Summary
Recent advances in genome sequencing efforts have revealed an abundance of novel putative lectins. Amongst these, many galectin-related proteins have been found in all corners of the eukaryotic superkingdom, characterized by many conserved residues but intriguingly lacking critical amino acids. Here we present a structural and biochemical analysis of one representative, the galectin-related lectin CGL3 found in the inky cap mushroom Coprinopsis cinerea. This protein contains all but one conserved residues known to be involved in β-galactoside binding in galectins. A Trp residue strictly conserved among galectins is changed to an Arg in CGL3 (R81). Accordingly, the galectin-related protein is not able to bind lactose. Screening of a glycan array revealed that CGL3 displays preference for oligomers of β1-4 linked N-acetyl-glucosamines (chitooligosaccharides) and GalNAcβ1-4GlcNAc (LacdiNAc). Carbohydrate-binding affinity of this novel lectin was quantified using isothermal titration calorimetry and its mode of chitooligosaccharide coordination, not involving any aromatic amino acid residues, was studied by x-ray crystallography. The structural information was used to alter the carbohydrate-binding specificity and substrate affinity of CGL3. The importance of residue R81 in determining the carbohydrate-binding specificity was demonstrated by replacing this Arg by a Trp residue (R81W). This single amino acid change led to a lectin that failed to bind chitooligosaccarides but gained lactose-binding. Our results demonstrate that, similar to the legume lectin fold, the galectin fold represents a conserved structural framework upon which dramatically altered specificities can be grafted by few alterations in the binding site and that, in consequence, many metazoan galectin-related proteins may represent lectins with novel carbohydrate-binding specificities.
Keywords: galectin, glycan array, chitooligosaccharide, LacDiNAc, mushroom
Introduction
Lectins are non-immunoglobulin carbohydrate-binding proteins widely distributed in nature. This group of proteins is highly diverse both in structure and specificity and plays a key role in the recognition of the vast arrays of glycocodes presented by a living cell. The galectin family of lectins is characterized by a conserved 130 amino acid carbohydrate recognition domain (CRD) and specificity for β-galactoside-containing oligosaccharides 1. The family is subdivided according to the multiplicity of CRDs and the presence of other domains as well as the preference of the individual CRDs for specific extensions of the core β-galactoside 2; 3; 4. The occurrence of galectins is restricted to multicellular eukaryotic organisms, excluding plants, and they are usually found as large protein families within a given species. Their expression is often regulated in response to both internal (developmental) and external cues 5; 6. Galectins have been implicated in a wide variety of biological processes such as cell adhesion 7, innate immunity 8; 9; 10, cell differentiation and development 8, signal transduction 11, regulation of cell proliferation and cell death 12; 13 and pre-mRNA splicing 14. A role of galectins in vesicular trafficking of glycoproteins may further contribute to such multiple functions 15. The lack of a signal sequence for classical secretion and the absence of glycosylation suggests that galectins are synthesized in the cytoplasm and secreted by alternative secretory pathways 16.
The prototype galectins CGL1 and CGL2 of the homobasidiomycete Coprinopsis cinerea were the first fungal members of the galectin family identified 17. Subsequently, representatives from the homobasidiomycetes Agrocybe cylindracea 18 and Agrocybe aegerita 19 were described. The crystal structures of the proteins from C. cinerea and A. cylindraceae were determined in ligand-free state as well as in complex with β-galactoside-containing oligosaccharides and are basically superimposable with the known structures of the mammalian galectins 20; 21. It is remarkable, that the mushroom-forming homobasidiomycetes seem to be the only representatives of the fungal kingdom in which galectins occur. The function of the fungal orthologs remains unclear. Since the onset of their expression in the case of C. cinerea coincides with the onset of fruiting body development, it was hypothesized that they play a role in fruiting body formation 17. However, recent studies in which the expression of C. cinerea galectins CGL1 and CGL2 was silenced could not support this hypothesis 22.
The sequenced genome of C. cinerea revealed a third putative galectin, CGL3. The protein sequence displays all β-galactoside-coordinating residues of galectins except for a critical Trp which is replaced by an Arg. Similar galectin-related proteins with changes in the galectin signature have been described in metazoans; intriguingly, for none of them has carbohydrate-binding been reported 5. Examples are mammalian galectin-related inter-fiber protein (GRIFIN), mammalian galectin-related protein (GRP; also referred to as HSPC159), mammalian Charcot-Leyden crystal protein (CLC) and GALE5 from the mosquito Anopheles gambiae. The CLC structure was reported in its ligand-free form and with a mannose molecule in its CRD 23. However, this binding appears to be unspecific since Man-binding of CLC could not be demonstrated biochemically 24.
Here, we show that CGL3 is expressed in C. cinerea fruiting bodies and that, in contrast to CGL2, neither endogenous nor recombinant CGL3 is able to bind lactose. In contrast, CGL3 bound specifically to LacdiNAc and, most remarkably, to chitooligosaccharides, oligomers of β1-4 linked GlcNAc. These carbohydrates are not ligands for CGL2. As a molecular basis for the difference in carbohydrate-binding specificity between CGL2 and CGL3, we present the crystal structure of the ligand-free and the chitotetraose-bound forms of CGL3. This is the first functional and structural characterization of the specific interaction between a galectin-related lectin and an oligosaccharide not containing any galactose.
Results
Primary Structure and expression of CGL3
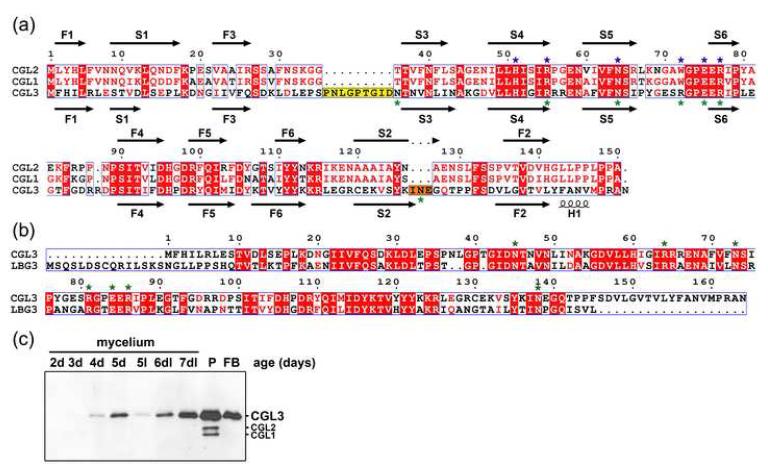
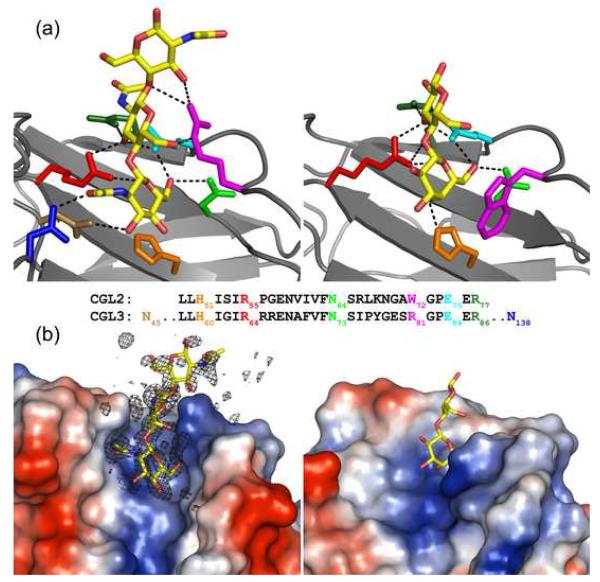
A BLAST search in the genome sequence of C. cinerea strain Okayama 7 (http://www.broad.mit.edu/annotation/genome/coprinus_cinereus/Home.html) using the amino acid sequences of the well-characterized C. cinerea galectins CGL1 and CGL2 (Genbank Acc. AF130360) revealed the presence of a third member of the galectin family in this model organism. The corresponding gene, which was termed cgl3, does, in contrast to the cgl1 and cgl2 genes, not contain any predicted introns and was cloned from genomic DNA of our laboratory strain AmutBmut 25; 26 and sequenced (Genbank Acc. DQ408306). The coding sequence shows 54 % identity on DNA level and 35/70 % identity/similarity on amino acid level to the coding sequence of AmutBmut CGL2. A primary structure alignment of the two known galectins and the third putative galectin (CGL3) of C. cinerea is shown in Figure 1a. Despite the rather low overall sequence identity, the signature of 7 residues directly involved in sugar coordination of galectins is almost completely conserved in CGL3 (6 out of 7 residues). Surprisingly, the Trp residue at position 72 in CGL2, which was shown to be essential for sugar binding 20, is replaced by an Arg residue at the corresponding position 81 in CGL3. In addition, the CGL3 protein has, compared to CGL1 and CGL2, two short insertions at positions 36-44 and 141-143. A similar galectin-family consisting of two putative galectin-orthologs and one putative CGL3 ortholog was found in the genome of the ectomycorrhizal homobasidiomycete Laccaria bicolor (alignment of the putative CGL3 ortholog is shown in Fig. 1b).
