Abstract
Winter cress (Barbarea vulgaris) is resistant to a range of insect species. Some B. vulgaris genotypes are resistant, whereas others are susceptible, to herbivory by flea beetle larvae (Phyllotreta nemorum). Metabolites involved in resistance to herbivory by flea beetles were identified using an ecometabolomic approach. An F2 population representing the whole range from full susceptibility to full resistance to flea beetle larvae was generated by a cross between a susceptible and a resistant B. vulgaris plant. This F2 offspring was evaluated with a bioassay measuring the ability of susceptible flea beetle larvae to survive on each plant. Metabolites that correlated negatively with larvae survival were identified through correlation, cluster, and principal component analyses. Two main clusters of metabolites that correlate negatively with larvae survival were identified. Principal component analysis grouped resistant and susceptible plants as well as correlated metabolites. Known saponins, such as hederagenin cellobioside and oleanolic acid cellobioside, as well as two other saponins correlated significantly with plant resistance. This study shows the potential of metabolomics to identify bioactive compounds involved in plant defense.
Plants are sessile organisms that have developed various strategies to adapt to or counteract abiotic and biotic stress. The ability to accumulate low-molecular-weight bioactive compounds, often referred to as allelochemicals, secondary metabolites, or bioactive natural products, provides a chemical defense against herbivorous insects used by plants. As a result of natural selection, insects often develop mechanisms to adapt to such compounds and eventually manage to break the resistance.
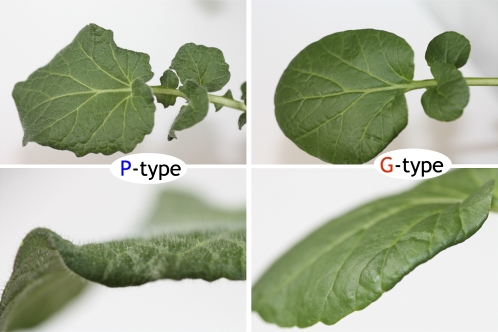
The interaction between Barbarea vulgaris (Brassicaceae) and the flea beetle Phyllotreta nemorum (Coleoptera: Chrysomelidae) is a unique model system to study chemical defenses in plants and counteradaptations in insects (Nielsen, 1997a; de Jong et al., 2000; Agerbirk et al., 2001, 2003b; Nielsen and de Jong, 2005). B. vulgaris, a biennial or short-lived perennial wild crucifer (MacDonald and Cavers, 1991), is polymorphic with respect to insect resistance: the pubescent P-type is susceptible to all known flea beetle genotypes, whereas the glabrous G-type is resistant to most common genotypes of the insect (Nielsen, 1997a, 1997b; Agerbirk et al., 2003a). In contrast, P. nemorum is polymorphic with respect to plant defenses (Breuker et al., 2005; Nielsen and de Jong, 2005).
B. vulgaris has a potential as an oil crop for use at northern latitudes (Börjesdotter, 1999) and is considered to be an important genetic resource for food and agriculture (International Treaty on Plant Genetic Resources for Food and Agriculture; ftp://ftp.fao.org/ag/cgrfa/it/ITPGRe.pdf). It has been used for salads and garnishes as well as a medicinal plant (Senatore et al., 2000). B. vulgaris has a wide native distribution area (Eurasia) and is furthermore naturalized in North America, Africa, Australia, New Zealand, and Japan as a weed (Hegi, 1958; MacDonald and Cavers, 1991; Tachibana et al., 2002). The subspecies arcuata is by far the most common Barbarea taxon in Denmark and comprises two morphologically, biochemically, and cytologically deviating genotypes, P and G, which differ by glucosinolate profiles, flea beetle resistance, and leaf pubescence (Agerbirk et al., 2003b; Fig. 1). B. vulgaris is a diploid; the G-type has 2n = 16 chromosomes, while the P-type has 2n = 16 or 2n = 18 chromosomes (Ørgaard and Linde-Laursen, 2008). B. vulgaris is phylogenetically positioned between Arabidopsis (Arabidopsis thaliana) and allopolyploid oil seed rape (Brassica napus; Bailey et al., 2006). Accordingly, research on plant-insect interaction in B. vulgaris may be applied to B. napus.
Figure 1.
Rosette leaves of P- and G-types of B. vulgaris subspecies arcuata. The P-type has hairs, while the G-type does not.
Glucosinolates constitute a group of defense compounds present in crucifers and play a key role in host selection by crucifer specialists (Renwick, 2002). These compounds are feeding and oviposition stimulants for a number of specialist insects, which have become adapted to such compounds as an outcome of long-standing coevolutionary interactions with host plants containing them (Renwick, 2002; Thompson, 2005). Therefore, glucosinolates no longer offer efficient protection against many specialist insects, and the relationship between glucosinolate profiles of plants and their suitability as food for insects is not simple (Nielsen et al., 2001; Poelman et al., 2008; van Leur et al., 2008). The P-type B. vulgaris contains the R-isomer of 2-hydroxy-2-phenylethylglucosinolate, whereas the G-type contains the S-isomer. However, the differences in glucosinolate profiles between the P- and G-types are not related to resistance to flea beetles (Agerbirk et al., 2003b).
As a putative response to renewed selection pressure from herbivorous insects, a number of crucifers have evolved a second generation of defense secondary compounds (e.g. cucurbitacins in Iberis species, cardenolides in Cheirantus and Erysimum species, and saponins in B. vulgaris). These compounds are feeding deterrents for a number of insect species (Nielsen, 1978; Renwick, 2002; Shinoda et al., 2002; Agerbirk et al., 2003a). Until now, Barbarea is the only crucifer known to contain saponins. Two saponins, oleanolic acid cellobioside (3-O-β-cellobiosyloleanolic acid) and hederagenin cellobioside (3-O-β-cellobiosylhederagenin), have been identified in B. vulgaris (Shinoda et al., 2002; Agerbirk et al., 2003a). The restricted distribution of such saponins in crucifers suggests that they originated later than the glucosinolates, which have a much wider distribution in the family.
Saponins are triterpenoid glycosides widely distributed in higher plants (Hostettmann and Marston, 1995; Sparg et al., 2004; Vincken et al., 2007). They are constituents of many plant drugs and folk medicines and possess a wide range of biological activities, including antifungal, antibacterial, molluscicidal, and insecticidal activities (Hostettmann and Marston, 1995; Sparg et al., 2004; Chwalek et al., 2006; Güçlü-Ustündağ and Mazza, 2007; Gauthier et al., 2009). The toxicity of saponins to fungi and insects is thought to be a result of their ability to form complexes with sterols in the plasma membrane, thus destroying the cellular semipermeability and leading to cell death. Although saponins are toxic to cold-blooded animals, their oral toxicity to mammals is low (for review, see Hostettmann and Marston, 1995; Sparg et al., 2004; Güçlü-Ustündağ and Mazza, 2007).
Hederagenin cellobioside has been identified as an active defense compound of B. vulgaris against the world-wide pest diamondback moth (Shinoda et al., 2002), which has become resistant to most insecticides. Oleanolic acid cellobioside concentration has been shown to correlate with resistance of B. vulgaris to the diamondback moth (Agerbirk et al., 2003a). This compound is present in the resistant G-type plant, and its concentration declines in autumn at the same time as the decline in resistance toward diamondback moth (Agerbirk et al., 2001, 2003b). The impact of the two saponins on defense against flea beetles, a major pest in oil seed rape, has not been reported previously.
The objective of this study was to develop an unbiased strategy to identify metabolites that correlate with resistance to flea beetle larvae in B. vulgaris and to provide knowledge that may facilitate more efficient and sustainable breeding for resistance toward insect pests. The results presented in this study are significant for understanding chemical plant defense against insects and may be utilized in future crop protection breeding by screening for the presence of similar bioactive compounds, biosynthetic enzymes, and genetic markers or transfer of resistance components to crop plants.
