Summary
Histone modifications have been implicated in stem cell maintenance and differentiation. We have analyzed genome-wide changes in gene expression and histone modifications during differentiation of multipotent human primary hematopoietic stem/progenitor cells (HSCs/HPCs) into erythrocyte precursors. Our data indicate that H3K4me1, H3K9me1 and H3K27me1 associate with enhancers of differentiation genes prior to their activation and correlate with basal expression, suggesting that these mono-methylations are involved in the maintenance of activation potential required for differentiation. In addition, although the majority of genes associated with both H3K4me3 and H3K27me3 in HSCs/HPCs become silent and lose H3K4me3 after differentiation, those that lose H3K27me3 and become activated after differentiation are associated with increased levels of H2A.Z, H3K4me1, H3K9me1, H4K20me1 and RNA polymerase II in HSCs/HPCs. Thus, our results suggest that gene expression changes during differentiation are programmed by chromatin modifications present at the HSC/HPC stage and we provide a resource for enhancer and promoter identification.
Introduction
The adult hematopoietic system consists of multiple distinct blood cell lineages and is continuously regenerated from common hematopoietic stem cells (HSCs) under normal conditions or following bone marrow transplantation (Morrison et al., 1995). While a stable pool is maintained by self-renewal, the multipotent HSCs continuously differentiate to produce a large number of blood cells. It remains unclear how the balance between self-renewal and differentiation is controlled and how a decision for differentiation is specified at molecular levels. However, it is clear that transcription programs, which include both activation of genes involved in the target lineage and repression of genes involved in non-target lineages, play essential roles during this process of fate determination (Surani et al., 2007). These specific transcription programs are controlled by a close coordination between transcription factors and chromatin states, both of which are regulated by extracellular signals. Appropriate chromatin modifications, including histone modifications, can help to maintain a relatively stable expression pattern of either activation or repression in stem cells or terminally differentiated cells. Indeed, enzymes that modulate chromatin structure, including Brg1 and Ezh2, have been implicated in regulating embryonic development and embryonic stem (ES) cell function (reviewed by (Surani et al., 2007)) and changes in chromatin structure have been reported during differentiation of ES cells (Bernstein et al., 2005; Chambeyron and Bickmore, 2004; Mikkelsen et al., 2007; Mohn et al., 2008) and other cells such as T cells and red blood cells (Ansel et al., 2006; de Laat et al., 2008).
Previous studies have provided genome-wide maps of histone modifications H3K4me3 and H3K27me3 in ES cells (Bernstein et al., 2006; Mikkelsen et al., 2007; Pan et al., 2007; Zhao et al., 2007). Many critical regions involved in pluripotency and differentiation of ES cells are associated with both H3K4me3 and H3K27me3 modifications and are termed ‘bivalent domains’ (Bernstein et al., 2006). The relative levels of these two modifications can effectively discriminate genes that are expressed or repressed in ES cells (Mikkelsen et al., 2007) and human CD4+ T cells (Barski et al., 2007; Roh et al., 2006). Regions with both H3K4me3 and H3K27me3 modifications have been proposed to play critical roles and can be resolved to monovalent modification in ES cell differentiation (Azuara et al., 2006; Bernstein et al., 2006; Mohn et al., 2008). However, it is not clear what controls the fate choice of bivalent genes.
CD34+ or CD133+ cells from human bone marrow or periphery blood contain hematopoietic stem cells that can sustain long-term hematopoiesis after transplantation (Morrison et al., 1995; Yin et al., 1997). These cells can be differentiated into particular cell types in vitro under defined conditions. The best characterized pathways for in vitro differentiation of the CD34+ or CD133+ cells are the production of erythrocyte precursor cells, which can be further induced to mature red blood cells (Giarratana et al., 2005). Even though the mechanisms of hematopoietic differentiation are still not fully clear, it is known that extensive reorganization of chromatin structure at critical loci occurs during the process (de Laat et al., 2008; Litt et al., 2001), which is regulated by a complex interplay between cis elements and trans factors including both transcription factors and chromatin-modifying enzymes. Only limited chromatin regions have been analyzed during the differentiation of hematopoietic stem cells into erythrocyte cells and it is therefore important to investigate the global changes of chromatin modifications during such process. However, almost any stem cell populations isolated from humans, including the CD34+ or CD133+ hematopoietic stem cells, are complex and composed of multiple progenitors with different differentiation potentials. While considerable efforts have been put in developing techniques to prospectively isolate sub-populations, the best procedures yield relatively complex mixture of cells and a very low number of cells. Another challenge is that these cells do not self renew in culture and are therefore in limiting quantities. Even with these limitations, the CD4+ and CD133+ cells isolated from human bone marrow or periphery blood have demonstrated clinical efficacy in transplantation therapies (Bitan et al., 2005) and are therefore widely used in treatment of various hematological disorders. It is therefore crucial to understand the epigenetic states of these important cells. Investigation of the global patterns of chromatin modification in these cells can provide valuable information, help understand the differences between HSCs/HPCs with hESCs and more differentiated hematopoietic cells and may provide clues for designing better therapeutic strategies. We have therefore examined the genome-wide distribution of chromatin modifications in the human HSCs/HPCs and differentiated erythrocyte precursor cells. Our data indicate that differentiation of CD133+ cells into CD36+ cells is accompanied by dramatic changes of histone modifications at critical genetic regions. Even though both H3K27me3 and H3K9me3 have been implicated in gene repression, we find that they are correlated with silencing different subsets of genes. Our data indicate that while a small fraction of bivalent genes in HSCs/HPCs lost H3K27me3 after differentiation, the majority of the bivalent genes lost H3K4me3. The association of increased levels of H3K4me1, H3K9me1, H2A.Z and RNA polymerase II with the bivalent genes indicates the potential of their losing H3K27me3 and becoming transcriptionally active during differentiation. These results suggest that the choice of resolution of bivalent modifications during differentiation is already programmed at the HSC/HPC stage.
Results
Experimental design
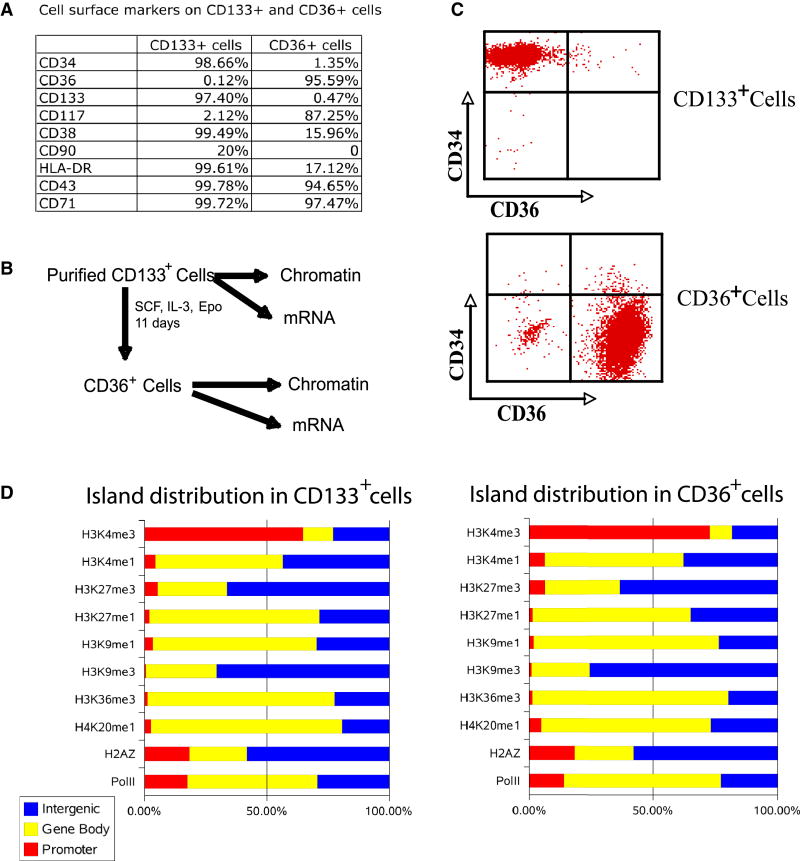
Mobilized CD133+ cells were purified from human periphery blood lymphocytes (PBLs) using established protocols (Migliaccio et al., 2002). To better characterize the composition of the cell populations, we stained the cells for the presence of various surface markers. Human HSCs are characterized by high levels of CD34 and CD133 expression, intermediate levels of CD117 (c-kit) and CD90 (Thy-1), and no or low levels of CD38, HLA-DR and CD71 (Szilvassy, 2003). Indeed, we found high levels of CD34 and CD133 expression in more than 98% of the cells and CD117 and CD90 expression in a small fraction (2 to 20%) of the cells, consistent with these cells being stem cells or progenitor cells (Figure 1A). However, more than 99% of the cells were CD38, HLA-DR and CD71-positive, indicating that the vast majority of these cells were early progenitor cells.
