Abstract
Plant architecture is determined by genetic and developmental programs as well as by environmental factors. Sessile plants have evolved a subtle adaptive mechanism that allows them to alter their growth and development during periods of stress. Phytohormones play a central role in this process; however, the molecules responsible for integrating growth- and stress-related signals are unknown. Here, we report a gain-of-function rice (Oryza sativa) mutant, tld1-D, characterized by (and named for) an increased number of tillers, enlarged leaf angles, and dwarfism. TLD1 is a rice GH3.13 gene that encodes indole-3-acetic acid (IAA)-amido synthetase, which is suppressed in aboveground tissues under normal conditions but which is dramatically induced by drought stress. The activation of TLD1 reduced the IAA maxima at the lamina joint, shoot base, and nodes, resulting in subsequent alterations in plant architecture and tissue patterning but enhancing drought tolerance. Accordingly, the decreased level of free IAA in tld1-D due to the conjugation of IAA with amino acids greatly facilitated the accumulation of late-embryogenesis abundant mRNA compared with the wild type. The direct regulation of such drought-inducible genes by changes in the concentration of IAA provides a model for changes in plant architecture via the process of drought adaptation, which occurs frequently in nature.
Plant architecture is vitally important for rice (Oryza sativa), as it is closely related to yield potential. Plants with a desirable structural form are capable of increased grain production in resource-limited fields compared with plants with less desirable builds. The three fundamental determinants of rice architecture, tiller number, leaf/tiller angle, and plant height, which are also important agronomic traits, are largely under the control of genetic and developmental programs; however, they are also influenced by environmental factors.
The effects of environmental conditions on plant architecture are derived from the fact that increased planting density inhibits the production of vegetative branches by grasses due to shade and nutrient deficiencies (Doust, 2007). In addition, sessile plants are frequently subjected to abiotic stresses, such as water deficiency, that can decrease the number of tillers and plant height.
The genetic and environmental interactions involved in determining plant architecture are thought to be mediated by plant hormones. The fact that some of the genes cloned from morphologically defective mutants encode components involved in hormone biosynthesis, perception, and signaling suggests that the phytohormone orchestra plays a crucial role in the creation of plant architecture, and auxin appears to be the central player. As seen in the maintenance of apical dominance, basipetal polar auxin transport in the shoot suppresses axillary bud outgrowth at the shoot base, likely through interplay with other messengers, including cytokinin and strigolactone, a newly discovered branching hormone (for review, see McSteen, 2009).
The establishment of an auxin gradient is prerequisite for its functions in plant morphogenesis, which include effects on root patterning, vascular tissue differentiation, axillary bud formation, flower organ development, and tropistic growth (Benková et al., 2003; Friml et al., 2003; Esmon et al., 2006). The auxin maxima in the plant body determine the location of primordia outgrowth, which involves the creation of distinct cell types that will give rise to various organs (Reinhardt et al., 2000; Grieneisen et al., 2007). Thus, the presence of stable local auxin maxima is a deciding factor in the ability of a plant to adopt an appropriate structural design. The asymmetric accumulation of auxin in certain cells results mainly from directional transport as well as from the dynamic biosynthesis, degradation, and conjugation of free indole-3-acetic acid (IAA), the main form of active auxin in plants (Woodward and Bartel, 2005; Paciorek and Friml, 2006). In rice, the perturbation of auxin homeostasis causes pleiotropic abnormalities leading to dramatic changes in plant architecture. For example, enhanced or reduced IAA biosynthesis via the overexpression or repression of OsYUCCA1 results in fluctuations in the level of IAA. These fluctuations produce severe abnormalities in shoot, root, and stem development, leading to dwarfism in transgenic rice plants (Yamamoto et al., 2007).
The maintenance of IAA homeostasis through the conversion of free IAA to a conjugated form is a conserved mechanism in monocots and dicots. Several gene families have been identified that are involved in the conjugation of free IAA with sugars, amino acids, or methyl groups (Qin et al., 2005; Woodward and Bartel, 2005). Proteins belonging to the GH3 family are responsible for converting active IAA to its inactive form via the conjugation of IAA with amino acids (Staswick et al., 2005). GH3 was first identified in Glycine max as an early auxin-responsive gene (Hagen and Guilfoyle, 1985). GH3 functions in the negative feedback regulation of IAA concentration, in that excess IAA up-regulates GH3 expression, causing the IAA conjugated to amino acids to be either stored or degraded. It has been shown that members of this gene family in Arabidopsis (Arabidopsis thaliana) are also regulated by hormones and environmental factors, including salicylic acid (SA), abscisic acid (ABA), pathogen infection, and light (Takase et al., 2004; Park et al., 2007a, 2007b; Zhang et al., 2007). Therefore, in addition to functioning in growth and development under normal conditions, GH3 genes also participate in plant resistance to biotic and abiotic stress. Similarly, OsGH3.8 and OsGH3.1 in rice reportedly play dual roles in development and bacterial resistance through the regulation of auxin signaling (Ding et al., 2008; Domingo et al., 2009). However, no other OsGH3 members have been reported that function in abiotic stress adaptation in rice.
Drought stress triggers the production of ABA and induces the expression of numerous genes via ABA-dependent and -independent pathways. Synchronized changes in plant architecture during drought-stress adaptation have been observed; however, no molecular mechanism has been reported. Here, we describe the cloning of OsGH3.13, a new member of the GH3 gene family, from a gain-of-function mutant, tld1-D (for increased number of tillers, enlarged leaf angles, and dwarfism). TLD1/OsGH3.13 is suppressed in aboveground tissues in rice under normal growth conditions in order to maintain a reasonable structural design; however, it is strongly induced in rice seedlings subjected to drought stress. The activation of TLD1/OsGH3.13 in tld1-D mutant rice results in IAA deficiency and dramatic changes in architecture; however, it also enhances drought tolerance. The loss-of-function mutant tld1 does not show visible differences from wild-type plants in normal growth and drought conditions. Here, we provide evidence that the down-regulation of IAA facilitates the accumulation of late-embryogenesis abundant (LEA) mRNA and switches the focus of rice plants from growth to stress adaptation at the expense of plant architecture and tissue patterning. Our results demonstrate the crucial role of IAA in integrating diverse developmental and environmental cues.