RESULTS
Crossing of P- and G-Type Plants
The P-type plants are susceptible to flea beetles, while the G-type plants are resistant. To generate a segregating hybrid population, independent reciprocal crosses between 11 different G-type and eight P-type parent plants resulted in 21 different F1 combinations. F1 plants from all hybrid combinations were selfed by bud pollination to obtain F2 seeds. While most F1 plants produced only a few seeds upon bud pollination of 25 to 30 flower buds, one hybrid combination (female × male) produced approximately 160 F2 seeds. This F2 population was used for bioassays and untargeted metabolite profiling. Accordingly, the F2 plant segregating population was derived from a single F1 plant.
Flea Beetle Larvae Survival
The parental P- and G-type B. vulgaris plants, the F1, and the segregating F2 hybrids were tested for resistance to flea beetle larvae. Five larvae were placed together with one leaf, and the number of larvae that survived was determined after 72 h.
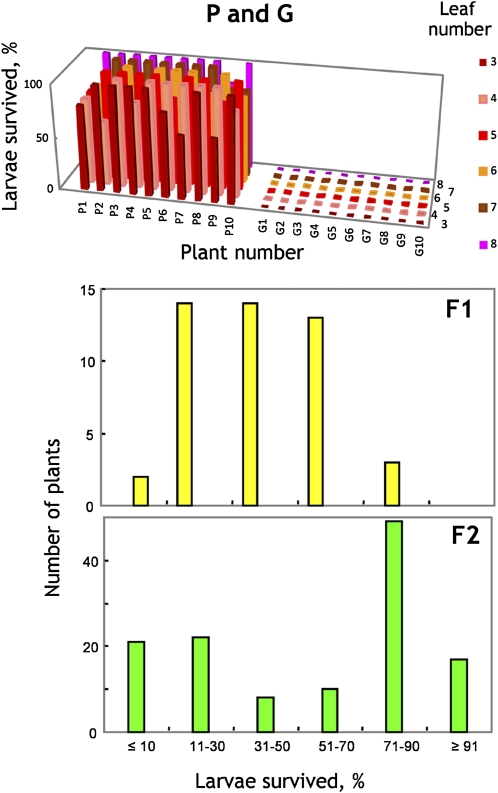
None of the flea beetle larvae survived on the parental G-type leaves, while 91% (4.6 ± 0.6 larvae) survived on the P-type leaves. Survival was independent of rosette leaf number (Fig. 2). Larvae survival on the F1 plants was variable but tended to be intermediate between those found on the parental P- and G-types. Twenty-eight percent of the larvae survived on the F1 plant used to produce the F2 population. The F2 population expressed the whole range from full susceptibility to full resistance (Fig. 2).
Figure 2.
Survival of flea beetle larvae on 10 individual parental P-type (P1–P10) and G-type (G1–G10), F1, and F2 B. vulgaris plants. Each leaf (nos. three to eight) of an individual plant was incubated with five flea beetle larvae for 3 d. Twenty-eight percent of larvae survived on the F1 plant used to generate the F2 population.
While the P-type plant carries hairs, the G-type does not (Fig. 1). The segregating F2 population contained the full range of hair numbers (data not shown). Correlation analysis showed that there was no association between hair number and resistance (r2 = 0.0026, slope −0.0016); thus, hairiness does not seem to be related to susceptibility. Segregation of the F2 for resistance is quantitative, with some tendency for grouping at both ends of the scale (Fig. 2). A 3:1 susceptible-to-resistant ratio may be proposed, with susceptibility being dominant. Higher order gene models are also possible but highly speculative.
Untargeted Metabolite Profiling by Liquid Chromatography-Mass Spectrometry
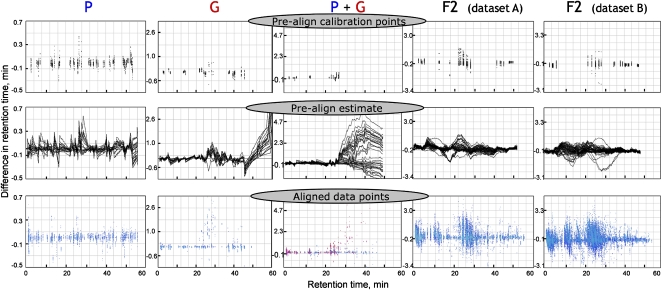
To identify metabolites that confer resistance in B. vulgaris, methanol extracts of the parental P- and G-type plants were analyzed by untargeted metabolite profiling by liquid chromatography-mass spectrometry (LC-MS). The LC-MS chromatograms of the P- and G-types were quite different (Fig. 3, A and E), and it was not obvious which metabolites were involved in resistance. To identify metabolites that differ between the parental P- and G-types, alignment of the chromatograms using the MetAlign software for untargeted profiling of metabolites (de Vos et al., 2007; Lommen, 2009) was pursued. MetAlign performs automated peak alignment, local noise calculation, baseline correction, and extraction of all relevant mass-to-charge ratio (m/z) signals from all LC-MS data sets. It further normalizes profiles, aligns chromatographic peaks to correct for retention time shifts, and produces a multivariate compatible data output. Retention time drifts in LC-MS profiles between samples is not linear and is a major obstacle in aligning LC-MS data sets. MetAlign deals with this obstacle by first identifying prealign calibration points and subsequently in an iterative process aligns the chromatographic peaks.
Figure 3.
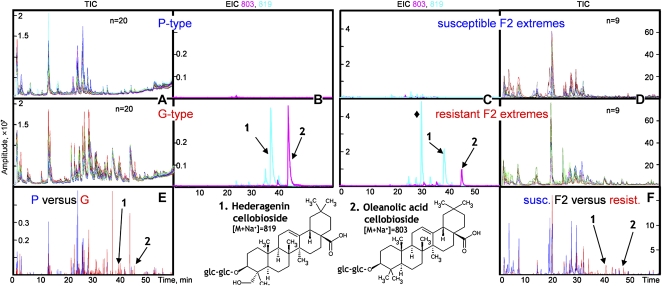
Metabolite profiling of B. vulgaris by LC-MS. A and D, Overlapped total ion chromatograms (TIC) of 20 P-type and 20 G-type plants (A) and of the nine most susceptible and nine most resistant F2 extremes (D). B and C, Extracted ion chromatograms (EIC) of m/z 803 (in pink; [M+Na]+ for oleanolic acid cellobioside) and m/z 819 (in cyan; [M+Na]+ for hederagenin cellobioside) of a P-type and a G-type (B) and susceptible and resistant F2 extremes (C). E and F, Example of a baseline-corrected and normalized profile of a P-type (marked in blue) versus a G-type (marked in red; E) and of susceptible (marked in blue) and resistant (marked in red) F2 extremes (F).
It was only possible to partially align the parental P- and G-type LC-MS profiles, mainly due to the many m/z signals from approximately 30 to 50 min, which were present in the resistant G-type but absent in the susceptible P-type (Figs. 3, A and E, and 4). This resulted in the lack of prealign calibration points beyond approximately 30-min retention time and thus impeded reliable alignments of the profiles with MetAlign (Fig. 4).
Figure 4.
Differential retention display of F2 plant data sets (A and B) and P and G data sets aligned separately or together, generated by MetAlign, which shows differences in retention between all files referenced with regard to the first data set in the group. When two data sets are aligned together, blue and red points are from data sets of group 1 (P-type) and group 2 (G-type), respectively. Black lines represent “prealign estimates” of retention differences (=final shift correction profiles) between the data sets and data set 1 of group 1. These prealign estimates are calculated by interpolation and extrapolation using prealign calibration points (black points). Prealign calibration points were calculated from chromatographic peaks adhering to restriction parameters of the alignment options and criteria that were selected.
To circumvent the inability to align the parental P and G genotypes, we took advantage of the F2 population. Based on resistance and susceptibility to flea beetle larvae, the nine most susceptible (larvae survival more than 93.3%) and the nine most resistant (larvae survival less than 6.7%) F2 lines were selected. These plant lines were designated as the most extreme lines. As a result of recombination between the two parental genomes, the genetic background in the segregating F2 population was randomized, and accordingly this reduced the diversity of LC-MS profiles (Fig. 3, D and F). This enabled reliable alignment of the LC-MS profiles of the most extreme F2 lines. Consequently, we used the t test function of MetAlign to point out metabolites that are significantly different between the extreme lines (Supplemental Table S1). The analysis of these 18 extreme lines showed that the randomization of the genetic background enabled reliable alignment and normalization of the F2 population. For the subsequent metabolite profiling of the entire F2 population consisting of 127 plants, the F2 lines were randomly divided into two groups: group A consisting of 58 F2 offspring, and group B consisting of 69 F2 offspring. These two groups represented two biological replicates and were analyzed by two separate LC-MS runs. Subsequently, MetAlign was used to align the LC-MS profiles of the group A and group B F2 lines (Fig. 4). The number of prealign calibration points within the first approximately 50 min show that it indeed was possible to align the two data sets (Fig. 4).
