.D.), relative to reporter gene activity in cells transfected with PG-luc reporter gene and expressing wild-type p53. Inset expands Y-axis to illustrate low level luciferase reporter responses. Asterisk (*) indicates genotypes where activities of p53 C275S allele and empty vector differed significantly (p < 0.05, by t-test).
DISCUSSION
Yeast studies indicate that deletion of thioredoxin reductase is not inhibiting p53 activity via an effect on the overall redox state of the cell, as defined by the redox state of glutathione (13). Although the Δtrr1 mutation results in a 2.5-fold increase in the GSSG:GSH ratio, restoration of the normal GSSG:GSH ratio in Δtrr1 cells by overexpressing the GLR1 gene encoding glutathione reductase does not restore p53 activity (13). Furthermore, deletion of the GLR1 gene, which results in more than a fivefold increase in the GSSG:GSH ratio (17, 31), does not inhibit p53 activity (13). Thus, glutathione oxidation is neither sufficient nor necessary for p53 inhibition. Rather, p53 specifically depends on an intact thioredoxin system. We have previously shown that oxidized thioredoxin accumulates in yeast lacking thioredoxin reductase (13). Given the current observation that thioredoxin deletion does not inhibit p53 activity and, in fact, suppresses the inhibitory effect of thioredoxin reductase deletion on p53 activity, our results suggest that it is the presence of oxidized thioredoxin rather than the absence of reduced thioredoxin that inhibits p53 activity. Oxidized thioredoxin may inhibit p53 activity by removing electrons from one or more cysteine residues on p53. Alternatively, it is possible that oxidized thioredoxin affects p53 activity by a mechanism that does not involve disulfide/dithiol exchange between the two proteins. For example, NADP+ and NADPH have been shown to have reciprocal allosteric effects on the DNA binding activity of circadium rhythm transcription factors (32). Also, reduced, but not oxidized, thioredoxin binds and allosterically inhibits the apoptosis signaling kinase 1 (ASK1) (33). Thus, it was possible that oxidized thioredoxin affected p53 activity by an allosteric mechanism rather than a mechanism involving disulfide-dithiol exchange. Our analyses using p53 cysteine substitution mutants were an attempt to obtain evidence for a mechanism involving disulfide-dithiol exchange between p53 and thioredoxin.
In mammalian cells, one mechanism for redox control of p53 is thought to be mediated by the dual function protein Ref1. Ref1 was originally purified and cloned by virtue of its ability to stimulate the DNA binding activity of the Fos/Jun transcription factor AP1 (34) and was subsequently shown to possess apurinic/apyrimidinic endonuclease (APE) activity. The redox active domain and APE domain functionally map to the N-terminal and C-terminal halves of the protein, respectively (35). Ref1 was purified a second time by virtue of its ability to stimulate the transcription-inducing activity of p53 (6). Consistent with a role for Ref1 in controlling p53, p53 cysteine reduction and increased p53 reporter gene transactivation activity in the presence of selenomethionine are suppressed in cells co-transfected with a dominant-negative allele of the ref1 gene (27). However, it is also clear that the thioredoxin system can affect the activity of p53 in the absence of Ref1. Thioredoxin enhances p53 binding to DNA in vitro in the absence of Ref1 (1). Furthermore, thioredoxin reductase mutations affect p53 activity in yeast (7, 8), even though yeast contains no close homolog of Ref1, and the most similar yeast protein, the apurinic/apyrimidinic endonuclease APN2 (orf YBL019W), lacks the N-terminal domain and Cys65 residue that is essential for the redox activity of human Ref1 (35). Finally, our current results showing that purified thioredoxin and p53 physically interact are consistent with a direct effect of thioredoxin on p53 activity. Although it remains possible that, in vivo, oxidized thioredoxin affects p53 activity via a primary effect on an intermediary protein analogous to Ref1, the simplest explanation compatible with all observations is that thioredoxin directly interacts with p53 and thereby affects the ability of p53 to bind and transactivate target genes.
