Abstract
Plant roots and microorganisms interact and compete for nutrients within the rhizosphere, which is considered one of the most biologically complex systems on Earth. Unraveling the nitrogen (N) cycle is key to understanding and managing nutrient flows in terrestrial ecosystems, yet to date it has proved impossible to analyze and image N transfer in situ within such a complex system at a scale relevant to soil-microbe-plant interactions. Linking the physical heterogeneity of soil to biological processes marks a current frontier in plant and soil sciences. Here we present a new and widely applicable approach that allows imaging of the spatial and temporal dynamics of the stable isotope 15N assimilated within the rhizosphere. This approach allows visualization and measurement of nutrient resource capture between competing plant cells and microorganisms. For confirmation we show the correlative use of nanoscale secondary ion mass spectrometry, and transmission electron microscopy, to image differential partitioning of 15NH4+ between plant roots and native soil microbial communities at the submicron scale. It is shown that 15N compounds can be detected and imaged in situ in individual microorganisms in the soil matrix and intracellularly within the root. Nanoscale secondary ion mass spectrometry has potential to allow the study of assimilatory processes at the submicron level in a wide range of applications involving plants, microorganisms, and animals.
Nitrogen (N) is the primary nutrient limiting plant production worldwide and is consequently a major regulator of ecosystem functioning. Understanding N and carbon (C) cycling at the plant-soil interface remains one of the greatest challenges limiting our ability to predict and manage nutrient flows in terrestrial systems. For example, it is well established that plants lose large amounts of C from their roots into the rhizosphere (approximately 20%–40% of total net primary production) and that this stimulates microbial proliferation around the root (Foster, 1988; Jones et al., 2004). In the case of N, and organic forms in particular, there seems to be strong competition between soil microorganisms and plant roots and the flow of N is intrinsically linked to the flow of C in the rhizosphere (Jones et al., 2009). Understanding ways to manipulate and manage rhizosphere C flow to maximize nutrient uptake, prevent metal toxicity, and prevent pathogen attack remains a priority for designing sustainable agricultural systems (Jones and Hinsinger, 2008).
In complex environments such as the rhizosphere, it has previously been impossible to differentiate nutrient assimilation by competing organisms, as it is difficult to spatially and temporally separate these organisms for individual analysis due to their size and their close physical and biological interactions. Understanding the link between the heterogeneity of the soil's physical and chemical environment and its impact on biological processes marks a current frontier in soil science (Young and Ritz, 1998; O'Donnell et al., 2007). Until now we have relied largely on mathematical modeling to describe nutrient flow in such complex biological systems (Schnepf and Roose, 2006; Falconer et al., 2008). However, much of our knowledge of functioning in the rhizosphere has been gained from simple model systems (e.g. hydroponics; Jones et al., 2004) under sterile conditions or where nutrients are applied in excess. This does not necessarily reflect the level of competition for C and nutrients that exists between plants and microorganisms within the soil environment.
Furthering our understanding is limited by the lack of suitable methods for reliably detecting, imaging, and measuring rhizosphere processes in situ at the submillimeter scale. Spatial mapping of nutrient flows within the rhizosphere has hitherto not been attained. Using isotopic tracers in time-series experiments can be a powerful technique for measuring assimilatory processes and nutrient transfer between soil, plant, and microorganisms (Prosser et al., 2006; Baggs, 2008). Traditional bulk mass spectrometry techniques, however, lose the spatially resolved information necessary to determine nutrient pathways, by averaging the signal between the different components in the system (Cliff et al., 2007). Secondary ion mass spectrometry (SIMS) now provides opportunities for the detection and, more importantly, direct visualization of nutrient flow within structures in situ. SIMS combines mass spectrometry with imaging at biologically meaningful spatial scales, allowing elemental mapping and the measurement of stable isotope ratios (e.g. 15N/14N). SIMS has been used to image C and N assimilation in soil microorganisms in the past; however, these studies were hampered by instrument limitations and were performed ex situ (Cliff et al., 2002; Pumphrey et al., 2008). Recent developments in SIMS instrumentation have led to the NanoSIMS 50 (for nanoscale SIMS; Cameca), a dynamic SIMS instrument combining ultrahigh lateral resolution, high mass resolution, and high sensitivity, thereby improving the possibility to undertake spatially discrete studies of nutrient flow at submicron length scales (Lechene et al., 2007; Eybe et al., 2008).