Figure 1. Differentiation of CD133+ cells into CD36+ cells.
A. Cell surface markers on CD133+ and CD36+ cells. The cells were stained using specific antibodies indicated on the left column and analyzed by flow cytometry. The fractions of cells exhibited positive staining for each antibody are indicated.
B. Experimental scheme.
C. FACS analysis of the CD34 and CD36 expression levels before (upper panel) and after (lower panel) differentiation.
D. The genome-wide distribution of different modifications. Total numbers of islands in promoter, gene body and intergenic regions were identified for each modification (see Experimental Procedures for details). The graph shows the fraction of islands in each different region for each modification in CD133+ and CD36+ cells.
We differentiated the CD133+ cells to CD36-expressing cells, which are precursor cells of erythrocytes (Figure 1B), using an established protocol (Wong et al., 2008). FACS analysis confirmed that more than 98% of the CD133+ cells expressed high levels of CD34 and no detectible levels of CD36 (Figure 1C). After differentiation, more than 95% of the cells expressed CD36 with the concurrent disappearance of CD34 expression (Figures 1A and 1C). At the same time, CD117 increased from 2.12% in CD133+ cells to 87.25% in CD36+ cells, whereas CD38 and HLA-DR decreased to 15.96% and 17.12%, respectively after differentiation (Figure 1A).
To examine the changes of chromatin modifications before and after differentiation, we fragmented the chromatin from CD133+ and CD36+ cells using micrococcal nuclease (MNase) digestion and performed ChIP-Seq analysis for histone methylation (H3K4me1, H3K4me3, H3K9me1, H3K9me3, H3K27me1, H3K27me3, H3K36me3 and H4K20me1) and histone variant H2A.Z. as described previously (Barski et al., 2007). The distribution of RNA polymerase II (Pol II) was also determined using ChIP-Seq. To test the reproducibility of the modification patterns, we repeated the ChIP-Seq analyses for H3K4me1 and H3K27me1 in both CD133+ and CD36+ cells. Our results indicate that all the major peaks of these modifications are reproducible (Figure S1).
Global chromatin modification patterns in CD133+ and CD36+ cells
To identify genomic regions associated with various histone modifications, we searched for regions enriched for modifications using a statistical model (see Supplemental Information for details). Our data indicate that the distribution of modification islands varied dramatically among different modifications. For example, 65% and 73% of the H3K4me3 islands were found in the promoter regions in CD133+ and CD36+ cells, respectively, whereas 71% to 76% of the H3K9me3 islands were detected in the intergenic regions with less than 1% in promoter regions (Figure 1C; Table S1). Other modifications including H3K4me1, H3K9me1, H3K27me1, H3K36me3 and H4K20me1 were mainly detected in the transcribed regions (Figure 1C; Table S1). We then compared directly the distribution profiles of the modification islands for 20,446 known genes and found that the overall patterns of modifications were very similar between CD133+ and CD36+ cells (Figure S2). Interestingly, the number of promoters associated with H3K9me1 decreased from 52% in CD133+ cells to 25% in CD36+ cells; the promoters associated with H3K27me1 decreased from 27% to 8%, while the association with other modifications was similar (Figure S3).
Correlation between chromatin modification and gene expression in each cell type
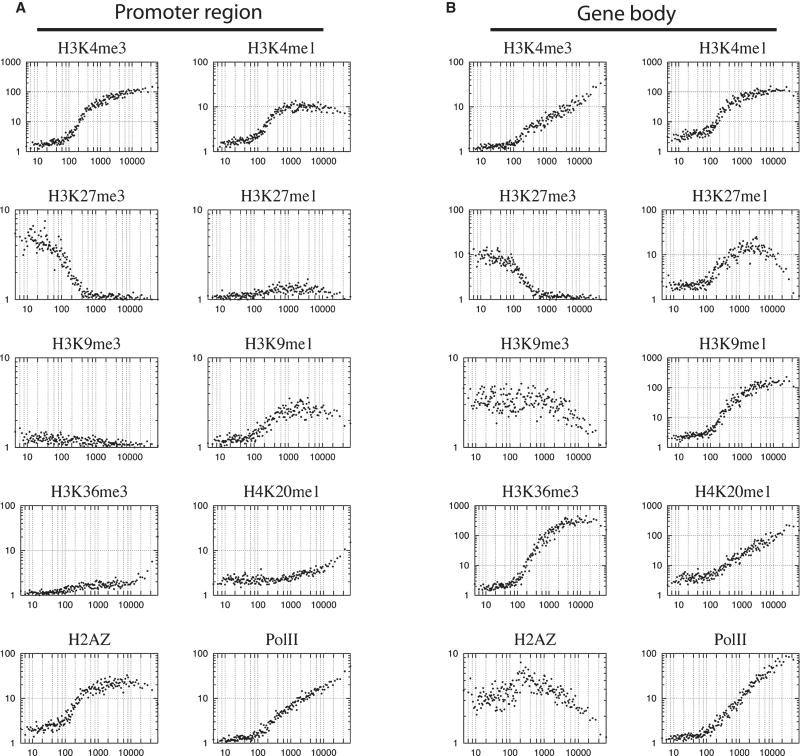
To analyze the relationship between chromatin modification and gene expression in each cell type, we examined the mRNA expression profiles in CD133+ and CD36+ cells, respectively, using gene expression microarrays. From a total of 20,446 known genes with corresponding probes on the arrays, 10,130 (50%) and 9,502 (46%) were unambiguously expressed in CD133+ and CD36+ cells, respectively. To test how the modification level, which is indicated by the number of reads in a region, correlates with gene expression level, we plotted the modification levels against the expression levels in the promoter and gene body regions, respectively, for CD36+ cells (Figure 2). This analysis indicates that a positive correlation exists between gene expression and H3K4me3, H3K4me1, H3K9me1, H3K36me3 and H4K20me1 for both promoters and gene body regions, which is consistent with our observations in CD4+ T cells (Barski et al., 2007). It appears that H2A.Z in the gene body region positively correlated with expression for less active genes but negatively correlated with expression for more active genes. H3K27me1 showed a similar trend with H2A.Z but the turning point for correlation was much higher. Similar results were obtained for CD133+ cells (data not shown).
Figure 2. Correlation between histone modification and gene expression in CD36+ cells.
Genes were grouped to 100-gene (one dot in the figure) sets according to expression level. The histone modification levels in promoter (A) and gene body (B) regions were calculated for the same 100-gene sets (see Experimental Procedure for detail). The y-axis indicates the histone modification level and x-axis indicates the expression level.