Figure 1. Sequence comparisons.
(a) Sequence alignment of C. cinerea galectins CGL2, CGL1 and the galectin-related protein CGL3. Secondary structure elements derived from the crystal structure of CGL2 (1UL9) and CGL3 (2R0F) are shown on top and bottom of the sequences, respectively. Strands are labeled to indicate the order and position in the β-sandwich. Identical amino acid residues conserved in all three proteins are shown in white letters on red background, and similar residues are shown in red on white background. The conserved β-galactoside-coordinating residues of the galectin carbohydrate recognition domain are marked with blue asterisks. Amino acid residues involved in chitooligosaccharide coordination by CGL3 are highlighted with green asterisks. The two prolonged regions of CGL3 are accented with yellow and orange backgrounds, in Figure 3d these regions are colored accordingly. (b) Sequence alignment of C. cinerea galectin-related protein CGL3 with the putative orthologous protein found in L. bicolor (LBG3) (http://genome.jgi-psf.org/Lacbi1/Lacbi1.home.html). (c) Analysis of CGL3 expression in C. cinerea. Equal amounts of proteins extracted from fungal tissues of C. cinerea strain AmutBmut were separated by SDS-PAGE, blotted to nitrocellulose and probed with anti-CGL3 antibodies. The age of the mycelial samples is indicated, P refers to primordia (fruiting structures ≤ 5 mm), FB stands for immature fruiting bodies (10-20 mm). Except for one mycelial sample cultivated in continuous light at 37 °C for 5 days (5l), all cultures were initially grown in complete darkness (d) at 37 °C for the indicated number of days and some exposed to an alternating 12 h dark/light (dl) cycle at 25 °C after 5 days (6dl, 7dl, P, FB). The additional bands in the primordia sample are due to crossreaction of the anti-CGL3 antiserum with CGL1 and CGL2.
Expression of CGL3 in C. cinerea was verified by immunoblotting of protein extracts from different developmental stages using an antiserum raised against pure N-terminally His-tagged CGL3 (His8-CGL3). Similar to CGL2 17, CGL3 expression was induced upon initiation of fruiting body formation with maximal expression in primordia and was repressed by exposure to constant light (Fig. 1c).
Carbohydrate-Binding Specificity of CGL3 and CGL2
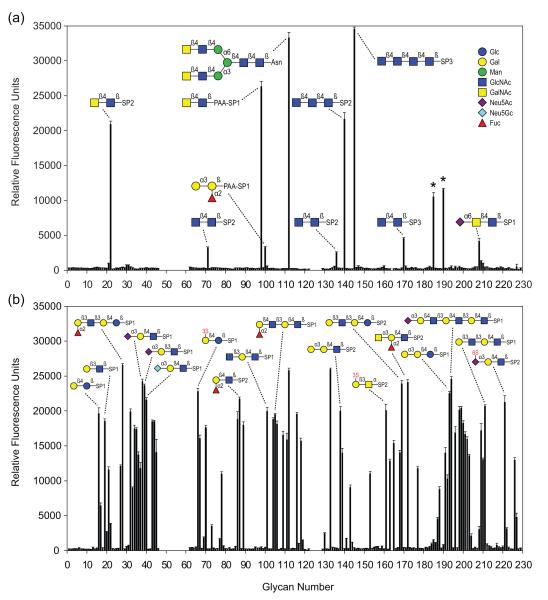
His8-CGL3 as well as CGL2 were expressed in E. coli and purified using metal affinity resin and lactosyl-sepharose, respectively. Since affinity-chromatography using lactosyl-sepharose indicated that neither endogenous nor recombinant CGL3 was able to bind lactose (data not shown; see below), lectin activity of CGL3 was assessed by exposing the protein to a broad variety of glycans. For this purpose binding of recombinant His8-CGL3, and recombinant CGL2 as control, to a defined glycan array containing biotinylated glycans captured on streptavidin-coated microtiter plates was monitored using the respective antisera. The entire list of glycans tested and the results of the binding assays can be found in the glycan screen raw data section on the homepage of the Protein-Carbohydrate Interaction Core H of the Consortium for Functional Glycomics (http://www.functionalglycomics.org/glycomics/publicdata/primaryscreen.jsp) and is summarized in supplementary Table 1. Oligosaccharide structures recognized by CGL3 are shown in Figure 2a and Table 1. In contrast to CGL2 and all known galectins, CGL3 did not bind to typical galectin ligands such as lactose or Galβ1-4GlcNAc (LacNAc), but exhibited a distinct specificity for highly N-acetylated glycans such as GalNAcβ-4GlcNAc (LacdiNAc) as well as di-, tri- and tetrasaccharides composed β1-4 linked N-acetyl-glucosamines (chitooligosaccharides). The fact that neither β-linked GlcNAc nor β-linked GalNAc alone were bound by CGL3 suggested that the disaccharides chitobiose and LacdiNAc represented the minimal ligands for CGL3. The presence of two 2′-acetamido groups seemed to be a prerequisite for disaccharide binding since this substitution was observed in all CGL3-bound disaccharides. Substitutions at the 4′-position of the ultimate GlcNAc of the chitobiose core found in all N-linked glycans e.g. Man3GlcNAc2 (Manα1-3(Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAc), glycan no. 146, were not tolerated by CGL3. In contrast, CGL2 did not show any affinity neither for chitooligosaccharides nor LacdiNAc in this assay (Fig. 2b). These results showed that CGL3 is a galectin-related protein with a novel oligosaccharide specificity including structures lacking Gal.
Figure 2. Sugar-binding specificities of CGLs.
(a) Binding of CGL3 to carbohydrates of the Consortium of Functional Glycomics (CFG) glycan array. Results shown are averages of triplicate measurements of fluorescence intensity. Error bars indicate the standard deviations of the mean. Glycans marked with an asterisk were not considered in the analysis due to heterogeneity. (b) CGL2 binding to the CFG glycan array. Raw data as well as the entire list of glycans with their respective spacers (SP) can be found on the CFG homepage (http://functionalglycomics.org) and in supplementary Table 1. S stands for sulphate.
Table 1.