Here, we present the application of NanoSIMS to image and measure competition for the uptake of 15N in situ within the rhizosphere. Imaging and analysis of the same sample by transmission electron microscopy (TEM) and NanoSIMS unequivocally confirms that 15N added to the rhizosphere can be readily detected as it is taken up from soil by individual microorganisms and a competing plant root. The ability to measure 15N assimilation in situ at this previously unattainable scale provides the first opportunity to simultaneously image and measure N flow pathways in complex biological systems at a scale appropriate to the size of the competing organisms.
RESULTS AND DISCUSSION
NanoSIMS Imaging of the Root-Soil Interface
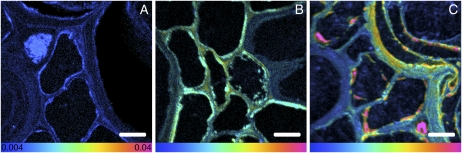
The in situ spatial relationship of the wheat (Triticum aestivum) roots to the soil matrix is clearly revealed by scanning electron microscopy. Figure 1 shows cross sections of three roots, with mineral grains (predominantly quartz in this system) pushed aside as the roots have expanded. From these preparations, levels of 15N enrichment within the cell walls and cytoplasm of the roots were imaged using NanoSIMS. Expression of these NanoSIMS 15N/14N ratio data as hue saturation intensity (HSI) images reveals the N pathway into the roots at the subcellular level (Fig. 2). In control samples, where there was no 15N enrichment, abundance levels are represented by blue (15N/14N = 0.004; Fig. 2A), which is equivalent to the terrestrial 15N/14N value (=0.00368). In samples exposed to 15N enrichment for 0.5 h, increased 15N was observed within the intercellular spaces and cell walls (Fig. 2B). In samples exposed to 15N enrichment for 24 h, the cell cytoplasm had become an order of magnitude more 15N enriched (15N/14N = 0.04, Fig. 2C) than the cytoplasm of control samples. Monitoring the uptake of N into plant roots at the subcellular level has not been achieved before. This type of NanoSIMS analysis could help to address a number of plant and soil science problems (for review, see Herrmann et al., 2007b). In particular we see application in (1) visualizing nutrient flows between plants and fungi (both symbionts and pathogens), (2) studying plant and microbial competition for nutrients and low Mr organic molecules, (3) defining how active and inactive microbial populations are spatially organized within the rhizosphere and how this is affected by plant species and mineralogical associations, (4) studying interactions between microorganisms including determination of the microsite conditions conducive to horizontal gene transfer, (5) determining the mechanisms by which soil organic matter is stabilized, (6) visualizing signal exchange in the rhizosphere, and (7) understanding the nature and spatial location of fixation sites (e.g. phosphorus) in soil.
Figure 1.
Plant roots within the soil matrix. Scanning electron micrograph of a polished soil plug, showing the in situ location of roots (arrows) within the soil matrix. Bar = 100 μm.
Figure 2.
HSI images showing 15N enrichment in root cells. A, Control with no 15N enrichment. B, 15N enrichment for 0.5 h. C, 15N enrichment for 24 h. The color scale indicates the 15N/14N ratio, with natural abundance levels (no enrichment) represented as blue (0.004), changing to pink with increasing 15N levels (0.04). Bars = 5 μm.
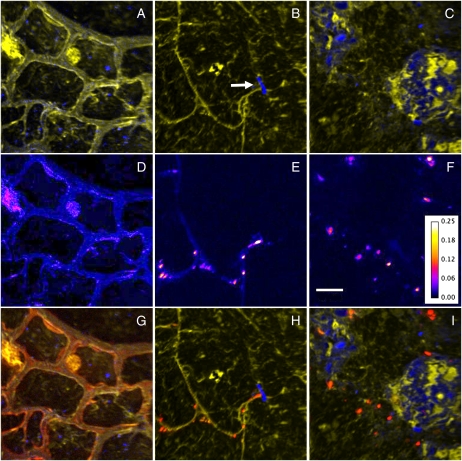
Visualization of the competitive uptake of 15N within the rhizosphere was achieved by acquiring NanoSIMS ion images of 12C14N and 12C15N from a total area several hundred microns in size, incorporating the three major regions of interest: plant root, rhizosphere, and soil matrix. These maps reveal both the spatial relationships of the organic material and the soil quartz grains, and the isotopic variations (15N/14N) between the different organic components. The plant cell structure (Fig. 3A), soil organic matter, and mineral particles (Fig. 3, B and C) can be distinguished clearly. Fine-scale detail, such as a root hair that has come into contact with a soil particle and subsequently diverged, is visible (Fig. 3B, arrow). Areas of low signal strength depict void spaces in the root (i.e. vacuoles) and soil (i.e. pore spaces). Images illustrating 15N/14N ratios reveal the level of assimilated 15NH4+ throughout the root cells (Fig. 3D), within microorganisms associated with root hairs (Fig. 3E), and within microorganisms throughout the soil matrix (Fig. 3F), thus allowing for the level of 15N enrichment within any given component to be determined. Combining the three ion images as a color montage further illustrates the direct in situ spatial relationship of the different system components and links this to the levels of 15N enrichment (Fig. 3, G–I).