RESULTS
Isolation and Phenotypic Characterization of tld1-D, a Rice Mutant
During a large-scale screen of rice receptor-like kinase (RLK) knockdown mutants transformed with RNA interference constructs (PTCK303-RLKs; Wang et al., 2004), we identified a transgenic plant that was unique at the T0 generation, in that it had an abnormally high number of tillers, larger leaf angles, and a dwarf-like appearance; therefore, it was named tld1 (Fig. 1, A–C). Analysis by reverse transcription (RT)-PCR did not reveal the reduced expression of AK103598 (http://cdna01.dna.affrc.go.jp/cDNA), the sequence against which the construct was created, compared with wild-type rice (data not shown). After self-pollination, the ratio of tld1 plants to wild-type plants in the T1 population was 52:18, or nearly 3:1 (χ2 = 0.019), suggesting the existence of a dominant, gain-of-function mutation caused by a single T-DNA insertion. Southern blotting confirmed the presence of a single insertion in the tld1 mutant plants (Supplemental Fig. S1); thus, the mutants were renamed tld1-D.
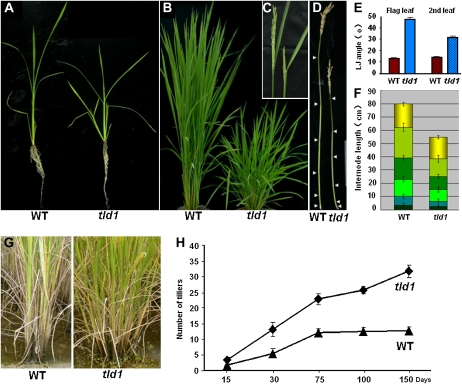
Figure 1.
Phenotypic comparison of wild-type (WT) and homozygous tld1-D rice plants at various stages of development. A, Seedling stage. B, Tillering stage. tld1-D displayed a dwarf-like phenotype with excess tillering. C, Heading stage. The shorter panicle and increased flag leaf angle in tld1-D are shown (right). D, Primary culms at the filling stage. Arrowheads indicate the nodes. E, Statistics for the flag and second leaf angles at the stage shown in C (n = 15). F, Statistics for the length of each internode and panicle shown in D (n = 15). G, Mature rice plants at the filling stage, showing the increased number of tillers in tld1-D. H, Statistics for the number of tillers in the wild-type and tld1-D plants at different stages.
The tld1-D plants tillered earlier than their wild-type counterparts at the seedling stage. The tillers in tld1-D were first visible at about 21 d after germination (Fig. 1A), whereas no tillers were visible in the wild-type plants until 28 d after germination under natural conditions. At the tillering stage (about 75 d after germination), the average tiller number per tld1-D plant was 22.67 ± 1.15, twice the average number per wild-type plant (11.66 ± 0.58; Fig. 1B). The effective tiller number per tld1-D plant increased to 31 ± 1.00 at the filling stage; the wild-type plants showed no such increase at the same stage (only about 12.67 ± 1.15 per plant; Fig. 1, G and H). Obviously expanded leaf angles were observed throughout the life span of tld1-D (Fig. 1, A–C). The average inclination angles for the flag (first) and second leaf blades were 47.73° ± 4.76° and 31.55° ± 3.91°, respectively, in tld1-D, whereas they were 13.09° ± 1.70° and 14.27° ± 1.79°, respectively, in the wild-type plants (Fig. 1, C and E). The dwarf-like stature of tld1-D was conferred by a reduction in the length of each internode (Fig. 1, D and F), and the short inflorescences in tld1-D caused a reduction in the number of spikelets (Supplemental Fig. S2, A and C). Most of the tld1-D seeds were abnormally thin due to developmental defects (Supplemental Fig. S2B). As a result of these defects, the productivity of tld1-D was not greatly enhanced, despite the fact that each plant had nearly 20 more tillers than the wild-type plants.
The Cloning of TLD1
The tld1-D phenotype cosegregated with the hygromycin resistance gene at the T1 generation as identified by PCR (data not shown), and Southern blotting suggested the presence of a single T-DNA insertion in the mutant; thus, thermal asymmetric interlaced-PCR was used to amplify the sequence flanking the T-DNA border (Liu et al., 2004). The T-DNA was found to be inserted into the first exon of Os11g32530 (Fig. 2A), which is predicted to encode a retrotransposon. All of the tld1-D mutant plants harbored the flanking sequence (data not shown). Based on these results, we conclude that the phenotype of the tld1-D plants was caused by this single-locus insertion.
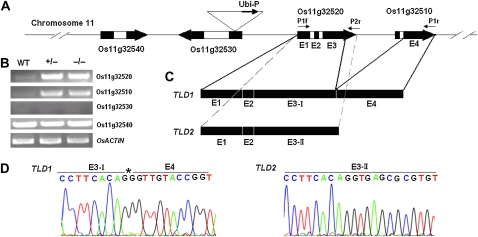
Figure 2.
The cloning of TLD1 and TLD2. A, Schematic representation of the T-DNA insertion site on rice chromosome 11. The ubiquitin promoter in the T-DNA was inserted into Os11g32530. White boxes, Introns; black boxes, exons; E1 to E4, the four exons in Os11g32520 and Os11g32510. The primers used for cloning are indicated above the exons. B, RT-PCR analysis of the transcript levels in wild-type (WT), heterozygous tld1-D (+/−), and homozygous tld1-D (−/−) seedlings. OsACTIN was used as an internal control. C, The selective splicing of Os11g32520 and Os11g32510 produces two splice variants: TLD1 and TLD2. The accession number for TLD2 mRNA in the National Center for Biotechnology Information database is NM 001074537. D, Nucleotide sequences of the TLD1 and TLD2 cDNAs. The left panel shows the junction between E3-I and E4 in TLD1. The asterisk indicates the cleavage site at the end of E3 that produces E3-I. The right panel shows the E3 region shown in the left panel without cleavage, which produces E3-II of TLD2.
However, no Os11g32530 mRNA was detected in the wild-type or heterozygous tld1-D plants by RT-PCR (Fig. 2B). On the other hand, the RNA expression of Os11g32520 and Os11g32510, located 7.8 and 13.6 kb downstream of the insertion site, respectively, was dramatically increased both in the heterozygous and homozygous tld1-D plants compared with the wild-type plants. The transcription of another neighboring gene, Os11g32540, was unaffected (Fig. 2B).
Our results indicate that the tld1-D phenotype is a result of the activation of Os11g32520 and Os11g32510 expression. Os11g32510 and Os11g32520 are both predicted to encode GH3-like proteins (http://signal.salk.edu/cgi-bin/RiceGE); however, neither of them encodes a complete GH3 protein. Os11g32520 contains the first two of three conserved motifs in GH3 proteins, while Os11g32510 includes only the third conserved motif, as indicated by an alignment of their amino acid sequences with those of other GH3 proteins.