In vitro results suggest that p53 is extremely prone to oxidation. Binding of p53 to target DNA in vitro requires the presence of reductant in the binding buffer (2, 3, 6), and incubation of p53 in the absence of reductant leads to rapid changes in immunoreactivity with conformation-specific antibodies (2). Alkylation studies with [14C]-iodoacetamide indicate that p53 rapidly becomes refractive to labeling when the protein is incubated or stored in the absence of reductant (2). Other studies suggest that p53 is prone to oxidation in vivo. Treatment of human cells with the copper chelator pyrrolidine dithiocarbamate inhibits p53 activity, and the pattern of p53 reactivity with alkylating reagents suggests that an undetermined number of p53 cysteines become oxidized (36, 37). Furthermore, essentially no p53 cysteines are accessible to thiol-directed alkylating reagents unless the cells are pre-treated with the micronutrient selenomethionine (27).
Recent biochemical and pharmacological evidence suggests that the link between thioredoxin reductase and p53 activity suggested by our yeast results also exists in mammalian cells. Using an approach involving sequential treatment of cell lysates with N-ethylmaleimide, DTT and 3-(maleimidopropioryl)-biocytin, it was demonstrated that transiently-expressed p53 in human H1299 cells contain at least one and possibly several oxidized cysteines (27). Significantly, if cells are pre-incubated with selenomethionine, all of the p53 cysteines that were previously oxidized become reduced. Similarly, in cells expressing a p53 fragment containing only Cys275 and Cys277, no cysteines are reactive with an alkylating reagent unless the cells are pre-incubated with selenomethionine (27). In addition to affecting the redox state of p53, pre-incubation with selenomethionine also modestly induces a p53-dependent reporter gene in transfected H1299 cells (27). Thioredoxin reductase is one of twenty-five selenoproteins identified in the mammalian proteome (38). Its penultimate selenocysteine residue is essential for catalytic activity (39), and incubation with selenium has been shown to boost thioredoxin reductase activity levels in several cells lines (40). Although the evidence is indirect, the effects of selenomethionine on the redox state and activity of p53 (27) are consistent with the idea that p53 is prone to oxidation in certain mammalian cell lines and that thioredoxin reductase helps to maintain p53 in the reduced and active state.
Further support for the control of p53 activity through redox communication with the thioredoxin reductase system comes from a recent study that shows high concentrations of prostaglandins PGA1, PGA2, and certain other electrophillic lipids inhibit both thioredoxin reductase activity and p53-dependent reporter gene transactivation in transfected mammalian cells (41). In addition, a biotinylated PGA1-derivative forms covalent adducts with several cellular proteins, including thioredoxin reductase and thioredoxin, but does not form adducts with p53. Furthermore, pre-incubation of cells with selenite partially suppresses the inhibitory effect of PGA2 on p53-dependent reporter gene transactivation (41). Although prostaglandin and selenite treatment may affect the activity of other proteins in the cell, the results are consistent with the conclusion that efficient target gene transactivation by p53 in mammalian cells requires thioredoxin reductase. Proof of a linkage between thioredoxin reductase activity and p53 activity in mammalian cells will require the development of methods to specifically inhibit thioredoxin reductase in higher eukaryotes.
We proposed and tested a model for regulation of p53 transactivation activity by direct cysteine redox communication between p53 and thioredoxin in yeast. No single replacement of cysteine with serine relieved the dependence of p53 on thioredoxin reductase, nor did any combination of replacements involving the six nonessential residues (C124S, C135S, C141S, C182, C229S and C277S). We could not assess thioredoxin reductase dependence of the p53 C135/229S allele due to extremely low levels of reporter gene transactivation in both TRR1 and Δtrr1 cells. However, the C135/141A-HEX mutant, which contains substitutions at codon 135 and 229, was both active and regulated by thioredoxin reductase. For these reasons, we consider it unlikely that oxidation of any of the six nonessential cysteine residues of p53 was responsible for thioredoxin reductase dependence.