Glycans of the glycan array recognized by CGL3
| Avg. S/Na | Glycanb | Trivial name | Glycan no.c |
|---|---|---|---|
| 228.43 | GlcNAcβ1-4GlcNAcβ1-4GlcNAcβ1-4GlcNAcβ–Sp3 | Chitotetraose | 145 |
| 220.09 | GalNAcβ1-4GlcNAcβ1-4Manα1-6(GalNAcβ1-4GlcNAcβ1-4Manα1- 3)Manβ1-4GlcNAcβ1-4GlcNAcβ-N–Sp1 |
Bi-LDN (remodeled from human fibrinogen) | 112 |
| 173.86 | GalNAcβ1-4GlcNAcβ–PAA–Sp1 | LacdiNAc | 98 |
| 143.16 | GlcNAcβ1-4GlcNAcβ1-4GlcNAcβ–Sp2 | Chitotriose | 140 |
| 138.06 | GalNAcβ1-4GlcNAcβ–Sp2 | LacdiNAc | 22 |
| 76.35 | Mixed glycans-N–Sp1 | KLH (Keyhole Limpet Hemocyanin N-glycans) | 190 |
| 69.31 | GalNAcβ1-4(Fucα1-3)GlcNAcβ1-4Manα1-3(GalNAcβ1-4(Fucα1- 3)GlcNAcβ1-4Manα1-6)Manα1-4GlcNAcβ1-4(Fucα1-3)GlcNAcβ-N–Sp1 |
Bi-LDNF (remodeled from human fibrinogen) | 185 |
| 29.91 | GlcNAcβ1-4GlcNAcβ–Sp3 | Chitobiose | 170 |
| 27.26 | Neu5Acα2-6GalNAcβ1-4GlcNAcβ–Sp1 | 6SLDN | 208 |
| 21.62 | Galα1-3(Fucα1-2)Galβ–PAA–Sp1 | Btri | 100 |
| 21.37 | GlcNAcβ1-4GlcNAcβ–Sp2 | Chitobiose | 71 |
| 16.46 | GlcNAcβ1-4GlcNAcβ–Sp2 | Chitobiose | 136 |
S/N is the signal to noise ratio, where the signal represents the average signal measured in relative fluorescence units of triplicate determinations over background (noise) of the assay
The entire list of glycans with their respective spacers (Sp) can be found on the homepage of the Consortium for Functional Glycomics (http://www.functionalglycomics.org)
Glycan number refers to the designated number of the biotinylated glycan in the glycan array (plate array version 3.7) of the Consortium for Functional Glycomics
Thermodynamics of CGL3-Chitooligosaccharide Interaction
Isothermal titration calorimetry (ITC) was used to characterize the interaction between recombinant His8-CGL3 and chitooligosaccharides. ITC measurements using recombinant CGL2 and lactose were performed as a reference. The calculated dissociation constants of His8-CGL3 and chitooligosaccharides (310-351 μM) were in a similar range as measured for the reference pair CGL2 and lactose (271 μM) confirming a specific interaction (Table 2). A representative thermogram of CGL3-binding to chitotriose is shown in supplementary Figure 1. The Kd value for CGL2 and lactose were slightly higher than the one previously determined by surface plasmon resonance (85.4 μM) 20. The binding between CGL3 and chitotriose was strongly enthalpy-driven, reflected by an enthalpy contribution (ΔH) of −55.9 kJ/mol counterbalanced by an unfavorable entropy contribution (TΔS) of −35.9 kJ/mol and resulting in the free energy of binding of −20.0 kJ/mol. This observation holds true for all combinations of lectins and carbohydrates measured and is in accordance with numerous other lectins where such an entropy barrier is characteristic 27. The increase in affinity from chitobiose to chitotriose but not from chitotriose to chitotetraose suggested that three GlcNAc residues were coordinated by CGL3.
Table 2.
Binding constants and thermodynamic parameters determined by ITC at 298° K
| Protein | Carbohydrate | Ka (103 M−1) | Kd (10−6 M) | ΔG (kJ/mol) | ΔH (kJ/mol) | TΔS (kJ/mol) |
|---|---|---|---|---|---|---|
| CGL3 | chitobiose | 2.85±0.044 | 351±5.5 | −19.7 | −47.5±0.24 | −27.8 |
| CGL3 | chitotriose | 3.23±0.034 | 310±4.3 | −20.0 | −55.9±0.23 | −35.9 |
| CGL3 | chitotetraose | 3.17±0.043 | 315±4.3 | −20.0 | −54.7±0.27 | −34.7 |
| CGL3 R81A | chitotriose | 1.12±0.020 | 893±15.7 | −17.4 | −50.3±1.46 | −32.9 |
| CGL2 | lactose | 3.69±0.07 | 271±3.3 | −20.4 | −51.2±0.23 | −30.8 |
Deviations represent experimental errors of single measurements. A representative thermogram showing chitotriose binding to CGL3 is depicted in supplementary Figure 1.
Overall Structure of CGL3
The structures of ligand-free protein (both untagged and His8-tagged) as well as of the untagged protein in complex with chitotetraose were determined. Details of data collection and structural refinement are reported in Table 3 and Materials and Methods. A tetrameric arrangement of the lectin was observed in all three structures. Size-exclusion chromatography experiments confirmed the tetrameric state in solution (data not shown).
Table 3.
Summary of structure determination and refinement
| Diffraction data | |||
|---|---|---|---|
| Data set | Ligand free | Chitotriose | His8-ligand free |
| Space group | P6122 | P1 | P6122 |
| Cell constants (Å) | a, b = 112.38 c = 175.66 |
a = 47.90 b = 59.70 c = 60.20 α = 73.0° β = 90.7° γ = 76.4° |
a, b = 109.86 c = 173.097 |
| Resolution limit (Å) | 20 – 2.0 (2.0 – 2.1) | 20 – 1.9 (1.9 – 2.0) | 50 – 2.3 (2.3 – 2.4) |
| Measured Reflections | 201733 (24219) | 114167 (18462) | 416130 (50104) |
| Unique reflections | 41642 (4561) | 46020 (7188) | 27885 (3231) |
| Redundancy | 4.8 (5.3) | 2.5 (2.6) | 14.9 (15.5) |
| Mean I/σ (I) | 9.8 (8.0) | 11.0 (8.0) | 21.2 (4.4) |
| Rsym (I) | 0.112 (0.240) | 0.075 (0.164) | 0.112 (0.75) |
| Refinement statistics | |||
|---|---|---|---|
| Resolution range used for refinement (Å) |
15 – 2.0 (2.0 – 2.05) | 15 – 1.9 (1.9 – 1.95) | 46 – 2.3 (2.3 – 2.36) |
| No. reflections used | 41187 (2836) | 43261 (3210) | 26580 (1926) |
| Completeness % | 97.09 (93.37) | 93.8 (95.26) | 99.58 (99.16) |
| Rcryst | 0.242 (0.406) | 0.195 (0.198) | 0.233 (0.282) |
| No. reflection for Rfree | 2260 (150) | 2325 (170) | 1407 (83) |
| Rfree | 0.250 (0.371) | 0.254 (0.306) | 0.269 (0.375) |
| Model contents (resid.) | |||
|---|---|---|---|
| Amino acid | 326 | 636 | 325 |
| H2O | 299 | 316 | 48 |
| Carbohydrate | - | 12 | - |
| Average B factor (Å2) | 32.15 | 23.31 | 34.22 |
| Estimated coordinate error (Rfree / Å) |
0.135 | 0.181 | 0.198 |
| Rmsd from ideal bond length (Å) |
0.006 | 0.013 | 0.015 |
| Rmsd from ideal bond angle (°) |
1.4 | 2.1 | 2.1 |
| Ramachandran geometry | |||
|---|---|---|---|
| Most favoured | 86.1 % (236) | 89 % (477) | 85.7% (233) |
| Allowed | 12.8 % (35) | 10.6 % (57) | 13.6 % (37) |
| Generously allowed | 0.4 % (1) | 0.4% (2) | 0.4% (1) |
| Disallowed | 0.7 % (2) | 0 | 0.4 % (1) |
| PDB ID | 2R0F | 2R0H | - |
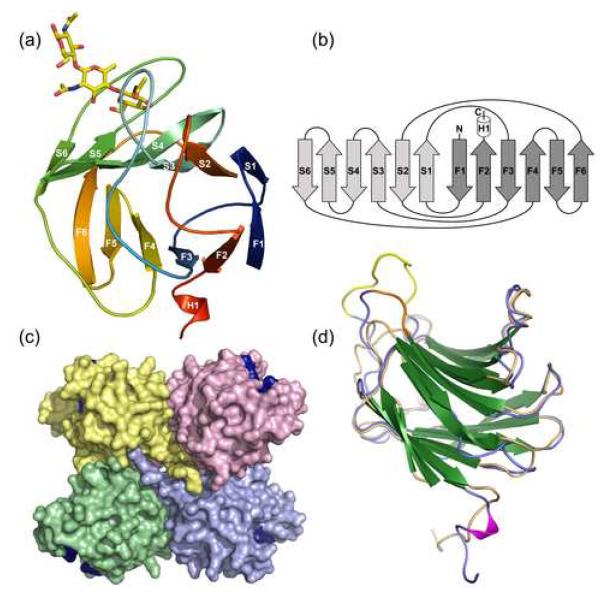
The protein exhibited a typical galectin fold with the CRD contained in a single globular domain formed by a sandwich of two anti-parallel beta sheets (Fig. 3). Four CGL3 molecules assembled into a tetramer that contained the CRDs in alternating, “trans” orientation (Fig. 3c). As a consequence and unlike the canonical galectin dimer found in mammals (which have both CRDs facing the same plane relative to the dimer axis), adjacent CRDs of CGL3 are oriented towards opposing sides of the tetramer.