Both H3K9me3 and H3K27me3 have been implicated in gene repression. Indeed, both of them were associated with genes expressed low levels. H3K27me3 negatively correlated with the genes showing low and intermediate levels of expression. In contrast, H3K9me3 negatively correlated with expression for genes with higher expression levels (Figure 2B). Interestingly, these genes were not associated with H3K27me3, suggesting that H3K9me3 and H3K27me3 may modulate expression of different subsets of genes (Figure 2B).
Genome-wide gene expression changes reveal differentiation to erythrocyte lineage and inhibition of other lineages
Differentiation of mouse HSCs to specific hematopoietic lineages is associated with distinct gene expression patterns (Chambers et al., 2007). To identify the specific expression patterns in human HSCs and the differentiated erythrocyte precursor cells, we examined their expression profiles and found 1,041 genes up-regulated and 1,697 genes down-regulated more than 4-fold upon differentiation into CD36+ cells (Table S2). Gene ontology analysis of down-regulated genes indicates that the top biological processes associated with these genes were related with immune responses or cellular defenses against pathogens (Figure S4A). For example, among the down-regulated genes, IFITM1, IFITM2, IFITM3, GBP1, OAS1, OAS2 and IFNGR1 are involved in interferon-mediated cellular defense systems, while IL16, LCK, IL15, LAT2, LY75, CIITA, IL6R and BCL2 contribute to the functions of T, B or NK cells. In contrast, many genes strongly induced in CD36+ cells were involved in oxygen transport (Figure S4A), which are among the functions of erythrocytes, consistent with CD36+ cells being precursors of erythrocytes. For example, several hemoglobin genes including HBD, HBZ, HBE1, HBM, HBB, HBG1 and HBA2 were among these up-regulated genes.
Transcription factors play essential roles in the maintenance and differentiation of stem cells. Our gene profiling analysis revealed that 1,328 and 1,290 transcription factors (see Experimental Procedures for definition) were expressed in CD133+ and CD36+ cells, respectively. Among these, 218 transcription factors were unique to CD133+ and 38 unique to CD36+ cells (Table S3). In addition, although expressed in both cell types, 38 transcription factors were up-regulated and 53 were down-regulated in CD36+ cells more than 3-fold. Among these transcription factors, the members of Hox gene family, that encode homeodomain-containing transcription factors and play critical roles during development (Krumlauf, 1994), exhibited the most dynamic regulation. Our analysis indicates that five HoxA genes including HoxA3, HoxA5, HoxA7, HoxA9 and HoxA10 were expressed in CD133+ cells and were down-regulated 15 to 137-fold upon differentiation into CD36+ cells as measured by DNA microarray analysis (Figure S4B), which was also confirmed using qPCR assays (Figure S4D). Among the five HoxB genes expressed in CD133+ cells, HoxB2 was up-regulated, whereas HoxB6 was silenced during the differentiation (Figure S4C). GATA3, a zinc-finger transcription factor known to be required for T cell development and differentiation (Ting et al., 1996), was silenced in CD36+ cells. Several transcription factors including IRF1, STAT1 and STAT2 involved in interferon-mediated cellular defense system were also down-regulated. The expression of PBX1, a homeodomain transcription factor required for NK and B cell development (Sanyal et al., 2007), decreased approximately 7-fold in CD36+ cells. In contrast, the erythroid Kruppel-like factor (EKLF or KLF1) and GATA1, which control the development and differentiation of erythroid lineage (Donze et al., 1995; Takahashi et al., 1997), were up-regulated 28 and 19-fold, respectively. These data collectively indicate that the HSCs/HPCs were efficiently differentiated into the erythrocyte-committed cells.
Dynamic changes of chromatin modifications at critical loci encoding transcription factors
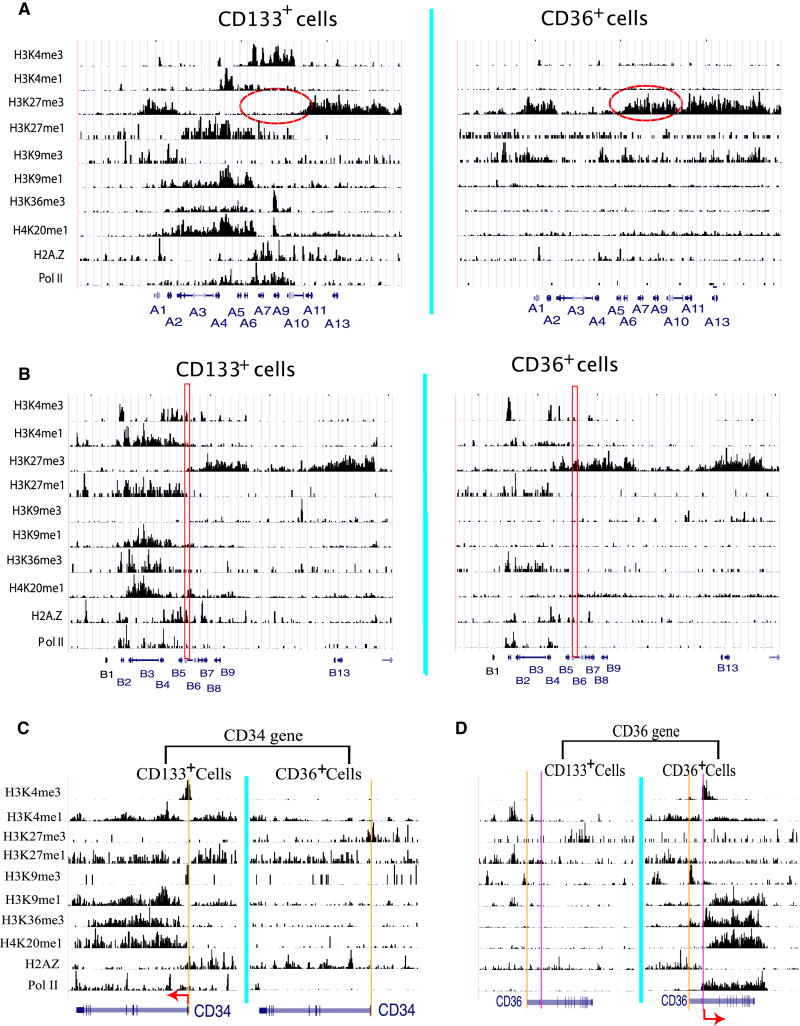
Our analysis of the histone modification patterns indicates that differentiation of the HSCs/HPCs into CD36+ cells induced a broad change in histone modifications in the Hox gene clusters (Figures 3A and 3B and Figure S5). The HoxA genomic locus was characterized by high levels of H3K4me3, H3K4me1, H3K9me1, H3K36me3, H4K20me1 and H2A.Z in the center region containing A5, A6, A7, A9 and A10, bracketed by high levels of H3K27me3 at both sides of the locus in CD133+ cells (Figure 3A, left panels). This region was associated with high levels of expression of A5, A7, A9 and A10 (Figure S4B). In CD36+ cells, the H3K27me3 modification spread to the center region (highlighted region in Figure 3A, right panel) (see Figure S6 for higher resolution display) with increased H3K9me3 signals in the right half of the domain, while other “active” modifications disappeared. These epigenetic changes were associated with silencing of all HoxA genes (Figure S4B). The HoxB locus behaved differently (Figure 3B). The region containing HoxB2 to B6 was associated with H3K4me3 and other marks indicative of active transcription in CD133+ cells, whereas the region containing HoxB7 to B13 associated with H3K27me3. However, the H3K27me3 signal spread to the HoxB5 and HoxB6 genes (B6 promoter highlighted in red) with concurrent decrease of H3K4me3 signals in the same region upon differentiation into CD36+ cells, which is consistent with the repression of HoxB5 and HoxB6 genes (see Figure S7 for higher resolution display). Unexpectedly, we also detected a striking decrease of H3K4me1, H3K27me1, H3K9me1 and H4K20me1 signals at HoxB2, B3 and B4 genes, even though the H3K4me3 and Pol II signals did not show much change in the same region. The HoxC and HoxD genomic loci associated with high levels of H3K27me3 and almost no active modifications in both CD133+ and CD36+ cells (data not shown), consistent with the silent state of these two loci. The KLF1 (EKLF) and GATA1 genes also exhibited dramatic changes of chromatin modification (Figure S8), which is consistent with their strong induction in CD36+ cells.
