Abstract
The early phase of the interaction between tree roots and ectomycorrhizal fungi, prior to symbiosis establishment, is accompanied by a stimulation of lateral root (LR) development. We aimed to identify gene networks that regulate LR development during the early signal exchanges between poplar (Populus tremula × Populus alba) and the ectomycorrhizal fungus Laccaria bicolor with a focus on auxin transport and signaling pathways. Our data demonstrated that increased LR development in poplar and Arabidopsis (Arabidopsis thaliana) interacting with L. bicolor is not dependent on the ability of the plant to form ectomycorrhizae. LR stimulation paralleled an increase in auxin accumulation at root apices. Blocking plant polar auxin transport with 1-naphthylphthalamic acid inhibited LR development and auxin accumulation. An oligoarray-based transcript profile of poplar roots exposed to molecules released by L. bicolor revealed the differential expression of 2,945 genes, including several components of polar auxin transport (PtaPIN and PtaAUX genes), auxin conjugation (PtaGH3 genes), and auxin signaling (PtaIAA genes). Transcripts of PtaPIN9, the homolog of Arabidopsis AtPIN2, and several PtaIAAs accumulated specifically during the early interaction phase. Expression of these rapidly induced genes was repressed by 1-naphthylphthalamic acid. Accordingly, LR stimulation upon contact with L. bicolor in Arabidopsis transgenic plants defective in homologs of these genes was decreased or absent. Furthermore, in Arabidopsis pin2, the root apical auxin increase during contact with the fungus was modified. We propose a model in which fungus-induced auxin accumulation at the root apex stimulates LR formation through a mechanism involving PtaPIN9-dependent auxin redistribution together with PtaIAA-based auxin signaling.
Most temperate forest trees develop a mutualistic root symbiosis with ectomycorrhizal soil fungi. During the establishment of ectomycorrhiza (ECM), fungal hyphae invade the root from root cap cells in and upwards to the epidermis (Horan et al., 1988). After attachment to epidermal cells, hyphae multiply to form a series of layers that differentiate to establish a mantle structure around the root (Horan et al., 1988). An internal network of hyphae between the epidermis and root cortex cells forms the Hartig net (Blasius et al., 1986), while extraradical hyphae prospect throughout the surrounding soil and gather nutrients. Morphological observations of forming ECMs have shown that in the vicinity of the fungus the root architecture of the host plant is profoundly modified. Interaction with hyphae stimulates lateral root (LR) formation, dichotomy of the root apical meristem in conifer species, and cytodifferentiation of root cells (radial elongation and root hair decay; Horan et al., 1988; Dexheimer and Pargney, 1991; Ditengou et al., 2000). Even though several studies have focused on LR stimulation during ECM formation, it still remains unclear what the molecular mechanisms are that modify root development during contact with the fungus.
In the herbaceous model plant Arabidopsis (Arabidopsis thaliana), LR development is well described (Malamy and Benfey, 1997), and the analysis of its molecular regulation is ongoing. LRs in Arabidopsis are derived from a subset of pericycle cells, termed pericycle founder cells, which are adjacent to the two xylem poles (for review, see Casimiro et al., 2003; De Smet et al., 2006). LR initiation (LRI) is thought to go through two independent checkpoints (De Smet et al., 2007; Dubrovsky et al., 2008). First, priming of pericycle founder cells occurs in the basal meristem, which is a region at the transition between the meristem and the elongation zone of the root. Second, LRI itself, comprising cell cycle reactivation of the founder cells and subsequent further cell divisions, occurs in more proximal regions of the root and leads to the formation of a LR primordium (Malamy and Benfey, 1997). Finally, the new LR emerges after having grown through the cortex and epidermis of the parental root.
The phytohormone auxin (indole-3-acetic acid [IAA]) is considered to be one of the main triggers regulating all of the different steps of LR formation (Bainbridge et al., 2008; Ditengou et al., 2008; Laskowski et al., 2008; Nibau et al., 2008). For instance, the acquisition of founder cell identity, cell cycle reactivation, and LR emergence all correlate with and require local auxin accumulation in specific cell types (Dubrovsky et al., 2008; Fukaki and Tasaka, 2009). Recent studies have suggested that the coordination of polar auxin transport by auxin influx carriers (AUX/LAX) and auxin efflux carriers (PIN) is responsible for establishing an auxin gradient along the root with specific local maxima that regulate LR development (Bainbridge et al., 2008; Ditengou et al., 2008; Laskowski et al., 2008). In combination with polar auxin transport, local auxin biosynthesis in specific root cells was proposed to contribute to the formation of the auxin gradient (Petersson et al., 2009). Lastly, one should not neglect that the localized expression of auxin homeostasis-regulating genes like IAA-amido-synthetase GH3 may significantly interfere with the apparition of auxin maxima (Brady et al., 2007) and thereby influence LR development (Nakazawa et al., 2001; Khan and Stone, 2007). GH3 IAA-amido-synthetase conjugates auxin to amino acids or sugars (Staswick et al., 2005) and thereby controls through catabolism and storage free (active) auxin levels in the cell (Hagen and Guilfoyle, 1985; Ljung et al., 2002; Staswick et al., 2005). During LRI, once an auxin maximum is formed in pericycle founder cells, auxin triggers its signaling cascade by binding to its receptors (Badescu and Napier, 2006). This activates auxin-dependent transcription factors (AUX/IAA and ARF) that act on cell cycle-regulating targets (Fukaki et al., 2005; Dreher et al., 2006) to elicit cell cycle reactivation. The mechanism that confers founder cell identity to pericycle cells in the basal meristem is distinct from LRI. Recent results reveal its independence from the basic auxin signaling pathway. A mechanism where the asymmetric distribution of auxin itself acts as a morphogenic trigger to prime pericycle cells was proposed (Dubrovsky et al., 2008). Together, these data point out the crucial role of auxin gradients in root development.
Recently, evidence has been reported for an ectomycorrhizal fungus-induced alteration of the endogenous auxin balance in the root apex of Arabidopsis plants (Splivallo et al., 2009). Even though identification of key molecular factors that could explain this alteration is still lacking, earlier results on tree roots suggest an impact of the fungus on root polar auxin transport and auxin conjugation, which could be involved in auxin level changes. For instance, inhibitors of polar auxin transport, such as 2,3,5-triiodobenzoic acid, restrict the stimulation of LR formation in conifer seedlings by ectomycorrhizal fungi (Karabaghli et al., 1998). Moreover, differential expression of a GH3 auxin-amido-synthetase during ECM formation has been reported in Pinus pinaster (Charvet-Candela et al., 2002; Reddy et al., 2006) and implied that the presence of the fungus also alters auxin homeostasis in the root via a mechanism involving enzymatic activities. Fungus-impacted polar auxin transport and auxin conjugation may thus be possible pathways responding to or causing an altered root auxin balance. How they are integrated in the perception of fungal signals and then trigger LR formation on a molecular basis during the interaction is still an enigma.
A recent study with two species of ectomycorrhizal truffle fungi (Tuber borchii and Tuber melanosporum) has suggested that fungal IAA and ethylene impact LR branching in Arabidopsis and Cistus incanus during the early phase of interaction (Splivallo et al., 2009). Interestingly, only a combined exogenous IAA/ethylene treatment, but not each phytohormone alone, was able to fully mimic the effect of fungal signals on root development. A multiplicity of other identified signals (auxins, alkaloids, cytokinins, flavonols, polyamines) have been found to act in either a synergistic (rutine/zeatine) or antagonistic (IAA/hypaphorine) manner during ECM formation, but to date evidence for their molecular impact on root developmental programs is lacking (Ditengou and Lapeyrie, 2000; Martin et al., 2001; Jambois et al., 2005; Martin and Nehls, 2009).
All these data show that significant knowledge on two levels is still lacking. First, which fungal signaling molecules precisely influence plant root development and how they cross talk has never been clearly demonstrated. Second, how fungus-induced auxin level alterations are connected to root development modification is another question that needs to be addressed to complete the molecular understanding of symbiotic fungus-root interactions.
The aim of this study was to identify gene networks inside the root, with special focus on auxin transport, conjugation, and signaling, that bridge the gap between fungus-induced root endogenous auxin alteration and LR formation in the early phase of the Laccaria-Populus interaction. The availability of the genomic sequences of Populus trichocarpa (Tuskan et al., 2006) and Laccaria bicolor (Martin et al., 2008) has facilitated the molecular analysis of this interaction. Moreover, in recent years, several components of the auxin signaling pathways in poplar have been identified (Moyle et al., 2002; Schrader et al., 2003; Kalluri et al., 2007; Teichmann et al., 2008).
We characterized the symbiotic interaction between Populus tremula × Populus alba (hereafter termed poplar) and L. bicolor at both the physiological and molecular levels and also explored the nonmycorrhizal Arabidopsis-L. bicolor interaction. We showed that in both species interacting with L. bicolor, LR development was due to diffusible, non-host-specific signaling molecules. In this context, LR induction required polar auxin transport through PtaPIN9 (highly homologous to AtPIN2) as well as auxin signaling through poplar Aux/IAA proteins. Based on these results, we propose a model involving these genes as important regulators during fungus-induced LR development.