Figure 3. Overall structure of CGL3.
(a) Schematic representation of one monomer of chitotriose-bound CGL3. Strands of the two β-sheets are labeled as in Figure 1a. Bound chitotriose is shown in stick representation. (b) topology diagram of CGL3 with the two anti-parallel beta sheets colored in light gray and dark gray, respectively. Termini are indicated with N and C. (c) surface representation of the ligandfree CGL3 tetramer. Location of the four sugar-binding pockets is marked by coloring the amino acids I43, N45 and R81 in dark blue. (d) the structure of one chain of ligandfree CGL3 is overlayed with one monomer of CGL2. CGL3 is colored in blue and the insertions are highlighted in yellow and orange (gaps in the alignment in Fig. 1a). CGL2 is colored in beige. Beta-strands of both structures are colored in green.
The structure showed very high similarity to CGL2 20, in both tertiary and quaternary structure (Fig. 3d). Despite moderate to low sequence identity, both structures could be accurately aligned with an average root mean square deviation (r.m.s.d.) of only 1.42 Å with regard to Cα positions (Fig. 3d; C-terminal segments were not aligned for r.m.s.d. calculation). R.m.s.d. greater than 5.0 Å were found at residues 31 to 44 (which contains a nine residue insertion in the region connecting strands F3 and S3 compared with CGL2), residues 94 to 96 (loop connecting S6 and F4), residues 115 to 119 (connecting F5 and F6), residues 132 to 140 (in S2 and connecting to F2, another insertion compared to CGL2) and the C-termini which appear completely dissimilar to the CGL2 C-terminal assembly (data not shown). The C-terminus of CGL3 contained a very short α-helical segment (H1) at the end of strand F2 that continued further in an extended conformation. The electron density of this region was poorly defined, impeding complete model building in case of the ligand-bound structure where the C-terminal four to six residues were not observed in the electron density. We therefore believe the very C-terminal region to contain some intrinsic disorder.
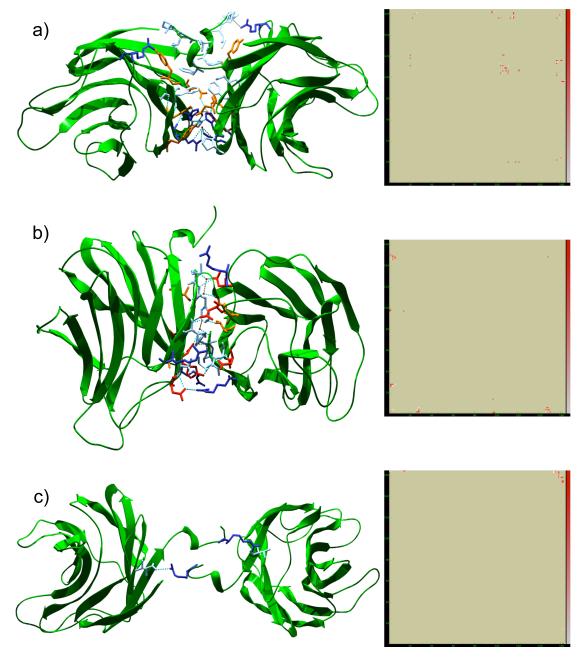
The interface area created by the tetramer was about 2′740 Å2. In the ligand-free structure, there were twenty water molecules involved in the assembly. However, merely six of these were buried in the interface and therefore bone fide excluded from exchange with the bulk solvent. The majority of solvent atoms involved in the assembly thus mediate hydrogen bonds along the periphery of the interface (data not shown). Hence the interface was relatively “dry” with just over two solvent molecules per 1′000 Å2. In accordance, the vast majority of residues involved in interface formation were non-bonded interactions between apolar residues. Interactions between the individual monomers of CGL3 leading to the tetrameric quarternary structure are depicted in Figure 4.
Figure 4. Multimerization interfaces and contact maps for the CGL3 tetramer.
The tetramer interface is illustrated for each unique interface within the tetramer. Cartoon representations are shown on the left with the corresponding contact maps on the right. In the interest of simplicity, only residues involved in typical bonding are depicted, i.e. polar residues and charged residues taking part in electrostatic interactions and hydrophobic residues in non-bonded interactions, respectively. Accordingly, residues are colored by type (pale blue: hydrophobic; orange: polar; blue: basic; red: acidic). Further, only water molecules buried within the interface are shown (red spheres). Dashed lines: turquoise, strong hydrogen bonds; gray, weak hydrogen bonds. Contact maps to the right represent heat-maps for the sum of non-bonded contacts between residues with an arbitrary scale given to the right (red, strong contacting; white, weak contacting). Subunit A is represented on the ordinate of the heat maps. (a) interface A-B. (b) interface A-C. (c) interface A-D.
Carbohydrate Coordination
While the overall sequence identity with the Coprinopsis galectins was moderate, most of the strictly conserved residues involved in coordination of carbohydrate ligands by galectins were conserved in CGL3, even at the level of rotamers (Fig. 5a). The most prominent exception was the substitution of the pivotal Trp residue of galectins with Arg at the equivalent position 81 in CGL3. The architecture of the binding site was equally conserved within the concave face of the beta sandwich fold. The residues coordinating the carbohydrate ligand were invariably found in strands S4 to S6 and the adjacent loop regions, respectively. In the CGL3-chitotetraose cocrystal the electron density of the carbohydrate ligand was well defined, bar the GlcNAc at the reducing end of the glycan, for which no electron density was observed. Hence the ligand was modelled as a chitotriose molecule. The electron density for the carbohydrate is shown in Figure 5b, representing a simulated annealing omit map. These results were in accordance with the ITC results where no increase in affinity from chitotriose to chitotetraose was observed (Table 2).
Figure 5. Comparison between carbohydrate coordination by CGL3 and CGL2.
(a) chitotriose coordination of CGL3 (left; PDB ID: 1R0H) and lactose coordination of CGL2 (right; PDB ID: 1ULC). Important amino acid side chains are displayed as sticks and colored according to the local sequence alignment in the middle of the figure. H-bonds are depicted as dashed black lines. (b) surface representation of the sugar binding pockets of the two lectins with the bound glycans shown as sticks. Protein electrostatics have been mapped onto the Connolly surface (vacuum electrostatics: red negatively, blue positively charged). The electron density for chitotriose is shown in the left panel as a simulated annealing omit map (Fo-Fc) contoured at 3.0 sigma.
Figure 5 illustrates the coordination of chitotetraose by CGL3 via direct hydrogen bonding. Direct as well as indirect hydrogen bonding via bridging water molecules is depicted in supplementary Figure 2. A summary of all interactions is found in Table 4. The GlcNAc moiety at the non-reducing end was most deeply buried in the binding site, in strikingly the same orientation as the beta-galactosides in galectins. The 2′-acetamido group of the non-reducing sugar was coordinated by direct hydrogen bonding of the carbonyl oxygen to Asn138 and water-mediated hydrogen bonding of the amide nitrogen to the backbone amide of Arg81. The 3′-OH of the sugar at the non-reducing end formed a hydrogen bond with the amide of Asn45 and was further coordinated by water molecules bridging to Asn47 and Asn138. Similarly, the 4′-hydroxyl was extensively water networked stretching as far as Ser134 and Lys136 residues located in the far end of the groove within beta-sheet S2. The exocyclic 6′-hydroxyl donated a hydrogen bond to Glu84 and received a bond from Asn73, both highly conserved interactions amongst galectins. Arg64 coordinated both the cyclic 5′-oxygen of the non-reducing GlcNAc and the 3′-OH of the second GlcNAc moiety, the latter hydroxyl group in turn donating a bond to Glu84 and accepting a further bond from Arg86. This too, represented a mode of coordination that is shared with some galectins. The amide group of the central GlcNAc was indirectly hydrogen bonded to Glu67 at the very entrance to the binding groove. As in CGL2, there was an identical quartet of salt-bridged Glu and Arg residues forming a cluster at the site of entrance to the binding groove (Arg64/Glu67/Glu84/Arg86). Arg81 donated a hydrogen bond to the exocyclic 6′-hydroxyl of the penultimate GlcNAc and further coordinated the 3′-OH of the GlcNAc at the reducing end, which was the sole direct interaction with this moiety and probably accounts for the observed difference in affinity between chitobiose and chitotriose/chitotetraose (Table 2). Furthermore, the glycosidic ether oxygen at the reducing end of the penultimate GlcNAc was hydrogen bonded to the the Nη of Arg81. Interestingly, the conserved His60 was not involved in any hydrogen bonding with the ligand, but appeared to be engaged in hydrophobic interaction with C4′ and C6′ of the non-reducing GlcNAc.
