Abstract
As a genetic platform, tomato (Solanum lycopersicum) benefits from rich germplasm collections and ease of cultivation and transformation that enable the analysis of biological processes impossible to investigate in other model species. To facilitate the assembly of an open genetic toolbox designed to study Solanaceae, we initiated a joint collection of publicly available gene manipulation tools. We focused on the characterization of promoters expressed at defined time windows during fruit development, for the regulated expression or silencing of genes of interest. Five promoter sequences were captured as entry clones compatible with the versatile MultiSite Gateway format: PPC2, PG, TPRP, and IMA from tomato and CRC from Arabidopsis (Arabidopsis thaliana). Corresponding transcriptional fusions were made with the GUS gene, a nuclear-localized GUS-GFP reporter, and the chimeric LhG4 transcription factor. The activity of the promoters during fruit development and in fruit tissues was confirmed in transgenic tomato lines. Novel Gateway destination vectors were generated for the transcription of artificial microRNA (amiRNA) precursors and hairpin RNAs under the control of these promoters, with schemes only involving Gateway BP and LR Clonase reactions. Efficient silencing of the endogenous phytoene desaturase gene was demonstrated in transgenic tomato lines producing a matching amiRNA under the cauliflower mosaic virus 35S or PPC2 promoter. Lastly, taking advantage of the pOP/LhG4 two-component system, we found that well-characterized flower-specific Arabidopsis promoters drive the expression of reporters in patterns generally compatible with heterologous expression. Tomato lines and plasmids will be distributed through a new Nottingham Arabidopsis Stock Centre service unit dedicated to Solanaceae resources.
Solanaceae provide the world's most important vegetable crops, including tomato (Solanum lycopersicum), potato (Solanum tuberosum), and pepper (Capsicum annuum), and are a model eukaryote family for evolutionary, genetic, and genomic studies. More than 3,000 Solanaceous species grow in habitats ranging from rain forests to deserts and mountains with regular snowfall. Thanks to conserved genome organization, this family is an excellent subject to explore the genetic and molecular basis of adaptation to diverse environments. Within the Solanaceae, tomato is a broadly used model system for studying plant-microbe interactions and the biology of fleshy fruits, owing to its simple diploid genetics, the short generation time, the routine protocols for production of transgenic plants, and its exceptional publicly available genetic and genomic resources. These resources include large collections of single-gene mutations, an extensive EST database, a high-density genetic map, microarrays, an emerging genome sequence, and well-characterized populations designed for genetic mapping of simple and quantitative trait loci, and they are used by an active Solanaceae research community involving research laboratories in more than 30 countries focusing on both basic and strategic research (Knapp, 2002; Mueller et al., 2005; Wu et al., 2006).
The first draft sequence of the entire euchromatic portion of the tomato genome will soon be available (Mueller et al., 2009). As already witnessed for other research communities dedicated to key species, the availability of a well-annotated genome will mark the onset of new lines of investigations and will greatly facilitate the characterization of genetic functions. Taking advantage of the EU-SOL European integrated project (http://www.eu-sol.net/), we initiated a collaborative effort for the construction of publicly available tools enabling gene manipulation at specific stages of tomato fruit development and in well-defined tissues and cell types. Tomato is a berry with a thick pericarp that encloses many seeds. The fruit results from the fusion of a number of carpels, the ovary wall becoming the fleshy pericarp. Pollination triggers cell proliferation, followed by extensive cell expansion, resulting in a mature fruit that ranges from 1 to 1,000 g (Gillaspy et al., 1993; Lemaire-Chamley et al., 2005; Gonzalez et al., 2007). Fruit development and ripening are also characterized by dramatic metabolic shifts, including the conversion of starch to sugars, chloroplasts to chromoplasts, and extensive cell wall disassembly, all under tight genetic control (Giovannoni, 2004). Changes in the spatial or temporal expression of regulatory genes or downstream effectors controlling these processes will substantially affect fruit tissue properties and the ripening process (Manning et al., 2006; Giovannoni, 2007). Indeed, the ability to alter subtly the timing and location of expression of key regulators is an important goal for crop biotechnology but is often constrained by lack of suitable promoters.
To this end, a core set of versatile resources designed for temporal and tissue-specific manipulation of gene expression in the tomato fruit has been created by independent laboratories. All clones and vectors were constructed with the flexible Gateway recombinational cloning technology (for review, see Karimi et al., 2007a). Promoters that displayed tissue-specific transcriptional activity have been captured as Gateway entry clones. These regulatory sequences can be combined at will with any gene, hairpin RNA (hpRNA), expression cassette, or artificial microRNA (amiRNA) via MultiSite Gateway protocols for the creation of novel binary T-DNA vectors. Expression of reporter genes in specific cell types illustrated the functionality of such vectors. Silencing of the endogenous gene coding for phytoene desaturase with amiRNA in stable transgenic tomato plants demonstrated that such vectors can be used to efficiently control transcript abundance in fruit tissues. In addition, tomato driver lines were generated for the transcriptional activation of any responder transgene locus, based on the LhG4 two-component system. Expression of several reporters and phenotypes induced within these lines facilitated their communal use for genetic manipulations. The assembly of a Solanaceae genetic toolbox is an important asset for future functional analysis in tomato and related crops.
RESULTS AND DISCUSSION
Cloning Strategy and Structure of the Vector Collection
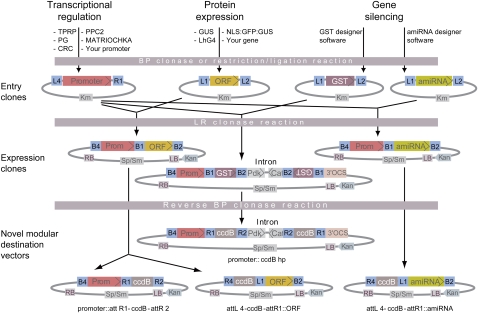
The Gateway MultiSite recombinational cloning framework is ideally suited for constructing versatile collections of genetic elements to be assembled in predetermined order, orientation, and open reading frame (ORF) registry (Karimi et al., 2005, 2007a, 2007b). The overall strategy underlying our vector resources is illustrated in Figure 1. Three types of elements have been considered so far: plant promoters, to direct the transcription of any gene of interest in well-defined tomato tissues; ORFs, to produce the encoded proteins; and gene-specific sequence tags (GSTs) or amiRNA precursors, to silence targeted genes.
Figure 1.
Assemblage of different genetic elements captured into entry clones via LR reaction to facilitate genetic studies in tomato. B1, B2, B4, L1, L2, L4, and R1: corresponding att Gateway recombination sites. Sp, spectinomycin; Sm, streptomycin; Km and Kan, kanamycin bacterial and plant selectable markers, respectively; LB, left T-DNA border; RB, right T-DNA border; Prom, promoter; pdk-cat intron, pHELLSGATE12-derived intron spacer; ccdB, counter-selectable bacterial marker; and 3′OCS, octopine synthase terminator.