RESULTS
Poplar-L. bicolor Ectomycorrhiza Development in Vitro
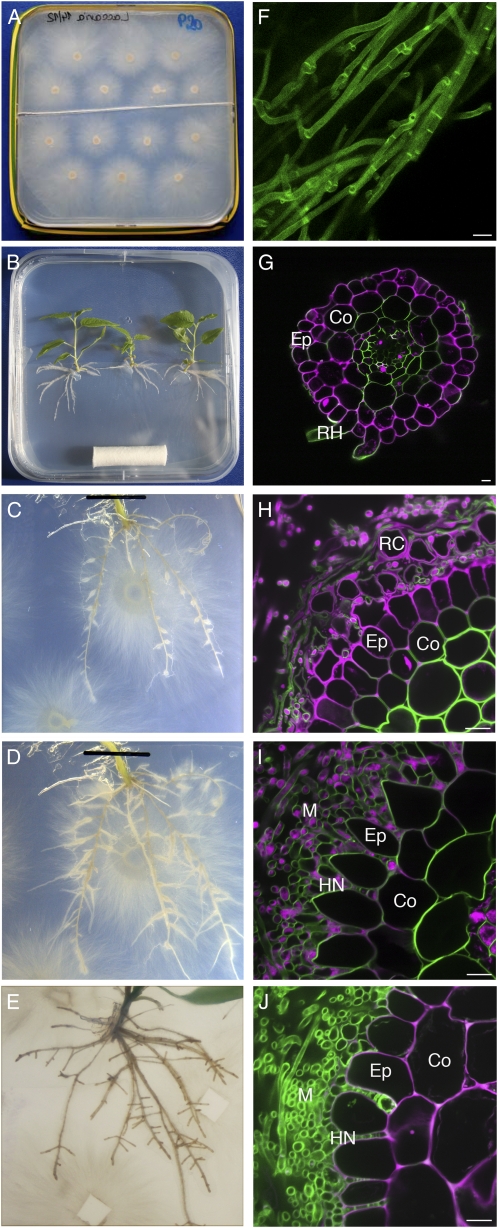
An in vitro sandwich culture system for the generation of poplar-L. bicolor ECMs under controlled conditions was adapted from previously described methods (Chilvers et al., 1986; Horan et al., 1988; Fig. 1, A and B). The time course of ECM development was assessed by observing root development (Fig. 1, C–E) and dual fluorescence-stained transverse root sections (Fig. 1, H–J). Two fluorescent markers were used, UVitex 2B for detection of fungal cell walls (green in Fig. 1) and propidium iodide for visualization of plant cell walls (magenta in Fig. 1). At 3 d of direct interaction (DODI), no change in root system morphology was visible (Fig. 1C). A discontinuous mycelium was attached to the root surface, and the rectangular epidermis cells were still tightly connected to one another (Fig. 1H). After 10 DODI, numerous LRs had developed and had a swollen appearance at their basis (Fig. 1D). At this stage, confocal microscopic observation showed the presence of a dense mycelium sheath (mantle) surrounding the roots and the start of Hartig net development: epidermis cells had an oval shape and were spaced and hyphae penetrated between them (Fig. 1I). After 30 DODI, we observed numerous short LRs, swollen from their basis up to the tip (Fig. 1E). Again, a Hartig net restricted to the epidermal cell layer was visible that suggests functional symbiosis (Fig. 1J).
Figure 1.
Time course of colonization of poplar roots by L. bicolor in an in vitro sandwich culture system. A and B, L. bicolor and poplar precultures, respectively. C to E, Root development of poplar at 3 DODI (C), 10 DODI (D), and 30 DODI (E). Note the increasing LR number starting at 10 DODI. Root swelling at the LR basis started at 10 DODI, and LR arrest was observed at 30 DODI. F, L. bicolor hyphae from precultures after UVitex staining. G, Transverse root section after propidium iodide UVitex dual staining (green, UVitex; magenta, propidium iodide). H to J, Dual fluorescence-stained transverse root sections at 3 DODI (H), 10 DODI (I), and 30 DODI (J). Co, Cortex; Ep, epidermis; HN, Hartig net; M, mantle; RC, root cap cells; RH, root hair. Note hyphae attachment at 3 DODI and mantle and Hartig net development from 10 to 30 DODI. In H, single, magenta-colored cells surrounding the epidermis are detached root cap cells. Images shown are representative from a series of three experiments. Bars = 10 μm.
LR Development in the Presence of L. bicolor
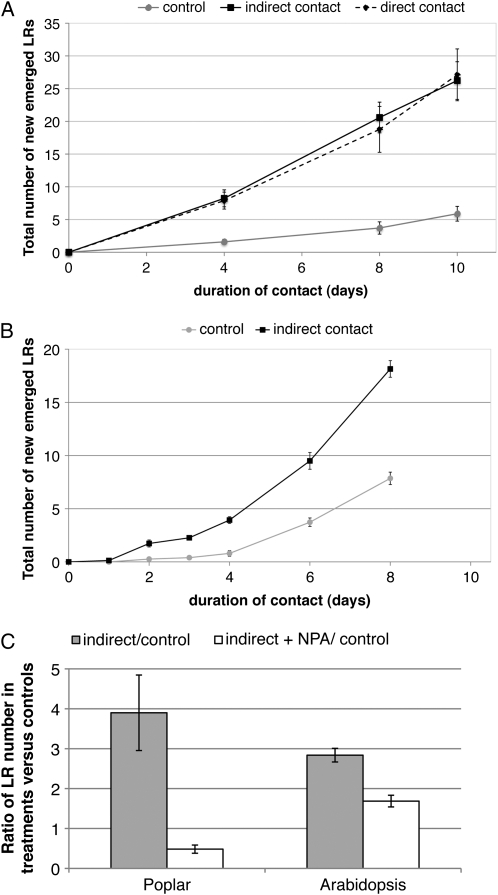
Analysis of the time course of LR induction in poplar interacting with L. bicolor under in vitro conditions showed that 4 DODI between the symbiotic partners was sufficient to increase the LR number by 3-fold compared with control plants without fungus (Fig. 2A). At 10 DODI, control plants had formed approximately five new LRs, whereas plants interacting with L. bicolor developed about 25 LRs. In the in vitro sandwich culture system, we also demonstrated an indirect interaction between the partners. This was achieved by placing a cellophane membrane between the two partners, which prohibits root colonization by hyphae but allows the exchange of signaling molecules between plant and fungus. Comparison of the degree of LR development in roots directly and indirectly interacting with L. bicolor showed that under both conditions, the number of LRs established over the induction time course was identical (Fig. 2A).
Figure 2.
Time course of early LR development during the interaction of L. bicolor with mycorrhizal poplar and nonmycorrhizal Arabidopsis. A and B, Time course of LR development in poplar in response to L. bicolor (A) and in Arabidopsis (B). Beginning at 4 d (A) and 2 d (B) of poplar and Arabidopsis-L. bicolor interaction, a significant (Student's t test P < 0.05) LR stimulation was observed. LR stimulation was similar in indirect and direct contact in poplar. C, Effect of the polar auxin transport inhibitor NPA on LR increase in poplar (10 μm NPA) after 8 DOII and in Arabidopsis (5 μm NPA) after 6 DOII. LR ratio of plants in indirect contact versus control plants without fungus in the presence and absence of NPA is shown. LR stimulation was significantly (Student's t test P < 0.01) reduced by the NPA treatment. In each experiment and each condition, 10 to 15 individual poplar plants or 15 Arabidopsis seedlings were observed. Error bars indicate se. If not visible, they were smaller than the symbol at the data point.
Next, we asked whether L. bicolor signals were able to induce LR formation in the nonmycorrhizal plant Arabidopsis. Similar to poplar, LR development was also induced in Arabidopsis during indirect contact (Fig. 2B). The time course of LR emergence was similar to that observed in poplar, with the first significant increase in LR development at 2 d of indirect interaction (DOII). After 8 DOII with L. bicolor, Arabidopsis seedlings had formed about 18 new LRs, whereas control plants had only formed eight.
Taken together, the results from the colonization and LR development experiments demonstrated that LR stimulation is characteristic for the early phase of plant-fungus interaction and is thus temporally distinguished from mature ECM formation, typified by Hartig net formation, root swelling, and root growth arrest.
Inhibition of Polar Auxin Transport during Plant-L. bicolor Interactions
The impact of polar auxin transport on fungus-induced LR stimulation was investigated. Therefore, we quantified LRs formed when L. bicolor interacted indirectly with poplar or Arabidopsis in the presence of the polar auxin transport inhibitor 1-naphthylphthalamic acid (NPA) at time points where we had observed a strong difference between control conditions and indirect contacts (8 and 6 DOII, respectively). The NPA treatment dramatically reduced fungal LR induction in both nonmycorrhizal and mycorrhizal plant species (Fig. 2C). In poplar, 10 μm NPA completely inhibited LR induction by the fungus. In Arabidopsis, the presence of 5 μm NPA reduced LR induction by 40% compared with controls. This severe reduction of LR induction is a first argument for the involvement of polar auxin transport in LR development during the early phase of the L. bicolor-plant interaction.