Figure 3. Changes in histone modifications accompany changes in gene expression during differentiation.
A. Histone modification profiles in the HoxA locus (chr7:27,054,903-27,243,612) in CD133+ and CD36+ cells. The data are displayed as custom tracks on the UCSC genome browser. The positions of the HoxA genes are indicated below the panel. Y-axis shows the number of sequence reads detected in 200 bp-windows and x-axis shows the chromosome coordinates in the genome.
B. Histone modification profiles in the HoxB locus (chr17:43,931,826-44,203,425) in CD133+ and CD36+ cells. The positions of the HoxB genes are indicated below the panel. The promoter region of the HoxB6 gene is highlighted in red.
C. Histone modification profiles in the CD34 gene locus (chr1:206,110,000-206,170,000) in CD133+ and CD36+ cells. The position and direction of transcription of the CD34 gene are indicated below the panel. The coordinate of the annotated TSS is indicated by the vertical orange line.
D. Histone modification profiles in the CD36 gene locus (chr7:80,090,000-80,170,000) in CD133+ and CD36+ cells. The position and direction of transcription of the CD34 gene are indicated below the panel. The vertical orange line indicates the annotated TSS and the vertical pink line indicates the actual TSS.
Dynamic changes of chromatin modifications of cell surface marker genes
As shown in Figure 1, the CD34 gene was highly expressed in CD133+ cells and became silent in CD36+ cells. Our data indicate that in CD133+ cells, the active CD34 gene promoter associated with high levels of H3K4me3 and H2A.Z, while the gene body region associated with widespread signals of H3K4me1, H3K9me1, H3K27me1, H3K36me3 and H4K20me1 modifications (Figure 3C, left panel). High levels of Pol II were also detected throughout the gene. In contrast, these signals either decreased (H3K4me1 and H3K27me1) or disappeared (H3K4me3, H3K9me1, H3K36me3 and H4K20me1) in CD36+ cells, while H3K27me3 was elevated in the promoter region (Figure 3C, right panel). Interestingly, H2A.Z signals decreased in the promoter region and increased in the gene body of the CD34 gene when it became silent in CD36+ cells. The histone modifications in the CD133 gene exhibited similar changes to that in the CD34 gene during differentiation (Figure S9). In contrast to the CD34 and CD133 genes, the CD36 gene was silent in CD133+ cells and became highly active in CD36+ cells. The silent CD36 gene promoter associated with high levels of H3K4me1 and H3K27me1, but not any detectible levels of H3K4me3 in CD133+ cells (Figure 3D, left panel). As expected, high levels of H3K4me3 were detected in the promoter region and high levels of H3K4me1, H3K9me1, H3K36me3 and H4K20me1 were detected in the transcribed region in CD36+ cells (Figure 3D, right panel). It is interesting to note that although H3K4me1, H3K9me1 and H3K27me1 marked the promoter region upstream of the first exon of the CD36 gene in CD133+ cells, the actual transcription may initiate from the second exon as suggested by the H3K4me3 and PolII binding signals as well as the distribution of H3K9me1, H3K36me3 and H4K20me1 modifications (Figure 3D, right panel). These data indicate that dramatic epigenetic changes accompanied the changes in expression states of these marker genes during differentiation of HSCs/HPCs to erythrocyte precursor cells.
Correlation between the dynamic changes of chromatin modification and gene expression during differentiation
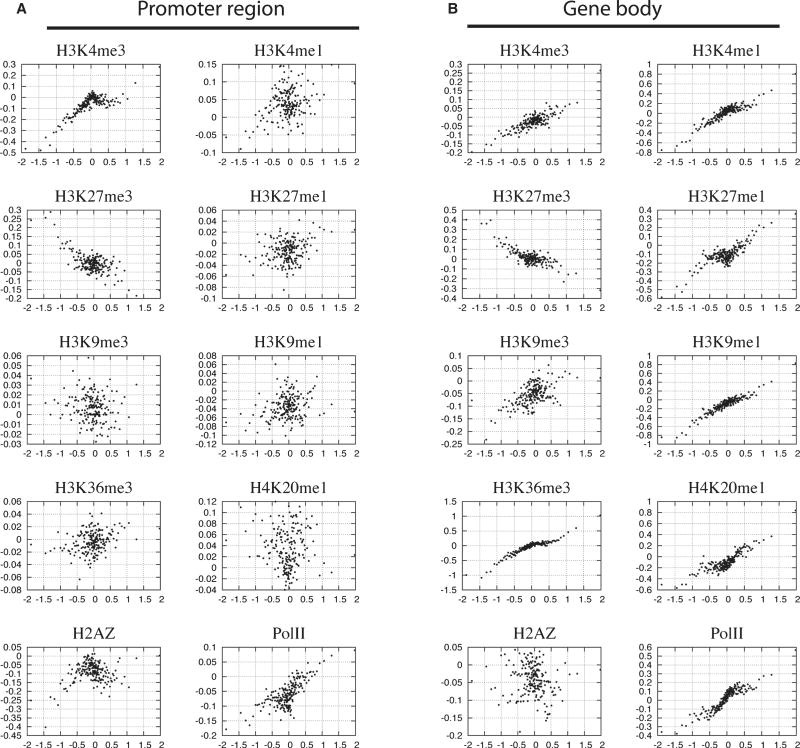
To test how the dynamic changes of these modifications correlate with gene expression change on a global level, we compared directly the changes of expression with the changes of histone modifications in the promoter and gene body regions, respectively, during differentiation of CD133+ cells to CD36+ cells (Figure 4). The x-axis indicates the changes of gene expression level and the y-axis represents the changes of the number of reads on the histone modification islands detected in the promoter (Figure 4A) and transcribed regions (Figure 4B). In the promoter region, the H3K4me3 modification change positively correlated with gene expression change, whereas H3K27me3 inversely correlated with gene expression and no clear correlation was observed for other modifications (Figures 4A). However, in the gene body region, in addition to the positive correlation with H3K4me3 and negative correlation with H3K27me3, gene expression showed positive correlation with all other modifications except H3K9me3 and H2A.Z (Figure 4B). A positive correlation was detected between gene expression change and changes in Pol II levels in both the promoter and gene body regions.
Figure 4. Correlation between changes in histone modification and gene expression during differentiation from CD133+ to CD36+ cells.
The fold changes in both expression level and histone modification level were calculated for each gene during differentiation. The genes were grouped into 100-gene sets according to their expression changes and the changes were averaged for each set of 100 genes (one dot in the figure); the histone modification changes were then averaged for the same sets of 100-genes. The y-axis indicates the fold change (log10 scale) of histone modification in the promoter region (A) or gene body region (B). The x-axis indicates that the fold change in expression levels. The clear gaps in some figures are due to cut-off thresholds of histone modification islands.