Each element was first captured in an entry clone, either via restriction/ligation cloning in a plasmid carrying multiple restriction sites flanked by attL and attR recombination sites or via PCR amplification and addition of flanking attB sites, followed by a BP Clonase reaction. We chose to clone all promoters in attL4-promoter-attR1 cassettes and all other elements in attL1-sequence-attL2 cassettes. The resulting entry clones can be recombined with various Gateway destination vectors in LR Clonase reactions to create expression vectors. Interestingly, two or more elements available as entry clones can be assembled via a single MultiSite LR Clonase reaction in the order and orientation defined by the position of the attL and attR sites flanking the recombined sequences (Cheo et al., 2004; Karimi et al., 2007b). In our configuration, any promoter can be positioned easily upstream of an ORF, a GST, or an amiRNA precursor and drive its tissue-specific transcription in transgenic plants. Lastly, novel modular destination vectors can be constructed from any expression clone via reverse BP Clonase reactions, further expanding the possible cloning schemes (Karimi et al., 2007b).
Fruit-Specific Promoters
Flowers and fruits are the main targets for genetic improvement of fruit crops. However, these organs are the last to form, and their manipulation with broadly expressed promoters is therefore restricted to nonpleiotropic genetic perturbations. Promoters active solely in these organs bypass this limitation and permit the functional analysis of all genes involved in fruit development. Such sequences facilitate the engineering of fruit quality, either directly or by providing proof-of-concept information from which strategies can be developed to harness natural variation.
As an initial set of regulatory sequences of interest, we chose four promoters from tomato genes shown previously to be transcribed specifically at different stages of fruit growth and maturation. In addition, we included a well-characterized Arabidopsis (Arabidopsis thaliana) promoter of which the activity is restricted to specific carpel and nectary tissues during flower development (Table I). The five promoter sequences are documented in Supplemental Table S1. The anticipated temporal and spatial transcriptional activity in the fruit for each cloned promoter is summarized below. The size of each captured sequence is also indicated in parentheses. They all end in a window of 1 to 34 nucleotides upstream of their cognate ORF.
Table I.
Genetic elements in entry clones
| Class | Element(s) | Recipient pDONR | Entry Clone | att Sites |
|---|---|---|---|---|
| Promoter | PPC2 | pDONR P4-P1R | pEN-L4-PPC2-R1 | attL4–attR1 |
| Promoter | TPRP | pDONR P4-P1R | pEN-L4-TPRP-R1 | attL4–attR1 |
| Promoter | IMA | pDONR P4-P1R | pEN-L4-IMA-R1 | attL4–attR1 |
| Promoter | CRC | pEN-L4-R1 | pEN-L4-CRC-R1 | attL4–attR1 |
| Promoter | PG | pEN-L4-R1 | pEN-L4-PG-R1 | attL4–attR1 |
| Reporter enzyme | SI | pDONR221 | pEN-L1-SI-L2 | attL1–attL2 |
| amiRNA | PDS | pDONR221 | pEN-L1-miRpds-L2 | attL1–attL2 |
| Two component | LhG4AtO | pDONR221 | pEN-L1-LhG4ATO4-L2 | attL1–attL2 |
INHIBITOR OF MERISTEM ACTIVITY (0.5 kb)
INHIBITOR OF MERISTEM ACTIVITY (IMA; accession number AM261628) encodes a 90-amino-acid tomato protein that harbors a putative central zinc finger domain similar to the recently characterized MINI ZINC FINGER proteins (Hu and Ma, 2006; Sicard et al., 2008). While absent from vegetative organs, IMA expression is detected in all floral organs and to a greater extent in carpels and petals. However, the highest expression was observed in the developing fruit. IMA transcripts are detected as early as the preanthesis stage corresponding to isolated carpels, accumulate gradually and strongly in fruit to reach a maximum at 10 d after anthesis (daa), and then decrease to reach basal levels at the mature green stage. IMA acts as an inhibitor of cell proliferation during flower termination (Sicard et al., 2008). Its promoter sequence was identified via the Solanaceae Genome Network (SGN) database in a BAC clone encompassing the IMA gene sequence (accession number AC122544).
CRABS CLAW (3.7 kb)
The Arabidopsis CRABS CLAW (CRC) gene belongs to the YABBY transcription factor family (Bowman and Smyth, 1999). It is required for nectary development in Arabidopsis flowers and for abaxial-adaxial polarity specification of carpels. The CRC promoter is active throughout carpel primordium initiation but becomes restricted to the valve abaxial domain upon anthesis. In addition, it is expressed in central placental domains and in nectaries throughout their development (Lee et al., 2005). This promoter was chosen because it marks the earliest stages of carpel initiation, a pattern not presently available from endogenous tomato promoters.
TPRP (2.6 kb)
TPRP is a cell wall tomato Pro-rich protein (Salts et al., 1991). TPRP expression levels are high in all tissues of young tomato fruits, corresponding with cell division phases II and III of fruit development (Gillaspy et al., 1993). Promoter activity is dramatically reduced in mature green and ripe fruits and completely absent in all other parts of the plant (Salts et al., 1991; Carmi et al., 2003). The TPRP promoter has been used successfully to down-regulate the expression of the DE-ETIOLATED1 gene, resulting in an increase of the carotenoid and flavonoid content restricted to the fruit (Davuluri et al., 2005).
PPC2 (2.0 kb)
The PPC2 gene codes for a phosphoenolpyruvate carboxylase isoform present in developing tomato fruits that is presumably involved in organic acid accumulation and CO2 fixation. LYCes;PPC2 is highly and specifically expressed during the phase of rapid fruit growth, corresponding to cell expansion. Expression increases initially at the end of cell division and decreases at the beginning of ripening. In young growing fruits, LYCes;PPC2 mRNA has been specifically located by in situ hybridization in the pericarp and the gel surrounding the seeds (Guillet et al., 2002). Accordingly, the 2.0-kb promoter fragment has recently been shown to direct fruit-specific GUS expression during the cell expansion phase in the placenta, gel, and later in the pericarp tissues in transgenic tomato plants (C. Guillet and C. Rothan, unpublished data).
Polygalacturonase (4.8 kb)
Polygalacturonase (PG) is a cell wall hydrolase abundantly secreted during fruit ripening and contributing to fruit softening. PG expression is tightly related to the ripening process. Transcriptional activation at the onset of ripening results in a high level of PG mRNA (Biggs and Handa, 1989; DellaPenna et al., 1989). Within the ripening fruit, a 1.4-kb promoter fragment has been shown to direct GUS expression mostly to the outer region of the pericarp and the columella tissue in transgenic tomato plants (Montgomery et al., 1993). However, a longer (4.8 kb) PG promoter was preferred because it yields higher levels of chloramphenicol acetyl transferase reporter activity in ripening transgenic tomato fruits (Nicholass et al., 1995) and has been successfully used to engineer metabolite fruit content (Davidovich-Rikanati et al., 2007; Kovacs et al., 2007).