Since bound zinc is required for p53 DNA binding activity and zinc is lost from p53 under oxidizing conditions in vitro (2, 4, 5), it is possible that p53 activity is controlled in vivo through redox regulation of the zinc-coordinating cysteine residues C176, C238 and C242. X-ray crystallography of murine p53 shows zinc-free protein containing a disulfide between C173 and C239 (homologs of human p53 C176 and C242, respectively) (42). The structure is superimposable on the structure of zinc-containing reduced p53, with the exception of the L3 loop containing C235, C239 and R245 (homologs of human C238, C242 and R248). This loop is rotated nearly ninety degrees from the normal position in the oxidized structure, thereby moving R245 away from the minor groove of DNA. We speculate that disulfide formation between C176 and C242 may serve as a mechanism for preventing gross unfolding and degradation of of the apoprotein when zinc is lost due to either stochastic or regulated events.
The possibility of C275 serving as a redox control point is intriguing. The residue lies in the major groove when p53 binds DNA. Mutation of the residue to either serine or alanine — shape and dipole mimics, respectively — uniformly inactivated p53 transactivation in the yeast model system. The incompatibility of both shape and dipole analogs indicated that a cysteine thiolate anion may either be required for DNA contact or for maintaining local structure such that an essential DNA contact by a nearby residue is made. For example, the essential DNA-contacting residue R273 is in the same sheet-loop structure (24), and may be dependent on a C275 thiolate for proper orientation. In addition, mutation of C275S strongly impaired the ability of p53 to transactivate a panel of response elements. Finally, it was previously shown that at least one unidentified p53 cysteine is oxidized under standard cell culture conditions, that the residue is fully reduced in response to selenomethionine treatment, and that a truncated p53 peptide containing only cysteines 275 and 277 exhibits the same pattern (27). Our result, showing that mutation of C277 has no effect on thioredoxin reductase dependence, leaves C275 as the remaining available site for redox chemistry.
Supplementary Material
ACKNOWLEDGEMENT
We thank Affi Agbivade for help in deriving yeast strains, and Michael Schimerlik and the OSU EHSC CIA core for help with binding data analyses.
This work was supported by NIH/NCI grant (R01 CA82633), NIH/NIEHS Center Pilot Project grant (P30 ES00210), a Medical Research Foundation of Oregon grant, and a Linus Pauling Institute Pilot Project grant to G.F.M.
Abbreviations
- DTT
dithiothreitol
- EMSA
electrophoretic mobility shift assay
- F/D
50% Dulbecco’s modified Eagle’s medium and 50% Hamm’s F12 medium
- GSH
glutathione
- GSSG
glutathione disulfide
- IPTG
isopropyl-β-D-1-thiogalactopyranoside
- LacZ
E. coli β-galactosidase
- LB
Luria broth
- ONPG
o-nitrophenyl-β-galactopyranoside
- PAPS
3'-phosphoadenosine-5'-phosphosulfate reductase
- PDTC
pyrrolidine dithiocarbamate
- PMSF
phenylmethanesulphonylfluoride
- TBST
tris-buffered saline containing 0.1 % Tween 20
- TRR1
thioredoxin reductase 1
- TRX1
thioredoxin 1
- TRX2
thioredoxin 2
- XGAL
5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside
- YEPD
yeast extract/peptone/dextrose medium
- YNB
yeast nitrogen base medium.