Modification of Root Auxin Gradients during Interaction
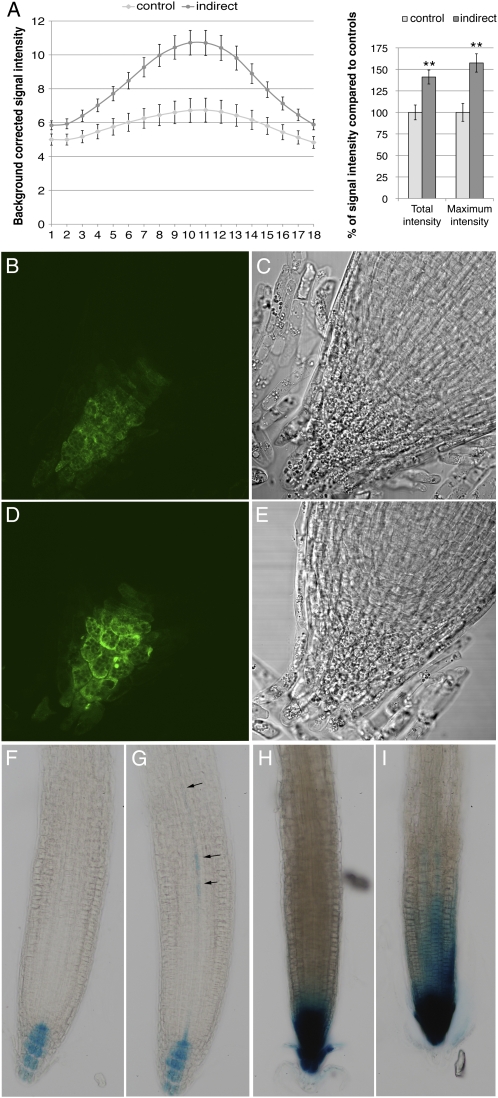
Auxin response in roots during contact with the fungus was assessed using the synthetic DR5 auxin-inducible promoter (Ulmasov et al., 1995) fused either to a GUS (Arabidopsis) or GFP (P. tremula × P. tremuloides) reporter gene. While DR5 reports on auxin levels in Arabidopsis (Petersson et al., 2009), it is primarily used as an auxin response promoter in poplar. GFP fluorescence intensity was observed on entire LR apices of DR5:GFP plants using confocal laser scanning microscopy. For each root, the signal intensity from each of 18 optical sections in a Z-stack (distance between slices was 1 μm) was measured to generate a profile of the auxin accumulation over the root apex (Fig. 3A). The result of this profiling showed that the presence of the fungus increased the total fluorescence in the root apex by 42% compared with control roots. The overall profile in roots was altered as well by the fungus: a strong auxin maximum was observed in the central part of the root at 3 DOII (57% increase compared with controls), and the profile showed a higher amplitude at the apex center than in the rather equally distributed signal in control roots. No difference was observed concerning the localization of the GFP signal (Fig. 3, B and D). In both conditions, the quiescent center zone was rather weakly stained and a stronger signal was observed in columella initials. As for Arabidopsis DR5:GUS, the presence of the fungus stimulated the appearance of a weak GUS signal in the provasculature within the basal meristem in 28 out of 32 analyzed plants (arrows in Fig. 3G). This signal was absent in 32 out of 35 control plants (Fig. 3F). In poplar, no provascular fluorescence was observed, probably due to high tissue thickness in this region. Interestingly, the provascular signal in Arabidopsis was absent at 3 DOII when the contact was made in the presence of 1 or 10 μm NPA (Supplemental Fig. S1).
Figure 3.
Modification of the auxin gradient in P. tremula × P. tremuloides and Arabidopsis upon interaction with L. bicolor. A, Left, distribution of average, background-corrected GFP fluorescence intensity in the 18 optical sections of the Z-stack through the LR apices of P. tremula × P. tremuloides DR5:GFP plants in control conditions or at 3 DOII. Right, quantification of the signal intensity change as percentage of fluorescence in control root apices. Total intensity (sum of 18 sections) as well as maximum intensity (one section, around Z-stack section 9–11) increased significantly. Quantification from at least seven root apices per condition is shown (** Student's t test P < 0.05). B to E, P. tremula × P. tremuloides DR5:GFP LR apex under control conditions (B and C) and at 3 DOII (D and E). F and G, Arabidopsis DR5:GUS in control conditions (F) and at 3 DOII (G). Note the signal in provasculature at 3 DOII (arrows in G). Examples shown are out of more than 30 biological replicates. H and I, Arabidopsis pin2 DR5:GUS in control conditions (H) and at 3 DOII (I). A strong auxin accumulation at the root apex in control plants is observed, and the presence of the fungus enlarged this signal upwards. Examples shown are out of 15 biological replicates.
Thus, L. bicolor-stimulated LR induction is paralleled by an NPA-sensitive auxin response increase at the root apex and in provascular tissues in Arabidopsis.
Transcript Profiling of Poplar Roots during Early Interaction with L. bicolor
To identify which molecular processes are induced in poplar roots by signals released by the symbiont, an oligoarray-based transcript profile of poplar roots at 3 DOII was generated. This corresponds to the time point where LR induction has begun (Figs. 1 and 2). Indirect contact was chosen because it limits possible root colonization-related transcriptome changes.
Of the 39,303 genes that were represented by specific oligonucleotides on the poplar array, 26,125 (66%) were expressed in control roots. In roots interacting with L. bicolor, 7% (2,945) of all genes were differentially expressed (>2-fold; Student's t test P < 0.05). Among these genes, 71% (2,095) were up-regulated and 29% (850) were down-regulated. Of the up-regulated genes, 17% (351) were induced de novo, as their expression could only be detected upon contact. Fifteen percent (132) of the down-regulated genes were completely repressed, as their transcripts were no longer detected in roots (Supplemental Table S1). Among the genes showing homology to known genes in the databases, most were involved in cell wall-related functions, such as laccases, carbohydrate-acting hydrolases, cell wall-related kinases, and phenylpropanoid metabolism enzymes (Supplemental Table S1). The second most represented gene class was transcription factors, and these included SCARECROW and PLETHORA (PLT). Together, these genes are known to control root apical meristem cell identity (Sabatini et al., 2003; Aida et al., 2004). PLT furthermore acts on PIN gene expression at the root apex (Blilou et al., 2005; Galinha et al., 2007).
In order to identify within the differentially expressed genes those that may be connected to LR initiation, we compared our results to LRI genes analyzed in Arabidopsis (Vanneste et al., 2005). These authors had identified 913 LRI genes in a comparative, microarray-based approach with wild-type and auxin signaling mutant plants during chemical stimulation of LRI. A total of 46 differentially expressed poplar genes were overlapping with LRI genes in Arabidopsis (Supplemental Table S2). These contained early auxin-responsive genes involved in transcriptional regulation (AUX/IAA) and auxin conjugation (GH3) as well as cell cycle regulators. A focus was made on the regulation of members of auxin signaling-related (PtaIAA) and homeostasis-related (PtaGH3) gene families.
Six out of the 35 poplar auxin-responsive transcription factors Aux/IAA were identified as being differentially expressed: PtaIAA28.1, PtaIAA33.2, PtaIAA3.3, PtaIAA3.4, PtaIAA19.3, and PtaIAA34 (names refer to Kalluri et al., 2007). Within these six PtaIAA family members, transcripts of PtIAA28.1 were the most abundant in control roots, and PtIAA28.1 was the only family member to be down-regulated upon contact with L. bicolor. PtaIAA34 was induced de novo in the presence of L. bicolor.
As for the 12 poplar homologs of known GH3 IAA-amido-synthetase genes that code for proteins regulating auxin homeostasis (Hagen and Guilfoyle, 1985; Ljung et al., 2002; Staswick et al., 2005), four transcripts were induced: PtaGH3-1 and PtaGH3-2 as well as PtaGH3-7 and PtaGH3-8, which respectively can be considered as duplicates based on their homology to each other and to Arabidopsis homologs (Supplemental Fig. S2). Referring to the phylogenetic analysis, these four targets are part of group II GH3 proteins that are active on auxin in other plant species (Staswick et al., 2005).
As the auxin gradient was affected in the plant during contact with the fungus, we also screened the microarray data for differentially expressed genes involved in polar auxin transport. Among the eight annotated members of the auxin influx carrier gene family (PtaAUX), two (PtaAUX3 and PtaAUX6) were differentially expressed (Table I; phylogenetic tree in Supplemental Fig. S3). Furthermore, four of the 16 members of the auxin efflux carrier gene family (Supplemental Fig. S4), PtaPIN2, PtaPIN4, PtaPIN9, and PtaPIN12, were induced in roots in interaction with the fungus (Table I).
Table I.
Microarray-based results of differentially expressed auxin-related genes in poplar roots after 3 DOII with the ectomycorrhizal fungus L. bicolor
Genes are listed by gene family and, within the gene family, by increasing transcript accumulation. From top to bottom: auxin-responsive transcription factors (PtaIAA), IAA-amido-synthetases (PtaGH3), auxin influx carriers (PtaAUX), and auxin efflux carriers (PtaPIN).
| Gene Namea | Gene Modelb | Expression in Control Rootsc | Fold Changed | Pe |
|---|---|---|---|---|
| PtaIAA28.1 | gw1.XVIII.808.1 | 4,470 | −1.9 | 0.0027 |
| PtaIAA33.2 | gw1.121.83.1 | 500 | 2.3 | 0.0001 |
| PtaIAA3.4 | estExt_fgenesh4_pm.C_LG_V0528 | 756 | 2.4 | 0.0001 |
| PtaIAA3.3 | fgenesh4_pm.C_LG_II000215 | 58 | 2.6 | 0.0026 |
| PtaIAA19.3 | estExt_fgenesh4_pm.C_LG_III0099 | 572 | 3.2 | 0.0004 |
| PtaIAA34 | gw1.X.53.1 | nd | 3.4 | 0.0023 |
| PtaGH3-8 | gw1.III.363.1 | nd | 2.8 | 0.0051 |
| PtaGH3-7 | fgenesh4_pg.C_LG_I000598 | 106 | 2.9 | 0.0151 |
| PtaGH3-2 | estExt_fgenesh4_pg.C_LG_IX0695 | 414 | 6.2 | 0.0000 |
| PtaGH3-1 | eugene3.02050011 | 2,288 | 8.3 | 0.0000 |
| PtaAUX3 | estExt_fgenesh4_pg.C_LG_X1704 | 1,029 | −2.0 | 0.0003 |
| PtaAUX6 | grail3.0001031001 | 1,022 | 2.9 | 0.0003 |
| PtaPIN12 | fgenesh4_pg.C_LG_XIX000547 | 3,975 | 2.2 | 0.0204 |
| PtaPIN2 | estExt_Genewise1_v1.C_LG_XVI1213 | 608 | 2.5 | 0.0267 |
| PtaPIN4 | estExt_fgenesh4_pm.C_LG_V0399 | 171 | 3.0 | 0.0330 |
| PtaPIN9 | fgenesh4_pm.C_LG_XVIII000434 | 294 | 3.9 | 0.0019 |
Gene name referring to the Joint Genome Institute Populus genome version 1.1 (PtaGH3, PtaPIN, PtaAUX) or to Kalluri et al. (2007; PtaIAA).