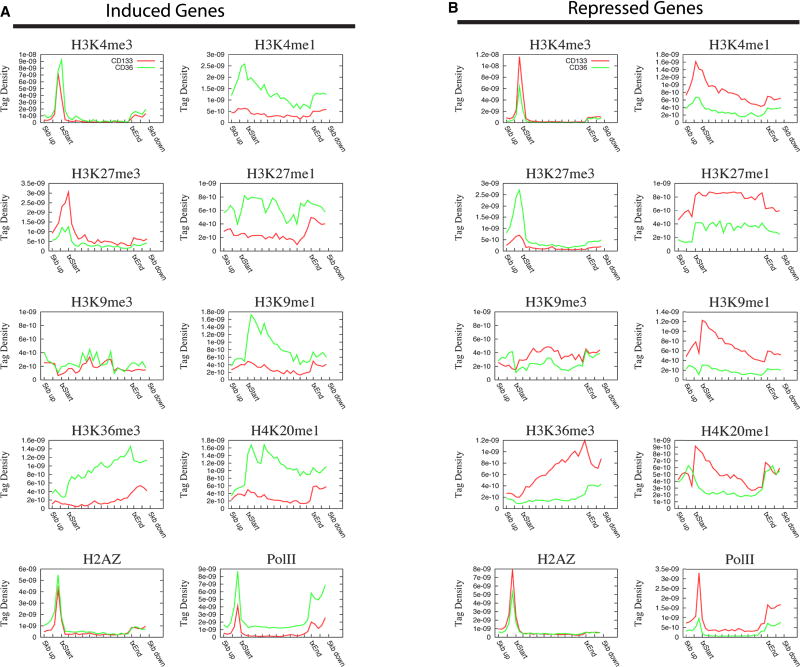
To examine the changes of histone modifications in specific gene groups, we separated the genes into four groups depending on their expression patterns during differentiation from CD133+ to CD36+ cells: (1) always-expressed (9,196 genes); (2) induced (306 genes); (3) repressed (934 genes); and (4) always-silent (7,420 genes). The modification profiles aligned relative to TSSs demonstrated similar patterns for always-expressed genes between CD133+ and CD36+ cells for all modifications (Figure S10A). The most interesting changes of modification profiles were observed for the induced and repressed gene groups. The active marks including H3K4me1, H3K9me1, H3K27me1, H3K36me3 and H4K20me1 were markedly increased, whereas the repressive mark H3K27me3 was decreased in the induced genes in CD36+ cells (Figure 5A). In contrast, opposite changes of the profiles were observed for the repressed genes (Figure 5B). H3K4me3 and H2A.Z levels, mainly in the promoter region, exhibited modest changes according to expression patterns, whereas Pol II showed significant changes in both promoter and gene body regions in correspondence to expression patterns. We also observed interesting changes in the always-silent genes. Even though these genes were not expressed in either CD133+ or CD36+ cells, the levels of H3K4me3, H2A.Z and Pol II in promoter regions and H3K4me1, H3K9me1 and H3K36me3 in either promoter or gene body regions were decreased after differentiation (Figure S10B). It is noteworthy that the level of H4K20me1 increased, which behaved differently from other active marks. These data suggest that the non-expressed genes become stably silenced by losing active modifications in differentiated cells.
Figure 5. Histone modification profiles in induced (A) and repressed (B) genes during differentiation of CD133+ cells (red) to CD36+ cells (green).
The tag density for modifications (see Supplementary Information for detail) is shown across the gene bodies as well as 5 kb 5′ and 3′ of the gene bodies.
Distinct chromatin modification patterns of bivalent genes in CD133+ and CD36+ cells
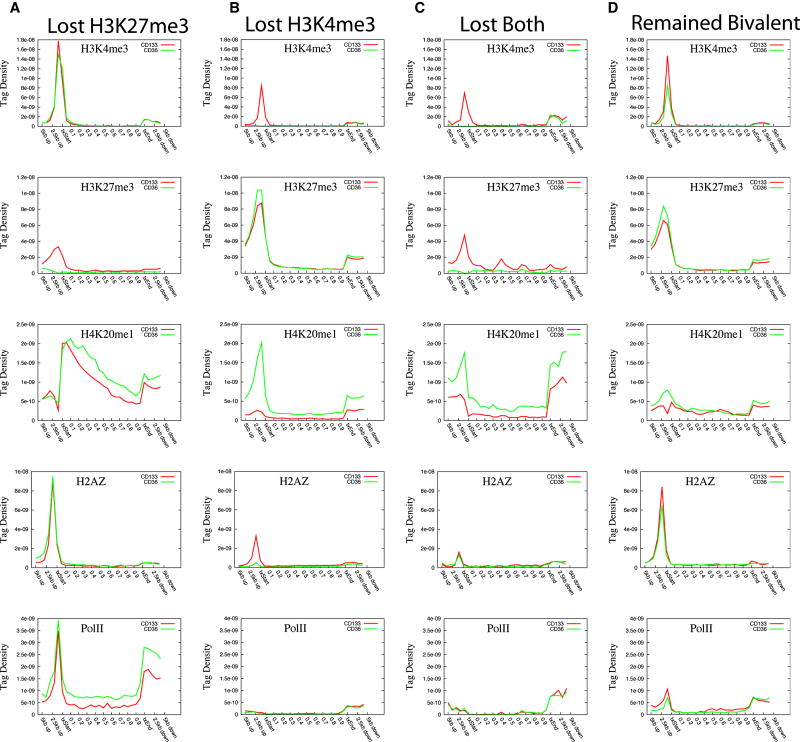
Although bivalent domains identified in ES cells can be resolved to monovalent modification after differentiation (Azuara et al., 2006; Bernstein et al., 2006; Mikkelsen et al., 2007), very little is known about the underlying mechanism controlling the fate of the bivalent modifications. To address this question, we identified 2,910 bivalent promoters in CD133+ cells and examined the presence of other modifications and Pol II at these promoters. The analysis revealed that many of them were also associated with H2A.Z (2,072; 71%), H3K4me1 (574; 20%), H3K9me1 (750; 26%) and H4K20me1 (886; 30%) (Table S4). Comparison of the histone modification profiles of the bivalent promoters between CD133+ and CD36+ cells indicates that H3K4me3 markedly decreased in CD36+ cells (Figure S11). Interestingly, we also observed a remarkable increase of H4K20me1 (Figure S11H) and modest increase of H3K9me3, H3K27me1 and H3K4me1 (Figures S11E, S11D, S11B) as well as modest decrease of H2A.Z (Figure S11I) in CD36+ cells. However, we did not observe a significant global change in H3K27me3 levels (Figure S11C). To reveal gene-specific changes, we examined the modifications of each bivalent gene before and after differentiation. This analysis revealed that among the 2,910 bivalent genes in HSCs/HPCs, 693 (24%) genes remained bivalent, 1,549 (53%) genes lost H3K4me3, 541 (19%) genes lost H3K27me3 and 127 (4%) genes lost both H3K4me3 and H3K27me3 after differentiation into CD36+ cells. Next, we examined changes of other modifications in these four groups of bivalent genes upon differentiation of CD133+ into CD36+ cells. For the genes that lost H3K27me3, increased levels of H3K4me1, H3K9me1, H3K27me1, H3K36me3 and H4K20me1 were detected in the gene body region, while no significant increases in H3K4me3 and H2A.Z were detected (Figure 6A, Figure S12A). Increased Pol II levels in the transcribed region indicated activation of these genes in CD36+ cells. Indeed, we detected 196 genes that showed more than 2-fold expression in CD36+ cells, whereas only 53 showed decreased expression. For the genes that lost H3K4me3 alone, we detected a remarkable increase of H4K20me1; modest decrease of H3K4me1 and H3K9me1; and almost complete loss of H2A.Z, while no significant change in H3K27me3 (Figure 6B, Figure S12B). A significant increase of H4K20me1 was also observed for the genes that lost both H3K4me3 and H3K27me3 (Figure 6C, Figure S12C). However, we did not find a correlation between the H4K20me1 level in these gene promoters and gene expression (data not shown), even though H4K20me1 in gene body regions demonstrated a certain degree of correlation with gene expression change (Figure S13). For the genes that remained bivalent in CD36+ cells, only modest changes of chromatin modifications were observed (Figure 6D, Figure S12D).