Each metabolite in the LC-MS chromatograms was represented by one chromatographic peak, which was represented by several m/z signals, including intact molecule ions, fragmentation products, and different adduct ions, as well as isotope ions thereof. The LC-MS profiles of the group A and B F2 plants were deconvoluted by MetAlign into approximately 7,000 m/z signals each. As many of these m/z signals represented isotope ions, various adduct ions, and fragments of the most intense peaks in the spectrum, the data sets were reduced by removing m/z signals that corresponded to isotope ions ([M + 1], [M + 2]) and m/z signals aligned inconsistently by MetAlign. This conservative approach reduced the B data set to 306 m/z signals for each of the 69 plants and the A data set to 319 m/z signals for each of the 58 plants.
Targeted Profiling for Saponins
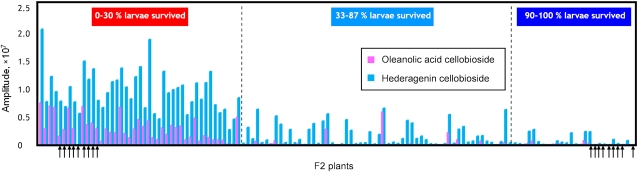
The parental G-type is known to contain the saponin oleanolic acid cellobioside, while the P-type does not (Agerbirk et al., 2003a). In addition to oleanolic acid cellobioside, we detected the saponin hederagenin cellobioside (Fig. 3B), which has previously been identified in a Japanese B. vulgaris genotype (Shinoda et al., 2002). To facilitate detection of glucosides, we added NaCl and formic acid to the solvents used for LC-MS profiling (Tattersall et al., 2001; Kristensen et al., 2005), and accordingly, hederagenin cellobioside and oleanolic acid cellobioside were detected as their sodium adducts ([M+Na]+ at m/z 819 and 803, with retention times 39.6 and 46.1 min, respectively). Based on the intensity of the peaks, oleanolic acid cellobioside and hederagenin cellobioside were the most abundant metabolites among a number of other m/z 803 and 819 peaks, but with different retention times. These minor additional 803 and 819 m/z peaks could only be detected in the resistant G-type plants (Fig. 3B). The F2 plants differed in their relative content of hederagenin cellobioside and oleanolic acid cellobioside. In contrast to the resistant extremes, most of the susceptible F2 extremes contained hederagenin cellobioside and oleanolic acid cellobioside in minor amounts (Fig. 5). The ability to align hederagenin cellobioside and oleanolic acid cellobioside across the F2 lines also serves to substantiate that the alignment generated by MetAlign was valid. Oleanolic acid cellobioside were not always the most abundant among the m/z 803 peaks. An m/z 819 chromatographic peak at 31.0 min was the strongest among m/z 819 peaks in many resistant and susceptible F2 extremes (Fig. 3C). This compound was not detected in the parental G-type plant; thus, its presence is the result of complementation between the P- and G-types. There were several m/z 803 and 819 chromatographic peaks at different retention times in the F2 population, which were not detected in the parental P- and G-type plants (Fig. 3, B and C). This phenomenon has been observed with Arabidopsis recombinant lines (Keurentjes et al., 2006).
Figure 5.
Distribution of hederagenin cellobioside and oleanolic acid cellobioside in 127 F2 plants with different resistance to flea beetle larvae. Intensities of [M+Na]+ for the two compounds (m/z 819 for hederagenin cellobioside and m/z 803 for oleanolic acid cellobioside) were used. The resistant and susceptible extremes used in the alignment of the LC-MS profiles by MetAlign are marked by arrows.
Correlation between Metabolites and Resistance
The original, unbiased data sets A and B contained approximately 7,000 m/z signals each. We used correlation analysis to filter out relevant m/z signals. This further served to discard m/z signals that aligned inconsistently as well as outliers. The observed ion intensity can be used as a relative measure of metabolite concentration (Skoog, 1985; Zacarias et al., 2002). Accordingly, we performed correlation and regression analysis between ion intensity and larvae survival with the aim to identify plant metabolites that correlate with resistance to flea beetle larvae in the F2 population. Permutation analysis with 2,000 permutations of the complete set of m/z signals was used to estimate a 95% significance level for correlation (r2) between relative metabolite level (intensity of m/z signal) and observed larvae survival on F2 plants.
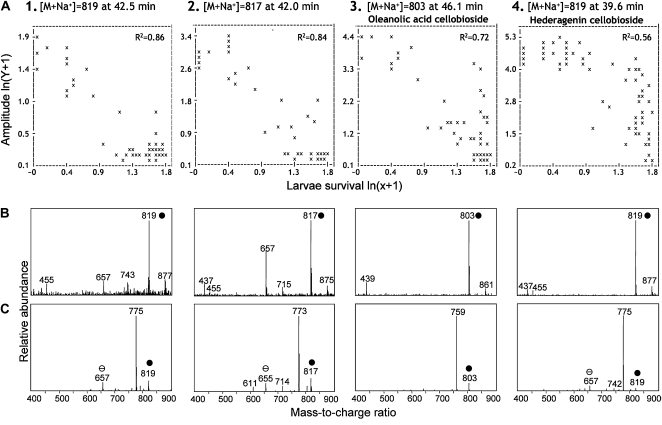
We detected 94 m/z signals in data set A and 51 in data set B that significantly correlated (r2 = 0.86–0.43 in A, r2 = 0.83–0.38 in B) with larvae survival (Supplemental Table S1). As expected, no m/z signals showed significant positive correlations with larvae survival. As many of the m/z signals present in both data sets were identical, the two data sets were manually merged to one data set consisting of 117 unique m/z signals. Two-thirds of the m/z signals represented fragments, various adducts, and isotope ions of the four compounds represented in Figure 6. The first compound most negatively correlated with the larvae survival was compound 1, with m/z 819 and r2 of 0.86 (data set A) or 0.83 (data set B). The second most negatively correlated compound was compound 2, with m/z 817 and r2 of 0.84 (data set A) or 0.77 (data set B). Oleanolic acid cellobioside (m/z 803) was identified as compound 3, with an r2 of 0.72 (data set A) or 0.61 (data set B). Hederagenin cellobioside (m/z 819) was identified as compound 4, with an r2 of 0.56 (data set A) or 0.64 (data set B). In the F2 population, compound 4 (hederagenin cellobioside, m/z 819) was the most abundant among the four compounds most correlated with the resistance. In the nine resistant F2 extremes, the average relative proportion of compounds 1 to 4 was 5:7:22:66, respectively. Resistant F2 extremes contained on average 36, 59, 17, and 11 times more of the compounds 1 to 4 than the susceptible F2 extremes, respectively. Of the 117 m/z signals, 66 signals could be directly related to the four compounds, being various adduct, fragment, and isotope ions. The remaining 51 m/z signals appeared to originate from 12 minor compounds, seven of which were probably saponins or saponin-like compounds, based on their characteristic fragmentation patterns (Supplemental Table S1). For the remaining five compounds, it was difficult to comment on the structure, primarily due to their low abundance.
Figure 6.