Gene model names from the Joint Genome Institute Populus genome version 1.1.
Background-corrected expression level as units of Cy3 fluorescence on microarray. nd, Not detected.
Ratio of expression levels in roots 3 DOII with L. bicolor versus expression levels in control roots only covered by a cellophane membrane.
Student's t test with false discovery rate (Benjamini-Hochberg) multiple testing correction.
Taken together, we identified 16 auxin-related genes that were differentially expressed in poplar roots during the interaction with L. bicolor (Table I) and that are likely to be involved in LRI and/or in modifying auxin gradients within the root. We have likely underestimated the expression ratios of several targets, as transcript analysis was realized on entire root systems, thereby diluting events with restricted, specific tissue localization.
Time Course of Auxin-Related Gene Expression during Poplar-L. bicolor Interaction
Using quantitative real-time PCR of cDNA synthesized from total RNA of root samples collected at 1, 3, and 10 DOII as well as from 4-week-old ectomycorrhizal roots (30 DODI), we assessed the transcript levels of the 16 identified auxin-related genes (Table I) over time during the poplar-L. bicolor interaction. Real-time PCR data from three independent biological replicates of 3 DOII were used to validate microarray data.
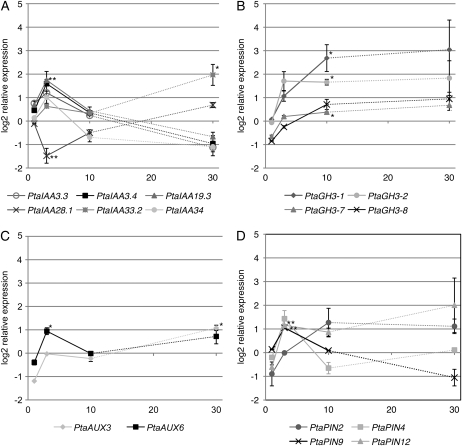
The expression profiles within the PtaIAA gene family members varied (Fig. 4A). During indirect contact, PtaIAA19.3 transcripts showed a significant increase at 3 DOII before leveling off back to constitutive levels. The transcript level increased 3.3-fold at 3 DOII (nonlogarithmic scale). The expression profile of PtaIAA28.1 was the inverse of the aforementioned PtaIAA. Its transcript level reached the lowest level (2.7-fold decrease) at 3 DOII and then increased slowly back to the level measured in control plants between 10 and 30 DODI. PtaIAA33.2 displayed yet another expression profile. It was significantly up-regulated up to 4-fold by 30 DODI and was the only late-induced member of the PtaIAA gene family. Taken together, PtaIAA19.3 and PtaIAA28.1 showed transient induction, paralleling fungus-stimulated LRI, whereas PtaIAA33.2 accumulation was activated during late symbiosis.
Figure 4.
Expression profiles of auxin-related target genes in poplar roots at 1, 3, and 10 DOII as well as 30 DODI (horizontal axis). Log2-transformed relative expression compared with control roots is shown. A, PtaIAA19.3 transcripts were significantly up-regulated with an accumulation maximum at 3 DOII. PtaIAA28.1 was down-regulated but bottomed out at the same time point. With the exception of PtaIAA33.2, these members of this gene family were only induced during the early phase of contact. B, PtaGH3 genes were slowly up-regulated only at 10 DOII. C, PtaAUX6 was early induced at 3 DOII, whereas PtaAUX3 was only induced in the late phase (30 DODI). D, PtaPIN transcript profiles differed from one another. PtaPIN9 and PtaPIN12 were induced early (3 DOII) but PtaPIN9 levels decreased in the late phase (30 DODI). * Student's t test P < 0.05; ** Student's t test P < 0.01.
The PtaGH3 gene family, involved in auxin conjugation, showed a homogeneous transcriptional pattern during the poplar-L. bicolor interaction (Fig. 4B). PtaGH3-1, PtaGH3-2, and PtaGH3-7 transcripts slowly accumulated, and their levels became significantly different from the controls after 10 DOII. PtaGH3-1 was the family member displaying the highest up-regulation, up to 6.4-fold higher than in control plants. By comparison, the induction of the other members, PtaGH3-2 and PtaGH3-7, was 3.4- and 1.3-fold, respectively. Even if transcript accumulation at 30 DODI was variable between different biological replicates, a persistent profile is indicated for all four PtaGH3 genes.
The two analyzed members of the auxin influx carrier family PtaAUX (Fig. 4C) showed a distinct regulation pattern. PtaAUX6 was significantly and transiently up-regulated at 3 DOII. The transcript levels of PtaAUX3 were not altered during the early phase of the interaction but became significantly up-regulated quite late, at 30 DODI.
Members of the auxin efflux carrier family PIN showed a very heterogeneous regulation profile (Fig. 4D). Only PtaPIN9 and PtaPIN12 were significantly elevated by 2.0- and 2.2-fold, respectively, at 3 DOII. Subsequently, PtaPIN9 levels decreased rapidly and reached constitutive levels at 10 DOII. PtaPIN12 levels varied between biological replicates at further time points but indicated a persisting induction up to 30 DODI.
In summary, L. bicolor caused the transient induction of a series of genes of the auxin signaling and polar auxin transport family, including PtaIAA19.3, PtaIAA28.1, PtaAUX6, and PtaPIN9. Members of the IAA-amido-synthetases family, PtaGH3-1, PtaGH3-2, and PtaGH3-7, accumulated slowly and their levels remained elevated throughout the contact. Lastly, PtaAUX3 and PtaIAA33.2 were specifically and only induced at late stages of the interaction.
NPA Effect on Gene Expression during Poplar-L. bicolor Interaction
We analyzed how the polar auxin transport inhibitor NPA, which inhibited LR stimulation by the fungus (Fig. 2C), influenced the expression of L. bicolor-regulated auxin-related genes. We generated cDNAs from poplar roots at 3 or 10 DOII in the presence or absence of NPA and subjected them to quantitative real-time PCR.
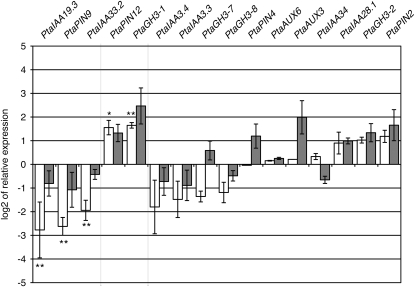
The expression profiles of certain members of the PtaIAA family were inverted by the presence of NPA (Fig. 5). The presence of NPA during the contact with L. bicolor abolished fungus-induced transcript accumulation of PtaIAA19.3, PtaPIN9, and PtaIAA33.2. A repression of 2.0 (PtaIAA19.3), 2.0 (PtaPIN9), and 2.5 (PtaIAA33.2) times compared with controls without fungus and NPA was observed instead of the previously observed significant induction of 3.3 and 2.0 times for PtaIAA19.3 and PtaPIN9, respectively (Figs. 4 and 5). In contrast, NPA-based inhibition of auxin polar transport amplified by 3-fold the induction of PtaPIN12 and PtaGH3-1 over levels induced by the fungus alone, thus leading to transcript levels 6- and 33-fold higher, respectively, than in control plants without fungus and NPA. Interestingly, for all five mentioned genes, NPA impacted transcript levels at 3 d of poplar-L. bicolor interaction, but at later stages (10 d), its effects on plants in interaction with the fungus were no longer statistically relevant. The differential expression upon contact with the fungus of all other gene targets was not significantly influenced by the presence of NPA at either 3 or 10 DOII.
Figure 5.
Influence of the polar auxin transport inhibitor NPA on gene expression after 3 DOII (white bars) or 10 DOII (gray bars) of poplar with L. bicolor. Ratios of the transcript levels in roots in the presence versus the absence of NPA during indirect interaction with L. bicolor are presented. The graph is separated into three parts. The left part presents genes whose fungal induction was significantly abolished by NPA (in order of magnitude referring to 3 DOII). A drastic reduction in PtaIAA19.3, PtaPIN9, and PtaIAA33.2 transcripts by the presence of NPA at 3 DOII was observed. The middle part presents genes whose fungal induction was significantly increased by the presence of NPA. Note the high up-regulation of PtaPIN12 and PtaGH3-1. The right part refers to genes whose expression was not statistically affected by NPA. * Student's t test P < 0.05; ** Student's t test P < 0.01.
These results indicate that the repression of fungus-induced LR induction by NPA is mirrored by dramatic changes in transcript abundance, especially that of early, transiently regulated auxin signaling and transport genes such as PtaPIN9 and PtaIAA19.3.