Table 4.
Chitotetraose coordination by CGL3
| Direct hydrogen bonds | ||
|---|---|---|
| DONOR | ACCEPTOR | Distance ± stddev (n=4) |
| Asn45 Nδ2 | O31 | 3.04 ± 0.15 |
| Arg64 Nη1 | O32 | 2.95 ± 0.06 |
| Arg64 Nη2 | O51 | 2.91 ± 0.12 |
| Asn73 Nδ2 | O61 | 3.01 ± 0.12 |
| Arg81 Nη2 | O33 | 3.00 ± 0.21* |
| Arg81 Nε | O62 | 2.82 ± 0.19 |
| Arg81 Nη2 | O43 | 3.02 ± 0.03* |
| O32 | Glu84 Oε1 | 2.52 ± 0.09 |
| O61 | Glu84 Oε1 | 2.67 ± 0.06 |
| Arg86 Nη1 | O32 | 2.94 ± 0.19 |
| Asn138 Nδ2 | O71 | 2.85 ± 0.12 |
| Water mediated hydrogen bonds | |
|---|---|
| Asn 47 Nδε2 | O31/O41* |
| Glu67 Oε2 | N22 |
| Arg81 N | O33/N21 |
| Ser134 Oγ | O41* |
| Lys136 N | O41* |
| Asn138 Oδ1 | O31 (O71/O41*) |
Distances are given in Å.
Numbering of chitotetraose atoms according to PDB Chemical Component Dictionary entry for chitotriose (CTO).
not observed in all four subunits.
The Arg residue at position 81 deserved particular attention since it took the place of the essential Trp of galectins (see below for functional investigations). Comparison of the complexed and the ligand-free binding site, revealed that this residue exhibited a significantly different conformation in the non-occupied binding cleft with a r.m.s.d. of 2.4 Å to the residue in the complexed protein, while the rest of the binding site remained quite unchanged (not shown). The temperature factor for this residue in the complexed structure was well below the average B-factor, whereas it was almost double the average B-factor when not complexed to the ligand. In addition, comparison of the uncomplexed and complexed state of the binding site revealed that water molecules were present at the positions corresponding to the 3′- and 6′-OH and the carbonyl oxygen of the 2′-acetamide positions of the non-reducing GlcNAc in the chitotetraose-bound crystal structure (not shown).
Functional Analysis of Carbohydrate-Coordinating Residues
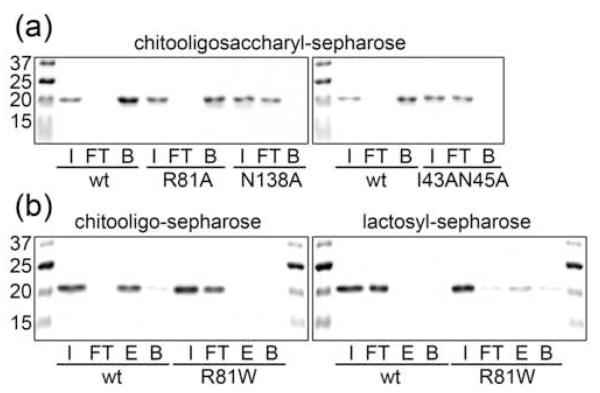
The functional significance of the above structural information for carbohydrate-binding by CGL3 was verified by changing critical carbohydrate-coordinating residues to Ala and assaying the chitooligosaccharide-binding of these CGL3 variants. Changed residues included I43 and N45 (double mutation), R81 and N138. In accordance with the CGL3-chitotetraose structure, changes of I43 and N45 as well as of N138 led to complete abrogation of chitooligosaccharide-binding as assayed by affinity chromatography (Fig. 6a). In contrast, change of R81 resulted in only a slight decrease in chitooligosaccharide-binding using the same assay. ITC measurements of the same mutant revealed an approximately three-fold reduced affinity towards chitotriose (Table 2). These results suggested that the contribution of R81 to the carbohydrate affinity of CGL3 was small. This was in contrast to the corresponding residue W72 in CGL2 where an analogous mutation abrogated binding 20. Interestingly, change of the R81 residue to Trp led to complete loss of chitotetraose binding probably due to steric hindrance (Fig. 6b). In summary, N45 (and/or I43) and N138 appear to be crucial for the affinity whereas R81 determines the specificity of carbohydrate-binding by CGL3.
Figure 6. Solid-phase binding assay using WT and mutated CGL3.
Coomassie-stained SDS-PAGE gels of proteins tested with chitooligosaccharyl- and lactosyl sepharose are shown. Abbreviations in the figure stand for the following terms: I (input), FT (flow through), E (elution) bound fraction specifically released using the respective sugars in solution, B (beads) residual protein on column detached by boiling the sepharose beads in protein sample buffer. Equivalent amounts of each fraction were loaded on the gel. (a) binding of WT and R81A, N138A and I43AN45A mutant forms of CGL3 to chitooligosaccharyl-sepharose. (b) binding of WT and R81W mutant form of CGL3 to chitooligosaccharyl- and lactosyl-sepharose.
Role of R81 for Carbohydrate-Binding Specificity of CGL3
The structure of the CGL3 carbohydrate-recognition groove suggested that all residues except a conserved Trp residue are in place for the coordination of lactose. We therefore tested the R81W variant of CGL3 for lactose coordination. Carbohydrate-binding was assessed using carbohydrate-affinity matrices (Fig. 6). In accordance with the results of the glycan array, CGL2 bound to lactose but not chitooligosaccharides (data not shown) whereas WT CGL3 bound to chitooligosaccharides but not to lactose. In contrast, CGL3 (R81W) was no longer able to bind to chitooligosaccharides but was partially retained on lactosyl-sepharose and specifically eluted from this matrix with lactose (Fig. 6b). However, the affinity of CGL3 (R81W) towards lactose or LacNAc was too low to be reliably quantified by ITC (data not shown). On the one hand, these results confirmed the different carbohydrate-binding specificities of CGL2 and CGL3 as determined by the glycan array. On the other hand, they demonstrated that the specificity of a galectin-related protein can be dramatically altered by change of a single amino acid residue.
Discussion
A characteristic of carbohydrate-recognition by lectins is the apparent discrepancy between the limited number of lectin folds and the comparably large variety of recognized carbohydrates. High-resolution structures of various lectin-carbohydrate complexes revealed that the carbohydrate-binding groove within a given lectin fold is highly variable in that changes in few carbohydrate-coordinating amino acid residues can result in significantly altered specificities. One of the best examples for this variability is the legume lectin fold with specificities ranging from monosaccharides such as Glc/Man, Gal/GalNAc, Fuc to GlcNAc/chitobiose to more complex oligosaccharides 28. This concept is less accepted for the galectin fold whose specificity seemed to be restricted to oligosaccharides harboring a core Gal in beta-glycosidic linkage to either Glc or GlcNAc and to vary just in the position and the nature of additional substituents on the Gal or Glc/GlcNAc moiety. However, it is known that there are a number of so-called galectin-like proteins which are homologous to galectins but display alterations in conserved Lac and LacNAc-coordinating residues and fail to bind to these minimal galectin ligands 5. These proteins are likely lectin candidates of the galectin fold family with altered carbohydrate-binding specificity. However, to our knowledge, there has hitherto been no experimental evidence to support this hypothesis.