The m/z signals most negatively correlated with flea beetle larvae survival, and their MS and MS/MS spectra. Each column represents one of the four compounds, most negatively correlated with the larvae survival. A, Correlation between ion intensity and larvae survival. Each plant is represented by an x. B, MS spectra of compounds 1 to 4 (chromatographic peaks at 42.5, 42.0, 46.1, and 39.6 min, respectively). C, MS/MS spectra showing fragmentation patterns of the base peaks of the four compounds: m/z 819, 817, 803, and 819, respectively. M/z 803 (compound 3) and 819 (compound 4) correspond to sodium adduct ions of oleanolic acid cellobioside and hederagenin cellobioside, respectively; m/z 819 (compound 1) is likely to correspond to a saponin with a structure similar to hederagenin cellobioside; m/z 817 (compound 2) is likely to correspond to the sodium adduct ion of hederagenin cellobioside or its isomer or to a saponin with a similar structure with an additional double bond in the aglycone. Loss of m/z 44 reflects the loss of a carboxylic group, and loss of m/z 160 reflects the loss of a glycose moiety. Thus, m/z 655 and 657 correspond to the monoglucosides; m/z 611 corresponds to the monoglycoside by cleaving of a carboxylic group; m/z 775, 773, and 759 correspond to the corresponding cellobioside by cleaving of a carboxylic group; m/z 714 corresponds to the monoglucoside minus Na2Cl; and m/z 742 corresponds to the cellobioside minus Na2Cl and water. For m/z, white circles correspond to monoglucoside and black circles correspond to cellobioside.
To facilitate fragmentation of metabolites, a quadrupole ion trap was employed. As metabolites that are structurally related break down in a similar pattern, the fragmentation pattern can be used as a tool to identify the putative chemical structure of a metabolite (Matsuda et al., 2009). Based on the characteristic fragmentation pattern, compound 1 (m/z of the base peak 819) has a structure similar to hederagenin cellobioside (compound 4), probably with some rearranged groups in the skeleton, as its fragments were represented by the loss of a carboxylic group (loss of m/z 44) and a glycose moiety (loss of m/z 160; Fig. 6, B and C). The fact that compound 2 has an m/z that is 2 mass units lower than that of hederagenin cellobioside and has the same fragmentation pattern is strongly indicative of the presence of a double bond in the aglycone of compound 2. Based on its fragmentation pattern, the m/z 819 compound at retention time 31.0 (Fig. 3B) is structurally related to hederagenin cellobioside but was not within the 150 most correlated m/z signals (r2 < 0.43 in data set A and r2 < 0.38 in data set B), indicating that this saponin is not related to resistance. The fragmentation pattern of this compound is similar to hederagenin cellobioside, as its fragments were represented by the loss of a carboxylic group and a glycose moiety. Accordingly, this compound has the same overall structure, but the decoration of the aglycone backbone, the hexose moiety, and linkage differ.
Clustering of Metabolites that Correlate with Resistance
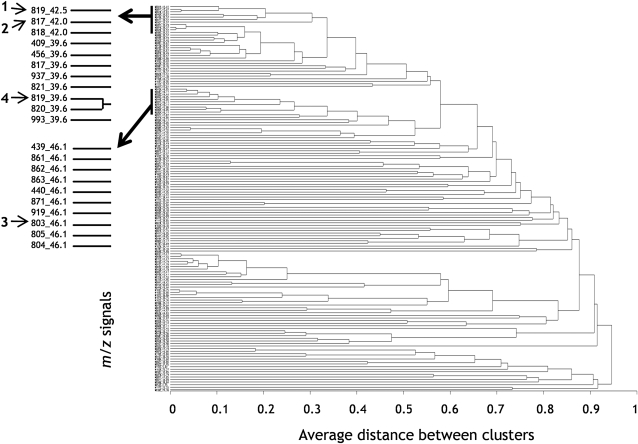
Clustering was used to build groups of metabolites (represented by m/z signals) with related accumulation patterns. Such groups are expected to contain functionally related metabolites, such as metabolites for a specific pathway or metabolites that are coregulated. Arbitrarily selected, the 150 m/z signals that correlated most with larvae survival were visualized in a dendrogram (Fig. 7). The dendrogram is based on UPGMA (for unweighted pair group method with arithmetic mean) clustering with distances 1 – r2 and was used to visualize how the m/z signal intensities covary among plants in the F2 population (Fig. 7). For data set A, the r2 range of the 150 m/z signals was from 0.86 to 0.30 (95% fractile = 0.43), and for data set B, the range was from 0.83 to 0.22 (95% fractile = 0.38). The dendrogram highlights the obstacle that many m/z signals correspond to a single metabolite. An example of this is the coclustering of m/z of 819, 820, and 821, corresponding to the base peak and two isotope ions ([M + 1], [M + 2]) of compound 4 (Fig. 7). The fact that the m/z signals corresponding to the same metabolite covary in offspring serves to substantiate that the correlation analysis is applicable.
Figure 7.
UPGMA dendrogram relating the 150 m/z signals of the F2 offspring most correlated with larvae survival. The m/z signals are represented by their m/z values followed by retention times. Base peaks of compounds 1 to 4 are as in Figure 6.
Compound 2 (m/z 817) from Figure 6, with a corresponding isotope ion m/z 818, and compound 1 (m/z 819) from Figure 6 covaried to a high extent (Fig. 7), indicating that compounds 1 and 2 co-occurred in the F2 plants. This, together with the structural similarity of the two compounds (Fig. 6, B and C), indicates that the two compounds belong to the same biosynthetic pathway. They were not clustered as closely as their isotope ions and fragments, possibly because compounds 1 and 2 were present in relatively small amounts, and thus the coefficient of variation in detection was relatively high. As expected, hederagenin cellobioside (compound 4, base peak m/z 819) covaried most with its two isotope ions, m/z 820 and 821. The m/z signals situated in the cluster between compounds 1 and 2 and 4 represented fragments and adduct ions of compound 4. Likewise, oleanolic acid cellobioside (compound 3, base peak m/z 803) covaried most with its two isotope ions, m/z 804 and 805, and clustered together with its fragments and adduct ions. The m/z signals situated in the cluster between base peaks of compounds 4 and 3 represented fragments and adduct ions of the compounds 1, 2, 3, and 4. One structural difference between oleanolic acid cellobioside and hederagenin cellobioside is the presence of a hydroxyl group at position 23 in hederagenin cellobioside. Although the biosynthetic pathways of hederagenin cellobioside and oleanolic acid cellobioside have not been elucidated, it is expected that the difference of the hydroxyl groups is due to the presence of an enzyme that either hydroxylates oleanolic acid cellobioside or dehydrates hederagenin cellobioside. The evident structural relatedness between compounds 1, 2, 3, and 4 (Fig. 6, B and C) and the fact that they covaried in offspring is an indication that they have a common biosynthetic origin.
Covariation between Metabolite Composition, Metabolite Profile, and Resistance
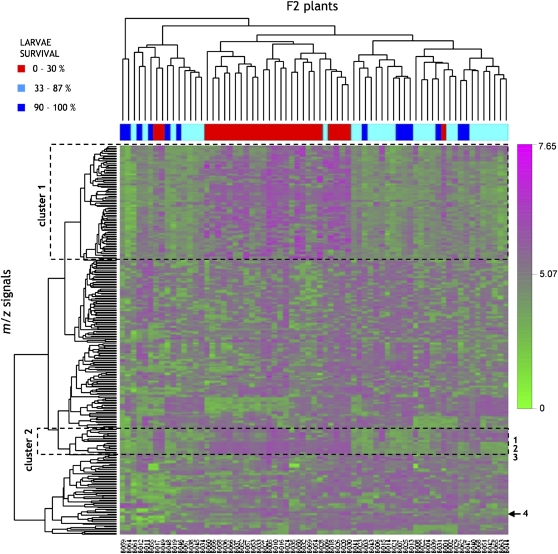
Two-way hierarchical cluster analysis (HCA) was used to visualize how the metabolites in the chromatograms and how the metabolite profiles of the F2 plants covary. In contrast to the cluster analysis depicted in Figure 7, which was based on the 150 m/z signals most correlated with resistance, we clustered 319 m/z signals of data set A or 306 m/z signals of data set B. To extend this analysis with the clustering of the metabolite profiles of the F2 plants, we performed two-way hierarchical clustering. Two-way HCA created a hierarchy of clusters where metabolites (represented by m/z signals) as well as plants were divided into clusters of similar patterns. Metabolite levels (represented by ion intensities) were represented by a vector. Similarity scores were calculated for all possible pairs of metabolites (angle between vectors), and the most similar vectors were then joined in a tree (Fig. 8). Clustering of m/z signals according to their metabolite profiles grouped the plants into clusters. Subsequently, we manually overlaid the clustering of the F2 plants with larvae survival The majority of F2 plants that were resistant (larvae survival 0%–30%) clustered together in one large cluster, while the more susceptible F2 plants clustered in two to three distinct clusters. Only four F2 plants deviated from this clustering (Fig. 8).