Footnotes
Supporting Information Available: Yeast strains used in this study are listed in Supplemental Table 1. Oligonucleotide primers used for p53 PCR amplification and site-directed mutagenesis are listed in Supplemental Table 2. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Ueno M, Masutani H, Arai RJ, Yamauchi A, Hirota K, Sakai T, Inamoto T, Yamaoka Y, Yodoi J, Nikaido T. Thioredoxin-dependent redox regulation of p53-mediated p21 activation. J Biol Chem. 1999;274:35809–35815. doi: 10.1074/jbc.274.50.35809. [DOI] [PubMed] [Google Scholar]
- 2.Hainaut P, Milner J. Redox modulation of p53 conformation and sequence-specific DNA binding in vitro. Cancer Res. 1993;53:4469–4473. [PubMed] [Google Scholar]
- 3.Rainwater R, Parks D, Anderson ME, Tegtmeyer P, Mann K. Role of cysteine residues in regulation of p53 function. Mol Cell Biol. 1995;15:3892–3903. doi: 10.1128/mcb.15.7.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fojta M, Kubicárová T, Vojtĕsek B, Palecek E. Effect of p53 protein redox states on binding to supercoiled and linear DNA. J Biol Chem. 1999;74:25749–25755. doi: 10.1074/jbc.274.36.25749. [DOI] [PubMed] [Google Scholar]
- 5.Hupp TR, Meek DW, Midgley CA, Lane DP. Activation of the cryptic DNA binding function of mutant forms of p53. Nucleic Acids Res. 1993;21:3167–3174. doi: 10.1093/nar/21.14.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jayaraman L, Murthy KG, Zhu C, Curran T, Xanthoudakis S, Prives C. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 1997;11:558–570. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 7.Casso D, Beach D. A mutation in a thioredoxin reductase homolog suppresses p53-induced growth inhibition in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1996;252:518–529. doi: 10.1007/BF02172398. [DOI] [PubMed] [Google Scholar]
- 8.Pearson GD, Merrill GF. Deletion of the Saccharomyces cerevisiae TRR1 gene encoding thioredoxin reductase inhibits p53-dependent reporter gene expression. J Biol Chem. 1998;273:5431–5434. doi: 10.1074/jbc.273.10.5431. [DOI] [PubMed] [Google Scholar]
- 9.Merrill GF, Dowell P, Pearson GD. The human p53 negative regulatory domain mediates inhibition of reporter gene transactivation in yeast lacking thioredoxin reductase. Cancer Res. 1999;59:3175–3179. [PubMed] [Google Scholar]
- 10.Matthews JR, Wakasugi N, Virelizier JL, Yodoi J, Hay RT. Thioredoxin regulates the DNA binding activity of NF-kappa B by reduction of a disulphide bond involving cysteine 62. Nucleic Acids Res. 1992;20:3821–3830. doi: 10.1093/nar/20.15.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xanthoudakis S, Miao G, Wang F, Pan YC, Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benezra R. An intermolecular disulfide bond stabilizes E2A homodimers and is required for DNA binding at physiological temperatures. Cell. 1994;79:1057–1067. doi: 10.1016/0092-8674(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 13.Merwin JR, Mustacich DJ, Muller EG, Pearson GD, Merrill GF. Reporter gene transactivation by human p53 is inhibited in thioredoxin reductase null yeast by a mechanism associated with thioredoxin oxidation and independent of changes in the redox state of glutathione. Carcinogenesis. 2002;23:1609–1615. doi: 10.1093/carcin/23.10.1609. [DOI] [PubMed] [Google Scholar]
- 14.Wallis JW, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 15.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. New York: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 16.Stotz A, Linder P. The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene. 1990;95:91–98. doi: 10.1016/0378-1119(90)90418-q. [DOI] [PubMed] [Google Scholar]
- 17.Muller EG. Thioredoxin deficiency in yeast prolongs S phase and shortens the G1 interval of the cell cycle. J Biol Chem. 1991;266:9194–9202. [PubMed] [Google Scholar]
- 18.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 19.Thiagalingam S, Kinzler KW, Vogelstein B. PAK1, a gene that can regulate p53 activity in yeast. Proc Natl Acad Sci U S A. 1995;92:6062–6066. doi: 10.1073/pnas.92.13.6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leid M, Kastner P, Lyons R, Nakshatri H, Saunders M, Zacharewski T, Chen JY, Staub A, Garnier JM, Mader S, Chambon P. Purification, cloning, and RXR identity of the HeLa cell factor with which RAR or TR heterodimerizes to bind target sequences efficiently. Cell. 1992;68:377–395. doi: 10.1016/0092-8674(92)90478-u. [DOI] [PubMed] [Google Scholar]