Figure 3.
NanoSIMS ion images highlighting in situ spatial relationships, coupled with levels of 15N enrichment. A to C, Overlays of 12C14N (yellow) and 28Si (blue) ion images representative of key regions: plant root cells (A), rhizosphere (B), and soil matrix (C). A root hair that has encountered a soil particle and diverged is seen in B (arrow). D to F, 15N/14N ratio images from same regions, showing areas of 15N enrichment. The color depicts the 15N/14N ratio, with the ratio scale shown in F. Bar = 5 μm. G to I, Overlay of 12C14N (yellow), 28Si (blue), and 12C15N/12C14N (red) images from each region.
NanoSIMS and TEM Imaging of Individual Microbial Cells
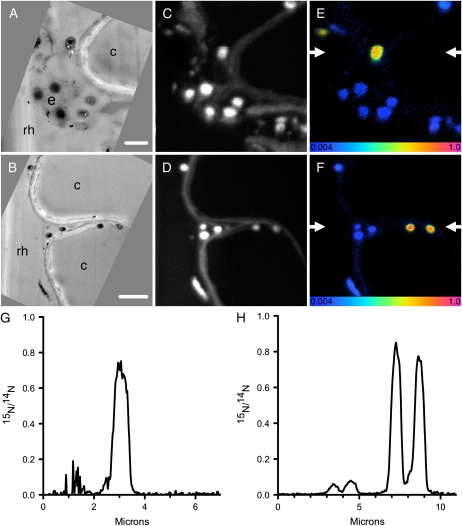
It was necessary to confirm that the 15N-enriched hot spots seen within the rhizosphere region corresponded to microorganisms. These images do appear very similar to the other reported SIMS images of isotopically labeled microorganisms in soil (Cliff et al., 2002; Herrmann et al., 2007a; Pumphrey et al., 2008). Nevertheless, by using a novel approach of correlative TEM and NanoSIMS analyses, we verified that these 15N hot spots within the rhizosphere are enriched microorganisms associated with plant roots. TEM imaging of extracted roots confirmed that individual microorganisms are adjacent to the root surface (Fig. 4, A and B), while corresponding NanoSIMS analyses of these regions (Fig. 4, C and D) identified that some, but not necessarily all, of these microorganisms were 15N enriched (Fig. 4, E and F).
Figure 4.
Correlative TEM imaging and NanoSIMS isotopic analyses of samples exposed to 15N for 24 h. A and B, TEM images confirm the presence of microorganisms in the rhizosphere (rh) and extracellular mulicage matrix (e) adjacent to the root cells (c). Bars = 1 μm (A) and 2 μm (B). C and D, NanoSIMS images of the same regions, visualized using 12C14N. E and F, 15N/14N NanoSIMS ratio images of the same regions, confirming that some of these microorganisms are 15N enriched. These 15N-enrichment images are shown as HSI images, where the color scale indicates the 15N/14N ratio, with natural abundance levels (0.004) changing to pink (1.0) with increasing 15N levels. G, Linescan from the region between the arrows indicated in E. H, Linescan from the region between the arrows indicated in F.