LR Development in Arabidopsis Auxin Mutants during Interaction with L. bicolor
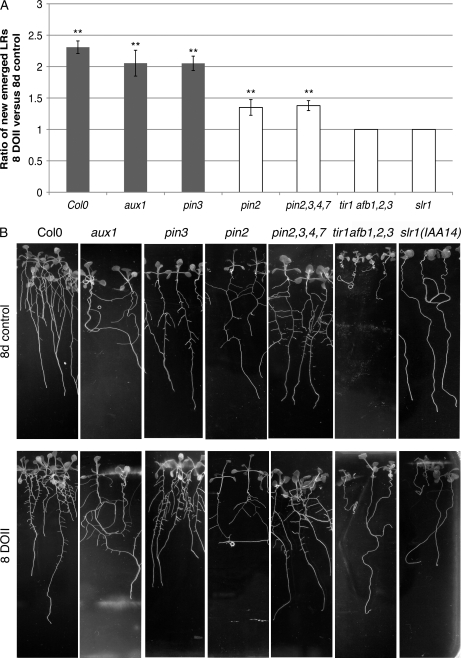
To gain additional insights into the crucial role of auxin transport and signaling during fungus-induced LR development, we assessed LR development in response to L. bicolor in different Arabidopsis mutants altered in auxin perception (tir1afb1,2,3), signaling (slr1 [IAA14]), influx (aux1), and efflux (pin2, pin2,3,4,7). The Arabidopsis mutants chosen were either those known to have an important LR phenotype (reduction or absence of LR development in slr1 and tir1afb1,2,3) or those genes most closely related to genes in poplar that were found to be regulated (pin2 [PtaPIN9], aux1 [PtaAUX3]) during L. bicolor-induced LR development. The effect of L. bicolor on plants defective in PIN3 or additionally also PIN4 and PIN7 was examined because of their role together with PIN2 in the auxin reflux loop at the root apex (Blilou et al., 2005). At 5 d after germination, mutants were placed into contact with L. bicolor, and LR development was observed after 8 DOII (Fig. 6). The auxin perception quadruple mutant tir1afb1,2,3 and the IAA14 protein-stabilized mutant slr1, which both did not form LRs under control conditions (Fig. 6B), also did not develop LRs in response to L. bicolor (Fig. 6). As for auxin transport mutants, we observed that in the presence of L. bicolor, aux1 was still able to develop LRs, as was pin3. The magnitude of LR stimulation in both mutants was comparable to that in the wild type. In both the single pin2 mutant and the quadruple pin2,3,4,7 mutants, a significant increase in LR during contact with the fungus was observed. LR stimulation in these mutants was 1.37 times higher as compared with controls of the same lines in the absence of fungus, whereas wild-type plants developed up to 2.4 times more roots during contact. LR stimulation in the quadruple mutant pin2,3,4,7 was not further inhibited than in the single pin2 mutant. We further analyzed auxin distribution in the pin2 mutant harboring the DR5:GUS reporter (Fig. 3, H and I). Whereas in wild-type plants the presence of the fungus stimulated auxin accumulation in specific cells of provascular tissues, in pin2 a strong and diffuse increase of DR5-directed GUS activity was observed in the epidermis and LR cap, and the provascular signal was visible only in two out of 13 analyzed plants at 3 DOII.
Figure 6.
LR development in Arabidopsis auxin mutants in the presence of L. bicolor after 8 DOII. A, Ratio of LR number that developed in plants in contact with L. bicolor versus control plants without fungus. Significant LR increases are marked by asterisks (** Student's t test P < 0.01). Ratios in mutants that differed significantly from ectoype Columbia (Col0) are represented as white bars. LR development was stimulated by L. bicolor in Columbia, pin3, and aux1 to a similar extent and significantly less in pin2 and pin2,3,4,7. The quadruple auxin receptor mutant tir1afb1,2,3 and the IAA14 gain-of-function mutant slr1 were completely insensitive to L. bicolor in terms of LR stimulation. B, Root development of control plants and plants after 8 DOII. Note the absence of LRs in the slr1 and tir1afb1,2,3 negative control plants and at 8 DOII. For each treatment and mutant line, 10 to 15 biological replicates were analyzed.
Together, these data imply a prominent role of auxin transport through AtPIN2 in fungus-regulated auxin gradient modification and LR initiation.
DISCUSSION
Although LR stimulation during ECM formation is a recognized phenomenon, the molecular mechanisms that are activated in roots by the presence of the fungus and that regulate LR stimulation are so far unknown. Here, we investigated the early interaction of L. bicolor with roots of a mycorrhizal and a nonmycorrhizal plant. Our molecular and functional analyses revealed that auxin transport and signaling are key mechanisms regulating LR development in response to fungal signaling molecules.
In interaction with L. bicolor, the early response of poplar resulted in an increase in LRs paralleled by an enhanced auxin response at the root apex, as revealed by an increase in DR5-driven GFP expression. Indirect contact experiments showed that these modifications did not require physical contact between hyphae and root cells, suggesting a role for diffusible signaling molecules. Using the nonmycorrhizal plant Arabidopsis, we further demonstrated that these diffusible signaling molecules induced LRs and root apex auxin response increase in a host plant-independent manner and that both did not rely on the mycorrhizal capacity of the plant partner. Equivalent results have been reported recently for the truffle-Arabidopsis and truffle-C. incanus interaction (Splivallo et al., 2009). Thus, LR-inducing signaling molecules from various fungi have rather broad functions in plant development, just as phytohormones would be expected to have.
Auxin Homeostasis Is Altered in the Root during Interaction with L. bicolor
The increase in auxin accumulation at the root apex during contact with the ectomycorrhizal fungus has only recently been reported by Splivallo et al. (2009), and our data confirm and extend this finding to another ectomycorrhizal fungus and to poplar as a mycorrhizal host plant. Two hypothetical scenarios can explain this auxin accumulation. First, auxin secreted by the ectomycorrhizal fungus may be taken up into the root and accumulate in the tip. The root tip is usually the first part of the root colonized by the fungus (Horan et al., 1988), which would explain this local auxin increase at the apex. Splivallo et al. (2009) have shown that exogenous auxin alone was not able to mimic the effect of the fungus, which also suggests that fungal auxin alone would not be sufficient to induce the observed DR5:GFP signal increase. For instance, Splivallo and coworkers (2009) have suggested a combined action of fungal auxin and ethylene on the root. As ethylene is known to impact endogenous auxin biosynthesis in roots (Stepanova et al., 2007, 2008), a second scenario may include an activation of endogenous auxin biosynthesis at the root apex by fungal ethylene, leading to the observed auxin accumulation. A recent study had shown that artificial induction of auxin biosynthesis at the quiescent center leads to enhanced DR5:GFP expression first in the columella region, then in the LR cap, and finally also in the epidermis and provascular strands (Blilou et al., 2005). Even if a lot weaker, results from Splivallo et al. (2009) and our data indicate an increase of the signal at the same sites. Auxin measurements will be necessary to confirm that the increased auxin response is actually due to an increased auxin quantity (Petersson et al., 2009) that might be caused by activation of auxin biosynthesis at the root apex.
Results from the microarray-based transcript profile have shown an increase in PtaPIN12 and PtaGH3 transcripts, whose Arabidopsis homologs are thought to be involved in auxin homeostasis (Staswick et al., 2005; Mravec et al., 2009). AtPIN5 (homolog of PtaPIN12) has been suggested to import auxin into the endoplasmic reticulum and thereby to decrease free auxin levels in the cell. Its overexpression was accompanied by an increase in IAA conjugates with amino acids, suggesting a cross talk with IAA-amido-synthetases of the GH3 family (Mravec et al., 2009). We found strong induction of PtaGH3-1 and PtaPIN12 during the combined NPA/fungus treatment, which coincided with a strong decrease of the DR5:GUS signal at the Arabidopsis root apex. Together, these results suggest that PtaGH3-1 and PtaPIN12 actively decrease the auxin maximum in the root apex. We hypothesize that both genes are involved in tuning auxin homeostasis during fungus-induced auxin accumulation in the root. Their impact may also explain why only a weak difference in auxin accumulation is observed at the root apex, especially as certain AtGH3 are expressed in these tissues (Brady et al., 2007). It will be necessary to investigate in more detail the spatial and temporal aspects of auxin accumulation in the root during interaction with the fungus and to connect the results to protein localization of auxin homeostasis regulators.
