Abstract
To analyze HIV-1 subtype distribution, sequence analysis was performed on serum specimens obtained in 1994 from the Rakai Health Sciences community cohort in Uganda. Portions of gag-p24 and env-gp41 were sequenced and HIV subtype was determined for 773 subjects residing in 10 community clusters in rural Uganda. Subtypes A (17%) and D (70%) were the most common strains in the population. Subtype distribution varied by geographic region with significantly more subtype A in northern community clusters compared with southern clusters (21% vs. 8%, p < 0.001) and more subtype D in southern clusters compared with northern clusters (78% vs. 65%, p < 0.008). These data illustrate the geographic complexity of subtype variation, which has important implications for HIV-1 vaccine design.
A distinct characteristic of HIV-1 is its high degree of genetic diversity, which is demonstrated in the multiple subtypes and circulating unique recombinant forms of the virus. HIV-1 is divided into nine different subtypes, identified through their genetic compositions, which can differ in their biological characteristics and geographic distribution.1,2 The predominant subtypes in Uganda are HIV-1 A, D, and recombinant forms of A and D.3,4
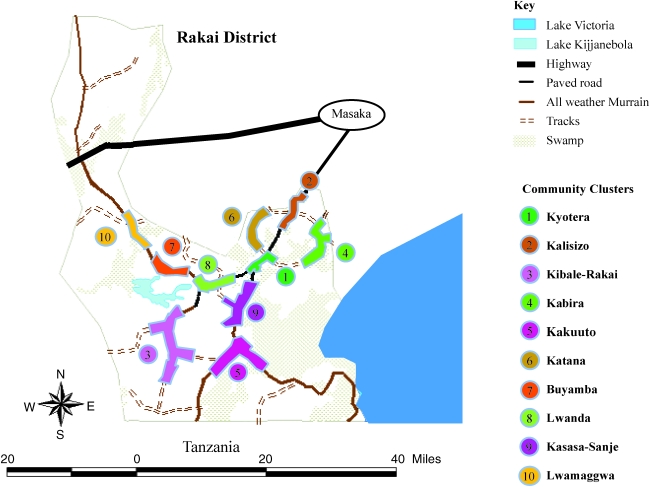
In this study, we analyzed the geographic subtype distribution in Rakai district, Uganda, where AIDS was first identified in East Africa.5 There were 12,164 participants aged 15–59 years enrolled in a community-randomized trial of STD Control for HIV Prevention who provided interview information on sociodemographic, behavioral and health information, and a serum during a baseline survey in 1994. Participants resided in 50 rural communities that were aggregated into 10 clusters, which are separated into 10 community clusters within Rakai: Kyotera, Kalisizo, Rakai, Kabira, Kakuuto, Katana, Buyamba, Lwanda, Kasasa-Sanje, and Lwamaggwa.6 Enrollment and study details have been previously described.2 Institutional Review Board approvals were obtained from Uganda Virus Research Institute's Science and Ethics Committee, Uganda National Council for Science and Technology, and from the Institution Review Boards (IRBs) of collaborating U.S. institutions (Columbia University and Johns Hopkins University).
Viral RNA was extracted from all available samples of HIV-1-positive subjects using a QIAmp Viral RNA Mini Kit (Qiagen, Valencia, CA). The eluted RNA was used for RT-PCR and nested PCR in two separate reactions to amplify portions of gag (HXB2 nt 1249 to 1704) and gp41 (HBX2 nt 7858 to 8260).7,8 Amplified samples were purified using ExoSAP-IT (USB, Cleveland, OH), and then sequenced using the Applied Biosystems 3730xl DNA Analyzer. Subtype assignments of the sequence data were generated using the NCBI genotyping database. Those subjects with both gag and gp41 sequences that were uniformly subtype A, C, or D were considered infected with that subtype.7,8 If the genomic regions differed by subtype, the subject was classified as infected with a recombinant strain.
Subtype distribution was analyzed by community cluster and then more broadly according to northern, central, and southern regions of Rakai district. Southern clusters were Kibale-Rakai, Kakuuto, and Kasasa-Sanje, northern clusters were Lwamaggwa, Buyamba, Katana, and Kalisizo, and Kabira, Kyotera, and Lwanda were designated as centrally located (Fig. 1). Chi-square tests were used for statistical inference.
FIG. 1.
Percent Subtype and Genetic Diversity of the Community Clusters Grouped by Geographic Region
There were 2058 HIV-positive subjects identified in 1994, of whom 89% (1837/2058) had sufficient serum volume for PCR and 42% (773/1837) of samples successfully generated interpretable sequence data for gag and gp41. The overall subtype distribution was 70% (543/773) HIV-1D, 17% (129/773) subtype A, and 12% (94/773) recombinant strains.
In the southern region, 78% (122/157) of individuals were infected with HIV-1D. This was significantly higher than HIV-1D infections in the northern region (65%, 238/364, p < 0.007) (Table 1). In the northern region, 21% (77/364) were infected with HIV-1A, compared to only 16% (39/252, p < 0.05) in the central region. The northern region communities also had a significantly higher percentage of HIV-1A infections compared to the southern region (8%, 13/157, p < 0.001).