The PPC2, TPRP, and IMA promoters were amplified by PCR with sense and antisense primers including the attB4 and attB1 sequences, respectively. The amplified fragments were recombined with pDONR P4-R1 to produce the corresponding entry clones. The larger PG and CRC promoters were introduced by conventional restriction/ligation cloning into pEN-L4-R1 in which a multicloning site is flanked by the attL4 and attR1 sequences.
Expression of the GUS Reporter in Tomato Fruits
In the context of MultiSite Gateway expression constructs, the described promoters (pEN-L4-promoter-R1) were fused to the GUS gene (pEN-L1-SI-L2; Karimi et al., 2007b) in the pK7m24GW destination vector via MultiSite LR Clonase reaction (Karimi et al., 2005; Table II). Initial validation of promoter activity was achieved with a previously described fruit transient expression assay (Orzaez et al., 2006). Briefly, the promoter-GUS binary expression vectors were transformed into Agrobacterium tumefaciens and injected into fruits of different developmental stages, from 20 daa to breaker + 11 d (Br+11). Several days after injection, fruits were sampled and thin sections were analyzed for GUS activity via classical histochemical assays (Supplemental Fig. S1 and Supplemental Materials and Methods S1). GUS staining consistent with previously published expression results were obtained for late stage promoters, such as in PGpro:GUS-injected ripening fruits, but the assessment of the promoter activity in the transient assay was restricted to more than 20-daa fruits and concurrent with wounding and stress. These technical constraints probably explain why temporal and spatial expression might be partly artifactual in agroinjected fruits. For example, the early-stage IMA promoter was active in ripening fruits, and higher level of GUS staining was observed in placenta and gel, presumably because of a better penetration of Agrobacterium in these tissues. Nevertheless, transient expression data provided the initial evidence that the Gateway cloning framework was compatible with gene expression in tomato, a prerequisite for in-depth analysis of promoter activity in stable transgenic plants.
Table II.
Expression vectors
| Destination and Modular Vector | Recombined Entry Clones | Resulting Expression Vector |
|---|---|---|
| pK7m24GW | pEN-L4-PPC2-R1 × pEN-L1-SI-L2 | pXK7SIPPC2 |
| pK7m24GW | pEN-L4-PG-R1 × pEN-L1-SI-L2 | pXK7SIPG |
| pK7m24GW | pEN-L4-TPRP-R1 × pEN-L1-SI-L2 | pXK7SITPRP |
| pK7m24GW | pEN-L4-IMA-R1 × pEN-L1-SI-L2 | pXK7SIIMA |
| pK7m24GW | pEN-L4-CRC-R1 × pEN-L1-SI-L2 | pXK7SICRC |
| pMK7S*NFm14GW | pEN-L4-PPC2-R1 | pXK7S*NFPPC2 |
| pMK7S*NFm14GW | pEN-L4-PG-R1 | pXK7S*NFPG |
| pMK7S*NFm14GW | pEN-L4-TPRP-R1 | pXK7S*NFTPRP |
| pMK7S*NFm14GW | pEN-L1-IMA-R1 | pXK7S*NFIMA |
| pMK7S*NFm14GW | pEN-L4-CRC-R1 | pXK7S*NFCRC |
| pK7GW2 | pEN-L4-miRpds-L2 | pXK7miRpds2 |
| pK7m24GW | pEN-L4-PPC2-R1 × pEN-L1-miRpds-L2 | pXK7miRpdsPPC2 |
| pK7m24GW | pEN-L4-PG-R1 × pEN-L1-miRpds-L2 | pXK7miRpdsPG |
| pK7m24GW | pEN-L4-TPRP-R1 × pEN-L1-miRpds-L2 | pXK7miRpdsTPRP |
| pK7m24GW | pEN-L4-IMA-R1 × pEN-L1-miRpds-L2 | pXK7miRpdsIMA |
| pK7m24GW | pEN-L4-CRC-R1 × pEN-L1-miRpds-L2 | pXK7miRpdsCRC |
| pK7m24GW | pEN-L4-PPC2-R1 × pEN-L1-LhGATO4-L2 | pXK7LhGATO4PPC2 |
| pK7m24GW | pEN-L4-PG-R1 × pEN-L1-LhGATO4-L2 | pXK7LhGATO4PG |
| pK7m24GW | pEN-L4-TPRP-R1 × pEN-L1-LhGATO4-L2 | pXK7LhGATO4TPRP |
| pK7m24GW | pEN-L4-IMA-R1 × pEN-L1-LhGATO4-L2 | pXK7LhGATO4IMA |
| pK7m24GW | pEN-L4-CRC-R1 × pEN-L1-LhGATO4-L2 | pXK7LhGATO4CRC |
To this end, the MicroTom cultivar was agroinfected with an improved protocol yielding transformation frequency consistently >80% (see “Materials and Methods”). The ploidy of regenerated plantlets was measured with flow cytometry, and polyploid plants were discarded. Between 12 and 25 independently transformed plants were generated for each promoter-GUS transgene (Table II). Following GUS staining and selection of plants with one or two transgene copies, two representative lines were selected for each construct, and the GUS staining pattern of 10 kanamycin-resistant T1 plants per original line was characterized at specific stages of development: flower, 6-mm buds (approximately stage 9 according to Baldet et al., 2006), and anthesis; fruit, 6 daa (cell division), 10 daa, 15 daa (cell expansion stage; Lemaire-Chamley et al., 2005), mature green (MG; transition to ripening), and Br+7 (ripe fruit). GUS staining of T1 leaves and entire young T2 plantlets completed the analysis.
As expected, strong GUS activity was observed in all 35Spro-GUS tissues, while no staining was detected in wild-type controls (Fig. 2). The five selected fruit promoters only resulted in fruit-specific expression, with no GUS staining in vegetative organs (Fig. 2). Flower buds showed weak GUS staining only in the IMApro:GUS, TPRPpro:GUS, and PPC2pro:GUS lines.
Figure 2.
GUS activity in tomato (cv MicroTom) stably transformed with promoter:GUS transgenes generated via MultiSite Gateway cloning. Bars = 10 mm.
In agreement with earlier reports (Sicard et al., 2008), IMApro:GUS fruits displayed weak but consistent GUS staining at early stages of development, particularly in placental and vascular tissues. Additional staining of seed tegument was observed in ripe fruit (Fig. 2). In CRCpro:GUS fruits, GUS staining was restricted to the outer carpel wall from early stages until MG (Fig. 2). This result is reminiscent of the CRC native expression that is largely restricted to the abaxial epidermis in the Arabidopsis carpel (from the base to the tip of the carpel, along its entire circumference), where it promotes abaxial cell fate (Eshed et al., 1999). In TPRPpro:GUS fruits, GUS staining peaked at the immature green stage (Fig. 2), consistent with earlier reports (Gillaspy et al., 1993). In PPC2pro:GUS fruits, strong GUS staining was limited to the fruit expansion phase (Fig. 2), in agreement with previous data (Guillet et al., 2002; C. Guillet and C. Rothan, unpublished data). In PGpro:GUS fruits, GUS activity was not detected in early stages, was weak at the onset of ripening (MG), and high in the ripe fruit (Br+7; Fig. 2), again as published previously (Montgomery et al., 1993).