We tried using several different pairs of primers to amplify the coding sequences of Os11g32510 and Os11g32520 using cDNA as the template. In the end, a large fragment about 2 kb in length was successfully amplified using the forward primer P1f and the reverse primer P1r; this fragment was named TLD1. A smaller fragment about 1.4 kb in length was also amplified using the P1f forward and P2r reverse primers (Fig. 2A); this fragment was designated as TLD2. Sequencing of the fragments revealed that TLD1 was alternatively spliced from four exons of the two genes. Exons 1 to 3 were from Os11g32520, while exon 4 was from Os11g32510 (Fig. 2, A and C). The splice site at the 3′ end of exon 3 (E3) in Os11g32520 is located 82 bp upstream of the stop codon (Fig. 2D, left panel, asterisk-labeled nucleotide [G]). Thus, the third exon (E3-I) in TLD1 is 82 bp shorter than the E3 from Os11g32520. As predicted, TLD2 was spliced from the three exons of Os11g32520 (Fig. 2, A, C, and D, right panel). Our results suggest that Os11g32520 and Os11g32510 together produce two splice variants, each possessing an open reading frame (ORF) with sequence identity at their 5′ ends; however, they are very different at their 3′ ends (Supplemental Fig. S3). Moreover, no mRNA is produced from Os11g32510, which contains an ORF.
Confirmation of the Functions of TLD1 and TLD2 in Rice and Arabidopsis
Protein sequence alignments revealed that TLD1 is closely related to GH3 proteins with known biological functions and includes three conserved motifs. TLD2 is a truncated version of TLD1, lacking 193 amino acids at its C-terminal end and containing only the first two conserved motifs (Supplemental Fig. S4).
To ascertain which variant is a functional protein and produces the tld1-D phenotype when activated, we generated transgenic rice plants harboring the vector 35S:TLD1/TLD2-cMyc to drive the overexpression of TLD1 and TLD2, respectively. Of the six lines produced, TLD1-6 was extremely short and died at the T0 generation without setting seed; in comparison, TLD1-3 and -5 exhibited varying degrees of dwarfism and had an increased number of tillers with a larger leaf angle, similar to the phenotype of tld1-D. In contrast, TLD1-1 and TLD2-3 looked like wild-type plants (Fig. 3, A–C). In agreement with the observed phenotypes, the greatest accumulation of the TLD1-cMyc fusion protein was noted in TLD1-5, which had a severe phenotype, whereas less accumulation was noted in TLD1-3, which had a milder phenotype; no overexpression was detected in the wild-type-like plant TLD1-1. In contrast, TLD2-cMyc fusion protein expression was detected in the wild-type-like plant TLD2-3 (Fig. 3E, left panel). Our results indicate that TLD1, but not TLD2, is capable of recapitulating the phenotype of tld1-D and is therefore a functional protein in rice.
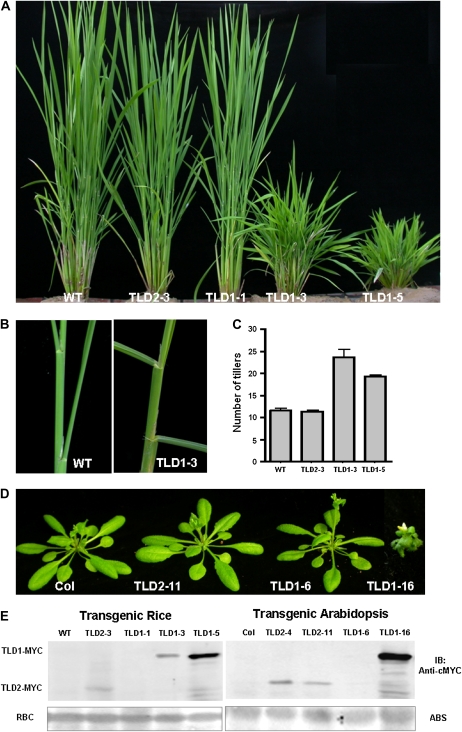
Figure 3.
Recapitulation of the tld1-D phenotype. A, Wild-type (WT), 35S:TLD1-MYC, and 35S:TLD2-MYC T1 rice plants at the tillering stage. Reappearance of the tld1-D phenotype was observed only in the TLD1-overexpressing lines. B, Enlarged view of the TLD1 overexpressers in A, showing the enlarged third and fourth leaf angles in TLD1-3 as compared with the wild type. C, Statistics for the number of tillers in the wild-type and transgenic lines shown in A (n = 6). D, The overexpression of TLD1 but not TLD2 in Arabidopsis causes severe morphologic alterations. E, Immunoblot assay of TLD1-cMyc and TLD2-cMyc protein expression in rice and Arabidopsis. ABS, Amido Black staining of Rubisco (RBC) was used as a loading reference. [See online article for color version of this figure.]
To further verify whether TLD1 functions as a GH3 protein in Arabidopsis, we transformed wild-type ecotype Columbia plants with the same overexpression vectors used in rice. A total of 16 out of 90 transgenic lines harboring 35S:TLD1-cMyc displayed a severe phenotype that included a dwarf-like appearance with downward-curving leaves, similar to GH3 activation mutants in Arabidopsis. Western blotting revealed the accumulation of the TLD1-cMyc fusion protein in all of the small plants, but not in the wild-type-like plants. However, not all of the 150 transgenic lines harboring the 35S:TLD2-cMyc construct exhibited an obvious phenotype, even though some of them overexpressed the TLD2-cMyc fusion protein, such as TLD2-11 (Fig. 3, D and E, right panel). These results confirm that TLD1, but not TLD2, functions as a GH3 protein genetically. In agreement with this, the in vitro biochemical assay demonstrates that TLD1, but not TLD2, has the IAA-conjugating activity (Fig. 4). Given these results, we next focus on functional studies of TLD1.
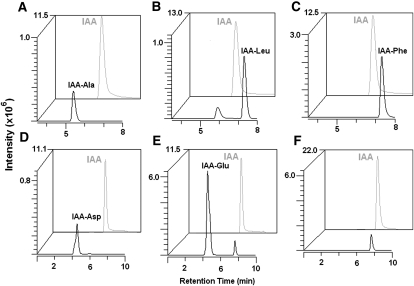
Figure 4.