Table 1.
Percent Subtype and Genetic Diversity of the Community Clusters Grouped by Geographic Region
| |
|
Subtype (N) (%) |
Diversity gag (%) |
Diversity gp41 (%) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Community cluster | N | A | D | C | R | A | D | A | D |
| North | |||||||||
| Kalisizo | 140 | 22.1 (31) | 66.4 (93) | 0.7 (1) | 10.7 (15) | 4.9 | 4.1 | 6.8 | 6.4 |
| Katana | 106 | 23.6 (25) | 60.4 (64) | 0.9 (1) | 15.1 (16) | 4.0 | 4.0 | 6.3 | 6.2 |
| Buyamba | 72 | 9.7 (7) | 77.8 (56) | 1.4 (1) | 11.1 (8) | 5.0 | 4.7 | 7.9 | 6.1 |
| Lwamaggwa | 46 | 30.4 (14) | 54.3 (25) | 0 | 15.2 (7) | 4.3 | 3.8 | 6.3 | 6.5 |
| All northern region | 364 | 21.2 (77) | 65.4 (238) | 0.8 (3) | 12.6 (46) | 4.6 | 4.2 | 6.8 | 6.3 |
| Central | |||||||||
| Kyotera | 78 | 15.4 (12) | 75.6 (59) | 0 | 9.0 (7) | 4.7 | 3.9 | 7.4 | 6.7 |
| Kabira | 87 | 13.8 (12) | 77.0 (67) | 0 | 9.2 (8) | 5.5 | 3.8 | 6.2 | 6.2 |
| Lwanda | 87 | 17.2 (15) | 65.5 (57) | 1.1 (1) | 16.1 (14) | 4.7 | 4.5 | 6.5 | 5.6 |
| All central region | 252 | 15.5 (39) | 72.6 (183) | 0.4 (1) | 11.5 (29) | 5.0 | 4.1 | 6.7 | 6.1 |
| South | |||||||||
| Kibale-Rakai | 42 | 9.5 (4) | 88.1 (37) | 2.4 (1) | 0 | 3.8 | 4.5 | 4.3 | 6.0 |
| Kakuuto | 36 | 13.9 (5) | 72.2 (26) | 5.6 (2) | 8.3 (3) | 8.2 | 5.3 | 6.3 | 6.3 |
| Kasasa-Sanje | 79 | 5.1 (4) | 74.7 (59) | 0 | 20.3 (16) | 5.8 | 3.5 | 8.0 | 6.1 |
| All southern region | 157 | 8.2 (13) | 77.7 (122) | 1.9 (3) | 12.1 (19) | 5.8 | 4.2 | 6.6 | 6.1 |
| Total | 773 | 16.7 (129) | 70.2 (543) | 0.9 (7) | 12.2 (94) | 5.0 | 4.2 | 6.7 | 6.2 |
Sequence diversity within subtypes A and D was calculated for both the gag and gp41 genomic regions and for both regions within each of the 10 community clusters. Overall sequence diversity of HIV-1A was 4.9% (±1.3) for gag and 6.8% (±1.1) for gp41 fragments, and 3.9% (±0.54) in the gag region and 6.2% (±0.30) in gp41 for HIV-1D fragments (Table 1). A t-test showed that diversity was not different between the two subtypes within gag (p = 0.056) or gp41 (0.277).
A previous study conducted between 1989 and 2000, examining portions of the envelope by sequencing and heteroduplex mobility assay, determined that the relative frequencies of subtypes among young pregnant women in 15 antenatal surveillance were spread evenly throughout Uganda centers. HIV-1A (45%) was the most prevalent, followed closely by subtype D (41%) in this study.9 Additionally, no significant change was found in relative frequencies of subtypes over the 11 sample years, nor were there significant differences in geographic distribution within Uganda. Reports from Kampala and Entebbe reported that HIV-1A (48% and 51%) was the most common subtype.10–12 A previous study based on full length sequencing of 46 HIV-1 samples from the Rakai district between 1998 and 1999 demonstrated that the predominant subtypes were D (54%), A/D (30%) recombinants, and A (15%). Compared with these full-length data from the same cohort, our data demonstrated a higher percentage of HIV-1D and a lower prevalence of recombinants.
Phylodynamic analyses of Ugandan viruses suggest that HIV-1A was introduced earlier than HIV-1D.13 In addition, HIV-1A appears to have originated in the Congo and spread from east to west, whereas HIV-1D appears to have originated from Tanzania and spread from south to north. These analyses are consistent with the findings of the present study in which HIV-1D was more common in the southern region located on the Tanzanian border compared to the northern region of Rakai district. Conversely, HIV-1A was least frequent in the south compared to the northern region.