In summary, the temporal regulation of the five promoters observed in stable transgenic tomato fruits was in agreement with the data reported in wild-type or stable transgenic plants. In addition to marking selected stages of fruit development, several tested promoters also offer the possibility to direct transgene expression to specific fruit tissues. For example, the CRC promoter can specifically target genes expressed in the epidermis of developing tomato fruit and involved in cuticle or phenylpropanoid synthesis (Mintz-Oron et al., 2008).
GFP Markers Specific to Fruit Cell Types
Promoter-GUS transgenes were useful to report tissue-specific transcription. Yet higher spatial resolution can be achieved via the transcriptionally regulated expression of a nuclear localization signal (NLS) fused to fluorescent proteins. Such markers enable the selection of particular cell types, as well as the tracking of cell proliferation and enlargement in the course of fruit development. Therefore, we generated a promoter:NLS-GFP-GUS construct for each of the five selected promoters via LR recombination into the modular destination vector pK7S*NFm14GW (Karimi et al., 2007b; Table II). Stable tomato (cv MicroTom) reporter lines carrying one of each promoter:NLS-GFP-GUS transgene (Table II) were generated, screened, and analyzed as described above. In addition, GFP production was verified in fresh 150-μm-thick pericarp sections with epifluorescence microscopy. Promoter:GUS and promoter:NLS-GFP-GUS lines displayed similar GUS staining patterns (data not shown).
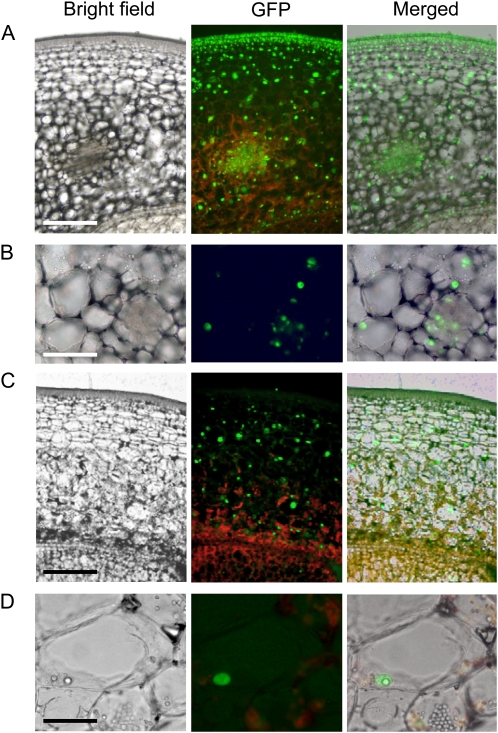
The NLS fused to GFP proved very effective because the fluorescence signal was detected in the nucleus of all cell types in the pericarp and gel of 35Spro:NLS-GFP-GUS fruits (Fig. 3, A and B). In the pericarp of PPC2pro:NLS-GFP-GUS fruits, GFP was absent from the fruit epidermis and from most underlying dividing cells and was essentially limited to the large expanding mesocarpic cells (Fig. 3, C and D). This observation is consistent with the hypothesis that malate synthesized via PECase serves as a counterion for potassium that accumulates in the vacuole, thus providing the driving force for fruit cell enlargement (Guillet et al., 2002).
Figure 3.
Detection of nuclear GFP in cells of tomato fruit pericarp. Sections were prepared from 8-daa fruits. A and B, 35Spro:NLS:GUS-GFP tissues showing GFP signal in all fruit tissues. B, Magnified image showing localization of GFP in the nucleus. C and D, Fruit pericarp cross section from 8-daa tomato fruits transformed with PPC2pro:NLS:GUS-GFP showing preferential GFP localization in expanding cells from fruit mesocarp. D, Magnified image of mesocarp showing localization of GFP in polyploid nucleus from large cell. Bars = 400 μm in A and C and 100 μm in B and D.
To conclude, in addition to the information provided by promoter-GUS transgenic lines about expression targeted at specific developmental stages and in particular tissues, promoter:NLS-GFP-GUS lines can be used to define the spatial distribution of a transgene driven by a selected promoter in tomato fruits with higher cellular resolution.
Vectors for Silencing Genes in Fruits
RNA-mediated gene silencing, or RNA interference, is a method of choice to generate loss-of-function phenotypes caused by partial to complete gene knockdown. amiRNAs have been shown to efficiently silence genes in multiple plant species, including tomato (Alvarez et al., 2006; Schwab et al., 2006; for review, see Ossowski et al., 2008). This approach entails the expression of an endogenous plant microRNA precursor engineered to yield a 21-nucleotide double-stranded amiRNA that is chosen to silence a gene, or several genes, of interest according to experimentally defined target selection parameters. The DNA sequence coding for an amiRNA can easily be synthesized by overlapping PCR amplification (Schwab et al., 2006) with the addition of the attB1 and attB2 recombination sites at their 5′ and 3′ ends, respectively, and subsequently captured as an amiRNA entry clone (attL1-amiRNA-attL2). Once in this format, any amiRNA precursor can be cloned downstream of any promoter of interest, including those described above, in a simple MultiSite LR Clonase reaction. The addition of attB-flanking sites to an amiRNA precursor (i.e. to the MIR319a backbone in this particular case) did not alter its ability to silence a target gene in Arabidopsis (Supplemental Fig. S2 and Supplemental Materials and Methods S1).
To confirm silencing in tomato, we generated MicroTom lines in which the PDS gene was silenced via the transcription of a matching amiR-pds precursor under the control of the cauliflower mosaic virus (CaMV) 35S or PPC2 promoter (Tables I and II). The PDS enzyme is involved in carotenoid biosynthesis and protects chlorophyll from photooxidation in developing tissues. PDS silencing causes photobleaching that can be readily recorded in the course of development. Bleaching was observed in all tissues of the 35Spro:amiR-pds tomato plants (Fig. 4, B and C). The most severely affected plantlets did not survive in the greenhouse (data not shown). In contrast, the 18 PPC2pro:amiR-pds primary transformants did not display any visual phenotype during vegetative growth. However, one-third of these T0 plants produced fruits that remained light orange during ripening and only turned light red when left on the plant (Fig. 4, D and E). Our observations are consistent with the PPC2 profile documented through reporter expression and indicating that the promoter activity is the highest at the early stages of fruit development (Fig. 2) and in the mesocarp (Fig. 3C), i.e. at developmental stages and in fruit tissues less sensitive to light than the exocarp where the 35S-driven expression is very high (Fig. 3A) and the photobleaching more likely to occur. Our results further illustrate how fruit promoters and constructs developed herein can be used to target specific fruit tissues and alter expression of candidate genes in tomato.