Here, we present a lectin whose primary, secondary and tertiary structure is highly similar to galectins but which displays an Arg residue at the position of the critical Trp residue. The structure of the chitotetraose-CGL3 complex revealed that this deviation allowed the coordination of a Glc pyranose ring at the position of the Gal pyranose ring in the carbohydrate-binding groove. Replacement of the Arg by Trp (R81W) abolishes the bindingof chitooligosaccharides (binding of LacdiNAc could not be tested due to the unavailability of this sugar) suggesting steric incompatibility between coordination of the acetamido group (see below) and stacking with the pyranose ring of the sugar at the non-reducing end. On the other hand, the same variant showed weak but significant binding of lactose suggesting that the carbohydrate-binding groove of CGL3 was capable of accommodating bona fide galectin ligands albeit with very low affinity. Interestingly, the Arg residue in CGL3 coordinated the two penultimate carbohydrates of the oligosaccharide rather than the sugar at the non-reducing end. However, in contrast to the corresponding W72 residue in CGL2, these coordinations by R81 appeared to contribute rather to the carbohydrate-binding specificity than to the affinity of CGL3 since changing this residue to an Ala (R81A) reduces chitotriose-binding only slightly. More critical in this respect were residues N45 and N138 coordinating the 2′-acetamido group of the sugar at the non-reducing end and leading to specific binding of per-N-acetylated oligosaccharides such as LacdiNAc and chitooligosaccharides. This finding was in agreement with other GlcNAc or GalNAc-binding lectins where the acetamido group of acetylated sugars is often a dominant carbohydrate recognition element 29. However, the apparent lack of stacking or hydrophobic interactions with aromatic amino acid residues is, to the best of our knowledge, unique among chitooligosaccharide-coordinating lectins 30; 31; 32. The only other example of a change of an aromatic by an aliphatic residue in a carbohydrate-binding groove is found among legume lectins. In the homotetrameric Dolichos biflorus lectin (DBL), which prefers GalNAc over Gal, the Phe or Tyr residue (stacking with the Gal pyranose ring in other legume lectins) is replaced by a Leu residue 33; 34. However, in contrast to CGL3, this Leu interacts with the same sugar moieties as the Phe/Tyr residues in the other family members and, accordingly, changing this residue to a Phe increased affinity for both GalNAc and Gal but did not alter specificity 34.
The biological role of CGL3 is unclear at present. Since fungal cell walls contain chitin, CGL3 might interfere with fungal growth. However, exogenous CGL3 did not affect spore germination or vegetative growth of a representative panel of fungi (data not shown). Furthermore, despite its developmental regulation, CGL3 does not seem to play an essential role in fruiting body formation in C. cinerea, since silencing of the cgl3 gene, performed as described for cgl1/2 22 did not affect this developmental process (data not shown).
In summary, CGL3 is the first example of a lectin with a galectin fold for which binding of a non-Gal-containing ligand was demonstrated both at biochemical and structural level. In the light of this finding, we hypothesize that many galectin-related proteins from animals containing deviations at positions strictly conserved among β-galactoside-binding galectins represent actual lectins with novel substrate specificities.
Materials and Methods
Cloning and Expression
The cgl3 gene was amplified from C. cinerea Amut Bmut genomic DNA by PCR with forward NdeI-CGL3N and reverse BamHI-CGL3C primers, introducing 5′-NdeI and 3′-BamHI restriction sites. Alternatively, forward primer NdeI-His8-CGL3N harboring the coding sequence for a N-terminal His8-tag was used. PCR products were ligated into pET24a using the introduced restriction sites. The same technique was applied to generate the CGL2 expression plasmid. Expression plasmids for CGL3 mutant proteins were obtained using overlap PCR. Sequences of all primers used are found in Table 5. Expression was carried out in E. coli BL21(DE3) in terrific broth supplemented with kanamycin at 37 °C. Cells were grown to an OD600 of 2.0 and induced with 0.5 mM isopropyl-β-D-thiogalactoside for 6 h, then harvested, frozen and stored at −20 °C.
Table 5.
Oligonucleotide primers
| NdeI–CGL3N | 5′–GGGGGGGGCATATGTTCCACATCCTCAGACTCG–3′ |
| BamHI–CGL3C | 5′–GGGGGGGGATCCTCAATTGGCGCGCGGCATGACG–3′ |
| NdeI–His8–CGL3N | 5′–GGGGGGGGCATATGGGCCATCATCATCATCATCATCATCACAGCGGCTTCCACATCCTCAGACTCGAATC–3′ |
| CGL3–I43AN45A–fwd | 5′–CAACTTGGGACCCACCGGGGCCGACGCCACCAACGTCAACTTGATCAAT–3′ |
| CGL3–I43AN45A–rev | 5′–ATTGATCAAGTTGACGTTGGTGGCGTCGGCCCCGGTGGGTCCCAAGTTG–3′ |
| CGL3–R81W–fwd | 5′–TACGGCGAAAGCTGGGGCCCTGAGGAACGAATTCC–3′ |
| CGL3–R81W–rev | 5′–TCGTTCCTCAGGGCCCCAGCTTTCGCCGTACGGGA–3′ |
| CGL3–R81A–fwd | 5′–GTATCCCGTACGGCGAAAGCGCGGGCCCTGAGGAACGAATTCC–3′ |
| CGL3–R81A–rev | 5′–GGAATTCGTTCCTCAGGGCCCGCGCTTTCGCCGTACGGGATAC–3′ |
| CGL3–N138A–fwd | 5′–GAGAAAGTGTCCTACAAGATCGCCGAAGGGCAGACCCCGCCT–3′ |
| CGL3–N138A–rev | 5′–AGGCGGGGTCTGCCCTTCGGCGATCTTGTAGGACACTTTCTC–3′ |
| NdeI–CGL2N | 5′–GGGGGGCATATGCTCTACCACCTCTTCGTCAACAA–3′ |
| BamHI–CGL2C | 5′–GGGGGGGGATCCCTAAGCAGGGGGAAGTGGGGGG–3′ |
Purification and Crystallization
Cells were resuspended in ice-cold TBS (10 mM Tris-HCl pH 7.5, 150 mM NaCl) containing 1 mM PMSF and ruptured using a French press (SLM Aminco, SLM Instruments Inc., UK). All subsequent steps were carried out at 4 °C. Cell debris was pelleted by consecutive centrifugation steps at 4300 g for 10 min and 27000 g for 30 min. The resulting supernatant was applied to Talon metal affinity resin (BD Biosciences, USA) in the case of His-tagged CGL3 or chitooligosaccharyl-sepharose (see below) in the case of untagged CGL3. After washing, proteins were eluted at 25 °C in TBS containing 200 mM imidazol or 200 mM chitooligosaccharides, respectively. Eluted proteins were purified with a HiLoad 16/60 Superdex 75 column (GE Healthcare, USA) equilibrated in TBS and finally concentrated in an Amicon Ultra-4 centrifugal filter device (Millipore, USA) with a cutoff of 10 kDa. Protein concentrations were determined using the BCA protein assay (Pierce, USA).
Crystals were grown at 18 °C using the hanging-drop vapor diffusion technique. Crystals of ligand-free, N-terminally His-tagged CGL3 were obtained by mixing 2.5 μl protein solution (26 mg/ml in TBS) with 2.5 μl mother liquor composed of 100 mM sodium citrate (pH 5.1) and 47% 2-Methyl-2,4-pentanediol (MPD). Untagged, ligand-free CGL3 crystallized with a mother liquor of 100 mM sodium citrate (pH 4.8) and 55% MPD. Cocrystals of untagged CGL3 were obtained by mixing 2.5 μl protein solution (15 mg/ml) containing 2 mM chitotetraose with 2.5 μl mother liquor consisting of 100 mM sodium citrate (pH 5.6), 0.9 M lithium sulfate and 0.5 M ammonium sulfate. Crystals were cryostabilized after two weeks in their respective mother liquor supplemented with 25% ethylene glycol and flash frozen in liquid nitrogen. Crystals of ligand-free CGL3 were frozen without further cryostabilization.
Structure Solution, Refinement and Analysis
Data sets were collected at the Swiss Light Source beamline X06SA at 100 K and processed with XDS 35. Intensities were converted to amplitudes in TRUNCATE as part of the CCP4 suite (CCP4, 1994). The structure of the N-terminally His-tagged CGL3 was initially solved by molecular replacement with a truncated CGL2 20 as a search molecule using Phaser 36. Iterative model rebuilding and refinement were done using Refmac5.2 37, CNS 38 and Coot 39. Model statistics were obtained with Procheck/Sfcheck as part of the CCP4 suite together with the programmes stated above (CCP4, 1994). Hydrogen bonding, subunit and ligand contact networks were analyzed with HBPLUS 40 and Ligplot 41. Molecular visualizations were done using PyMol 42 and Swiss PDB Viewer 43. Subunit contact maps were created using NOC (http://noch.sourceforge.net).