- 21.Kippert F. A rapid permeabilization procedure for accurate quantitative determination of beta-galactosidase activity in yeast cells. FEMS Microbiol Lett. 1995;128:201–206. doi: 10.1111/j.1574-6968.1995.tb07523.x. [DOI] [PubMed] [Google Scholar]
- 22.Giaccone G, Battey J, Gazdar AF, Oie H, Draoui M, Moody TW. Neuromedin B is present in lung cancer cell lines. Cancer Res. 1992;52:2732s–2736s. [PubMed] [Google Scholar]
- 23.Carmel-Harel O, Stearman R, Gasch AP, Botstein D, Brown PO, Storz G. Role of thioredoxin reductase in the Yap1p-dependent response to oxidative stress in Saccharomyces cerevisiae. Mol Microbiol. 2001;39:595–605. doi: 10.1046/j.1365-2958.2001.02255.x. [DOI] [PubMed] [Google Scholar]
- 24.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 25.Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 26.Buzek J, Latonen L, Kurki S, Peltonen K, Laiho M. Redox state of tumor suppressor p53 regulates its sequence-specific DNA binding in DNA-damaged cells by cysteine 277. Nucleic Acids Res. 2002;30:2340–2348. doi: 10.1093/nar/30.11.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seo YR, Kelley MR, Smith ML. Selenomethionine regulation of p53 by a ref1-dependent redox mechanism. Proc Natl Acad Sci U S A. 2002;99:14548–14553. doi: 10.1073/pnas.212319799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kern SE, Kinzler KW, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B. Identification of p53 as a sequence-specific DNA-binding protein. Science. 1991;252:1708–1711. doi: 10.1126/science.2047879. [DOI] [PubMed] [Google Scholar]
- 29.Kern SE, Pietenpol JA, Thiagalingam S, Seymour A, Kinzler KW, Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 30.Chene P. Mutations at position 277 modify the DNA-binding specificity of human p53 in vitro. Biochem Biophys Res Commun. 1999;263:1–5. doi: 10.1006/bbrc.1999.1294. [DOI] [PubMed] [Google Scholar]
- 31.Grant CM, Collinson LP, Roe JH, Dawes IW. Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol Microbiol. 1996;21:171–179. doi: 10.1046/j.1365-2958.1996.6351340.x. [DOI] [PubMed] [Google Scholar]
- 32.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 33.Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, Kawabata M, Miyazono K, Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xanthoudakis S, Miao GG, Curran T. The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc Natl Acad Sci U S A. 1994;91:23–27. doi: 10.1073/pnas.91.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verhaegh GW, Richard MJ, Hainaut P. Regulation of p53 by metal ions and by antioxidants: dithiocarbamate down-regulates p53 DNA-binding activity by increasing the intracellular level of copper. Mol Cell Biol. 1997;17:5699–5706. doi: 10.1128/mcb.17.10.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu HH, Thomas JA, Momand J. p53 protein oxidation in cultured cells in response to pyrrolidine dithiocarbamate: a novel method for relating the amount of p53 oxidation in vivo to the regulation of p53-responsive genes. Biochem J. 2000;351:87–93. doi: 10.1042/0264-6021:3510087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gladyshev VN, Hatfield DL. Selenocysteine-containing proteins in mammals. J Biomed Sci. 1999;3:151–160. doi: 10.1007/BF02255899. [DOI] [PubMed] [Google Scholar]
- 39.Gladyshev VN, Jeang KT, Stadtman TC. Selenocysteine, identified as the penultimate C-terminal residue in human T-cell thioredoxin reductase, corresponds to TGA in the human placental gene. Proc Natl Acad Sci U S A. 1996;93:6146–6151. doi: 10.1073/pnas.93.12.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berggren M, Gallegos A, Gasdaska J, Powis G. Cellular thioredoxin reductase activity is regulated by selenium. Anticancer Res. 1997;17:3377–3380. [PubMed] [Google Scholar]
- 41.Moos PJ, Edes K, Cassidy P, Massuda E, Fitzpatrick FA. Electrophilic prostaglandins and lipid aldehydes repress redox-sensitive transcription factors p53 and hypoxia-inducible factor by impairing the selenoprotein thioredoxin reductase. J Biol Chem. 2003;278:745–750. doi: 10.1074/jbc.M211134200. [DOI] [PubMed] [Google Scholar]
- 42.Kwon E, Kim DY, Suh SW, Kim KK. Crystal structure of the mouse p53 core domain in zinc-free state. Proteins. 2008;70:280–283. doi: 10.1002/prot.21608. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.