Figure 6. Distinct histone modification changes are associated with different fates of the bivalent genes.
Histone modification profiles for the bivalent genes that lost H3K27me3 (A), H3K4me3 (B), both H3K27me3 and H3K4me3 (C), or remained as bivalent (D), during differentiation. H3K4me3, H3K27me3, H4K20me1, H2A.Z and Pol II profiles are shown here. The profiles for other modifications are shown in Figure S13.
The fate of bivalent modifications after differentiation is linked to distinct chromatin modification patterns in HSCs/HPCs
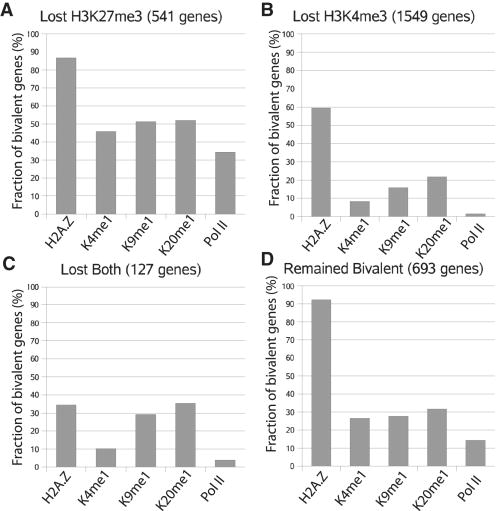
To identify the chromatin modification differences between the bivalent genes that lose either H3K4me3 or H3K27me3, we examined each of the bivalent promoters for association with other modifications and Pol II in CD133+ cells (Figure 7, Table S4). We find that H2A.Z was associated with 87% of the promoters that lost H3K27me3, whereas it was detected at only 59% of the bivalent promoters that lost H3K4me3 after differentiation. Similarly, H3K4me1 and H3K9me1 were detected at a much larger fraction of the bivalent promoters that lost H3K27me3 (46% and 51%, respectively) than at the promoters that lost H3K4me3 (8% and 15%, respectively). Interestingly, H3K4me1 and H3K9me1 were detected at 27% and 28%, respectively of the promoters that remained bivalent after differentiation. We also observed a striking difference for association of RNA Pol II with these different classes of bivalent genes (34%, 2% and 14% for the genes that lost H3K27me3, H3K4me3 and remained bivalent, respectively). These data suggest that the fate of bivalent modifications after differentiation is linked with chromatin modification patterns in the stem cell or progenitor cell stage.
Figure 7. The fate of bivalent genes after differentiation is linked to the chromatin modification patterns in HSCs/HPCs.
The fraction of the bivalent genes associated with H2A.Z, H3K4me1, H3K9me1, H4K20me1 and Pol II was indicated for those that lost H3K27me3 (A), lost H3K4me3 (B), lost both (C), or remained bivalent (D) after differentiation.
Functional enhancers of inducible genes are associated with various modifications before gene induction
Previous large-scale studies have found that potential transcriptional enhancers are associated with various histone modification patterns (Barski et al., 2007; Heintzman et al., 2007; Roh et al., 2005; Roh et al., 2007; Wang et al., 2008). In order to address whether enhancer regions associate with specific histone modifications prior to gene induction, we examined several genes that were induced during the differentiation of HSCs/HPCs to erythrocyte cells. As seen in Figure 3D, the silent CD36 gene was associated with H3K4me1, H3K27me1 and H3K9me1 in a region about 2 kb upstream of its TSS in CD133+ cells, suggesting that these modifications marked a potential enhancer element prior to gene induction.
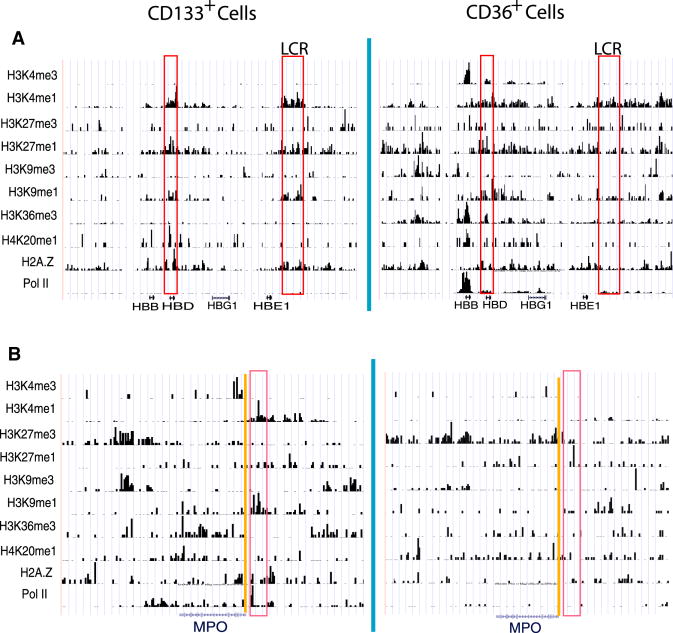
The locus control region (LCR) is a critical regulatory element of the globin genes (Grosveld et al., 1987), which are strongly induced in CD36+ cells. Indeed, we found that the LCR and the region upstream of the β–globin gene were associated with high levels of H3K4me1, H3K9me1, H3K27me1 and H2A.Z but not H3K4me3 and H3K27me3 modifications in CD133+ cells even though the globin genes were not expressed (Figure 8A). In contrast, the neutrophil-specific myeloperoxidase (Mpo), that was primed for expression in HSCs (Ford et al., 1996) and silenced in CD36+ cells, associated with H3K4me1 and H3K9me1 in a region upstream of its TSS in CD133+ cells; but these signals were erased in CD36+ cells (Figure 8B), suggesting a loss of activation potential after differentiation. These data indicate that the mono-methylations including H3K4me1, H3K27me1 and H3K9me1 are associated with critical regulatory elements in the cell stage before their target genes are expressed.
Figure 8. H3K4me1, H3K9me1 and H3K27me1 modifications mark critical regulatory regions before gene activation.
The histone modification profiles at the β-globin locus (chr11:5,171,954-5,279,020) (A) and MPO (chr17:53,682,809-53,732,673) (B) genomic regions in CD133+ and CD36+ cells are displayed. The critical promoter or enhancer regions are highlighted in red.
To examine the global changes of these mono-methylations during differentiation, we identified all genes associated with one of these mono-methylations (1,321 for H3K4me1, 1,475 for H3K9me1 and 1,106 for H3K27me1) but not with H3K4me3 in CD133+ cells (Table S5). Our data indicated that most of these modifications were correlated with each other (Figure S14). Interestingly, 53% of H3K4me1 promoters, 59% of H3K9me1 promoters and 71% of H3K27me1 promoters lost their respective methylation marks after differentiation into CD36+ cells. Among the promoters that lost mono-methylations, 31% to 40% demonstrated decreased expression, while only 10% to 14% showed increased expression. Together, these data suggest that these mono-methylations in HSCs/HPCs may maintain the activation potential of a subset of genes required for differentiation.
Discussion
We report here the generation of high-resolution genome-wide maps of eight histone modifications as well as histone variant H2A.Z and RNA Pol II, together with gene expression profiles in the human CD133+ HSCs/HPCs and CD36+ erythrocyte precursor cells. Our data provide information on the epigenetic states of each cell type and reveal interesting relationships between the fate of bivalent genes during differentiation and their association with several mono-methylations in the stem/progenitor cell stage.