These data demonstrate that HIV-1 subtypes were not randomly distributed in Rakai in 1994, with significantly more HIV-1D in the southern-most communities and significantly more HIV-1A in northern-most regions. Varying subtype distribution poses major implications for vaccine development, especially when, as in this study, variation and high frequency of recombination reflect the historical perspective.14 It is possible even in a small geographic region such as the Rakai district, Uganda that a broadly reactive vaccine would need to be developed to accommodate all subtypes and variations within those subtypes.
Acknowledgments
The authors would like to thank all the participants of the Rakai community cohort and the staff of the Rakai Health Sciences Group for their tireless work. Funding for this project was provided by the Laboratory of Immunoregulation, Division of Intramural Research, NIAID, NIH. GenBank accession numbers for these sequences are GQ332766–GQ334183, GQ253866–GQ253897, and GQ252692–GQ252793.
Disclosure Statement
No competing financial interests exist.
References
- 1.Taylor BS. Sobieszczyk ME. McCutchan FE. Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med. 2008;358:1590–1602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiwanuka N. Laeyendecker O. Robb M, et al. Effect of human immunodeficiency virus type 1 (HIV-1) subtype on disease progression in persons from Rakai, Uganda, with incident HIV-1 infection. J Infect Dis. 2008;197:707–713. doi: 10.1086/527416. [DOI] [PubMed] [Google Scholar]
- 3.Hu DJ. Baggs J. Downing RG, et al. Predominance of HIV-1 subtype A and D infections in Uganda. Emerg Infect Dis. 2000;6:609–615. doi: 10.3201/eid0606.000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yirrell DL. Kaleebu P. Morgan D, et al. Inter- and intra-genic intersubtype HIV-1 recombination in rural and semi-urban Uganda. AIDS. 2002;16:279–286. doi: 10.1097/00002030-200201250-00018. [DOI] [PubMed] [Google Scholar]
- 5.Serwadda D. Mugerwa RD. Sewankambo NK, et al. Slim disease: A new disease in Uganda and its association with HTLV-III infection. Lancet. 1985;2:849–852. doi: 10.1016/S0140-6736(85)90122-9. [DOI] [PubMed] [Google Scholar]
- 6.Kiwanuka N. Robb M. Kigozi G, et al. Knowledge about vaccines and willingness to participate in preventive HIV vaccine trials: A population-based study, Rakai, Uganda. J Acquir Immune Defic Syndr. 2004;36:721–725. doi: 10.1097/00126334-200406010-00009. [DOI] [PubMed] [Google Scholar]
- 7.Yang C. Dash BC. Simon F, et al. Detection of diverse variants of human immunodeficiency virus-1 groups M, N, and O and simian immunodeficiency viruses from chimpanzees by using generic pol and env primer pairs. J Infect Dis. 2000;181:1791–1795. doi: 10.1086/315439. [DOI] [PubMed] [Google Scholar]
- 8.Yang C. Dash B. Hanna SL, et al. Predominance of HIV type 1 subtype G among commercial sex workers from Kinshasa, Democratic Republic of Congo. AIDS Res Hum Retroviruses. 2001;17:361–365. doi: 10.1089/08892220150503726. [DOI] [PubMed] [Google Scholar]
- 9.Herbeck JT. Lyagoba F. Moore SW, et al. Prevalence and genetic diversity of HIV type 1 subtypes A and D in women attending antenatal clinics in Uganda. AIDS Res Hum Retroviruses. 2007;23:755–760. doi: 10.1089/aid.2006.0237.A. [DOI] [PubMed] [Google Scholar]
- 10.Harris ME. Serwadda D. Sewankambo N, et al. Among 46 near full length HIV type 1 genome sequences from Rakai district, Uganda, subtype D and AD recombinants predominate. AIDS Res Hum Retroviruses. 2002;18:1281–1290. doi: 10.1089/088922202320886325. [DOI] [PubMed] [Google Scholar]
- 11.Flys TS. Chen S. Jones DC, et al. Quantitative analysis of HIV-1 variants with the K103N resistance mutation after single-dose nevirapine in women with HIV-1 subtypes A, C, and D. J Acquir Immune Defic Syndr. 2006;42:610–613. doi: 10.1097/01.qai.0000221686.67810.20. [DOI] [PubMed] [Google Scholar]
- 12.Kaleebu P. French N. Mahe C, et al. Effect of human immunodeficiency virus (HIV) type 1 envelope subtypes A and D on disease progression in a large cohort of HIV-1-positive persons in Uganda. J Infect Dis. 2002;185:1244–1250. doi: 10.1086/340130. [DOI] [PubMed] [Google Scholar]
- 13.Gray RR, et al. Spatial phylodynamics of HIV-1 epidemic emergence in east Africa. AIDS. 2009;23(14):F9–F17. doi: 10.1097/QAD.0b013e32832faf61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peeters M. Sharp PM. Genetic diversity of HIV-1: The moving target. AIDS. 2000;14(Suppl. 3):S129–140. [PubMed] [Google Scholar]