Interestingly, these correlative TEM and NanoSIMS images highlight how the 15N/14N ratio varies between microorganisms (Fig. 4, E–H). Further, when the microorganisms in Figure 3 were analyzed for quantitative 15N analyses, individual organisms varied by as much as 110% relative to one another (more than 5 sdmean). These differences reflect real differences in the incorporation of the 15NH4+, and true heterogeneity in rhizosphere microbial assimilation. Variation clearly exists between endophyte microorganisms and those that appear to reside within an extracellular mucilage matrix (Fig. 4, F and H; Alvarez et al., 2004). Such substances at the surface of colonies can develop in response to moisture stress and as such can influence the rate of nutrient exchange between water films and microorganisms. Alternatively, the variation in 15N enrichment of individual bacterial cells could also simply reflect variable N assimilation by different species, or level of cellular activity at time of sampling. Future development of halogenated DNA probes that allow for correlative species identification using NanoSIMS will represent a major advance in understanding the roles and activities of microorganisms in the rhizosphere at this scale. While these have been developed successfully for microbial cultures (Behrens et al., 2008; Li et al., 2008; Musat et al., 2008), their application to three-dimensional structures, such as an intact soil core remains challenging, due to difficulties with nonspecific labeling and diffusion into, and removal of, chemical solutions from soil pores.
NanoSIMS Imaging of Plant-Microbial N Uptake
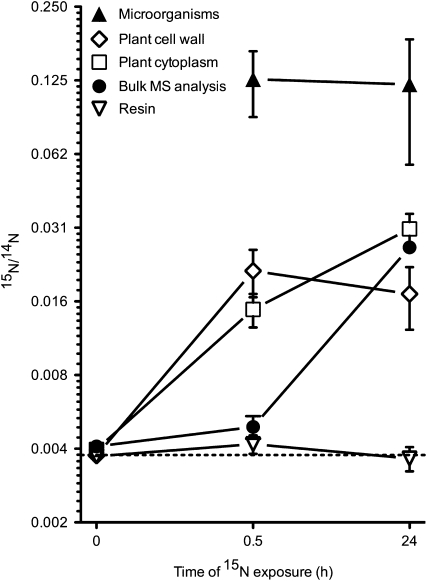
In addition to ion images, which visually depict variation in 15N enrichment, numerical 15N/14N ratio data were also extracted from image data. Numerical data were obtained from microorganisms, root cell walls, and root cell cytoplasm. Measurement of 15N/14N ratios from resin-only regions, which can be used as an internal reference standard, gave a mean value of 0.00374 ± 0.00005 (n = 58 regions). Although the uncertainty of these resin analyses does not incorporate propagated instrumental counting precision, the reproducibility of the measurements and agreement with the expected terrestrial 15N/14N value of 0.00368, confirm the validity of these 15N data.
Analyses of control samples with no 15N enrichment revealed that the 15N/14N ratio in the root cell walls and cytoplasm was also, as expected, highly similar to the terrestrial and resin values (Figs. 2A and 5). In samples exposed for 0.5 h to 15N enrichment, the root cell walls and cytoplasm were enriched by a factor of approximately 6 and 3, respectively (Figs. 2B and 5). In samples exposed to 15N for 24 h, the 15N/14N ratio of the cell walls showed no further net increase, yet enrichment in the cell cytoplasm continued to increase as N was assimilated into the plant cell (Figs. 2C and 5).
Figure 5.
15N/14N ratio data from individual rhizosphere components. Samples were exposed to (1) no 15N (control), (2) 15N for 0.5 h, and (3) 15N for 24 h. Values from resin-only regions are given as an internal reference standard. Microorganisms cannot be visualized in in situ samples without 15N enrichment, thus no data is recorded for microorganisms in control samples. Terrestrial abundance (0.00368) is indicated by the dotted line. It is clear that NanoSIMS offers the ability to distinguish the 15N levels in different components of the rhizosphere, compared to traditional bulk mass spectral (MS) analyses, which incorporate each of these into a single, diluted, and biased result.
At natural abundance (i.e. without 15N enrichment), individual microorganisms cannot accurately be visualized or distinguished in polished soil plug samples (they can only be visualized in thin sections of extracted roots in the TEM—see “Materials and Methods” section for more information). As such, no microorganisms could be detected in control samples and therefore, no data is recorded. After 15N exposure for 0.5 h, the 15N/14N ratio of microorganisms had increased by a factor of almost 50, and this remained similar after 24 h exposure to 15N (Fig. 5). In highly enriched microorganisms, where up to 50% of the N was derived from the added 15N, these data indicate that N uptake and subsequent assimilation is extremely rapid. This is consistent with studies showing that the pseudo residence time of NH4+ in soil under wheat is less than 24 h (Murphy et al., 2003).