Polar Auxin Transport Regulates LR Induction during the Root-L. bicolor Interaction
Expression analysis of polar auxin transporters revealed that the expression of PtaPIN9, which was induced during the early root-fungus interaction, was negatively impacted by the presence of the polar auxin transport inhibitor NPA. Furthermore, the reduction in PtaPIN9 transcripts paralleled the absence of LR stimulation. These results suggest a role of PtaPIN9 during fungus-induced LR induction. PtaPIN9 is the homolog of Arabidopsis AtPIN2, a protein involved in basipetal auxin transport from the root apex upwards to the elongation zone and in creating an auxin reflux loop at the root apex (Muller et al., 1998; Blilou et al., 2005). Interestingly, L. bicolor-induced LR stimulation was dramatically decreased in pin2 but not in pin3 transgenic Arabidopsis plants. Furthermore, the absence of PIN3, PIN4, and PIN7 in the quadruple mutant pin2,3,4,7 did not further decrease LR stimulation compared with single pin2 mutants. This suggests that specifically AtPIN2-dependent polar auxin transport is required for fungus-induced LR stimulation. AtPIN2 transcription has been shown to be positively influenced by auxin and by PLT genes (Sieberer et al., 2000; Benkova et al., 2003; Blilou et al., 2005; Vieten et al., 2005). If we assume functional homology between AtPIN2 and PtaPIN9, the increasing auxin accumulation at the root apex, together with the PLT increase, may impact PtaPIN9 transcription. Whether this leads to an increase in PtaPIN9 protein and also to a change in localization still needs to be confirmed, as the exact localization is crucial for directing the auxin flow (Laskowski et al., 2008). Nonetheless, we hypothesize that a fungus-induced increased amount of PtaPIN9 protein, localized to apical and lateral cell membranes in the epidermis and the cortex, leads to higher accumulation of auxin at the pericycle within the elongation zone/differentiation zone in the root, a mechanism thought to prime a larger region for LR induction (Laskowski et al., 2008). When Blilou and coworkers (2005) expressed a bacterial auxin biosynthesis gene at the quiescent center of Arabidopsis roots either in a pin2 mutant background or in NPA-treated wild-type plants, the induction of the DR5:GFP signal was, in contrast to a wild-type background without NPA, absent or only weak in provascular strands. Accordingly, in our study during Arabidopsis-fungus contact, the inhibition of auxin polar transport in general through NPA or specifically in pin2 led to the absence of the provascular auxin signal and to the absence of LR stimulation. Nevertheless, in pin2, a strong and diffuse auxin accumulation was observed at the root apex during contact (Fig. 3, H and I) that was absent in control plants, implying a function of AtPIN2 in efficiently redistributing excess auxin that accumulates during interaction with the fungus. Furthermore, it has also been reported that in the Arabidopsis wav6-52 mutant, which has an increased amount of PIN2 protein, a strong DR5:GFP signal extends into the root elongation zone (Abas et al., 2006). Together, our data compared with the literature strengthen the hypothesis that provascular auxin accumulation and LR induction may result from an apical/lateral auxin transport related to AtPIN2 (PtaPIN9). Immunolocalization of PtaPIN9 in poplar and quantification of PtaPIN9 induction in these specific tissues will allow confirming and deepening the impact of PtaPIN9 during fungus-root cross talk.
Auxin Signaling Regulates LR Induction during the Root-L. bicolor Interaction
Our transcript profiling analyses revealed a differential expression of early auxin-responsive genes involved in auxin signaling (PtaIAA). Quantitative real-time PCR analysis of these differentially expressed targets at various time points during the poplar-L. bicolor interaction identified genes, such as PtaIAA19.3 and PtaIAA28.1, whose transcript levels were modified early but only transiently. When defective or absent, homologs of the Arabidopsis auxin transcription regulators PtaIAA19.3 and PtaIAA28.1 are known to cause a strong LR phenotype (Tian and Reed, 1999; Rogg et al., 2001; Tatematsu et al., 2004; Muto et al., 2007). Strikingly, when NPA was applied during the poplar-L. bicolor interaction, PtaIAA19.3 transcript accumulation was repressed in the presence of the fungus and no LR stimulation was observed. This suggests a connection between auxin signaling via PtaIAA19.3 and fungus-induced LR development. Experiments with Arabidopsis slr1 (a stabilized form of AtIAA14 conferring a gain-of-function mutation), which is blocked in auxin signaling (Fukaki et al., 2002), confirmed that the classical auxin signaling pathway is required for fungus-induced LR development in plants. In poplar, PtaIAA19.3 may be a key actor in the basic auxin signaling pathway in roots during LRI.
CONCLUSION
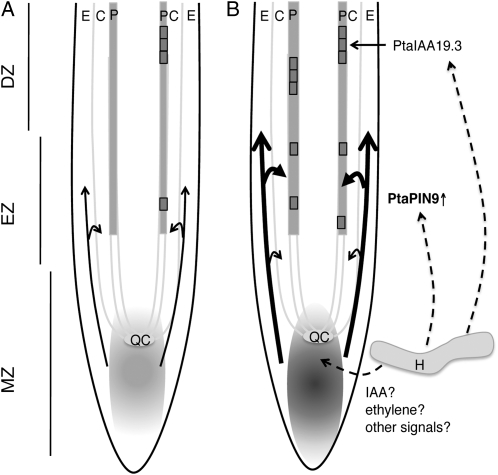
Our data suggested a model (Fig. 7) in which undefined fungal signals cause an auxin accumulation at the root apex and increase the apical/lateral polar auxin transport through PtaPIN9. Excess auxin that accumulates during contact is specifically transported to provascular pericycle cells, which are primed to LR founder cells. We assume that a stronger auxin accumulation in these tissues primes a higher number of cells. Priming of founder cells for LRI has been suggested to be independent of AUX/IAA signaling. However, LRI in the differentiation zone involved auxin accumulation and auxin signaling. Therefore, we propose that auxin signaling through PtaIAA19.3 in the differentiation zone is part of LRI downstream of the acquisition of founder cell specification in the basal meristem (meristem/elongation zone; Fig. 7).
Figure 7.
Hypothetical model of the molecular mechanism underlying fungus-induced LR development in poplar. A, PtaPIN9 directed basipetal polar auxin transport (arrows) before contact with the fungus. The auxin maximum around the quiescent center (QC) is indicated by gray shading in the meristematic zone (MZ). Pericycle founder cells (single gray box) are primed for LRI in the elongation zone (EZ), and LRs are initiated through subsequent cell divisions (multiple gray boxes) in the differentiation zone (DZ). B, The presence of the fungus stimulates auxin accumulation at the root apex (gray shading) through an unknown mechanism. The increased auxin level and/or other fungal signals stimulate PtaPIN9 expression. PtaPIN9 protein enhances basipetal auxin transport (thick arrows), which then primes more pericycle cells for LRI. LRI occurs through a PtaIAA19.3-dependent signaling mechanism in the differentiation zone. Whether fungal signals act directly on PtaIAA19.3 expression needs to be analyzed. C, Cortex; E, epidermis; H, hyphae; P, pericycle.
MATERIALS AND METHODS
Plant and Fungal Material and Growth Conditions
Experiments were performed with the hybrid poplar (Populus tremula × Populus alba; INRA clone 717-1-B4) and the transgenic hybrid Populus tremula × Populus tremuloides DR5:GFP. Plants were micropropagated in vitro and grown on half-strength Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) in glass culture tubes under a 16-h photoperiod at 24°C in a growth chamber.
The dikaryotic vegetative mycelium of the ectomycorrhizal fungus Laccaria bicolor strain S238N (Maire P.D. Orton) was maintained at 25°C on modified Pachlewski medium P5 (Deveau et al., 2007).
Seeds of Arabidopsis (Arabidopsis thaliana ectoype Columbia), aux1-t (also known as aux1-100; Bennett et al., 1996), pin2/eir1-1 (Roman et al., 1995), pin2 DR5:GUS (Sabatini et al., 1999), pin3 (salk_005544) and pin2,3,4,7 (http://signal.salk.edu/cgi-bin/tdnaexpress/; Blilou et al., 2005), slr1 (Fukaki et al., 2002), and tir1afb1,2,3 (Gray et al., 1999; Dharmasiri et al., 2005) were surface sterilized and sown on solid Arabidopsis medium (2.3 g L−1 MS salt, 1% Suc, 1.3% agar-agar [pH 6.0 adjusted with KOH], and 1 g L−1 MES sodium salt). After stratification for 1 to 2 d at 4°C, seeds were germinated under a long-day photoperiod (16 h of light, 8 h of darkness) at 21°C.
Coculturing in a Sandwich Culture System
For mycorrhiza formation on in vitro poplar by L. bicolor, we modified an existing sandwich coculture system initially developed for the Eucalyptus-Pisolithus interaction (Chilvers et al., 1986; Burgess et al., 1996; Fig. 1). Free-living mycelium of L. bicolor S238N was grown for 10 d on cellophane-covered agar (12 g L−1) plates containing sugar-reduced Pachlewski medium P20 (Deveau et al., 2007). In parallel, plant material was prepared. In order to synchronize rhizogenesis, stem cuttings from in vitro poplar plants were precultured on half-strength MS medium containing 2 mg L−1 indole butyric acid for 7 d. Rooted cuttings were then transferred to vertically arranged 12- × 12-cm square petri dishes half covered with a cellophane membrane and cultured for 3 weeks under a 16 h d−1 light photoperiod at 24°C.
For cocultures, plants were transferred to 12- × 12-cm petri dishes containing solidified (12 g L−1 agar) low-carbon Pachlewski medium (P20), pH 5.8, buffered with 1 g L−1 MES sodium salt covered by a 6- × 12-cm cellophane membrane. A mycelium-covered cellophane membrane was placed fungus side down (direct interaction) or fungus side up (indirect interaction) on the roots. Petri dishes were closed with Parafilm on the upper and lower sides and with Band-Aids (ensuring high gas permeability) on both lateral sides. Cultures were arranged vertically, and the lower part of the dish was covered with a small black plastic bag to prevent light from reaching the fungus and roots. The sandwich cultures were kept in the same conditions as the poplar plant cultures. When Arabidopsis was put into contact with L. bicolor, mycelium-covered cellophane membranes were directly laid on the plant roots grown on MS medium (with 1 g L−1 MES sodium salt). The medium used for NPA treatment and mutant-L. bicolor contact contained 1 g L−1 MES sodium salt. Control poplar and Arabidopsis plants were covered with a cellophane membrane without fungal mycelium.
Observation of Root Development
For LR quantification, 10 to 15 individual poplar plants (three per petri dish) or 10 to 15 Arabidopsis seedlings in the respective conditions were observed every 2 to 4 d using a Discovery V.8 stereomicroscope (Zeiss). LRs were counted and images were taken. To determine the extent of root colonization by L. bicolor, we observed root morphology every 10 d. Samples were taken for sectioning at each time point.