In the complexed structure, which contained a tetramer in the asymmetric unit, the tetramer gave rise to four-fold non-crystallographic symmetry (NCS), which was used throughout structure refinement as loose restraints for both side chain and main chain atoms. Carbohydrate molecules were not included in NCS restraints. The ligand-free structures contained a tetramer as a consequence of crystallographic symmetry relating the dimers in the asymmetric unit. Here, loose two-fold NCS restraints were used throughout refinement. No gross positional deviations were observed between subunits of the tetramer when NCS was not included in the refinement, suggesting that the use of loose NCS restraints was not forcing conformity that was not there to begin with (data not shown).
Analysis of the Φ/Ψ-torsion angles of the β1-4 linkages within the chitooligosaccharides revealed no significant conformational changes as compared to glycan structures that were energy minimized by molecular dynamics or when comparing against database values for glycan torsions in the PDB with similar resolution using the GlyTorsion tool44.
Sequence Alignments
Alignments were calculated with Multalin 45 using Blosum62-12-2 alignment parameters and figures were prepared with ESPript2.2 46.
Production of Antiserum against CGL3
Immunization of two rabbits with purified recombinant N-terminally His8-tagged CGL3 yielded two equally specific polyclonal antisera (Pineda Antikörper Service, Germany).
Glycan Array Analysis
Purified N-terminally His8-tagged CGL3 and untagged CGL2 were used at 30 μg/ml to probe the plate glycan array version 3.7 by Core H of the consortium for functional glycomics (http://www.functionalglycomics.org/static/index.shtml). Bound lectins were detected using the specific antisera and goat anti-rabbit IgG-Alexa Fluor 488 according to the consortium’s standard protocol.
Solid-Phase Carbohydrate-Binding Experiments
Lactosyl- and chitooligosaccharyl-sepharose were prepared by divinyl sulfone coupling 47. Chitooligosaccharide mixture, chitobiose, chitotriose and chitotetraose were purchased from Seikagaku (Japan), lactose from Sigma (USA). The experiments were performed using purified His8-CGL3 and the mutated versions thereof as well as untagged CGL2. CGL2 was purified as described above for untagged CGL3, using lactosyl-sepharose and lactose. For the binding experiments, equal amounts of pure protein were incubated with the respective matrices under agitation at 4 °C for 1 h. The bound proteins were eluted either with 200mM carbohydrates in solution and / or by boiling in SDS-PAGE sample buffer after washing. Protein samples were separated on a 15% SDS-PAGE and stained with Coomassie.
Isothermal Titration Microcalorimetry Measurements
Experiments were performed with a VP-ITC isothermal titration calorimeter (Microcal, MA, USA) at 25 °C using purified N-terminally His8-tagged CGL3 (1 mM) or untagged CGL2 (2.24 mM). 10 mM chitooligosaccharides and 20 mM lactose were dissolved in TBS. Chitooligosaccharides were titrated into a cell containing the protein solution in 48 injections of 6 μl preceeded by a single one of 2 μl. In the case of CGL2, 2 μl portions of lactose were injected 148 times. All injections were done at intervals of 3 min while stirring at 270 rpm. Experimental data were fitted to a theoretical titration curve using the MCS-ORIGIN software supplied by Microcal. Association constants (Ka and enthalpy change (ΔH) were obtained using a model of 1 ligand binding site per protein monomer. Dissociation constants (Kd), change in free energy (ΔG) and entropy of binding (TΔS) were calculated.
Supplementary Material
SI Figure 1: Isothermal titration calorimetry results for binding of CGL3 (1 mM) to chitotriose (10 mM) in TBS (pH 7.5) at 298° K. Top, data from 49 injections of 6 μl chitotriose into CGL3-containing cell. Bottom, plot of total heat released as a function of ligand concentration for the titration shown above. The best least-square fit for the obtained data is depicted by a solid line (hidden by the squares).
SI Figure 2: Chitotriose coordination by CGL3. Sugar binding pocket of ligand-bound CGL3 (PDB ID: 1R0H) is depicted as cartoon. Important amino acid residues are displayed as sticks and colored according to the partial CGL3 sequence on the bottom of the figure. Direct and indirect H-bonds are represented as dashed black and red lines, respectively. Water molecules are shown as blue spheres and chitotriose is colored in yellow.
Acknowledgments
The glycan-array analysis was conducted by the Protein-Carbohydrate Interaction Core of The Consortium for Functional Glycomics funded by the National Institute of General Medical Sciences grant GM62116. We thank Richard Alvarez and Angela Lee of the Consortium for Functional Glycomics for screening the glycan array as well as C. Schulze-Briese and S. Gutman for assistance with data collection at the Swiss Light Source (Villigen, Switzerland). We appreciate the collaboration with Nenad Ban of the Institute of Molecular Biology and Biophysics (ETH Zürich). This work was supported by funds of the ETH Zürich and the Swiss National Science Foundation (Grant No. 3100A0-116827 to M.K. and Fellowship PAOOA-109094 to P.J.W.).
Footnotes
Accession numbers Coordinates and structure factors have been deposited in the Protein Data Bank (PDB; New Brunswick, USA) under codes 2R0F and 2R0H.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barondes SH, Cooper DN, Gitt MA, Leffler H. Galectins. Structure and function of a large family of animal lectins. J Biol Chem. 1994;269:20807–10. [PubMed] [Google Scholar]
- 2.Kasai K, Hirabayashi J. Galectins: a family of animal lectins that decipher glycocodes. J Biochem (Tokyo) 1996;119:1–8. doi: 10.1093/oxfordjournals.jbchem.a021192. [DOI] [PubMed] [Google Scholar]
- 3.Leffler H, Carlsson S, Hedlund M, Qian Y, Poirier F. Introduction to galectins. Glycoconj J. 2004;19:433–40. doi: 10.1023/B:GLYC.0000014072.34840.04. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed H, Vasta GR. Galectins: conservation of functionally and structurally relevant amino acid residues defines two types of carbohydrate recognition domains. Glycobiology. 1994;4:545–8. doi: 10.1093/glycob/4.5.545. [DOI] [PubMed] [Google Scholar]
- 5.Cooper DN. Galectinomics: finding themes in complexity. Biochim Biophys Acta. 2002;1572:209–31. doi: 10.1016/s0304-4165(02)00310-0. [DOI] [PubMed] [Google Scholar]
- 6.Chiariotti L, Salvatore P, Frunzio R, Bruni CB. Galectin genes: regulation of expression. Glycoconj J. 2004;19:441–9. doi: 10.1023/B:GLYC.0000014073.23096.3a. [DOI] [PubMed] [Google Scholar]
- 7.He J, Baum LG. Galectin interactions with extracellular matrix and effects on cellular function. Methods Enzymol. 2006;417:247–56. doi: 10.1016/S0076-6879(06)17017-2. [DOI] [PubMed] [Google Scholar]
- 8.Pace KE, Baum LG. Insect galectins: roles in immunity and development. Glycoconj J. 2004;19:607–14. doi: 10.1023/B:GLYC.0000014092.86763.2f. [DOI] [PubMed] [Google Scholar]
- 9.Sato S, Nieminen J. Seeing strangers or announcing “danger”: galectin-3 in two models of innate immunity. Glycoconj J. 2004;19:583–91. doi: 10.1023/B:GLYC.0000014089.17121.cc. [DOI] [PubMed] [Google Scholar]
- 10.Kohatsu L, Hsu DK, Jegalian AG, Liu FT, Baum LG. Galectin-3 induces death of Candida species expressing specific beta-1,2-linked mannans. J Immunol. 2006;177:4718–26. doi: 10.4049/jimmunol.177.7.4718. [DOI] [PubMed] [Google Scholar]