While the HSI scale in Figure 4 suggests that the ratio values for the microorganisms in extracted, sectioned TEM samples are much higher than the ratio values for the microorganisms in the in situ plugs (Fig. 5), this is not the case. The HSI scale reflects the ratio from every individual pixel, whereas data from the soil plugs were obtained from selected regions of interest, thus giving an overall average of the 15N/14N ratio in the entire selected area (i.e. the whole microorganism). When microorganisms in TEM sections were analyzed using similar averaged, selected regions of interest, the levels of 15N enrichment (mean = 0.1378 ± 0.0724; minimum = 0.0410, maximum = 0.3230; n = 47 regions) were highly similar (P = 0.28) to that recorded for the plugs (0.1203 ± 0.0637; minimum = 0.0374, maximum = 0.3022; n = 30 regions). Therefore, ratio data from microorganisms are consistent between soil plug samples and TEM sections.
Advantages of NanoSIMS over Conventional Mass Spectrometry
The advantage of the spatial resolution of NanoSIMS, and the ability to distinguish individual components within the experimental system, is clearly evident. Comparison of the 15N/14N ratio values for the individual components in the rhizosphere acquired using NanoSIMS against the bulk mass spectrometric data reveals a stark contrast in results between the two techniques (Fig. 5). Bulk analysis includes all organic components in the system, large amounts of which have no 15N enrichment. This homogenizing effect produces a heavy bias and the component pathways of nutrient assimilation are completely obscured. Also, TEM imaging confirms that plant roots extracted from soil and cleaned still possess numerous bacteria attached to the root surface. Thus any traditional analysis of plant roots is actually reflective of the combined 15N/14N ratio from the plant cells and attached microorganisms. As shown from the NanoSIMS analyses, these different components can have orders-of-magnitude variations when individually resolved as separate isotopic analyses.
CONCLUSION
The multicomponent complexity of soil has previously made it impossible to differentiate biological processes at the scale at which they occur (i.e. micrometer scale). Our novel application of in situ sample preparation techniques, NanoSIMS analyses, and correlative TEM imaging has allowed for: (1) assimilation of 15N by individual microorganisms competing within the root (endorhizosphere), on the root surface (rhizoplane) and in the external soil (ectorhizosphere) to be spatially resolved; (2) visualization of the pathways of 15N uptake from soil into plant root components over time; (3) direct confirmation of the variability of 15N assimilation between bacterial cells, highlighting the close spatial arrangement between inactive and active cells; (4) confirmation that a region of interest within a sample can be structurally imaged using TEM, and subsequently isotopically analyzed using NanoSIMS; and (5) visualization and measurement of soil-microbe-plant interactions, which is unattainable at this scale using conventional mass spectrometry techniques.
MATERIALS AND METHODS
Preparation of Microcosms
A loamy-sand textured soil, dominated by quartz grains of sand and silt size, with 1.2% organic C was obtained from a freely draining soil located in Meckering, Western Australia (31°40′N, 117°00′E). Soil samples were collected from the Ap horizon (0–15 cm) using a stainless steel corer and stored in CO2 permeable polypropylene bags for transport back to the laboratory where they were sieved (<5 mm) and stored field moist at 4°C. Seeds of wheat (Triticum aestivum) were soaked for 24 h in water and then allowed to germinate on moistened filter paper at 20°C. After 3 d, each plant had one main root axis approximately 1 cm in length and two lateral roots 0.5 cm in length; the seedlings were then placed into individual soil-filled microcosms (Owen and Jones, 2001). The plant-soil microcosms were constructed from polyethylene tubing (30 cm long, 0.6 cm internal diameter). The microcosms were filled with soil to a bulk density of 1.25 g cm−3. After the addition of seedlings, the microcosms were maintained at 20°C, under a 12 h photoperiod. Microcosms were kept moist by the addition of deionized water daily. When the roots and associated root hairs had completely occupied the microcosm, making it essentially all rhizosphere soil (15 d after transplantation; shoots 12.4 ± 0.5 cm long, n = 12), 500 μL of 45 mg N L−1 (3 mm) (15NH4)2SO4 (0.99 15N/14N ratio; Isotec) was injected at three locations 3, 5, and 7 cm (total injected volume 1,500 μL) below the soil surface through premade holes. This ensured uniform distribution of 15N solution within specific microcosm regions for later NanoSIMS analysis. Control samples were injected with distilled water only.