Figure 4.
Tissue-specific amiRNA silencing in tomato fruits. Flowers at anthesis (top), mature leaves (middle), and ripe fruits (Breaker +7; bottom). A, The wild type (cv MicroTom). B and C, 35Spro:amiR-pds plants with strong to extreme photobleaching in all organs. D and E, PPC2pro:amiR-pds plants with moderate and transient photobleaching restricted to the fruit. Bars = 10 mm.
Because amiRNA target recognition relies on a short 21-nucleotide sequence, amiRNAs yield more specific silencing than the numerous small interfering RNAs derived from the long (hundreds of base pairs) double-stranded hpRNA molecules. However, because many genes encoded in Solanaceae have yet to be identified, hpRNA silencing is still valuable: for example, to avoid the degradation of a gene different from the target but serendipitously containing sequences matching a selected amiRNA or to silence simultaneously multiple closely homologous transcripts.
Therefore, we constructed a new series of hpRNA expression vectors with the aim to restrict gene silencing to specific tissues or organs (Supplemental Table S5). For this purpose, we implemented novel cloning schemes involving only BP and LR Clonase reactions to position any promoter available as an entry clone in front of a silencing transgene (Supplemental Figs. S3–S5 and Supplemental Materials and Methods S1). The constructs generated were derived from the pHELLSGATE12 vector (Wesley et al., 2001; Helliwell and Waterhouse, 2003; Hilson et al., 2004) in which the sequence targeted for silencing (GST), originally captured in an entry clone (attL1-GST-attL2), was transferred simultaneously in two independent Gateway cassettes separated by an intron spacer (attR1-ccdB-attR2-intron_spacer-attR2-ccdB-attR1). This spacer consisted of two head-to-head introns that enabled splicing of the encoded transcript regardless of its orientation. In this configuration, the attR1 and attR2 recombination sites were inverted with respect to one another, so that the two copies of the target sequence were inserted as inverted repeats into the resulting hpRNA expression clone (Helliwell and Waterhouse, 2003).
For both hpRNA and amiRNA silencing, software is available that selects the most likely specific target sites taking into consideration entire genome sequences: for example, SPADS for the design of GSTs cloned into hpRNA expression vectors (Thareau et al., 2003; Sclep et al., 2007) and Web MicroRNA Designer for the selection of amiRNA sequences introduced into the microRNA precursor backbone by overlapping PCR (http://wmd3.weigelworld.org/). Nevertheless, some amiRNAs and hpRNAs fail to yield efficient silencing, and current algorithms cannot accurately predict silencing efficiency.
Driver Constructs and Tomato Lines
In tomato, as in most crop plants, transformation protocols require tissue culture. However, because many tissue- and cell-type-specific promoters display extensive expression in callus, transgenes with deleterious effects might be selected against during tissue culture. A solution to this problem is to dissociate the target gene expression locus from its transcriptional regulator. For this purpose, we established reference tomato driver lines facilitating both gene misexpression and silencing. These lines were constructed with elements of the pOp/LhG4 system engineered for transcriptional activation into plant species (Moore et al., 1998; Rutherford et al., 2005; Wielopolska et al., 2005; Ori et al., 2007). Its main components are the LhG4 chimeric transcription factor, consisting of the bacterial LacI DNA-binding and yeast GAL4 activation domains, and the synthetic pOp promoters containing multiple copies of the lac operator bound by LhG4. The driver locus codes for the LhG4 gene under the control of a promoter with a particular spatio-temporal pattern. The responder locus contains a synthetic pOp promoter driving the transcription of a gene, hpRNA, or amiRNA of interest and is only active in the presence of LhG4. Both loci were combined in the same plant either by crossing or by transformation.
First, we created an array of reporter lines to validate the use of the pOp/LhG4 system in tomato, including pOP:GUS, pOP:ER-GFP, pOP:dsRFP, and pOP:KANADI1 (KAN1). The first three reporters were useful for the detailed examination of promoter activity. The pOP:KAN1 reporter drove the potent tomato KANADI1 gene that imposes abaxial identity and severe growth arrest whenever expressed ectopically outside of its normal domain (Eshed et al., 2001). This reporter provided a simple and sensitive test for spatial activity of any examined driver line because even low level of expression, possibly escaping detection with the other reporters, might result in clearly visible phenotypes. The KAN1 reporter is particularly relevant to reveal background activity within large organs, such as the tomato fruits, that are difficult to characterize via classical histological staining or microscopy analysis.
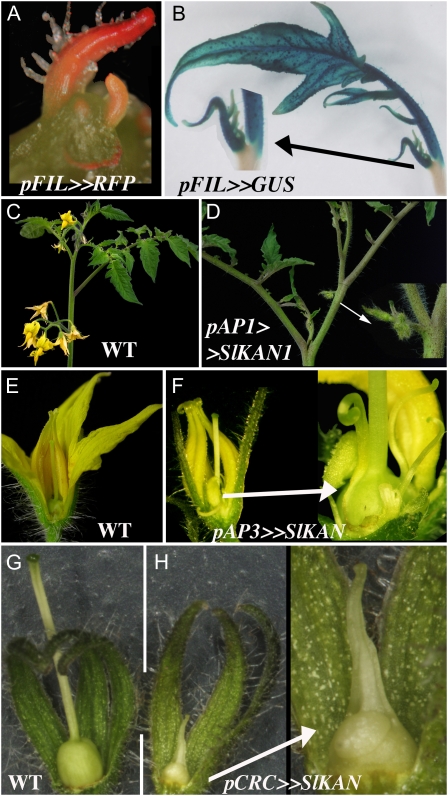
Second, we crossed these reporter lines with tomato driver lines in which the LhG4 transgene was under the control of one of five Arabidopsis flower-specific promoters (described in Pekker et al., 2005). Significantly, the Arabidopsis FIL, AP1, AP3, and CRC promoters maintained their specific expression domains in F1 tomato plants expressing both a driver and reporter construct. The FIL promoter stimulated expression in all organ primordia but not in the shoot apical meristem proper (Fig. 5, A and B). The AP1 promoter drove expression in the sepals, petals, and flower meristems because all were strongly inhibited upon transactivation of KAN1 (Fig. 5, C and D) and the AP3 promoter only in the petals and stamens, of which the petals became radialized upon transactivation of KAN1 (Fig. 5, E and F). The CRC promoter strongly inhibited carpel development upon KAN1 transactivation but nowhere else (Fig. 5, G and H). Only one promoter, FUL, failed to show the expected early expression in the pericarp (data not shown). The proportion of observed versus expected patterns was in the same range as that obtained when attempting to systematically recapitulate transcription pattern with cloned promoter sequences (Lee et al., 2006). To conclude, defined Arabidopsis promoters are a useful resource to misexpress or silence genes in other species, such as tomato.