Figure 8.
Heat map for two-way hierarchical clustering of F2 plants and their metabolites (represented as m/z signals). Base peaks of compounds 1 to 4 marked at the right of the heat map are as in Figure 6. Plants are marked with three different colors according to their level of resistance, as in Figure 5. Color scale indicates log10-transformed m/z signal intensities (relative metabolite levels).
Based on the heat map generated from the HCA, two distinct clusters of m/z signals related to resistance were identified (Fig. 8). Clusters 1 and 2 displayed very similar response patterns: m/z signals (log10-transformed relative metabolite levels) in clusters 1 and 2 were intense (shown in magenta) when present in resistant plants (shown in red) but weak (shown in green) in the susceptible plants (shown in blue). Cluster 2 contained base peaks of compounds 1, 2, and 3 (oleanolic acid cellobioside), while the base peak of compound 4 (hederagenin cellobioside) was not present in cluster 1 or 2. All of the metabolites detected by the correlation analysis as being significantly correlated with larvae survival (Fig. 6) were found in the two mentioned clusters, except for the base peak of hederagenin cellobioside (compound 4; m/z 819; Supplemental Table S1). However, while the base peak of hederagenin cellobioside did not cluster with compounds 1, 2, and 3, its fragments and adduct ions were present in clusters 1 and 2, indicating that the clustering of the base peak of compound 4 may be false. As compound 4 is approximately 30-fold more abundant than compound 1, the split behavior of the m/z signals of compound 4 is most likely a reflection of saturation of the detector.
Relationships between Plants and Metabolites
We used principal component analysis (PCA) to calculate the variance of each m/z signal and then rank the variance based on the magnitudes of the uncorrelated variables. PCA transforms the original data matrix into a smaller set of uncorrelated variables called principal components (PCs). Data are plotted as a swarm of points, and the first PC corresponds to the direction of the swarm, where the variance or spread is largest. PCA of m/z signals (log10-transformed relative metabolite levels) and plants of data set B showed that PC1 explained 20% of the total variation. PC2 contributed with 8.6%, PC3 with 7.7%, and PC4 with 5.2% of the total variance, respectively. Of the total variation, 90% was accounted for by 39 components.
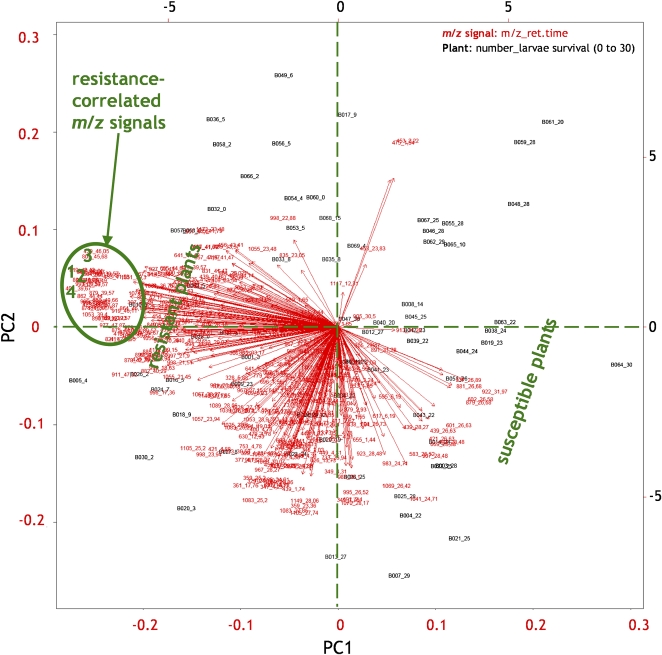
A biplot of PC1 versus PC2 (Fig. 9) provides a projection view of relationships between plants and their metabolites. The m/z signals that belong to the four compounds most correlated with the resistance (compounds 1, 2, 3, and 4 from Fig. 6) covaried to a great extent, since they grouped together (inside the green oval in Fig. 9). Base peaks of the four compounds had the largest negative scores for PC1, −0.18, −0.21, −0.24, and −0.18, respectively (Fig. 9; Supplemental Table S1); their values on the PC2 axis were close to 0. Susceptible plants had large positive loadings, while resistant plants had large negative loadings, on PC1. PC1 was associated with increasing susceptibility (decreasing resistance) in the plant. Higher PCs did not seem to reveal any further structure in the data set. It is worth noting that the larvae survival data were not included in this data matrix.
Figure 9.
Relationships between plants and metabolites (represented as m/z signals) identified by PCA. Red arrows represent m/z signals; the metabolites are represented by m/z values followed by retention times in minutes. Black points represent plants and their resistance toward insects; the letter B corresponds to data that are derived from data set B, followed by the plant line, and the larvae survival number out of 30 larvae is indicated after the underscore. Directions of arrows show the relative loadings of the metabolites on the first and second PCs. Base peaks of compounds 1 to 4 are as in Figure 6.
DISCUSSION
Saponins Confer Resistance to Flea Beetle Larvae
Metabolomics has attracted significant interest over the last few years (Tikunov et al., 2005; Hall, 2006; Keurentjes et al., 2006; de Vos et al., 2007; Khoo and Al-Rubeai, 2007; Sumner et al., 2007, 2008; Blow, 2008; Fiehn et al., 2008; Steinfath et al., 2008) and has contributed significantly to the discovery of bioactive compounds, improving food quality and cataloging metabolomes, as a new diagnostic and therapeutic approach and to study plant-herbivore interactions (Dixon et al., 2006; Ryan and Robards, 2006; Böttcher et al., 2008; Denkert et al., 2008; Jansen et al., 2009).
Our study demonstrated the power of untargeted metabolite profiling for identification of bioactive compounds involved in plant defense, using B. vulgaris and flea beetle larvae as a model system. The approach detected the two saponins hederagenin cellobioside and oleanolic acid cellobioside. Hederagenin cellobioside is known to confer resistance against diamondback moth (Shinoda et al., 2002), and oleanolic acid cellobioside is believed to have a similar effect on the diamondback moth (Agerbirk et al., 2003a), but the effect on flea beetles has not been reported. Thus, this work indicates that the two compounds are not only defense compounds of B. vulgaris against diamondback moth but also against flea beetles. In addition to oleanolic acid cellobioside and hederagenin cellobioside, we detected novel compounds that are possible defense compounds of B. vulgaris against flea beetles. Based on their characteristic fragmentation patterns, many of the novel compounds that relate to resistance appeared to be saponins. Compound 1 (m/z of the base peak 819) has a structure similar to hederagenin cellobioside, probably with some rearranged groups in the skeleton, and compound 2 (m/z of the base peak 817) probably has an additional double bond in the aglycone. Accordingly, our data have identified four saponins that may be defense compounds of B. vulgaris against a variety of insect species.
The F2 population accumulated the two known saponins hederagenin cellobioside and oleanolic acid cellobioside as well as several compounds likely to be saponins, some of which cosegregated with biological activity toward flea beetle larvae and others that did not correlate with resistance. Some of the predicted saponins in the F2 population were not present in the parental types, thus emphasizing the large potential for modification of metabolic composition through classical breeding.
The mode of action of saponins and their functionality are generally known to depend on their glycosylation pattern and secondary modifications of the saponin aglycone skeleton. Bidesmosidic saponins (with two sugar chains, often with one attached through a linkage at C-3 and the second attached through an ester linkage at C-28) are known to either lack or exhibit attenuated properties of the corresponding monodesmosides (with a single sugar, normally attached at C-3; Hostettmann and Marston, 1995). Saponin glycosides in monodesmoside form exhibit a broad spectrum of activity against yeast, fungi, mollusks, frogs, and fish, while those in bidesmoside form are generally inactive (Parkhurst et al., 1974; Schenkel et al., 1991; Favel et al., 1994; Lee et al., 2001; Sparg et al., 2004). In addition, the number, kind, and sequence of the sugar residues significantly modify antifungal effects of saponins (Mshvildadze et al., 2000; Zhang et al., 2005).