Preparation of Samples for NanoSIMS Analysis
After 0.5 and 24 h exposure to 15NH4+ a 2-cm midsection from the enriched zone of the microcosm containing intact plant roots and soil was quickly excised and the bottom of the core covered with a cotton (Gossypium hirsutum) membrane (BSN Medical), porous to water and solvents, which prevented soil movement during handling. Controls were only sampled at 24 h. The intact cores were immediately fixed with 2.5% glutaraldehyde in 0.1 m phosphate-buffered saline at 4°C. Samples were then rinsed in distilled water and dehydrated in a graded series of acetones. Infiltration of samples in acetone:araldite mixtures was conducted over a period of days, with the concentration of araldite gradually increased until 100%. For final embedding, samples were infiltrated and embedded in 100% Araldite 502 over several days, as described in detail for nonplanted soil cores by Herrmann et al. (2007a).
Previous studies indicate that the loss of nitrogenous compounds from samples using this process is negligible and that these types of compounds are generally well preserved (Peteranderl and Lechene, 2004; Herrmann et al., 2007a). Both plant and microorganisms are also structurally well preserved as evidenced in TEM images and by the retention of such material as the extracellular matrix. Additionally, removal of microorganisms from pores is likely to be minimal in large, undisturbed core samples where spaces are generally well isolated and not subject to significant fluid movement using this membrane-diffusion method. Low-temperature methods that are ideal for improved preservation of structure and cell chemistry, are generally only advantageous for small (<1 mm diameter) samples and are not suitable for use with large, soil core preparations.
Resin-embedded cores were cut into 3-mm thick slices using a diamond saw and reembedded into 10-mm diameter mounts. Samples were polished using increasing grades of silicon carbide paper followed by diamond pastes and coated with 5 nm gold. The location of roots within the soil matrix were identified using scanning electron microscopy (Zeiss, 1555 FESEM) at 10 kV, prior to NanoSIMS analysis (Fig. 1).
To confirm that 15N-enriched hot spots identified in these soil plugs using NanoSIMS were microorganisms associated with the root surface, roots were extracted from the soil so that complimentary TEM sections could be prepared for fine-scale structural and isotopic analyses. Following fixation in the polyethylene tubes, plant roots exposed to 15N for 24 h were extracted from the soil, sonicated for 5 min to remove large soil particles, dehydrated, and embedded in Araldite 502. Thin (approximately 100 nm) sections of extracted root material were cut transversely on glass knives and mounted on C-filmed copper grids. Sections were imaged at 120 kV by TEM (JEOL, 2100) using a digital camera (Gatan, SC1000 ORIUS), before being coated with 5 nm gold and transferred to the NanoSIMS for correlative isotopic analysis. It is important to note that TEM sections cannot be prepared from plant roots still embedded within soil cores, as the mineral grains within the soil matrix are too hard to permit cutting of sufficiently thin (<150 nm) sections.
NanoSIMS Analyses
All data were acquired using the NanoSIMS 50 (Cameca) at the University of Western Australia. A Cs+ primary ion probe, with an impact energy of approximately 16 kV, was rastered across the sample with a beam current of 1 to 2 pA. The primary ion beam was focused to a diameter of approximately 100 nm. The secondary ions 12C−, 12C14N−, 12C15N−, and 28Si− were recorded simultaneously on masses 12, 26, 27, and 28, respectively. The instrument was tuned to high mass resolution (6,000 mass resolving power) to minimize interferences from 13C2− and 12C13CH− on mass 26 and 13C14N− on mass 27. Ion images were acquired at a resolution of 256 × 256 pixels. Images are recorded as counts per pixel, with count times of 40 ms per pixel. All areas were presputtered with the primary ion beam prior to acquisition to remove surface contamination and to enhance the generation of secondary ions.
N is not easily ionized in SIMS and must be represented by the CN− ion, which is emitted with high contrast from organic material and is therefore particularly useful for showing structural features such as cell walls and organelles (Guerquin-Kern et al., 2005; McMahon et al., 2006). Images representing 15N/14N ratios were obtained by normalizing the 12C15N− counts to the 12C14N− counts for each pixel in the ion images. Numerical 15N/14N ratio data were extracted directly from the 12C15N−/12C14N− ion images by selecting regions of interest, pixels defining certain structural features, on the normalized image using the MIMS plugin for ImageJ (http://www.nrims.hms.harvard.edu/NRIMS_ImageJ.php). Background measurements were made from the resin infilling the natural soil pore space and cell vacuoles. Given the magnitude of 15N enrichment in these samples, corrections for dead time and quasi-simultaneous arrival (Slodzian et al., 2004) effects were not deemed necessary. Analyses of regions of interest of individual microbes from select images showed that not incorporating a dead time correction produced an error in estimates of 15N/14N ratios of less than 1% relative. Similarly, corrected useful ion yields (Kcor, Slodzian et al., 2004) measured from these microbes were less than 0.03, indicating a maximum error of a few percent in relative accuracy.