Microscopic Observation
In order to ascertain the formation of the intraradicular Hartig net, we subjected root sections to propidium iodide/UVitex double staining for plant and fungal structures, respectively. One-centimeter root tips were fixed in 4% (w/v) para-formaldehyde in phosphate-buffered saline (PBS; pH 7) overnight at 4°C. Roots were washed in PBS and embedded in 6% (w/v) agarose. Thirty-micrometer transverse sections were prepared using a Vibratome (Leica). Sections were stained in 1% (w/v) UVitex 2B (Polyscience) in PBS for 2 min, washed, and then counterstained with propidium iodide (1:100 dilution; Sigma-Aldrich; Moldenhauer et al., 2006; Xu et al., 2006). For DR5:GFP observation, fresh 2-cm root tips from P. tremula × P. tremuloides DR5:GFP were mounted in Slowfade Gold Antifade Reagent (Molecular Probes) and observed immediately. All samples were observed with a Radiance 2100 Rainbow confocal scanning laser microscope (Nikon-Bio-Rad) equipped with an Apochromat X60 (numerical aperture 1.4) oil objective. A wavelength of 405 nm was used for UVitex 2B excitation, and emission was detected between 500 and 560 nm. Propidium iodide was excited at 514 nm with an argon laser line, and the emitted fluorescence was detected above 550 nm. GFP was excited using the 488-nm argon laser line in conjunction with a 505- to 530-nm band-pass filter. Settings (laser intensity, gain, offset, magnification) for DR5:GFP observations were maintained equally between all samples. Histological detection of GUS was performed according to Scarpella et al. (2004). DR5:GUS plants were stained for 90 min, and pin2 DR5:GUS plants were stained for 120 min at 37°C. Samples were mounted in 50% (v/v) glycerol and observed using a Zeiss Axiovert 200M MOT device (Carl Zeiss Microimaging).
RNA Extraction and cDNA Synthesis
At each time point of quantitative real-time PCR analysis, three pools of six independent root systems for each control root and fungal treatment were harvested and frozen in liquid nitrogen. For microarray analysis, three additional pools of six root systems for each control and indirect interaction were harvested from a second experimental series. Total RNA was extracted from these samples using the RNAeasy kit (Qiagen) as per the manufacturer's instructions. An in-column digestion step with DNAse I (Qiagen) was part of the extraction. RNA quality was verified by Experion Standardsens Capillar gels (Bio-Rad). cDNA for NimbleGen microarrays was synthesized using the Smart cDNA Synthesis kit (Clontech) containing an amplification step on the cDNA level (Duplessis et al., 2005). RNA for real-time PCR was additionally subjected to a second DNA digestion step using DNAfree (Ambion) before synthesizing cDNA from 250 ng of total RNA using an iScript kit (Bio-Rad).
NimbleGen Microarray Transcript Profiling
The Populus whole genome expression array version 2.0 (S. DiFazio, A. Brunner, P. Dharmawardhana, and K. Munn, unpublished data) manufactured by NimbleGen Systems contains in duplicate three independent, nonidentical, 60-mer probes per whole gene model plus control probes and labeling controls. Included in the microarray are 65,965 probe sets corresponding to 55,970 gene models predicted on the P. trichocarpa genome sequence version 1.0 and 9,995 aspen cDNA sequences (P. tremula, P. tremuloides, and P. tremula × P. tremuloides). The Populus version 2.0 oligoarray is fully described in the platform Gene Expression Omnibus at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo). NimbleGen whole genome microarray analyses were performed in three biological replicates (independent from quantitative real-time PCR samples) with a technical replicate on each array as per the manufacturer's instructions. Expression data were processed in the following way. To ensure a high specificity, all independent 60-mer oligonucleotides for the 55,970 genes were blasted against the Populus genome version 1.1 available at http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html, and only probes with less than 10% homology to other gene models than the gene model they were designed for were retained for further analysis. Due to this stringent filtering, about 30% of the genes (16,667) were excluded, since all three independent oligonucleotides failed the given specificity. Fluorescence data were normalized between all different arrays using ARRAYSTAR software (DNASTAR). Average expression levels from all three biological repetitions were calculated for each gene from the specific independent probes and were used for further analyses. A Student's t test with false discovery rate (Benjamini-Hochberg) multiple testing correction was applied on the data using ARRAYSTAR software (DNASTAR). Transcripts with a significant P value (<0.05) and a more than 2.0 change in transcript level were considered as significantly differentially expressed in roots in contact with L. bicolor compared with control roots. The signal-to-noise threshold (background signal) was estimated as described by Martin et al. (2008). The complete expression data set is available (accession no. GSE16662) at the Gene Expression Omnibus at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/). Expression data for all genes that were represented by at least one specific 60-mer oligonucleotide on the array are also available in Supplemental Table S1.
Quantitative Real-Time PCR
Specific primer sequences can be found in Supplemental Table S3. We used ubiquitin (Kohler et al., 2004; P. trichocarpa gene model estExt_Genewise1_v1.C_LG_XV0407; GenBank accession no. CA825222) and a putative protein (Gutierrez et al., 2008; P. trichocarpa gene model estExt_fgenesh4_pm.C_LG_IX0344) as reference genes. Real-time PCR was performed using a Chromo4 Light Cycler and OpticonMonitor Software. Real-time PCR analyses were performed in three biological replicates (independent from microarray samples) with a technical replicate for each reaction. PCR was realized with a technical replicate for each cDNA from three biological repetitions using SYBRGreen Supermix following the manufacturer's instructions (Bio-Rad). Fold changes in gene expression between treated and control roots were based on ΔΔCt calculations according to Pfaffl (2001). The means of each of the three ΔΔCt values were presented as histograms. ΔΔCt values calculated with either of the reference genes were similar. Only ubiquitin-normalized data are presented. For each mean, ΔΔCt significance was calculated using Student's t test.
Phylogenetic Trees
Phylogenetic trees based on entire protein sequence alignments were constructed using MEGAlign (ClustalX alignment, pairwise deletion). A neighbor-joining algorithm (pairwise deletion, p-distance) with 5,000 bootstrap repetitions was chosen to establish the phylogenetic trees.
Image Analysis
For DR5:GFP fluorescence quantification, ImageJ software with the Stack-Measure Macro was used. Background intensity was equivalent for all images and was subtracted for further quantification. Image assembly of all figure panels was realized in Adobe Photoshop.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number GSE16662.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effect of NPA on auxin accumulation in AtDR5:GUS transgenic plant apices during contact with L. bicolor.
Supplemental Figure S2. Neighbor-joining cladogram of the eight members of the Populus auxin influx carrier protein family AUX/LAX and their four and 10 homologous protein sequences in Arabidopsis and rice (Oryza sativa), respectively.
Supplemental Figure S3. Neighbor-joining cladogram of the 16 members of the Populus auxin efflux carrier protein family PIN and their eight and 13 homologous protein sequences in Arabidopsis and rice, respectively.
Supplemental Figure S4. Neighbor-joining cladogram of the 12 members of the Populus IAA-amido-synthetase protein family GH3 and their 19 and 12 homologous protein sequences in Arabidopsis and rice, respectively.
Supplemental Table S1. Gene expression data of poplar roots after 3 DOII with L. bicolor.
Supplemental Table S2. List of Arabidopsis LRI genes (Vanneste et al., 2005) and expression data from their poplar homologs in control roots and roots after 3 DOII with L. bicolor.
Supplemental Table S3. Specific primers for auxin-related target genes in poplar used in quantitative real-time PCR.
Supplementary Material
Acknowledgments
We thank Antoine Larrieu, Malcolm Bennett, and Mark Estelle for sharing published material. We gratefully acknowledge the excellent technical support from Jean-Pierre Leclerc and Joëlle Gerard in Nancy and from Katja Rapp in Freiburg. We are also grateful to Benoit Hilselberger for microarray data processing and to Pierre Montpied for giving instructions for statistical analysis. We thank Stéphane Hacquard for providing Arabidopsis sequences for PIN protein alignments, Jonathan Plett for helpful comments on an early draft of the manuscript, and Krista Plett for proofreading the final version.
This work was supported by the European Commission within the Seventh Framework Program for Research, Project Energypoplar (grant no. FP7–211917), the French Space Agency, and the EVOLTREE network of excellence, by mobility grants from the German-French University and Egide/Procope, the German-French Research Cooperation, which enabled the cooperation between the two research groups, and by the French Ministry for Higher Education and Research (PhD scholarship to J.F.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Valérie Legué (valerie.legue@scbiol.uhp-nancy.fr).
The online version of this article contains Web-only data.