The compound with m/z 819 detected at 31.0 min was present in both susceptible and resistant F2 plants and had a similar fragmentation pattern to that of hederagenin cellobioside. This compound is very likely an isomer that does not possess biological activity against flea beetle larvae, at least at the occurring concentrations, since it did not correlate with resistance (r2 < 0.43 in data set A and r2 < 0.38 in data set B). These results indicate the importance of specific structural features as mediators of the activity against flea beetle larvae. It is thus of interest to unravel the relationship between chemical structure and biological activity of the different saponins present in B. vulgaris.
Multivariate Data Analysis
By applying two-way HCA, we identified two main clusters of metabolites that correlate with resistance to flee beetle larvae. The hederagenin cellobioside (compound 4) base peak was in a single distinct cluster, while its fragments, isotope ions, and adduct ions clustered together in clusters 1 and 2 (Fig. 8). The split behavior of the m/z signals of compound 4, and the apparent nonlinear correlation between larvae survival and ion intensity (Fig. 6), are most likely a reflection of saturation of the detector. As compound 4 is approximately 30-fold more abundant than compound 1, detector saturation is not an issue for compounds 1, 2, and 3. Hederagenin cellobioside was present in most of the F2 plants (Fig. 5), while compounds 1 and 2 were not detected in the susceptible F2 extremes. This is in contrast to the parental P- and G-types, where hederagenin cellobioside could only be detected in the G-type (Fig. 3). A different picture emerged when we applied PCA, in which hederagenin cellobioside (compound 4), including its base peak and most of its fragments and adduct ions, formed one group together with the three other most correlated with the resistance compounds. The base peaks of the four compounds contributed most to PC1, showing that they were strongly correlated with the resistance. Resistant and susceptible plants were distinguished by PCA into separate groups.
Efficiency of the Methods Used
Given the complexity of the data set, different data analysis tools were explored and correlation analysis was found to be the most informative. However, the correlation analysis did not provide information about the relationship between m/z signals and similarity between metabolites, while PCA, UPGMA cluster analysis, and HCA did. Particularly, HCA facilitated identification of patterns for each m/z signal by visualizing how m/z signal intensities change according to the resistance or susceptibility of the F2 plants. UPGMA clustered m/z signals according to cosegregation. It was difficult to see a clear separation of such clusters, which could indicate that most of the m/z signals used for the analysis belong to the same group of compounds and metabolic pathway and thus covary to some extent.
Regardless of the four statistical approaches applied, we obtained very similar results with the two biological replicates, data sets A and B, as the same four compounds were identified as being the most significant to resistance in B. vulgaris. Based on their fragmentation patterns, all four compounds were saponins. Most of the other compounds identified with the different approaches that correlated with resistance were saponins or saponin like, based on their fragmentation patterns, as represented by Supplemental Table S1. A few m/z signals related to resistance, as detected by PCA and HCA, were not identified as significantly correlated with resistance. This is probably due to their low abundance and thus relatively high coefficient of variation. The parental plant types, P and G, may differ in many genes that are not related to the resistance. Randomization of the background of genes and metabolites in the F2 offspring through meiotic recombination facilitates the detection of chemical compounds that truly correlate with resistance.
Perspectives
Insects remain the main pests to several major crops and stored products (Schoonhoven et al., 2005). Knowledge of bioactive plant compounds against herbivorous insects may be used to introduce natural resistance to cultivated plants and as an environmentally sustainable alternative to synthetic insecticides. Accordingly, there is an obvious need to better understand factors affecting interactions between insects and plants, as such knowledge is fundamental for the amelioration of future crop plants using biologically safe control strategies intended to control insect attacks.
This study demonstrates that untargeted metabolite profiling can be used to identify metabolites associated with resistance to flee beetle larvae in B. vulgaris. Our ecometabolomic approach has identified four main candidates that correlate with resistance. Future work is needed to purify the metabolites and to confirm the activity of each such candidate compound in bioassays, because inactive compounds may cosegregate with truly active compounds if they are metabolically or genetically closely linked.
Furthermore, it provides the foundation for future work focused on the isolation of key metabolites for structure elucidation by NMR and bioassays to reveal relationships between chemical structure and biological activity. At the genomic level, these results facilitate future studies of molecular mechanisms that confer resistance in B. vulgaris by demonstrating correlations between metabolites and gene expression.
The method developed based on untargeted metabolite profiling coupled to correlation and cluster studies and PCA can be applied to identify metabolites active in other systems, not only with regard to plant-insect interactions.
MATERIALS AND METHODS
Plants and Insects
The resistant G-type Barbarea vulgaris subspecies arcuata used in this work originated from Herlev, Denmark, whereas the susceptible P-type originated from Tissø, Denmark (de Jong et al., 2000; Agerbirk et al., 2003a). They were grown in a growth chamber at 20°C and an 18-h-light/6-h-dark photoperiod (de Jong et al., 2000). Light was supplied by 400-W HPI/T lamps, which produced 160 to 200 μmol quanta m−2 s−1 at the level of the leaf surface. Parental plants used in crossings were collected at natural growth sites in March and April 2003. F2 plants were offspring from one individual F1 plant from a cross between a G-type plant (no. 8) and a P-type plant (no. 6).
A stock of Phyllotreta nemorum from the ST line was maintained at 24°C and an 18-h-light/6-h-dark photoperiod as previously described (Nielsen, 1997a). The ST line is susceptible: it does not contain R genes that confer resistance to G-type and is unable to survive on the G-type of B. vulgaris arcuata (Nielsen, 1999).
Crossing of P- and G-Type Plants
Hybridization between G- and P-type B. vulgaris was performed under glasshouse conditions using manual emasculation of unopened female flower buds followed by paper bag isolation and pollination from a male 4 to 5 d later. Both P- and G-types were used as females to produce seeds of 21 different F1 combinations between 11 G-type and eight P-type parent plants. The following year, F1 plants from all hybrid combinations were bud pollinated to obtain F2 seeds. Unopened flower buds were opened with forceps and pollinated from opened flowers of the same plant. Hybrid combination P number 6 × G number 8 (female × male), which produced approximately 160 F2 seeds, was used subsequently for bioassays and metabolic profiling.
Bioassays
Levels of resistance of individual plants were determined in bioassays using freshly harvested leaves from 3- to 12-week-old plants grown in a growth chamber as described above. Leaves were placed in transparent plastic containers of suitable size (20 or 50 mL, according to the size of the leaf tested) together with a piece of moist filter paper. Newly emerged flea beetle larvae (less than 24 h old) were gently placed on the filter paper with a moist paint brush. Each container contained one leaf and five larvae. Containers were kept at 24°C for 72 h. After 72 h, larval survival was examined with a stereomicroscope. Larvae that were alive were close to their first molt and had produced a clear leaf mine, whereas dead larvae had not grown and were found dead either in a tiny leaf mine or on the leaf surface. From each individual plant, six leaves were tested; that is, survival rates on individual plants were determined using 30 larvae.
Metabolite Profiling
For the metabolite extraction, 8-mm leaf discs (about 15 mg fresh weight or 4 mg dry weight) from leaf 7 unless otherwise stated were frozen in liquid nitrogen and kept at −80°C. The samples were boiled with 500 μL of 85% methanol (60°C−70°C) in a water bath for 5 min and then cooled on ice. The extract was filtered with 45-μm Ultrafree-MC Durapore polyvinylidene difluoride filters (Millipore) and kept in glass containers at −20°C.
LC-MS analysis was carried out using an Agilent 1100 Series liquid chromatograph (Agilent Technologies) coupled to a Bruker Esquire 3000+ ion trap mass spectrometer (Bruker Daltonics). An XTerra MS C18 column (3.5 μm, 2.1 × 100 mm; Waters) with a flow rate of 0.2 mL min−1 was used. The mobile phases were as follows: A, formic acid (1 mL L−1) and NaCl (50 μm); B, acetonitrile (800 mL L−1) and formic acid (1 mL L−1). The gradient program was as follows: 0 to 3 min, isocratic 18% B; 3 to 60 min, linear gradient 18% to 80% B; 60 to 65 min, linear gradient 80% to 100% B; 65 to 70 min, isocratic 100% B; 71 to 85 min, isocratic 18% B. The mass spectrometer was run in positive ion mode. Total ion current and ion traces for specific [M+Na]+ adduct ions were used for locating compounds. Hederagenin cellobioside and oleanolic acid cellobioside were identified based on mass spectra and retention times for the pure compounds kindly provided by Dr. Tetsuro Shinoda (National Institute of Agrobiological Sciences, Ibaraki, Japan).