In vitro assay of IAA-amido synthetase activity. The indicated IAA-amino acid conjugate species in individual reactions catalyzed by TLD1 (A–E) or TLD2 (F) recombinant protein were detected by HPLC-ESI-MS/MS at different MRM spectra (for optimal parameters for the MRM, see Supplemental Table S1; for details of the biochemical reaction, see “Materials and Methods”). Conjugated IAA (black peaks) and free IAA (gray peaks) intensities are shown in arbitrary units. In F, no IAA-Glu peak was observed when TLD1 in E was replaced by TLD2.
tld1-D Is Deficient in Endogenous IAA and Shows Reduced Sensitivity to Exogenous IAA
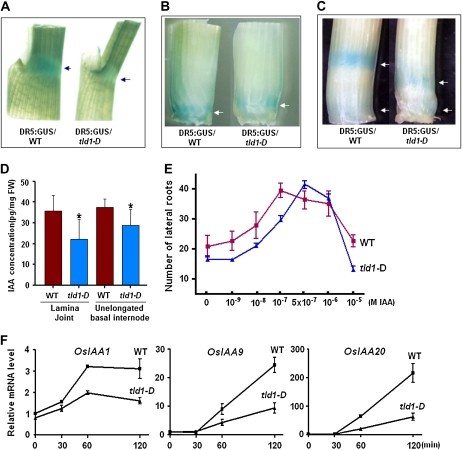
To determine whether the phenotypes we observed were caused by a deficiency in free IAA (active IAA), the auxin reporter DR5:GUS was introduced into wild-type and tld1-D plants and the level and distribution of active IAA were examined. In general, the level of GUS staining in the tld1-D seedlings was weaker than that detected in the wild type. GUS staining at the lamina joint, shoot apexes, and culm nodes were stronger in the wild type than in tld1-D, indicating that the local accumulated IAA maxima were reduced in the mutant plants (Fig. 5, A–C, arrows). We next collected tissue samples from the lamina joint and shoot apexes and measured the free IAA concentration by gas chromatography-mass spectrometry (GC-MS). Our results indicated lower free IAA concentrations in the tld1-D samples than in the wild-type samples (Fig. 5D), in agreement with our results using the DR5:GUS reporter. Additionally, three fewer vascular bundles were present in the culm, while fewer lateral and adventitious roots were observed in the tld1-D plants compared with wild-type plants, which is typical of IAA-deficient plants (Supplemental Fig. S5).
Figure 5.
The tld1-D mutant is deficient in endogenous IAA and shows reduced sensitivity to exogenous IAA. A to C, Auxin-responsive element DR5:GUS staining. The IAA maximum was reduced at the lamina joint in the second leaf (A, arrows), the unelongated basal internode of a 1-month-old seedling (B, arrows), and the nodes (C, top arrow, second node; bottom arrow, basal node) of tld1-D as compared with the wild type (WT). D, Quantification of free IAA in the lamina joint and unelongated basal internodes of 14-d-old seedlings by GC-MS. Bars with asterisks indicate P < 0.05. FW, Fresh weight. E, Effects of exogenous IAA on lateral root formation. The number of lateral roots per centimeter of adventitious root was counted (n = 60, from at least 20 seedlings). F, qRT-PCR analysis of auxin-responsive gene expression in 1-week-old seedlings treated with 10 μm IAA for the indicated times.
The decrease in endogenous IAA suggested increased IAA-conjugation activity in tld1-D. Therefore, we next investigated the ability of tld1-D to sequester exogenous IAA by analyzing the promotional effects of exogenous IAA on lateral root formation. The number of lateral roots was increased in the wild-type and tld1-D plants by IAA in a dose-dependent manner; however, the promotional effects of IAA on tld1-D, which peaked at 0.5 μm, were not obvious at concentrations below 1 nm. In comparison, in the wild-type plants, the number of lateral roots was highest at 0.1 μm IAA (Fig. 5E). These results suggest that tld1-D mutant plants are less sensitive than wild-type plants to exogenous IAA.
To further examine the reduced IAA sensitivity of tld1-D, the relative expression of several auxin rapid-response genes was analyzed at various time points following treatment with 10 μm IAA, as described previously (Jain et al., 2006). As shown in Figure 5F, OsIAA1, OsIAA9, and OsIAA20 were rapidly induced in both the wild-type and mutant plants; however, the level of induction was lower in tld1-D. These data demonstrate that TLD1/OsGH3.13 possesses IAA-conjugating activity in vivo. Moreover, the activation of TLD1 leads to reduced IAA maxima and the tld1-D phenotype.
Expression Pattern of TLD1 in Rice
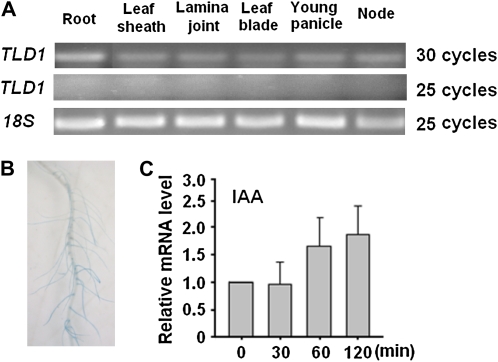
To understand the native roles of TLD1 in rice physiology and development, we analyzed the TLD1 expression pattern under normal conditions. Although TLD1 was expressed in all of the tissues examined, the level of transcription as determined by RT-PCR was low in the aboveground organs but relatively high in the roots (Fig. 6A). TLD1 promoter-driven GUS fusion protein expression was also used to examine the tissue-specific expression of TLD1 in greater detail. However, even with overnight staining, the GUS signal in most tissues was too weak to be visualized, except in the roots (Fig. 6B). TLD1 was rapidly induced by auxin, as indicated by quantitative (q)RT-PCR (Fig. 6C), indicating that it is an auxin early-response gene. The above results indicate that the promoter activity of TLD1 is extremely low in rice shoots and leads to a scarcity of TLD1 mRNA under normal growth conditions.
Figure 6.
Expression pattern of TLD1 in rice. A, RT-PCR analysis of the tissue-specific expression of TLD1. B, GUS staining of the roots of transgenic rice plants expressing ProTLD1:GUS. C, qRT-PCR analysis of TLD1 transcription in 1-week-old seedlings treated with 10 μm IAA.