The fate of bivalent domains during differentiation is linked to chromatin modification patterns at HSC/HPC stage
It has been suggested that HSCs and HPCs are primed for the potential of differentiation into multiple lineages by co-expression of various transcription factors, growth factors and their receptors involved in these lineages prior to the commitment to any specific lineage (Hu et al., 1997). Indeed, previous gene expression profiles have confirmed the expression of these critical factors in HSCs (Ivanova et al., 2002; Ramalho-Santos et al., 2002; Venezia et al., 2004). Although HSCs exhibit significant heterogeneity in gene expression between individual cells, co-expression of these factors in one single cell has been reported (Ramos et al., 2006; Warren et al., 2006). Therefore, the multipotentcy of HSCs and HPCs could be related with the expression potential, albeit at low levels, of many genes involved in the function and development of multiple lineages. One important question is what maintains the activation or silencing potential of these genes. H3K4me3 has been associated with active genes while H3K27me3 has been associated with silent genes in CD4+ T cells (Barski et al., 2007; Roh et al., 2006). We previously reported that the co-existence of H3K4me3 and H3K27me3 is generally correlated with reduced levels of gene expression in CD4+ T cells (Barski et al., 2007; Roh et al., 2006; Wang et al., 2008) and proposed that the co-existence of these two apparently counter-acting modifications provides a mechanism of either gene activation or repression through a shift in their balance (Roh et al., 2006; Wang et al., 2008). Studies using ES cells suggest that bivalent modifications maintain the activation or silencing potential of critical differentiation genes (Azuara et al., 2006; Bernstein et al., 2006). It is known that some of the bivalent genes can lose H3K4me3 while others can lose H3K27me3 during differentiation. However, it is not clear what controls the fate choice of the bivalent genes. The data we report here indicate that numerous genes involved in the development and differentiation of multiple lineages are associated with both the repressive mark H3K27me3 and the active mark H3K4me3 in HSCs/HPCs. We found that 19% of the bivalent genes lost H3K27me3 and became activated after differentiation into CD36+ cells. Interestingly, a large fraction of these genes were associated with H3K4me1 and H3K9me1 modifications in the HSC/HPC stage and also bound with RNA Pol II, suggesting that they were poised for activation. In contrast, the association of these mono-methylations with the bivalent promoters, which lost H3K4me3 after differentiation, was decreased dramatically in the HSC/HPC stage; in addition, almost no Pol II was detected in these promoters. These results are consistent with a model that the fate of bivalent genes during differentiation is controlled by epigenetic modifications including various mono-methylations that occur in the HSC/HPC stage.
Genome-wide silencing of the genes maintaining the multipotent potential of HSCs/HPCs
The loss of multipotency and commitment to differentiation is accompanied by the induction of genes involved in one particular lineage and the silencing of genes required for other lineages. We found that only 24% of the 2,910 bivalent genes remained bivalent after differentiation. Instead, most bivalent promoters (53%) lost H3K4me3 after differentiation. Even those that remained bivalent were associated with lower levels of H3K4me3 in CD36+ cells. These results indicate that the differentiation into CD36+ cells is accompanied by a genome-wide silencing of the genes involved in the development and differentiation of other lineages and that the silent states are locked off by a switch of epigenetic modifications.
Epigenetic signals on critical transcriptional regulatory elements
H3K4 methylation has been detected at silent loci in ES and differentiated cells (Barski et al., 2007; Bernstein et al., 2006; Guenther et al., 2007; Meissner et al., 2008; Mikkelsen et al., 2007; Roh et al., 2006; Wang et al., 2008; Weber et al., 2007). The presence of H3K4me3 at inactive genes has been proposed to be a memory for past transcriptional activity based on studies performed in yeast (Ng et al., 2003). However, we detected H3K4me3 signals at numerous genes, which are inactive in HSCs/HPCs but are required for functions of multiple lineages other than erythrocytes. Interestingly, upon commitment to the erythrocyte pathway, the H3K4me3 signals disappeared from these genes. These data suggest that the H3K4me3 signal present at the developmental genes in HSCs/HPCs may not be just a memory for past transcription, but instead may be part of an epigenetic state that maintains the activation potential of the genes required for specific differentiation pathways. In addition to H3K4me3 and H3K27me3 modifications, we also detected various mono-methylations as well as H2A.Z in these genes. H2A.Z typically co-localizes with H3K4me3 near the TSSs, whereas mono-methylations can be detected near TSSs or well within the gene body. A large scale analysis of histone modification patterns in 1% of the human genome has revealed that functional enhancers are marked by H3K4me1 (Heintzman et al., 2007). Our genome-wide analyses of histone modifications in human T cells have indicated that enhancers can be associated with histone acetylation (Roh et al., 2005; Roh et al., 2007), various histone methylations including H3K4me1, H3K4me3, H3K9me1 and H3K27me1 as well as histone variant H2A.Z (Barski et al., 2007; Wang et al., 2008). However, it is not clear whether the enhancers required for differentiation genes, which are not active in stem and progenitor cells, are marked with histone modifications prior to gene activation. Several lines of evidence suggest that this is the case. For example, the CD36 gene promoter is associated with H3K4me1, H3K9me1 and H3K27me1 in HSCs/HPCs, where it was not expressed. The LCR, which is required for efficient induction of the globin genes in erythrocytes, is also associated with these modifications. These data suggest that these critical regulatory elements are epigenetically modified long before the gene expression. Consistent with this, it was reported recently that H3K4me2 marks a subset of developmentally poised hematopoietic genes (Orford et al., 2008). It is curious that no H3K4me3 signals were detected at either the promoter or enhancer regions of these genes in HSCs/HPCs, suggesting that the mono-methylation of H3K4, H3K9 and H3K27 may be earlier epigenetic signals for the gene induction. This idea is also consistent with the observation that these modifications are detected in the paternal pronucleus at fertilization of an egg when the paternal genome is decondensed to prepare for transcriptional activation, whereas H3K4me3 and H3K27me3 only become detectible later after DNA replication (Lepikhov and Walter, 2004; Santos et al., 2005).
In summary, we have provided high-resolution genome-wide maps for eight histone modifications, histone variant H2A.Z and RNA polymerase II in both HSCs/HPCs and the differentiated CD36+ cells, an important resource for understanding the transcriptional regulation and epigenetic states of these clinically important human primary cells. Our data argue that the fate of bivalent genes during differentiation is programmed by chromatin modifications including H3K4me1, H3K9me1, H4K20me1 and H2A.Z at the HSC/HPC stage. In addition to bivalent genes, our data suggest that the mono-methylations are correlated with the basal level expression of a subset of differentiation genes in HSCs/HPCs and therefore may be involved in the maintenance of their activation potential required for differentiation.