Figure 5.
Tissue-specific transactivation in tomato with selected Arabidopsis promoters. A and B, Expression of dsRED or GUS, activated in trans by the organ primordia and abaxial domain pFIL:LhG4 driver. Abaxial expression is evident after leaf primordium has expanded. C and D, Flower-specific growth arrest stimulated by specific transactivation of a pOP:KAN1 reporter, with the pAP1:LhG4 driver. E and F, Petal-only radialization stimulated by specific transactivation of a pOP:KAN1 reporter, with the pAP3:LhG4 driver. G and H, Carpel-specific growth arrest stimulated by transactivation of the pOP:KAN1 reporter with the pCRC:LhG4 driver. WT, Wild type; SlKAN, KANADI1 gene of tomato.
Finally, we have expanded the repertoire of driver constructs by generating via MultiSite LR recombination all of the transcriptional fusions between each of the tomato fruit promoters described above and the LhG4AtO ORF, optimized for expression in plants (pEN-L1-LhATOG4-L2; Karimi et al., 2007b; Table II).
Perspectives
All plasmids with tomato sequences and tomato seed stocks described here are made available via the Nottingham Arabidopsis Stock Centre (NASC; http://Arabidopsis.info) to ensure long-term distribution and appropriate documentation of the materials. The centralization of resources and related information at the SGN Web site (Mueller et al., 2005) and complemented at NASC will also facilitate their integration into reference genome browsers as well as genotype and phenotype databases. As demonstrated by the Arabidopsis community, shared genetic tools promote rapid and significant scientific achievements, which is particularly relevant for a plant family of high commercial value. For example, the tomato promoters available as Gateway entry clones might be used to characterize genes involved in key processes taking place during the successive phases of fruit development, namely, cell proliferation (TPRP and CRC; mitotic cycle), cell expansion (PPC2; endoreduplication, starch synthesis and storage, and cell wall synthesis), and ripening (PG; cell wall and starch breakdown and soluble sugar and carotenoid accumulation), or in specific fruit tissues (IMA, vascular tissues; CRC, cuticle formation and secondary metabolism). Promoters only, or mainly, active in flower or fruit should avoid some of the common pitfalls associated with strong constitutive promoters (such as CaMV 35S) that also affect vegetative plant parts and therefore hamper the analysis of gene function, as clearly illustrated for PDS amiRNA silencing in tomato.
Finally, it is worth noting that the Gateway clones described here and in a recent companion article (Estornell et al., 2009) are compatible with a wide range of complementary entry clones (coding for alternative terminators, reporters, or activators) and destination vectors designed for various functional assays in plants or in other heterologous systems (for review, see Karimi et al., 2007a).
MATERIALS AND METHODS
Bacterial Strains
Host Escherichia coli strains were either DH5α (with entry and expression clones) or DB3.1 (with destination vectors; Invitrogen). Bacterial cultures were grown at 37°C in Luria broth medium with appropriate antibiotics.
BP and LR Clonase Reactions
Additional information about the basic Gateway site-specific recombinational cloning protocols is provided online by the manufacturer (http://www.invitrogen.com/). Entry clones were created in BP Clonase reactions with 50 to 100 ng of PCR product and 50 ng of donor vector. Expression clones were created in LR Clonase II reactions with 30 ng of each entry clones and 50 to 80 ng of destination vector. Reactions were done in 10 μL total volume containing 2 μL of BP or LR Clonase, incubated at 25°C overnight. The reaction was inactivated by addition of 1 μL of proteinase K, and 5 μL from each reaction was used to transform E. coli competent cells as described.
Promoter Entry Clones
Specific oligonucleotides were designed to amplify the PPC2, TPRP, and IMA promoters, including the attB4 and attB1 sites upstream of the 5′ and 3′ gene-specific priming sequences, respectively (Supplemental Table S2). Promoters were amplified by PCR with either tomato genomic or plasmid DNA as template. PCR mix contained 0.5 μm of each primer, 25 ng template DNA, 200 μm of each dinucleotide triphosphate, 2.5 units of Platinum HiFi DNA polymerase (Invitrogen), 1× PCR buffer, and 1 mm MgSO4. Programmed cycles were as follows: 5 min initial denaturing step at 94°C; 30 cycles of 30 s denaturation at 94°C, 30 s annealing at 50°C, 1 to 3 min extension (1 min per kb) at 68°C, and 5 min termination at 68°C. PCR products were purified with the High Pure PCR Purification Kit (Roche Diagnostics). BP Clonase recombination reactions were done with the purified PCR products and pDONR P4-P1R to generate the entry clones. After transformation into DH5α competent cells, colonies were screened by PCR for the correct insert size and clones were sequence validated.
Large DNA fragments are more difficult to capture into entry clones. Because the PG and CRC promoters were 4.8 and 3.7 kb, respectively, we chose conventional restriction/ligation techniques to build the corresponding entry clones. The PG promoter originally cloned into pBIN19 (Nicholass et al., 1995) was isolated as a fragment obtained by BamHI and partial HindIII digestion. The isolated fragment was ligated to pBlueScript (CLONTECH) plasmid linearized by BamHI-HindIII digestion to generate an intermediate clone. Two complementary PG fragments were ligated at once into SalI-XhoI-linearized pEN-L4-R1: from 5′ to 3′, a XhoI-XbaI fragment isolated from the PG intermediate clone, and an XbaI-XhoI fragment isolated from the original PG pBIN19 derivative. The plasmid pEN-L4-R1 carried a multiple cloning site flanked by the attL4 and attR1 recombination sites (attL4-XmnI-SalI-BamHI-KpnI-ccdB-XhoI-attR1; O. Nyabi, M. Karimi, P. Hilson, and J. Haigh, unpublished data). The CRC promoter originally cloned into pBS (Lee et al., 2005) was isolated as a SalI-XhoI fragment and cloned into the same sites within pEN-L4-R1. Digested fragments were separated by electrophoresis in agarose gels, extracted, and purified with the High Pure PCR Purification Kit (Roche Diagnostics). After transformation into DH5α competent cells, clones were screened by restriction analysis to identify plasmids with the expected insert in the correct orientation and subsequently sequence validated.