Bioinformatics and Statistics
The LC-MS profiles were aligned and normalized using MetAlign software (http://www.metalign.wur.nl/UK/). The t test function of MetAlign was used to reveal m/z signals significantly different between nine resistant and nine susceptible F2 extremes. Global normalization of the data was performed by autoscaling on total signal. That is, all amplitudes of m/z signals of the data set were summed together and used to normalize with regard to the first data set.
For correlation and regression analysis, a Java program, specifically coded for this type of data, was used. The data were normalized to adjust for average effects of each of the F2 plants. Correlation analysis measured r2 from correlation between the amount of each metabolite in each plant (m/z signal) and larvae survival on the same plant, both transformed with ln(Y + 1). The regression model was Y = μ + ax + bx × x, where Y = transformed amount of metabolite in plant, x = transformed survival of larvae on plant, and a and b are estimated regression parameters. Permutation analysis used 2,000 permutations of larvae survival rates to determine 95% significance threshold level for r2 to enable an argument for how many of the most correlated m/z signals to study. A binary distance matrix for the 150 m/z signals most highly correlated with larvae survival was generated. The distance between two m/z signals was calculated as 1 − r2, where r is the coefficient of correlation of levels of the two metabolites (m/z signals) over all F2 plants, with a regression model containing both a linear and a quadratic term and with both sides transformed with ln(Y + 1). This distance matrix was used to cluster the 150 selected m/z signals according to their coappearance in the F2 plant offspring with the SAS procedure proc Cluster using UPGMA (Sneath and Sokal, 1973) and subsequently draw the dendrogram with the SAS procedure proc Tree.
For PCA, R (R2.6.2; http://www.r-project.org/) was used. For two-way hierarchical clustering, the BioConductor package of R was used. The MetAlign output file containing relative metabolite levels (m/z signal intensities) in F2 plants was log10 transformed and used as input file for R.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. M/z signals identified with LC-MS, correlation, HCA, and PCA.
Supplementary Material
Acknowledgments
We are grateful to Susanne Bidstrup (Department of Plant Biology and Biotechnology, Faculty of Life Sciences, University of Copenhagen) for her help with extractions. We thank Dr. Tetsuro Shinoda (Invertebrate Gene Function Research Unit, Division of Insect Sciences, National Institute of Agrobiological Sciences, Ibaraki, Japan) for the kind gift of hederagenin cellobioside and oleanolic acid cellobioside and discussions and Nikoline Juul Nielsen (Department of Natural Sciences and Environment, Faculty of Life Sciences, University of Copenhagen) for discussions.
This work was supported by the Danish Agency for Science, Technology, and Innovation (grant no. 274–06–0370) and by the Villum Kann Rasmussen Foundation to “Pro-Active Plants.”
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Søren Bak (bak@life.ku.dk).
The online version of this article contains Web-only data.
References
- Agerbirk N, Olsen CE, Bibby BM, Frandsen HO, Brown LD, Nielsen JK, Renwick JAA (2003. a) A saponin correlated with variable resistance of Barbarea vulgaris to diamondback moth Plutella xylostella. J Chem Ecol 29 1417–1433 [DOI] [PubMed] [Google Scholar]
- Agerbirk N, Olsen CE, Nielsen JK (2001) Seasonal variation in leaf glucosinolates and insect resistance in two types of Barbarea vulgaris ssp. arcuata. Phytochemistry 58 91–100 [DOI] [PubMed] [Google Scholar]
- Agerbirk N, Ørgaard M, Nielsen JK (2003. b) Glucosinolates, flea beetle resistance, and leaf pubescence as taxonomic characters in the genus Barbarea (Brassicaceae). Phytochemistry 63 69–80 [DOI] [PubMed] [Google Scholar]
- Bailey CD, Koch MA, Mayer M, Mummenhoff K, O'Kane SL Jr, Warwick SI, Windham MD, Al-Shehbaz IA (2006) Toward a global phylogeny of the Brassicaceae. Mol Biol Evol 23 2142–2160 [DOI] [PubMed] [Google Scholar]
- Blow N (2008) Metabolomics: biochemistry's new look. Nature 455 697–700 [DOI] [PubMed] [Google Scholar]
- Börjesdotter D (1999) Potential oil crops: cultivation of Barbarea verna, Barbarea vulgaris and Lepidium campestre. PhD thesis. Acta Universitatis Agriculturae Sueciae, Uppsala
- Böttcher C, von Roepenack-Lahaye E, Schmidt J, Schmotz C, Neumann S, Scheel D, Clemens S (2008) Metabolome analysis of biosynthetic mutants reveals a diversity of metabolic changes and allows identification of a large number of new compounds in Arabidopsis. Plant Physiol 147 2107–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuker CJ, Victoir K, de Jong PW, van der Meijden E, Brakefield PM, Vrieling K (2005) AFLP markers for the R-gene in the flea beetle, Phyllotreta nemorum, conferring resistance to defenses in Barbarea vulgaris. J Insect Sci. 5 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwalek M, Lalun N, Bobichon H, Plé K, Voutquenne-Nazabadioko L (2006) Structure-activity relationships of some hederagenin diglycosides: haemolysis, cytotoxicity and apoptosis induction. Biochim Biophys Acta 1760 1418–1427 [DOI] [PubMed] [Google Scholar]
- de Jong PW, Frandsen HO, Rasmussen L, Nielsen JK (2000) Genetics of resistance against defences of the host plant Barbarea vulgaris in a Danish flea beetle population. Proc R Soc Lond B Biol Sci 267 1663–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denkert C, Budczies J, Weichert W, Wohlgemuth G, Scholz M, Kind T, Niesporek S, Noske A, Buckendahl A, Dietel M, et al (2008) Metabolite profiling of human colon carcinoma: deregulation of TCA cycle and amino acid turnover. Mol Cancer 7 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos CHR, Moco S, Lommen A, Keurentjes JJB, Bino RJ, Hall RD (2007) Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat Protoc 2 778–791 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Gang DR, Charlton AJ, Fiehn O, Kuiper HA, Reynolds TL, Tjeerdema RS, Jeffery EH, German JB, Ridley WP, et al (2006) Applications of metabolomics in agriculture. J Agric Food Chem 54 8984–8994 [DOI] [PubMed] [Google Scholar]
- Favel A, Steinmetz MD, Regli P, Vidal-Ollivier E, Elias R, Balansard G (1994) In vitro antifungal activity of triterpenoid saponins. Planta Med 60 50–53 [DOI] [PubMed] [Google Scholar]
- Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee do Y, Lu Y, Moon S, Nikolau B (2008) Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J 53 691–704 [DOI] [PubMed] [Google Scholar]
- Gauthier C, Legault J, Girard-Lalancette K, Mshvildadze V, Pichette A (2009) Haemolytic activity, cytotoxicity and membrane cell permeabilization of semi-synthetic and natural lupane- and oleanane-type saponins. Bioorg Med Chem 17 2002–2008 [DOI] [PubMed] [Google Scholar]
- Güçlü-Ustündağ O, Mazza G (2007) Saponins: properties, applications and processing. Crit Rev Food Sci Nutr 47 231–258 [DOI] [PubMed] [Google Scholar]
- Hall RD (2006) Plant metabolomics: from holistic hope, to hype, to hot topic. New Phytol 169 453–468 [DOI] [PubMed] [Google Scholar]
- Hegi G (1958) Illustrierte Flora von Mittel-Europa. Carl Hanser Verlag, Munich