Experimental Procedures
Isolation of CD133+ cells and differentiation into CD36+ cells
Multipotent HSCs were collected from the blood of healthy volunteers following mobilization with G-CSF and AMD3100 (NHLBI protocol 04-H-0179); CD133+ HSCs cells were isolated from apheresis products using immunomagnetic beads specific for CD133 (Miltenyi CliniMacs system) and stored in liquid nitrogen. For FACS analysis of cell surface markers and ChIP-Seq analysis of histone modifications, the cells were recovered in serum-free long-term culture medium (LTCM) containing 100 ng/ml Flt3-ligand (FL), 100 ng/ml megakaryocyte growth and development factor (MGDF), 100 ng/ml stem cell factor (SCF) and 10 ng/ml granulocyte colony-stimulating factor (G-CSF) at 37°C in 5% CO2 for 12 hrs. For differentiation into CD36+ cells, the CD133+ cells were suspended at 104 cells/ml in maintenance media that was prepared by 1:5 dilution of BIT 9500 (StemCell Technologies, Cat# 09500) in AMEM (Mediatech, Cat# 10-022-CV) for a final concentration of 10mg/ml BSA, 10μg/ml rh Insulin, 200μg/ml hTransferrin and supplemented with 900ng/ml ferrous sulfate (Sigma, Cat# F8633), 90ng/ml ferric nitrate (Sigma, Cat# F8508), 10-6M Hydrocortisone (Sigma, Cat# H6909), 100ng/ml rh SCF (PEPROTECH, Cat# 300-07), 5ng/ml rh IL-3 (PEPROTECH, Cat# 200-03), and 3IU/ml rh Epo (StemCell Technologies, Cat# 02625). The cells were incubated at 37°C in a 5% CO2/95% air atmosphere for 9 to 11 days, followed by FACS analysis for the presence of CD36+ cells.
Chromatin from CD133+ and CD36+ cells was prepared using micrococcal digestion as described (Barski et al., 2007). 10×106 cells were used for each ChIP-Seq analysis.
The flow cytometry analysis of CD133+ and CD36+ cells was performed using the following antibodies from BD Pharmingen: CD34-PE (#555822), CD36-PE (#555455), CD36-FITC (#555454), CD38-PE (#555460), CD177-APC (#341096), CD90-FITC (#555595), HLA-DR (#555812),CD43-APC (#560198), CD71-PE-Cy5 (#551143). CD133-APC (#130-090-854) was from MACS.
Gene expression analysis
Total RNAs from the CD133+ and CD36+ cells were isolated using the RNeasy kit (Qiagen, Cat# 74104) and subjected to DNA microarray analysis for gene expression profiles using the HG-U133 Plus 2.0 chip (Affymetrix, Cat# 900466). Two replicates were obtained and the average of the two values obtained for each probe ID were taken as the expression value of that probe ID. Gene expression categorization by the absent and present calls using MAS5.0 algorithm provided by Affymetrix was performed. Genes with conflicting calls in the two replicates were ignored. The expression data are provided in Table S6.
ChIP-Seq and data analysis
The ChIP-Seq analysis was performed as described previously (Barski et al., 2007). The sequence reads and expression data have been deposited in the NCBI Short Reads Archive (GSE12646). The BED files can be downloaded from the website (http://dir.nhlbi.nih.gov/papers/lmi/epigenomes/hghscmethylation.html). Detailed strategies for data analysis can be found in Supplemental Information.
Supplementary Material
Acknowledgments
We thank Dr. Susan Wong for helpful discussions and Dr. Warren Leonard for critical reading of the manuscript. The gene expression analysis using the Affymetrix microarrays was performed by the Gene Expression Core Facility of NHLBI. This work was supported by the Intramural Research Program of the NIH, National Heart, Lung and Blood Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bitan M, Shapira MY, Resnick IB, Zilberman I, Miron S, Samuel S, Ackerstein A, Elad S, Israel S, Amar A, et al. Successful transplantation of haploidentically mismatched peripheral blood stem cells using CD133+-purified stem cells. Exp Hematol. 2005;33:713–718. doi: 10.1016/j.exphem.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Boles NC, Lin KY, Tierney MP, Bowman TV, Bradfute SB, Chen AJ, Merchant AA, Sirin O, Weksberg DC, et al. Hematopoietic Fingerprints: An Expression Database of Stem Cells and Their Progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Klous P, Kooren J, Noordermeer D, Palstra RJ, Simonis M, Splinter E, Grosveld F. Three-dimensional organization of gene expression in erythroid cells. Curr Top Dev Biol. 2008;82:117–139. doi: 10.1016/S0070-2153(07)00005-1. [DOI] [PubMed] [Google Scholar]
- Donze D, Townes TM, Bieker JJ. Role of erythroid Kruppel-like factor in human gamma- to beta-globin gene switching. J Biol Chem. 1995;270:1955–1959. doi: 10.1074/jbc.270.4.1955. [DOI] [PubMed] [Google Scholar]
- Ford AM, Bennett CA, Healy LE, Towatari M, Greaves MF, Enver T. Regulation of the myeloperoxidase enhancer binding proteins Pu1, C-EBP alpha, -beta, and -delta during granulocyte-lineage specification. Proc Natl Acad Sci U S A. 1996;93:10838–10843. doi: 10.1073/pnas.93.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giarratana MC, Kobari L, Lapillonne H, Chalmers D, Kiger L, Cynober T, Marden MC, Wajcman H, Douay L. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat Biotechnol. 2005;23:69–74. doi: 10.1038/nbt1047. [DOI] [PubMed] [Google Scholar]
- Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, Enver T. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Lepikhov K, Walter J. Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC Dev Biol. 2004;4:12. doi: 10.1186/1471-213X-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliaccio G, Di Pietro R, di Giacomo V, Di Baldassarre A, Migliaccio AR, Maccioni L, Galanello R, Papayannopoulou T. In vitro mass production of human erythroid cells from the blood of normal donors and of thalassemic patients. Blood Cells Mol Dis. 2002;28:169–180. doi: 10.1006/bcmd.2002.0502. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Uchida N, Weissman IL. The biology of hematopoietic stem cells. Annu Rev Cell Dev Biol. 1995;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Orford K, Kharchenko P, Lai W, Dao MC, Worhunsky DJ, Ferro A, Janzen V, Park PJ, Scadden DT. Differential H3K4 methylation identifies developmentally poised hematopoietic genes. Dev Cell. 2008;14:798–809. doi: 10.1016/j.devcel.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-Genome Analysis of Histone H3 Lysine 4 and Lysine 27 Methylation in Human Embryonic Stem Cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Ramos CA, Bowman TA, Boles NC, Merchant AA, Zheng Y, Parra I, Fuqua SA, Shaw CA, Goodell MA. Evidence for diversity in transcriptional profiles of single hematopoietic stem cells. PLoS Genet. 2006;2:e159. doi: 10.1371/journal.pgen.0020159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Wei G, Farrell CM, Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17:74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol. 2005;280:225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Sanyal M, Tung JW, Karsunky H, Zeng H, Selleri L, Weissman IL, Herzenberg LA, Cleary ML. B-cell development fails in the absence of the Pbx1 proto-oncogene. Blood. 2007;109:4191–4199. doi: 10.1182/blood-2006-10-054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Szilvassy SJ. The biology of hematopoietic stem cells. Arch Med Res. 2003;34:446–460. doi: 10.1016/j.arcmed.2003.06.004. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Onodera K, Motohashi H, Suwabe N, Hayashi N, Yanai N, Nabesima Y, Yamamoto M. Arrest in primitive erythroid cell development caused by promoter-specific disruption of the GATA-1 gene. J Biol Chem. 1997;272:12611–12615. doi: 10.1074/jbc.272.19.12611. [DOI] [PubMed] [Google Scholar]
- Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- Venezia TA, Merchant AA, Ramos CA, Whitehouse NL, Young AS, Shaw CA, Goodell MA. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2004;2:e301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci U S A. 2006;103:17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Wong S, Zhi N, Filippone C, Keyvanfar K, Kajigaya S, Brown KE, Young NS. Ex vivo-generated CD36+ erythroid progenitors are highly permissive to human parvovirus B19 replication. J Virol. 2008;82:2470–2476. doi: 10.1128/JVI.02247-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J, Buck DW. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002–5012. [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, et al. Whole-Genome Mapping of Histone H3 Lys4 and 27 Trimethylations Reveals Distinct Genomic Compartments in Human Embryonic Stem Cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.