Creation of amiRNA Vectors
According to Schwab et al. (2006), the plasmid pRS300 was used as template to introduce an amiRNA sequence targeting the tomato (Solanum lycopersicum) and Arabidopsis (Arabidopsis thaliana) PDS gene (5′-TCCATGCAGCTACCTTTCCAC-3′) into the miR319a precursor by site-directed mutagenesis. Overlapping PCR amplification steps were done as described in the original cloning protocol except that, in the final PCR reaction, the oligonucleotides A and B (based on the pBSK backbone) were replaced by two alternative oligonucleotides, attB1-amiRNA-fw and attB2-amiRNA-rev, carrying the attB1 and attB2 sites, respectively. These two primers can be used for the amplification of any amiRNA derived from the miR319a precursor backbone (see Supplemental Materials and Methods S1 for details). The resulting PCR product containing the amiR-pds precursor was cloned into pDONR221 by BP Clonase reaction, resulting in the pEN-L1-miRpds-L2 entry clone (Table I) and then subcloned into the pK7WG2 or pK7m24GW destination vectors by LR Clonase reaction to yield the expression vectors driving amiR-pds silencing under the control of the CaMV 35S, PPC2, PG, TPRP, IMA, or CRC promoters (Table II).
Transactivation Clones
Reporter OP clones were sequence verified after restriction-ligation subcloning downstream of an OP array (10xOP-TATA-MCS-3′OCS in BJ36) and transferred into the NotI site of the pART27 binary vector. pOP:GUS and pOP:RFP were described previously (Lifschitz et al., 2006). pOP:KAN1 was generated by PCR amplification of the ORF of the tomato KAN1 ortholog, designated SGN-U321055. Driver lines were generated by transcriptional fusion of promoters in front of the chimeric LhG4 (MCS-LhG4-3′OCS in BJ36; Moore et al., 1998) and transferred into the NotI site of pART27. The Arabidopsis LhG4 promoter lines were described before (Pekker et al., 2005), but briefly, a 6.1-kb FILAMENTOUS FLOWER, a 1.7-kb APETALA1, a 0.5-kb APETALA3, and 3.5-kb CRC Arabidopsis promoter fragments were subcloned in front of LhG4 in pART27.
Tomato Transformation and Transactivation
MicroTom tomato lines were transformed with the Agrobacterium tumefaciens strain GV3101. Briefly, 8-d-old MicroTom cotyledons were cut at both extremities and in the middle, placed on solid KCMS in petri dishes, incubated at 25°C for 24 h, soaked for 30 min with shaking in Agrobacterium suspension (Agrobacterium was grown in Luria broth to OD = 1, pelleted, and resuspended in liquid KCMS to OD = 0.05–0.08), blotted dry on sterile Whatman paper, and incubated on solid KCMS in petri dishes in the dark for 48 h at 25°C. For regeneration, the cotyledons were laid on 2Z medium with 250 mg L−1 timentin (ticarcillin and clavulanate; GlaxoSmithKline) and 100 mg L−1 kanamycin and incubated during 15 d at 25°C in the light. Regeneration medium was changed every 15 d when necessary, with timentin concentration reduced to 150 mg L−1. In case of Agrobacterium overgrowth, timentin concentration was kept at 250 mg L−1. Regenerated plantlets were transferred to rooting medium supplemented with 100 mg L−1 kanamycin and 75 mg L−1 timentin. With this protocol and the vectors described herein, transformation frequencies observed using MicroTom cotyledons were consistently >80% and up to 90%. Less than 15% of the transgenic plantlets were polyploid according to flow cytometry analysis. Cultivation from cotyledon explants to transfer of transgenic plants into the greenhouse lasted 2 to 3 months, and 2 to 3 additional months were required to collect transgenic seeds. The composition of in vitro culture media is provided in Supplemental Tables S3 and S4.
Cotyledon transformation of M82 tomato and leaf discs were carried out according to McCormick (1991) with the Agrobacterium strain GV3101. promoter:LhG4 lines were crossed to 10xe lines to generate the F1 promoter≫reporter plants.
Upon request, all novel materials described will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
GUS Histochemical Staining
Tissues from stably transformed tomato plants were soaked, vacuum-infiltrated, and incubated for 30 min (PG), 1 h (35S, PPC2, and TPRP), 2 h (CRC), or overnight (IMA) at 37°C in GUS staining solution [0.5 mm X-gluc, 0.15 m NaH2PO4, pH 7, 2 mm K3Fe(CN)6, 2 mm K4Fe(CN)6, and 0.05% Triton X-100]. GUS-stained tissues were cleared overnight in 100% ethanol and stored in 70% ethanol.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. GUS activity in tomato fruits (cv MicroTom) agroinjected with promoter:GUS transgenes generated via MultiSite Gateway cloning.
Supplemental Figure S2. Albino phenotype resulting from PDS silencing in Arabidopsis.
Supplemental Figure S3. Creation of novel hpRNA destination vectors.
Supplemental Figure S4. Construction of the intron-spacer donor vector pDONR P1-R2-I-R2-P1.
Supplemental Figure S5. Alternative scheme to generate specific hpRNA destination vectors.
Supplemental Table S1. Theoretical promoter sequences.
Supplemental Table S2. Primers designed for PCR amplification of genetic elements and for validation of recombined plasmids.
Supplemental Table S3. Composition of cv MicroTom transformation media (per liter).
Supplemental Table S4. Vitamins for cv MicroTom transformation media.
Supplemental Table S5. Specific hpRNA destination vectors.
Supplemental Materials and Methods S1.
Supplementary Material
Acknowledgments
We thank Wilson Ardilez for sequencing and Martine De Cock for help in preparing the manuscript.