References
- Abas L, Benjamins R, Malenica N, Paciorek T, Wisniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C (2006) Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat Cell Biol 8 249–256 [DOI] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119 109–120 [DOI] [PubMed] [Google Scholar]
- Badescu GO, Napier RM (2006) Receptors for auxin: Will it all end in TIRs? Trends Plant Sci 11 217–223 [DOI] [PubMed] [Google Scholar]
- Bainbridge K, Guyomarc'h S, Bayer E, Swarup R, Bennett M, Mandel T, Kuhlemeier C (2008) Auxin influx carriers stabilize phyllotactic patterning. Genes Dev 22 810–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273 948–950 [DOI] [PubMed] [Google Scholar]
- Blasius D, Feil W, Kottke I, Oberwinkler F (1986) Hartig net structure and formation in fully ensheathed ectomycorrhizas. Nord J Bot 6 837–842 [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433 39–44 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318 801–806 [DOI] [PubMed] [Google Scholar]
- Burgess T, Dell B, Malajczuk N (1996) In vitro synthesis of Pisolithus-Eucalyptus ectomycorrhizae: synchronization of lateral tip emergence and ectomycorrhizal development. Mycorrhiza 6 189–196 [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ (2003) Dissecting Arabidopsis lateral root development. Trends Plant Sci 8 165–171 [DOI] [PubMed] [Google Scholar]
- Charvet-Candela V, Hitchin S, Reddy MS, Cournoyer B, Marmeisse R, Gay G (2002) Characterization of a Pinus pinaster cDNA encoding an auxin up-regulated putative peroxidase in roots. Tree Physiol 22 231–238 [DOI] [PubMed] [Google Scholar]
- Chilvers GA, Douglass PA, Lapeyrie F (1986) A paper-sandwich technique for rapid synthesis of ectomycorrhizas. New Phytol 103 397–402 [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frey NF, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al (2007) Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134 681–690 [DOI] [PubMed] [Google Scholar]
- De Smet I, Vanneste S, Inze D, Beeckman T (2006) Lateral root initiation or the birth of a new meristem. Plant Mol Biol 60 871–887 [DOI] [PubMed] [Google Scholar]
- Deveau A, Palin B, Delaruelle C, Peter M, Kohler A, Pierrat JC, Sarniguet A, Garbaye J, Martin F, Frey-Klett P (2007) The mycorrhiza helper Pseudomonas fluorescens BBc6R8 has a specific priming effect on the growth, morphology and gene expression of the ectomycorrhizal fungus Laccaria bicolor S238N. New Phytol 175 743–755 [DOI] [PubMed] [Google Scholar]
- Dexheimer J, Pargney JC (1991) Comparative anatomy of the host-fungus interface in mycorrhizas. Experientia 47 312–320 [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9 109–119 [DOI] [PubMed] [Google Scholar]
- Ditengou FA, Beguiristain T, Lapeyrie F (2000) Root hair elongation is inhibited by hypaphorine, the indole alkaloid from the ectomycorrhizal fungus Pisolithus tinctorius, and restored by indole-3-acetic acid. Planta 211 722–728 [DOI] [PubMed] [Google Scholar]
- Ditengou FA, Lapeyrie F (2000) Hypaphorine from the ectomycorrhizal fungus Pisolithus tinctorius counteracts activities of indole-3-acetic acid and ethylene but not synthetic auxins in eucalypt seedlings. Mol Plant Microbe Interact 13 151–158 [DOI] [PubMed] [Google Scholar]
- Ditengou FA, Teale WD, Kochersperger P, Flittner KA, Kneuper I, van der Graaff E, Nziengui H, Pinosa F, Li X, Nitschke R, et al (2008) Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc Natl Acad Sci USA 105 18818–18823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benkova E (2008) Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA 105 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplessis S, Courty PE, Tagu D, Martin F (2005) Transcript patterns associated with ectomycorrhiza development in Eucalyptus globulus and Pisolithus microcarpus. New Phytol 165 599–611 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M (2005) Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J 44 382–395 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29 153–168 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tasaka M (2009) Hormone interactions during lateral root formation. Plant Mol Biol 69 437–449 [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449 1053–1057 [DOI] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Mauriat M, Guenin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, et al (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6 609–618 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle TJ (1985) Rapid induction of selective transcription by auxins. Mol Cell Biol 5 1197–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan DP, Chilvers GA, Lapeyrie FF (1988) Time sequence of the infection process in eucalypt ectomycorrhizas. New Phytol 109 451–458 [Google Scholar]
- Jambois A, Dauphin A, Kawano T, Ditengou FA, Bouteau F, Legue V, Lapeyrie F (2005) Competitive antagonism between IAA and indole alkaloid hypaphorine must contribute to regulate ontogenesis. Physiol Plant 123 120–129 [Google Scholar]
- Kalluri UC, Difazio SP, Brunner AM, Tuskan GA (2007) Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol 7 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabaghli C, Frey-Klett P, Sotta B, Bonnet M, Le Tacon F (1998) In vitro effects of Laccaria bicolor S238N and Pseudomonas fluorescens strain BBc6 on rooting of de-rooted shoot hypocotyls of Norway spruce. Tree Physiol 18 103–111 [DOI] [PubMed] [Google Scholar]
- Khan S, Stone JM (2007) Arabidopsis thaliana GH3.9 influences primary root growth. Planta 226 21–34 [DOI] [PubMed] [Google Scholar]
- Kohler A, Blaudez D, Chalot M, Martin F (2004) Cloning and expression of multiple metallothioneins from hybrid poplar. New Phytol 164 83–93 [DOI] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Maree AF, Scheres B (2008) Root system architecture from coupling cell shape to auxin transport. PLoS Biol 6 e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Kowalczyk M, Marchant A, Celenza J, Cohen JD, Sandberg G (2002) Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol Biol 49 249–272 [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124 33–44 [DOI] [PubMed] [Google Scholar]
- Martin F, Aerts A, Ahren D, Brun A, Danchin EG, Duchaussoy F, Gibon J, Kohler A, Lindquist E, Pereda V, et al (2008) The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 452 88–92 [DOI] [PubMed] [Google Scholar]
- Martin F, Duplessis S, Ditengou F, Lagrange H, Voiblet C, Lapeyrie F (2001) Developmental cross talking in the ectomycorrhizal symbiosis: signals and communication genes. New Phytol 151 145–154 [DOI] [PubMed] [Google Scholar]
- Martin F, Nehls U (2009) Harnessing ectomycorrhizal genomics for ecological insights. Curr Opin Plant Biol 12 508–515 [DOI] [PubMed] [Google Scholar]
- Moldenhauer J, Moerschbacher BM, van der Westhuizen AJ (2006) Histological investigation of stripe rust (Puccinia striiformis f.sp. tritici) development in resistant and susceptible wheat cultivars. Plant Pathol 55 469–474 [Google Scholar]
- Moyle R, Schrader J, Stenberg A, Olsson O, Saxena S, Sandberg G, Bhalerao RP (2002) Environmental and auxin regulation of wood formation involves members of the Aux/IAA gene family in hybrid aspen. Plant J 31 675–685 [DOI] [PubMed] [Google Scholar]
- Mravec J, Skupa P, Bailly A, Hoyerova K, Krecek P, Bielach A, Petrasek J, Zhang J, Gaykova V, Stierhof YD, et al (2009) Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459 1136–1140 [DOI] [PubMed] [Google Scholar]
- Muller A, Guan C, Galweiler L, Tanzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K (1998) AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 17 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige M, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Muto H, Watahiki MK, Nakamoto D, Kinjo M, Yamamoto KT (2007) Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of Arabidopsis revealed by promoter-exchange experiments among MSG2/IAA19, AXR2/IAA7, and SLR/IAA14. Plant Physiol 144 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M (2001) DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J 25 213–221 [DOI] [PubMed] [Google Scholar]
- Nibau C, Gibbs DJ, Coates JC (2008) Branching out in new directions: the control of root architecture by lateral root formation. New Phytol 179 595–614 [DOI] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K (2009) An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SM, Hitchin S, Melayah D, Pandey AK, Raffier C, Henderson J, Marmeisse R, Gay G (2006) The auxin-inducible GH3 homologue Pp-GH3.16 is downregulated in Pinus pinaster root systems on ectomycorrhizal symbiosis establishment. New Phytol 170 391–400 [DOI] [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B (2001) A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman G, Lubarsky B, Kieber JJ, Rothenberg M, Ecker JR (1995) Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics 139 1393–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99 463–472 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E, Francis P, Berleth T (2004) Stage-specific markers define early steps of procambium development in Arabidopsis leaves and correlate termination of vein formation with mesophyll differentiation. Development 131 3445–3455 [DOI] [PubMed] [Google Scholar]
- Schrader J, Baba K, May ST, Palme K, Bennett M, Bhalerao RP, Sandberg G (2003) Polar auxin transport in the wood-forming tissues of hybrid aspen is under simultaneous control of developmental and environmental signals. Proc Natl Acad Sci USA 100 10096–10101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer T, Seifert GJ, Hauser MT, Grisafi P, Fink GR, Luschnig C (2000) Post-transcriptional control of the Arabidopsis auxin efflux carrier EIR1 requires AXR1. Curr Biol 10 1595–1598 [DOI] [PubMed] [Google Scholar]
- Splivallo R, Fischer U, Gobel C, Feussner I, Karlovsky P (2009) Truffles regulate plant root morphogenesis via the production of auxin and ethylene. Plant Physiol 150 2018–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jurgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133 177–191 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, Alonso JM (2007) Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19 2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann T, Bolu-Arianto WH, Olbrich A, Langenfeld-Heyser R, Gobel C, Grzeganek P, Feussner I, Hansch R, Polle A (2008) GH3:GUS reflects cell-specific developmental patterns and stress-induced changes in wood anatomy in the poplar stem. Tree Physiol 28 1305–1315 [DOI] [PubMed] [Google Scholar]
- Tian Q, Reed JW (1999) Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126 711–721 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ (1995) Composite structure of auxin response elements. Plant Cell 7 1611–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, De Rybel B, Beemster GT, Ljung K, De Smet I, Van Isterdael G, Naudts M, Iida R, Gruissem W, Tasaka M, et al (2005) Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17 3035–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benkova E, Benjamins R, Beeckman T, Luschnig C, Friml J (2005) Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132 4521–4531 [DOI] [PubMed] [Google Scholar]
- Xu B, Seong-Tae K, Dae-Sik L, Kastan MB (2006) Two molecularly distinct G2/M checkpoints are induced by ionizing irradiation. Mol Cell Biol 22 1049–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.