Unless otherwise stated, all results are presented as mean ± sd. The instrumental counting statistics for the background measurements typically had low internal precision due to the low number of counts. Conversely, the measurements of the highly enriched microorganisms had better internal precisions due to the high number of counts in those regions. The errors quoted in the text reflect the reproducibility of the different regions, which in this scenario is more relevant than the internal analytical precision. The 15N-labeled ammonium produces enrichment values that are orders of magnitude greater than the analytical precision, and so this error pales in significance.
HSIs (McMahon et al., 2006) were created using the MIMS plugin for ImageJ. Similarly, linescans were obtained from 12C15N/12C14N ratio images with a line width of 8 pixels, using the program ImageJ and plotted using GraphPad Prism (version 5.0b for Macintosh, GraphPad Software).
Conventional Mass Spectrometry
For comparison, traditional 15N/14N analysis of roots (i.e. plant root material with any attached microbial cells) was also performed. Plant roots were recovered from a separate 2-cm midsection from the enriched zone of the microcosm, washed in distilled water, and any soil particles that were attached to the plant roots were removed with the aid of tweezers and a stereo microscope. These samples were then dried at 70°C, weighed, and placed in a tin capsule for combustion and analysis by isotope ratio mass spectrometry (20/20 Europa Scientific).
Acknowledgments
We thank Frank Nemeth and Garry Cass for their technical support, and Karl Ritz and John Kuo for useful discussions. The authors acknowledge the facilities, scientific, and technical assistance of the Australian Microscopy and Microanalysis Research Facility at the Centre for Microscopy, Characterisation and Analysis, University of Western Australia, a facility funded by the university and state and commonwealth governments.
This work was supported by an Australian Research Council Discovery Project and a United Kingdom Natural Environmental Research Council Antarctic Funding Initiative grant. A.M.H. was funded through the European Commission (Marie Curie Outgoing International Fellowship Scheme FP6).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Daniel Murphy (daniel.murphy@uwa.edu.au).
Open Access articles can be viewed online without a subscription.
References
- Alvarez HM, Silva RA, Cesari AC, Zamit AL, Peressutti SR, Reichelt R, Keller U, Malkus U, Rasch C, Maskow T, et al (2004) Physiological and morphological responses of the soil bacterium Rhodococcus opacus strain PD630 to water stress. FEMS Microbiol Ecol 50 75–86 [DOI] [PubMed] [Google Scholar]
- Baggs EM (2008) A review of stable isotope techniques for N2O source partitioning in soils: recent progress, remaining challenges and future considerations. Rapid Commun Mass Spectrom 22 1664–1672 [DOI] [PubMed] [Google Scholar]
- Behrens S, Lösekann T, Pett-Ridge J, Weber PK, Ng WO, Stevenson BS, Hutcheon ID, Relman DA, Spormann AM (2008) Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl Environ Microbiol 74 3143–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliff JB, Bottomley PJ, Gaspar DJ, Myrold DD (2007) Nitrogen mineralization and assimiliation at millimetre scales. Soil Biol Biochem 39 823–826 [Google Scholar]
- Cliff JB, Gaspar DJ, Bottomley PJ, Myrold DD (2002) Exploration of inorganic C and N assimilation by soil microbes with time-of-flight secondary ion mass spectrometry. Appl Environ Microbiol 68 4067–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eybe T, Audinot JN, Bohn T, Guignard C, Migeon HN, Hoffmann L (2008) NanoSIMS 50 elucidation of the natural element composition in structures of cyanobacteria and their exposure to halogen compounds. J Appl Microbiol 105 1502–1510 [DOI] [PubMed] [Google Scholar]