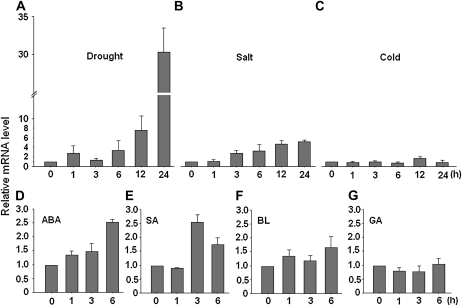
Based on the above results, we next considered the factors for TLD1 activation and the significance of that activation in rice. Wild-type seedlings subjected to various abiotic stresses showed a dramatic increase in the transcription of TLD1 compared with control seedlings: a nearly 30-fold increase was detected after 24 h of drought stress (Fig. 7A), whereas a 5-fold increase was observed following salt treatment (Fig. 7B). In contrast, cold stress exerted no obvious effect (Fig. 7C). We next analyzed the transcriptional activity of TLD1 in response to various plant hormones and found that TLD1 expression was induced by ABA and SA but not by brassinosteroids (BRs) or gibberellin (GA; Fig. 7, D–G). Based on these results, we conclude that the activation of TLD1/OsGH3.13 transcription might be positively correlated with drought or salt tolerance in rice.
Figure 7.
Induced expression of TLD1 in response to stress and phytohormone treatment. Two-week-old wild-type rice seedlings were treated for the indicated number of hours with drought (roots exposed to air without water; A), 200 mm NaCl (B), cold (4°C; C), 50 μm ABA (D), 500 μm SA (E), 10 μm 24-epibrassinolide (BL; F), or 50 μm GA (G). The mRNA expression of TLD1 relative to that at 0 h was determined by qRT-RCR.
Free IAA Is Decreased in tld1-D and Drought-Stressed Wild-Type Seedlings
The levels of free IAA, conjugated IAA species, and ABA, a hormonal marker of drought stress, were examined in whole tld1-D and wild-type seedlings with or without drought-stress treatment by HPLC/electrospray ionization (ESI)-MS/MS. As shown in Table I, free IAA was decreased while ABA was increased in tld1-D by about half compared with the control seedlings; the drought-stressed wild-type seedlings showed a similar response, although the magnitude of the change was greater. In addition, the levels of three types of IAA-amino acid conjugates, IAA-Ala, IAA-Phe, and IAA-Asp, but not IAA-Leu, were increased in the tld1-D seedlings. Among them, the contents of IAA-Ala and IAA-Phe were about twice that in the wild-type seedlings, while the levels of IAA-Asp were about three times the levels in the wild-type plants. Furthermore, during the drought treatment of the wild-type seedlings, the level of IAA-Ala and IAA-Asp were dramatically increased while the levels of IAA-Phe and IAA-Leu were also increased, but to a lesser extent. Overall, our results are in agreement with previous data showing that 24 h of drought stress dramatically activated the expression of TLD1/OsGH3.13.
Table I.
Quantification of the hormones in wild-type and tld1-D seedlings
Numbers in boldface are fold changes relative to wild-type/D0 control (untreated wild-type) seedlings; numbers in parentheses are amounts of hormone (ng g−1). D0 to D24, Drought treated for 0, 6, 12, or 24 h; n.d., not detected. The values are means ± sd of three to six independent samples.
| Hormone | Wild Type/D0 | tld1-D | Wild Type/D6 | Wild Type/D12 | Wild Type/D24 |
|---|---|---|---|---|---|
| IAA | 1 (14.932 ± 1.515) | 0.67 (10.021 ± 0.761) | 0.28 (4.175 ± 0.587) | 0.13 (1.899 ± 0.631) | n.d. |
| IAA-Ala | 1 (0.231 ± 0.117) | 2.58 (0.595 ± 0.096) | 10.04 (2.320 ± 0.177) | 16.66 (3.848 ± 0.330) | 33.58 (7.756 ± 1.253) |
| IAA-Phe | 1 (1.842 ± 0.431) | 2.00 (3.681 ± 0.663) | 1.44 (2.650 ± 0.845) | 1.95 (3.590 ± 0.213) | 5.13 (9.451 ± 1.590) |
| IAA-Leu | 1 (0.050 ± 0.017) | 0.76 (0.038 ± 0.009) | 0.92 (0.046 ± 0.015) | 1.20 (0.060 ± 0.020) | 4.02 (0.201 ± 0.057) |
| IAA-Asp | 1 (0.009 ± 0.003) | 3.11 (0.028 ± 0.005) | 4.88 (0.044 ± 0.006) | 9.11 (0.082 ± 0.007) | 27.78 (0.250 ± 0.012) |
| ABA | 1 (4.044 ± 0.923) | 1.49 (6.123 ± 0.033) | 94.21 (380.998 ± 1.737) | 46.70 (188.852 ± 5.580) | 33.35 (134.876 ± 3.829) |
tld1-D Shows Enhanced Drought Tolerance
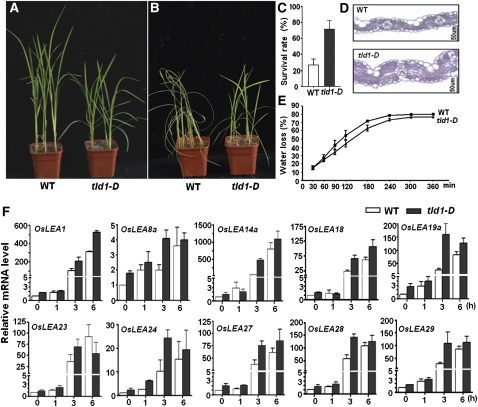
We next compared the drought and salt tolerance of 3-week-old tld1-D and wild-type rice seedlings and found that the tld1-D seedlings were more drought tolerant. About 70% of the tld1-D seedlings survived 7 d without watering, while only approximately 30% of the wild-type seedlings survived (Fig. 8, A–C). The rate of water loss was slower in tld1-D (s = 24) than in the wild-type plants (s = 28.8), especially within the first 3 h (Fig. 8E). The transcription of several drought-inducible LEA genes was dramatically increased in tld1-D compared with the wild type over the course of the treatment (Fig. 8F). Microscopic analysis of leaf blade cross sections revealed an increased number of cell layers in tld1-D (Fig. 8D), which may explain the reduced water loss in the mutant seedlings. On the other hand, no obvious improvement in salt tolerance was detected in tld1-D as compared with the wild-type seedlings (data not shown).
Figure 8.