- 11.Nakahara S, Raz A. On the role of galectins in signal transduction. Methods Enzymol. 2006;417:273–89. doi: 10.1016/S0076-6879(06)17019-6. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez JD, Baum LG. Ah, sweet mystery of death Galectins and control of cell fate. Glycobiology. 2002;12:127R–36R. doi: 10.1093/glycob/cwf081. [DOI] [PubMed] [Google Scholar]
- 13.Hsu DK, Yang RY, Liu FT. Galectins in apoptosis. Methods Enzymol. 2006;417:256–73. doi: 10.1016/S0076-6879(06)17018-4. [DOI] [PubMed] [Google Scholar]
- 14.Liu FT, Patterson RJ, Wang JL. Intracellular functions of galectins. Biochim Biophys Acta. 2002;1572:263–73. doi: 10.1016/s0304-4165(02)00313-6. [DOI] [PubMed] [Google Scholar]
- 15.Delacour D, Greb C, Koch A, Salomonsson E, Leffler H, Le Bivic A, Jacob R. Apical sorting by galectin-3-dependent glycoprotein clustering. Traffic. 2007;8:379–88. doi: 10.1111/j.1600-0854.2007.00539.x. [DOI] [PubMed] [Google Scholar]
- 16.Nickel W. The mystery of nonclassical protein secretion. A current view on cargo proteins and potential export routes. Eur J Biochem. 2003;270:2109–19. doi: 10.1046/j.1432-1033.2003.03577.x. [DOI] [PubMed] [Google Scholar]
- 17.Boulianne RP, Liu Y, Aebi M, Lu BC, Kues U. Fruiting body development in Coprinus cinereus: regulated expression of two galectins secreted by a non-classical pathway. Microbiology. 2000;146(Pt 8):1841–53. doi: 10.1099/00221287-146-8-1841. [DOI] [PubMed] [Google Scholar]
- 18.Yagi F, Hiroyama H, Kodama S. Agrocybe cylindracea lectin is a member of the galectin family. Glycoconj J. 2001;18:745–9. doi: 10.1023/a:1021193432577. [DOI] [PubMed] [Google Scholar]
- 19.Yang N, Tong X, Xiang Y, Zhang Y, Liang Y, Sun H, Wang DC. Molecular character of the recombinant antitumor lectin from the edible mushroom Agrocybe aegerita. J Biochem (Tokyo) 2005;138:145–50. doi: 10.1093/jb/mvi109. [DOI] [PubMed] [Google Scholar]
- 20.Walser PJ, Haebel PW, Kunzler M, Sargent D, Kues U, Aebi M, Ban N. Structure and functional analysis of the fungal galectin CGL2. Structure. 2004;12:689–702. doi: 10.1016/j.str.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Ban M, Yoon HJ, Demirkan E, Utsumi S, Mikami B, Yagi F. Structural basis of a fungal galectin from Agrocybe cylindracea for recognizing sialoconjugate. J Mol Biol. 2005;351:695–706. doi: 10.1016/j.jmb.2005.06.045. [DOI] [PubMed] [Google Scholar]
- 22.Walti MA, Villalba C, Buser RM, Grunler A, Aebi M, Kunzler M. Targeted gene silencing in the model mushroom Coprinopsis cinerea (Coprinus cinereus) by expression of homologous hairpin RNAs. Eukaryot Cell. 2006;5:732–44. doi: 10.1128/EC.5.4.732-744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swaminathan GJ, Leonidas DD, Savage MP, Ackerman SJ, Acharya KR. Selective recognition of mannose by the human eosinophil Charcot-Leyden crystal protein (galectin-10): a crystallographic study at 1.8 A resolution. Biochemistry. 1999;38:13837–43. doi: 10.1021/bi990756e. [DOI] [PubMed] [Google Scholar]
- 24.Leonidas DD, Elbert BL, Zhou Z, Leffler H, Ackerman SJ, Acharya KR. Crystal structure of human Charcot-Leyden crystal protein, an eosinophil lysophospholipase, identifies it as a new member of the carbohydrate-binding family of galectins. Structure. 1995;3:1379–93. doi: 10.1016/s0969-2126(01)00275-1. [DOI] [PubMed] [Google Scholar]
- 25.Swamy S, Uno I, Ishikawa T. Morphogenic effects of mutations at the A and B incompatibility factors in Coprinus cinereus. J Gen Microbiol. 1984;130:3219–3224. [Google Scholar]
- 26.May G, Le Chevanton L, Pukkila PJ. Molecular analysis of the Coprinus cinereus mating type A factor demonstrates an unexpectedly complex structure. Genetics. 1991;128:529–38. doi: 10.1093/genetics/128.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dam TK, Brewer CF. Thermodynamic studies of lectin-carbohydrate interactions by isothermal titration calorimetry. Chem Rev. 2002;102:387–429. doi: 10.1021/cr000401x. [DOI] [PubMed] [Google Scholar]
- 28.Loris R, Hamelryck T, Bouckaert J, Wyns L. Legume lectin structure. Biochim Biophys Acta. 1998;1383:9–36. doi: 10.1016/s0167-4838(97)00182-9. [DOI] [PubMed] [Google Scholar]
- 29.Weis WI, Drickamer K. Structural basis of lectin-carbohydrate recognition. Annu Rev Biochem. 1996;65:441–73. doi: 10.1146/annurev.bi.65.070196.002301. [DOI] [PubMed] [Google Scholar]
- 30.Harata K, Muraki M. Crystal structures of Urtica dioica agglutinin and its complex with tri-N-acetylchitotriose. J Mol Biol. 2000;297:673–81. doi: 10.1006/jmbi.2000.3594. [DOI] [PubMed] [Google Scholar]
- 31.Hayashida M, Fujii T, Hamasu M, Ishiguro M, Hata Y. Similarity between protein-protein and protein-carbohydrate interactions, revealed by two crystal structures of lectins from the roots of pokeweed. J Mol Biol. 2003;334:551–65. doi: 10.1016/j.jmb.2003.09.076. [DOI] [PubMed] [Google Scholar]
- 32.Loris R, De Greve H, Dao-Thi MH, Messens J, Imberty A, Wyns L. Structural basis of carbohydrate recognition by lectin II from Ulex europaeus, a protein with a promiscuous carbohydrate-binding site. J Mol Biol. 2000;301:987–1002. doi: 10.1006/jmbi.2000.4016. [DOI] [PubMed] [Google Scholar]
- 33.Bouckaert J, Hamelryck T, Wyns L, Loris R. Novel structures of plant lectins and their complexes with carbohydrates. Curr Opin Struct Biol. 1999;9:572–7. doi: 10.1016/s0959-440x(99)00007-x. [DOI] [PubMed] [Google Scholar]
- 34.Hamelryck TW, Loris R, Bouckaert J, Dao-Thi MH, Strecker G, Imberty A, Fernandez E, Wyns L, Etzler ME. Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus. J Mol Biol. 1999;286:1161–77. doi: 10.1006/jmbi.1998.2534. [DOI] [PubMed] [Google Scholar]
- 35.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appli. Cryst. 1993;26:795–800. [Google Scholar]
- 36.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–64. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–93. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 41.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–34. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 42.DeLano WL. The PyMOL Molecular Graphics System. 2006.
- 43.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 44.Lutteke T, Frank M, von der Lieth CW. Carbohydrate Structure Suite (CSS): analysis of carbohydrate 3D structures derived from the PDB. Nucleic Acids Res. 2005;33:D242–6. doi: 10.1093/nar/gki013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–90. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fornstedt N, Porath J. Characterization studies on a new lectin found in seeds of Vicia ervilia. FEBS Lett. 1975;57:187–91. doi: 10.1016/0014-5793(75)80713-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SI Figure 1: Isothermal titration calorimetry results for binding of CGL3 (1 mM) to chitotriose (10 mM) in TBS (pH 7.5) at 298° K. Top, data from 49 injections of 6 μl chitotriose into CGL3-containing cell. Bottom, plot of total heat released as a function of ligand concentration for the titration shown above. The best least-square fit for the obtained data is depicted by a solid line (hidden by the squares).
SI Figure 2: Chitotriose coordination by CGL3. Sugar binding pocket of ligand-bound CGL3 (PDB ID: 1R0H) is depicted as cartoon. Important amino acid residues are displayed as sticks and colored according to the partial CGL3 sequence on the bottom of the figure. Direct and indirect H-bonds are represented as dashed black and red lines, respectively. Water molecules are shown as blue spheres and chitotriose is colored in yellow.