- Hostettmann K, Marston A (1995) Chemistry and Pharmacology of Natural Products: Saponins. Cambridge University Press, Cambridge, UK
- Jansen JJ, Allwood JW, Marsden-Edwards E, van der Putten WH, Goodacre R, van Dam NM (2009) Metabolomic analysis of the interaction between plants and herbivores. Metabolomics 5 150–161 [Google Scholar]
- Keurentjes JJ, Fu J, de Vos CH, Lommen A, Hall RD, Bino RJ, van der Plas LH, Jansen RC, Vreugdenhil D, Koornneef M (2006) The genetics of plant metabolism. Nat Genet 38 842–849 [DOI] [PubMed] [Google Scholar]
- Khoo SHG, Al-Rubeai M (2007) Metabolomics: an emerging tool for understanding metabolic systems. In Systems Biology, Vol 5. Springer, Dordrecht, The Netherlands, pp 237–273
- Kristensen C, Morant M, Olsen CE, Ekstrøm CT, Galbraith DW, Møller BL, Bak S (2005) Metabolic engineering of dhurrin in transgenic Arabidopsis plants with marginal inadvertent effects on the metabolome and transcriptome. Proc Natl Acad Sci USA 102 1779–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MW, Kim SU, Hahn DR (2001) Antifungal activity of modified hederagenin glycosides from the leaves of Kalopanax pictum var. chinense. Biol Pharm Bull 24 718–719 [DOI] [PubMed] [Google Scholar]
- Lommen A (2009) MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal Chem 81 3079–3086 [DOI] [PubMed] [Google Scholar]
- MacDonald MA, Cavers PB (1991) The biology of Canadian weeds. 97. Barbarea vulgaris R. Br. Can J Plant Sci 71 149–166 [Google Scholar]
- Matsuda F, Yonekura-Sakakibara K, Niida R, Kuromori T, Shinozaki K, Saito K (2009) MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J 57 555–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mshvildadze V, Favel A, Delmas F, Elias R, Faure R, Decanosidze G, Kemertelidze E, Balansard G (2000) Antifungal and antiprotozoal activities of saponins from Hedera colchica. Pharmazie 55 325–326 [PubMed] [Google Scholar]
- Nielsen JK (1978) Host plant discrimination within Cruciferae: feeding responses of four leaf beetles (Coleoptera: Chrysomelidae) to glucosinolates, cucurbitacins and cardenolides. Entomol Exp Appl 24 41–54 [Google Scholar]
- Nielsen JK (1997. a) Variation in defences of the plant Barbarea vulgaris and in counteradaptations by the insect Phyllotreta nemorum. Entomol Exp Appl 82 25–35 [Google Scholar]
- Nielsen JK (1997. b) Genetics of the ability of Phyllotreta nemorum to survive in an atypical host plant Barbarea vulgaris ssp. arcuata. Entomol Exp Appl 82 37–44 [Google Scholar]
- Nielsen JK (1999) Specificity of a Y-linked gene in the flea beetle Phyllotreta nemorum for defences in Barbarea vulgaris. Entomol Exp Appl 91 359–368 [Google Scholar]
- Nielsen JK, de Jong PW (2005) Temporal and host-related variation in frequencies of genes that enable Phyllotreta nemorum to utilize a novel host plant, Barbarea vulgaris. Entomol Exp Appl 115 265–270 [Google Scholar]
- Nielsen JK, Hansen ML, Agerbirk N, Betersen BL, Halkier BA (2001) Responses of the flea beetles Phyllotreta nemorum and P. cruciferae to metabolically engineered Arabidopsis thaliana with an altered glucosinolate profile. Chemoecology 11 75–83 [Google Scholar]
- Ørgaard M, Linde-Laursen I (2008) Meiotic analysis of Danish species of Barbarea (Brassicaceae) using FISH: chromosome numbers and rDNA sites. Hereditas 145 215–219 [DOI] [PubMed] [Google Scholar]
- Parkhurst RM, Thomas DW, Skinner WA, Cary LW (1974) Molluscicidal saponins of Phytolacca dodecandra: lemmatoxin. Can J Chem 52 702–705 [Google Scholar]
- Poelman EH, Galiart RJFH, Raajimakers CE, van Loon JJA, van Dam NM (2008) Performance of specialist and generalist herbivores feeding on cabbage cultivars is not explained by glucosinolate profiles. Entomol Exp Appl 127 218–228 [Google Scholar]
- Renwick JAA (2002) The chemical world of crucivores: lures, treats and traps. Entomol Exp Appl 104 35–42 [Google Scholar]
- Ryan D, Robards K (2006) Metabolomics: the greatest omics of them all? Anal Chem 78 7954–7958 [DOI] [PubMed] [Google Scholar]
- Schenkel EP, Werner W, Schulte KE (1991) Die saponine aus Thinouia coriacea. Planta Med 57 463–467 [DOI] [PubMed] [Google Scholar]
- Schoonhoven LM, van Loon JJA, Dicke M (2005) Insect-Plant Biology. Oxford University Press, Oxford
- Senatore F, D'Agostino M, Dini I (2000) Flavonoid glycosides of Barbarea vulgaris L. (Brassicaceae). J Agric Food Chem 48 2659–2662 [DOI] [PubMed] [Google Scholar]
- Shinoda T, Nagao T, Nakayama M, Serizawa H, Koshioka M, Okabe H, Kawai A (2002) Identification of a triterpenoid saponin from a crucifer, Barbarea vulgaris, as a feeding deterrent to the diamond back moth, Plutella xylostella. J Chem Ecol 28 587–599 [DOI] [PubMed] [Google Scholar]
- Skoog DA (1985) Principles of Instrumental Analysis, Ed 3. Saunders College Publishing, Philadelphia
- Sneath PHA, Sokal RR (1973) Numerical Taxonomy. WH Freeman and Company, San Francisco, pp 230–234
- Sparg SG, Light ME, van Staden J (2004) Biological activities and distribution of plant saponins. J Ethnopharmacol 94 219–243 [DOI] [PubMed] [Google Scholar]
- Steinfath M, Groth D, Lisec J, Selbig J (2008) Metabolite profile analysis: from raw data to regression and classification. Physiol Plant 132 150–161 [DOI] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, Beger R, Beale MH, Daykin C, Fan TW, Fiehn O, Goodacre R, Griffin JL, et al (2007) Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, Urbanczyk-Wochniak E, Broeckling CD (2008) Metabolomics data analysis, visualization, and integration. In D Edwards, ed, Plant Bioinformatics: Methods and Protocols, Vol 406. Humana Press, Totowa, NJ, pp 409–436 [DOI] [PubMed]
- Tachibana M, Watanabe H, Itoh K (2002) Distribution of a naturalized weed, Barbarea vulgaris R. Br. in the Tohoku area of Japan. Journal of Weed Science and Technology 47 235–241 [Google Scholar]
- Tattersall DB, Bak S, Jones PR, Olsen CE, Nielsen JK, Hansen ML, Høj PB, Møller BL (2001) Resistance to an herbivore through engineered cyanogenic glucoside synthesis. Science 293 1826–1828 [DOI] [PubMed] [Google Scholar]
- Thompson JN (2005). The Geographic Mosaic of Coevolution. University of Chicago Press, Chicago
- Tikunov Y, Lommen A, de Vos CHR, Verhoeven HA, Bino RJ, Hall RD, Bovy AG (2005) A novel approach for nontargeted data analysis for metabolomics: large-scale profiling of tomato fruit volatiles. Plant Physiol 139 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leur H, Vet LEM, van der Putten WH, van Dam NM (2008) Barbarea vulgaris glucosinolate phenotypes differentially affect performance and preference of two different species of Lepidopteran herbivores. J Chem Ecol 34 121–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincken JP, Heng L, de Groot A, Gruppen H (2007) Saponins, classification and occurrence in the plant kingdom. Phytochemistry 68 275–297 [DOI] [PubMed] [Google Scholar]
- Zacarias A, Bolanowski D, Bhatnagar A (2002) Comparative measurements of multicomponent phospholipid mixtures by electrospray mass spectroscopy: relating ion intensity to concentration. Anal Biochem 308 152–159 [DOI] [PubMed] [Google Scholar]
- Zhang JD, Cao YB, Xu Z, Sun HH, An MM, Yan L, Chen HS, Gao PH, Wang Y, Jia XM, et al (2005) In vitro and in vivo antifungal activities of the eight steroid saponins from Tribulus terrestris L. with potent activity against fluconazole-resistant fungal pathogens. Biol Pharm Bull 28 2211–2215 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.