Enhanced drought tolerance of tld1-D. A and B, Three-week-old wild-type (WT) and tld1-D seedlings were grown on soil before (A) and after (B) being subjected to drought stress for 5 d. Two representative pots of seedlings are shown. C, The survival rates of the tld1-D and wild-type plants were determined after 7 d of drought and 7 d of recovery (n = 60). D, Cross sections of wild-type and tld1-D leaves by microscopy. E, Water loss from 3-week-old tld1-D and wild-type seedlings. F, qRT-PCR analysis of LEA expression in wild-type and tld1-D plants during drought-stress treatment.
tld1 Shows No Difference from Wild-Type Rice Plants in Normal Growth and Drought Stress
To clarify if TLD1/GH3.13 is necessary for the regulation of plant architecture and drought tolerance, the TLD1 loss-of-function mutant was analyzed as well. tld1 (PFG_2B-40268.L) harbored one T-DNA insertion at the third intron, which interfered with the splicing and led to little transcripts of TLD1 (Supplemental Fig. S7, B–D). In normal growth conditions, the aboveground organs of tld1 show almost the same architecture as those of wild-type plants (Supplemental Fig. S7A). In drought conditions, tld1 behaves like the wild type: both of them cannot survive a 7-d water deficit (Supplemental Fig. S8, A and B), and the water losses for both are almost the same (Supplemental Fig. S8C). Furthermore, the transcripts of LEAs in tld1 are not increased as much as in wild-type plants before and after drought stress (Supplemental Fig. S8D). As knockout of TLD1 does not cause the mutant to be more susceptible to drought stress than wild-type plants, there should be other GH3 members that function redundantly with TLD1/OsGH3.13. Indeed, TLD1 homologues OsGH3.1, -2, -4, and -8 are also induced by drought stress (Supplemental Fig. S9).
DISCUSSION
TLD1 Encodes OsGH3.13, a Functional IAA-Amido Synthetase Affecting Rice Architecture Establishment
In this study, we characterized tld1-D, a gain-of-function rice mutant characterized by an increased number of tillers, enlarged leaf angles, and dwarfism (Fig. 1), resulting from ubiquitin promoter insertion and the subsequent activation of TLD1 expression (Fig. 2). We cloned two cDNA splice variants, TLD1 and TLD2 (Fig. 2; Supplemental Fig. S3); however, only TLD1 was capable of recapitulating the tld1-D phenotype when overexpressed in wild-type rice plants (Fig. 3).
TLD1 encodes a complete OsGH3.13 protein, a new member of the GH3 family, which was identified through an extensive database search of the rice genome (Terol et al., 2006). Of the 13 GH3 genes known in rice, only OsGH3.8 and OsGH3.1 have been functionally characterized; both genes are involved in disease resistance to rice pathogens (Ding et al., 2008; Domingo et al., 2009). In this study, we verified that OsGH3.13 cDNA is processed from four exons located at two loci: Os11g32520 and Os11g32510 (http://signal.salk.edu/cgi-bin/RiceGE; Fig. 2). The sizes of exons 2, 3 (E3-I in TLD1), and 4 were as predicted by Terol et al. (2006), which confirms the presence of a splicing site between exons 3 and 4 (Fig. 2, C and D, asterisk in left panel). In contrast, exon 1 has 99 more bp than predicted at its N terminus; translation may be initiated at the first in-frame ATG (Supplemental Fig. S3, boxed). Thus, the ORF of TLD1 contains 1,986 nucleotides, which makes it 99 bp longer than the predicted length of OsGH3.13, encoding a protein with 662 amino acids. Overexpression of the full-length coding sequence of TLD1 as a 90-kD cMyc fusion protein in rice and Arabidopsis, which was confirmed by western blotting using anti-cMyc antibodies (Fig. 3E), revealed the GH3 function of the protein, suggesting that the longer ORF of OsGH3.13 might be correct.
TLD1 contains all of the conserved regions of GH3 proteins (Terol et al., 2006) and has adenylation activity both in vivo (Table I) and in vitro (Fig. 4, A–E). TLD2 lacks 193 amino acids at its C-terminal end, corresponding to the third consensus motif (Supplemental Fig. S4), has no adenylation activity (Fig. 4F), and cannot recapitulate the tld1-D phenotype (Fig. 3). We conclude that this C-terminal segment is essential for the function of TLD1/OsGH3.13 in determining the architecture of rice plants and that the presence of the third motif may be prerequisite for IAA-adenylation activity, as most known functional GH3 proteins show no sequence variation in this region (Supplemental Fig. S4).
The Alterations in Architecture in tld1-D Are Caused by Reduced IAA Maxima
The constitutive activation of GH3s results in severe dwarf plants, as has been shown in Arabidopsis mutants dfl1-D (GH3.6), ydk1-D (GH3.2), and wes1-D (GH3.5; Nakazawa et al., 2001; Takase et al., 2004; Park et al., 2007a; Zhang et al., 2007) and OsGH3.8 and OsGH3.1 overexpression rice mutants (Ding et al., 2008; Domingo et al., 2009); however, no in-depth analysis of the alterations in architecture caused by the overexpression of these genes has been reported. We produced several lines of evidence showing that the activation of TLD1/OsGH3.13 in rice results in endogenous IAA deficiency (Table I) and reduced sensitivity to exogenous IAA (Fig. 5). The reduced IAA maxima at the shoot apexes, lamina joint, and nodes in tld1-D (Fig. 5) led to an increased number of tillers, enlarged leaf angles, and dwarfism (Figs. 1 and 3, A–C).
The IAA maxima at the basal nodes in rice, which are the result of basipetal transport, contribute to the repression of tiller bud outgrowth at the seedling stage, as illustrated by the auxin efflux carrier knockdown mutant OsPIN1, in which reduced IAA transport leads to tiller outgrowth (Xu et al., 2005). In this study, the reduced IAA maxima observed at the shoot apexes or basal nodes due to the conversion of free IAA to its inactive form in the TLD1/OsGH3.13 overexpressers also resulted in excessive tillering (Figs. 1, 3, A and C, and 5, B and D). This finding supports the suggested role of IAA homeostasis in the control of rice tillering; axillary meristem initiation and tiller bud outgrowth both appeared to be enhanced in tld1-D compared with the wild-type plants.
Leaf angle is another agronomic trait affecting rice plant architecture. Erect leaves are advantageous for shade avoidance, particularly in response to high-density planting. The plant hormone BR is known to affect leaf angle. The BR-deficient and -insensitive rice mutants d2 and d61 have erect leaves (Yamamuro et al., 2000; Hong et al., 2003), and exogenously supplied BRs induce lamina bending. IAA has also been shown to induce lamina bending at a high concentration (Takeno and Pharis, 1982); for example, the large leaf angles observed in the mutant la1-ZF802 might be caused by enhanced auxin transport (Li et al., 2007). Here, we showed that a reduction in the IAA maximum at the lamina joint perturbs the distribution of IAA, as visualized by DR5:GUS staining (Fig. 5, A and D), and thus impairs tissue patterning (Supplemental Fig. S6), leading to lamina inclination (Figs. 1, C and E, and 3, A and B). Based on our results, we conclude that in addition to BRs, leaf angle is determined by the distribution of IAA at the lamina joint, but not simply by the quantity of IAA. Any perturbation in IAA homeostasis (i.e. either an increase or a decrease in the IAA concentration) will result in changes in leaf angle.