This work is supported by the European Solanaceae Integrated project, EU-SOL, through the 6th Framework Programme of the European Commission (FOOD–CT–2006–016214).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Pierre Hilson (pierre.hilson@psb.ugent.be).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y (2006) Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldet P, Hernould M, Laporte F, Mounet F, Just D, Mouras A, Chevalier C, Rothan C (2006) The expression of cell proliferation-related genes in early developing flowers is affected by a fruit load reduction in tomato plants. J Exp Bot 57 961–970 [DOI] [PubMed] [Google Scholar]
- Biggs MS, Handa AK (1989) Temporal regulation of polygalacturonase gene expression in fruits of normal, mutant, and heterozygous tomato genotypes. Plant Physiol 89 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR (1999) CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126 2387–2396 [DOI] [PubMed] [Google Scholar]
- Carmi N, Salts Y, Dedicova B, Shabtai S, Barg R (2003) Induction of parthenocarpy in tomato via specific expression of the rolB gene in the ovary. Planta 217 726–735 [DOI] [PubMed] [Google Scholar]
- Cheo DL, Titus SA, Byrd DR, Hartley JL, Temple GF, Brasch MA (2004) Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: functional analysis of multi-segment expression clones. Genome Res 14 2111–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich-Rikanati R, Sitrit Y, Tadmor Y, Iijima Y, Bilenko N, Bar E, Carmona B, Fallik E, Dudai N, Simon JE, et al (2007) Enrichment of tomato flavor by diversion of the early plastidial terpenoid pathway. Nat Biotechnol 25 899–901 [DOI] [PubMed] [Google Scholar]
- Davuluri GR, van Tuinen A, Fraser PD, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Uhlig J, et al (2005) Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat Biotechnol 23 890–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D, Lincoln JE, Fischer RL, Bennett AB (1989) Transcriptional analysis of polygalacturonase and other ripening associated genes in Rutgers, rin, nor, and Nr tomato fruit. Plant Physiol 90 1372–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL (1999) Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99 199–209 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11 1251–1260 [DOI] [PubMed] [Google Scholar]
- Estornell LH, Orzáez D, López-Peña L, Pineda B, Antón MT, Moreno V, Granell A (2009) A multisite gateway-based toolkit for targeted gene expression and hairpin RNA silencing in tomato fruits. Plant Biotechnol J 7 298–309 [DOI] [PubMed] [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W (1993) Fruits: a developmental perspective. Plant Cell 5 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ (2004) Genetic regulation of fruit development and ripening. Plant Cell 16 S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ (2007) Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol 10 283–289 [DOI] [PubMed] [Google Scholar]
- Gonzalez N, Gévaudant F, Hernould M, Chevalier C, Mouras A (2007) The cell cycle-associated protein kinase WEE1 regulates cell size in relation to endoreduplication in developing tomato fruit. Plant J 51 642–655 [DOI] [PubMed] [Google Scholar]
- Guillet C, Just D, Bénard N, Destrac-Irvine A, Baldet P, Hernould M, Causse M, Raymond P, Rothan C (2002) A fruit-specific phosphoenolpyruvate carboxylase is related to rapid growth of tomato fruit. Planta 214 717–726 [DOI] [PubMed] [Google Scholar]
- Helliwell C, Waterhouse P (2003) Constructs and methods for high-throughput gene silencing in plants. Methods 30 289–295 [DOI] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Ma H (2006) Characterization of a novel putative zinc finger gene MIF1: involvement in multiple hormonal regulation of Arabidopsis development. Plant J 45 399–422 [DOI] [PubMed] [Google Scholar]
- Karimi M, Bleys A, Vanderhaeghen R, Hilson P (2007. b) Building blocks for plant gene assembly. Plant Physiol 145 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10 103–105 [DOI] [PubMed] [Google Scholar]
- Karimi M, Depicker A, Hilson P (2007. a) Recombinational cloning with plant Gateway vectors. Plant Physiol 145 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S (2002) Tobacco to tomatoes: a phylogenetic perspective on fruit diversity in the Solanaceae. J Exp Bot 53 2001–2022 [DOI] [PubMed] [Google Scholar]
- Kovacs K, Zhang L, Linforth RST, Whittaker B, Hayes CJ, Fray RG (2007) Redirection of carotenoid metabolism for the efficient production of taxadiene [taxa-4(5),11(12)-diene] in transgenic tomato fruit. Transgenic Res 16 121–126 [DOI] [PubMed] [Google Scholar]
- Lee J-Y, Baum SF, Alvarez J, Patel A, Chitwood DH, Bowman JL (2005) Activation of CRABS CLAW in the nectaries and carpels of Arabidopsis. Plant Cell 17 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-Y, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN (2006) Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA 103 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire-Chamley M, Petit J, Garcia V, Just D, Baldet P, Germain V, Fagard M, Mouassite M, Cheniclet C, Rothan C (2005) Changes in transcriptional profiles are associated with early fruit tissue specialization in tomato. Plant Physiol 139 750–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y (2006) The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA 103 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38 948–952 [DOI] [PubMed] [Google Scholar]
- Mintz-Oron S, Mandel T, Rogachev I, Feldberg L, Lotan O, Yativ M, Wang Z, Jetter R, Venger I, Adato A, et al (2008) Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol 147 823–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S (1991) Transformation of tomato with Agrobacterium tumefaciens. In K Lindsey, ed, Plant Tissue Culture Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–9
- Montgomery J, Pollard V, Deikman J, Fischer RL (1993) Positive and negative regulatory regions control the spatial distribution of polygalacturonase transcription in tomato fruit pericarp. Plant Cell 5 1049–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore I, Gälweiler L, Grosskopf D, Schell J, Palme K (1998) A transcription activation system for regulated gene expression in transgenic plants. Proc Natl Acad Sci USA 95 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Klein Lankhorst R, Tanksley SD, Giovannoni JJ, White R, Vrebalov J, Fei Z, van Eck J, Buels R, Mills AA, et al (2009) A snapshot of the emerging tomato genome sequence. Plant Genome 2 78–92 [Google Scholar]
- Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin C, Wright MH, Ahrens R, Wang Y, et al (2005) The SOL Genomics Network. A comparative resource for Solanaceae biology and beyond. Plant Physiol 138 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholass FJ, Smith CJS, Schuch W, Bird CR, Grierson D (1995) High levels of ripening-specific reporter gene expression directed by tomato fruit polygalacturonase gene-flanking regions. Plant Mol Biol 28 423–435 [DOI] [PubMed] [Google Scholar]
- Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, et al (2007) Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39 787–791 [DOI] [PubMed] [Google Scholar]
- Orzaez D, Mirabel S, Wieland WH, Granell A (2006) Agroinjection of tomato fruits. A tool for rapid functional analysis of transgenes directly in fruit. Plant Physiol 140 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53 674–690 [DOI] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y (2005) Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S, Brandizzi F, Townley H, Craft J, Wang Y, Jepson I, Martinez A, Moore I (2005) Improved transcriptional activators and their use in mis-expression traps in Arabidopsis. Plant J 43 769–788 [DOI] [PubMed] [Google Scholar]
- Salts Y, Wachs R, Gruissem W, Barg R (1991) Sequence coding for a novel proline-rich protein preferentially expressed in young tomato fruit. Plant Mol Biol 17 149–150 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclep G, Allemeersch J, Liechti R, De Meyer B, Beynon J, Bhalerao RP, Moreau Y, Nietfeld W, Renou J-P, Reymond P, et al (2007) CATMA, a comprehensive genome-scale resource for silencing and transcript profiling of Arabidopsis genes. BMC Bioinformatics 18 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard A, Petit J, Mouras A, Chevalier C, Hernould M (2008) Meristem activity during flower and ovule development in tomato is controlled by the mini zinc finger gene INHIBITOR OF MERISTEM ACTIVITY. Plant J 55 415–427 [DOI] [PubMed] [Google Scholar]
- Thareau V, Déhais P, Serizet C, Hilson P, Rouzé P, Aubourg S (2003) Automatic design of gene-specific sequence tags for genome-wide functional studies. Bioinformatics 19 2191–2198 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang M, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27 581–590 [DOI] [PubMed] [Google Scholar]
- Wielopolska A, Townley H, Moore I, Waterhouse P, Helliwell C (2005) A high-throughput inducible RNAi vector for plants. Plant Biotechnol J 3 583–590 [DOI] [PubMed] [Google Scholar]
- Wu F, Mueller LA, Crouzillat D, Pétiard V, Tanksley SD (2006) Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade. Genetics 174 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.