- Falconer RE, Bown JL, White NA, Crawford JW (2008) Modelling interactions in fungi. J R Soc Interface 5 603–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster RC (1988) Microenvironments of soil organisms. Biol Fertil Soils 6 189–203 [Google Scholar]
- Guerquin-Kern JL, Wu TD, Quintana C, Croisy A (2005) Progress in analytical imaging of the cell by dynamic secondary ion mass spectrometry. Biochim Biophys Acta 1724 228–238 [DOI] [PubMed] [Google Scholar]
- Herrmann AM, Clode PL, Fletcher IR, Nunan N, Stockdale EA, O'Donnell AG, Murphy DV (2007. a) A novel method for the study of the biophysical interface in soils using nano-scale secondary ion mass spectrometry. Rapid Commun Mass Spectrom 21 29–34 [DOI] [PubMed] [Google Scholar]
- Herrmann AM, Ritz K, Nunan N, Clode PL, Pett-Ridge J, Kilburn MR, Murphy DV, O'Donnell AG, Stockdale EA (2007. b) Nano-scale secondary ion mass spectroscopy—a new analytical tool in biogeochemistry and soil ecology: a review article. Soil Biol Biochem 39 1835–1850 [Google Scholar]
- Jones DL, Hinsinger P (2008) The rhizosphere: complex by design. Plant Soil 312 1–6 [Google Scholar]
- Jones DL, Hodge A, Kuzyakov Y (2004) Plant and mycorrhizal regulation of rhizodeposition. New Phytol 163 459–480 [DOI] [PubMed] [Google Scholar]
- Jones DL, Nguyen C, Finlay RD (2009) Carbon flow in the rhizosphere: carbon trading at the soil-root interface. Plant Soil 321 5–33 [Google Scholar]
- Lechene CP, Luyten Y, McMahon G, Distel DL (2007) Quantitative imaging of nitrogen fixation by individual bacteria within animal cells. Science 317 1563–1566 [DOI] [PubMed] [Google Scholar]
- Li T, Wu TD, Mazéas L, Toffin L, Guerquin-Kern J-L, Leblon G, Bouchez T (2008) Simultaneous analysis of microbial identity and function using NanoSIMS. Environ Microbiol 10 580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon G, Glassner BJ, Lechene CP (2006) Quantitative imaging of cells with multi-isotope imaging mass spectrometry (MIMS)—nanoautography with stable isotope tracers. Appl Surf Sci 252 6895–6906 [Google Scholar]
- Murphy DV, Recous S, Stockdale EA, Fillery IRP, Jensen LS, Hatch DJ, Goulding KWT (2003) Gross nitrogen fluxes in soil: theory, measurement and application of 15N pool dilution techniques. Adv Agron 79 69–118 [Google Scholar]
- Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F, Horreard F, Amann R, Jørgensen BB, Kuypers MMM (2008) A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci USA 105 17861–17866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell AG, Young IM, Rushton SP, Shirley MD, Crawford JW (2007) Visualization, modelling and prediction in soil microbiology. Nat Rev Microbiol 5 689–699 [DOI] [PubMed] [Google Scholar]
- Owen AG, Jones DL (2001) Competition for amino acids between wheat roots and rhizosphere microorganisms and the role of amino acids in plant N acquisition. Soil Biol Biochem 33 651–657 [Google Scholar]
- Peteranderl R, Lechene CP (2004) Measure of carbon and nitrogen stable isotope ratios in cultured cells. J Am Soc Mass Spectrom 15 478–485 [DOI] [PubMed] [Google Scholar]
- Prosser JI, Rangel-Castro JI, Killham K (2006) Studying plant-microbe interactions using stable isotope technologies. Curr Opin Biotechnol 17 98–102 [DOI] [PubMed] [Google Scholar]
- Pumphrey GM, Hanson BT, Chandra S, Madsen EL (2008) Dynamic secondary ion mass spectrometry imaging of microbial populations utilizing 13C-labelled substrates in pure culture and in soil. Environ Microbiol 11 220–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepf A, Roose T (2006) Modelling the contribution of arbuscular mycorrhizal fungi to plant phosphate uptake. New Phytol 171 669–682 [DOI] [PubMed] [Google Scholar]
- Slodzian G, Hillion F, Stadermann FJ, Zinner E (2004) QSA influences on isotopic measurements. Appl Surf Sci 231: 874–877
- Young IM, Ritz K (1998) Can there be a contemporary ecological dimension to soil biology without a habitat? Soil Biol Biochem 30 1229–1232 [Google Scholar]