Dwarfism in tld1-D mutant plants results from the uniformly shortened length of each internode. The culms displayed a dn-type mutation (for reference, see Takeda, 1977), similar to GA-related dwarf mutants (Hong et al., 2004). Correlated with these shortened internodes, DR5:GUS staining revealed that the IAA maximum at each node in tld1-D was significantly reduced (Fig. 5C). Internode elongation depends on cellular proliferation at the node and intercalary meristem immediately above the node, in addition to cell elongation at the upper region of the zone of division (Uozu et al., 2000). Having the proper auxin maximum is required for cell division and thus meristem formation. We believe that the decreased nodal concentration of IAA in the culm of tld1-D impaired active cell division, resulting in impaired culm elongation.
In addition to the alterations listed above, tld1-D also exhibited a fine culm, slender seeds, short panicles, and a reduced number of lateral roots. Taken together, our data indicate that these pleiotropic abnormalities in development were the consequence of the reduced IAA maximum. Therefore, the expression of TLD1 in rice is greatly suppressed in aboveground organs in order to maintain a reasonable IAA concentration and distribution that will support normal growth and development. That is why the loss of function of TLD1 does not show any visible phenotype in aboveground organs (Supplemental Fig. S7).
Down-Regulation of the IAA Concentration by TLD1/OsGH3.13 Is an Important Event in Drought-Stress Adaptation
Although inactive under normal growth conditions (Fig. 6A), TLD1/OsGH3.13 was highly induced by drought stress and ABA treatment (Fig. 7, A and D). Accordingly, the level of free IAA declined rapidly while the level of IAA-amino acid conjugates was increased in response to drought stress (Table I). Therefore, TLD1/OsGH3.13 may function under drought stress to down-regulate the free IAA concentration. Drought tolerance testing revealed an increased survival rate and reduced water loss in tld1-D compared with the wild type (Fig. 8, A–C and E), and the constitutive activation of TLD1/OsGH3.13 in tld1-D lowered the free IAA level to about half that in the wild-type control (Table I). These findings confirm the positive role of TLD1/OsGH3.13 in drought-stress adaptation. Similarly, positive roles for Arabidopsis GH3.5 in abiotic stress have been described (Park et al., 2007a).
Drought-stress adaptation in plants involves molecular and cellular events as well as physiological and biochemical events. One group of drought-inducible gene products, the LEA proteins, may function as protective macromolecules in detoxification and the alleviation of cellular damage during dehydration (Shinozaki and Yamaguchi-Shinozaki, 2007). Expression of the barley (Hordeum vulgare) LEA gene HVA1 in rice confers tolerance to water deficiency (Xu et al., 1996). The increased mRNA levels of many LEAs in tld1-D provide a molecular hallmark for the mutant's enhanced drought tolerance (Fig. 8F; gene names follow Wang et al., 2007). Most of the LEAs tested showed an increase in transcription that occurred more rapidly in tld1-D than in the wild type over the course of drought stress, implying that down-regulation of the IAA concentration facilitates the accumulation of LEAs. Among them, OsLEA1, OsLEA14a, OsLEA19a, OsLEA27, and OsLEA29 have been shown by microarray analysis to be highly induced by drought stress (Xiang et al., 2008). OsLEA19a has also been shown to enhance drought tolerance when overexpressed in rice (Xiao et al., 2007). Together, these results suggest that dynamic changes in the IAA concentration directly regulate drought-responsive LEA gene expression.
Alterations in architecture and tissue patterning due to the decrease in IAA in tld1-D might be another kind of adaptation to drought tolerance. For example, the thickened leaf blades of the mutant were advantageous in terms of preventing water loss at the seedling stage (Fig. 8D). Although the grain yield may be affected, it could be worthwhile for rice plants to survive drought stress at the expense of growth and development. In Arabidopsis, DELLA-induced growth repression is beneficial for plant survival in the face of changes in the natural environment (Achard et al., 2006). In rice, drought induces IAA/AUX expression, as indicated by microarray analysis, and causes the redistribution of auxin, as shown by a DR5:GUS reporter assay (Song et al., 2008), suggesting that the simultaneous dampening of IAA signaling and changes in the IAA concentration constitute an appropriate response to drought stimuli. We believe that down-regulation of the IAA concentration in rice is another important event in the switch from normal growth to drought adaptation, with TLD1/OsGH3.13 as a key regulator. Water deficiency may trigger the activation of TLD1/OsGH3.13 and four other members, causing a decrease in free IAA, leading to the accumulation of drought-inducible functional genes, such as LEAs, and drought tolerance. Concomitantly, alterations in plant architecture or tissue patterning may also contribute to drought tolerance.
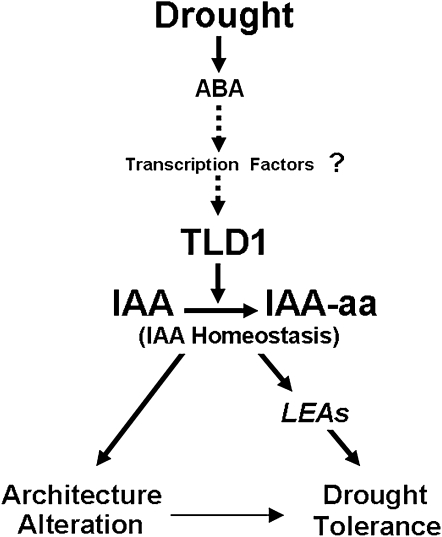
It is interesting that TLD1/OsGH3.13 is inactive under normal growth conditions but activated under drought stress. An analysis of the 2.3-kb promoter region of TLD1/OsGH3.13 revealed multiple cis-acting elements, including ARF-binding sites (TGTCTC), cytokinin-responsive elements (NGATT), and drought-responsive elements, including ABRE-like (for ABA-responsive element-like; ACGTG), MYBRS-like (WAACCA), and MYCRS (CNAATG; http://www.dna.affrc.go.jp/PLACE/) elements, which are thought to be the targets of ABA-inducible transcription factors (Shinozaki and Yamaguchi-Shinozaki, 2007). We suppose that endogenous ABA mainly contributes to the induction of TLD1/OsGH3.13. First, the 6-h drought stress dramatically induces endogenous ABA content (Table I); second, exogenous ABA treatment also induces the TLD1/OsGH3.13 mRNA level for 6 h (Fig. 7D); third, significant drought induction of TLD1/OsGH3.13 takes a long time (12–24 h; Fig. 7A). Considering the cis-elements analyzed above together, we proposed a working model in which an ABA-dependent system may govern drought-inducible TLD1/OsGH3.13 (Fig. 9). Recently, it was reported that ABA induction of GH3.3 and GH3.5 genes in Arabidopsis depends on the R2R3-type MYB transcription factor MYB96, which can act as a molecular link to mediate ABA-auxin cross talk in drought stress (Seo et al., 2009). This shed light on the upstream activators of TLD1/OsGH3.13 in rice. The identification of the upstream repressor and activator of TLD1/OsGH3.13 will help elucidate the dual roles of OsGH3.13 in growth and drought tolerance and will be useful for optimizing crop design in the future.
Figure 9.
Working model for TLD1 function in response to drought stress. Water deficiency triggers TLD1/OsGH3.13 expression through an ABA-dependent system, causing the alteration of IAA homeostasis, leading to the accumulation of drought-inducible functional genes, such as LEAs, and drought tolerance. Concomitantly, alterations in plant architecture or tissue patterning may also contribute to drought tolerance. aa, Amino acid.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The transgenic and wild-type rice (Oryza sativa ‘Nipponbare’) plants were grown in the field under natural conditions or in plant boxes containing a hydroponic culture solution in a greenhouse at 24°C to 30°C (16 h of light/8 h of darkness). The TLD1 T-DNA insertional mutant tld1 (PFG_2B-40268.L) is in the Hwayoung background and was obtained from the POSTECH Biotech Center. Arabidopsis (Arabidopsis thaliana) was grown in a growth room at 22°C under a 16-h/8-h light/dark cycle.
Vector Construction and Plant Transformation
The full-length ORFs of TLD1 and TLD2 were amplified from tld1-D cDNA, while the 2.3-kb promoter sequence of TLD1 was amplified from genomic DNA using Primer STAR HS DNA polymerase with GC buffer (Takara). The resulting fragments were cloned into pGEM-T Easy and sequenced. The ORFs of TLD1 and TLD2 were introduced into modified pCAMBIA1300 using XbaI and BamHI to generate 35S:TLD1/TLD2-7cMYC-6His. The TLD1 promoter was cloned into pCAMBIA1391Z using SalI and EcoRI to generate ProTLD1:GUS. The gene-specific primers used for cloning are listed in Supplemental Table S2. The DR5:GUS construct was generated as described previously (Scarpella et al., 2003). Transgenic rice plants were generated through Agrobacterium tumefaciens (strain EHA105)-mediated transformation (Yang et al., 2004).
RT-PCR and qRT-PCR
Total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer's instructions. RT-PCR analyses of TLD1, TLD2, Os11g32530, and Os11g32540 transcription were performed using the Takara RNA PCR (AMV) kit version 3.0 (Takara) with gene-specific primers (Supplemental Table S3). qRT-PCR analyses of gene expression were done using an ABI 7000 sequence detection system (Applied Biosystems) according to the manufacturer's protocol with SYBR Premix Ex Taq (Takara) and gene-specific primers (Supplemental Table S4). ACTIN (Os03g50890) was used as an internal control to normalize all data. Three independent experiments were performed, and representative results from a single experiment are shown. Each point is the mean ± sd of three parallel replicates.
Western-Blot Analysis
Total protein was extracted from rice or Arabidopsis seedlings with 2× SDS protein sample buffer and then separated by 10% SDS-PAGE. The blot carrying the transferred total proteins was probed with anti-cMyc monoclonal antibodies (Sigma-Aldrich). Anti-mouse IgG IRDye 800 conjugate (Rockland) was used as the secondary antibody.
Histological Analysis
Histochemical GUS analysis was performed as described (Jefferson, 1987). To create paraffin-embedded sections, the samples were fixed in formaldehyde-acetic acid at 4°C overnight, embedded in Paraplast Plus (Sigma-Aldrich) after dehydration in a graded series of ethanol, and sectioned at 8-μm intervals. The sections were stained with safranin and then photographed (Zeiss ImagerA1).
Plant Chemical and Abiotic Stress Treatments
Two-week-old rice seedlings were immersed in a hydroponic culture solution containing 10 μm IAA, 50 μm ABA, 500 μm SA, 10 μm 24-epibrassinolide, or 50 μm GA or exposed to cold (4°C), drought (no watering), or salt (200 mm NaCl) for the indicated times. The shoots of the plants were then collected for RNA isolation or hormone quantification.
To assess the effects of IAA on wild-type and tld1-D mutant rice, seeds were surface sterilized and sown on 0.5× Murashige and Skoog medium containing 0.25% phytagel and allowed to germinate for 1 d before being transferred to 0.5× Murashige and Skoog medium supplemented with various concentrations of IAA, as indicated in Figure 5E, for 6 d. The numbers of lateral roots were then calculated.
For drought-stress treatment, 3-week-old wild-type and tld1-D mutant plants grown in a mixture of sand and soil (1:1) were subjected to no watering for 7 d and then allowed to recover for 7 d before being analyzed for their survival rate. To measure the rate of water loss, excised leaves from five 3-week-old seedlings were exposed to air at room temperature (approximately 24°C) and weighed at the time points shown in Figure 8E.
Enzyme Assays
The coding sequences of TLD1 and TLD2 were amplified by PCR (for primers, see Supplemental Table S2) and cloned into pGEX2TK vector (Amersham Bioscience). Glutathione S-transferase (GST)-recombinant proteins were expressed in Escherichia coli BL21 codon plus strain with isopropylthio-β-galactoside induction and purified with Glutathione Sepharose 4B (Amersham Bioscience) according to the manufacturer's protocol. IAA-amido synthetase assay was performed as described earlier (Staswick et al., 2005). The reaction mixture (100 μL), containing 50 mm Tris-HCl, pH 8.7, 3 mm MgCl2, 3 mm ATP, 1 mm dithiothreitol, 1 mm IAA, 1 mm amino acid, and about 20 μg of purified GST-TLD1/GST-TLD2 fusion protein, was allowed to be incubated at 30°C for 12 h. The free IAA and conjugated IAA in a 10-μL mixture in each reaction were detected with HPLC-ESI-MS/MS at selected multiple reaction monitoring (MRM; for parameters, see Supplemental Table S1).
Hormone Quantification by HPLC-ESI-MS/MS and GC-MS
Two-week-old seedlings were used for hormone quantification according to the method of Matsuda et al. (2005). After extraction and purification, the dried samples were reconstituted in 200 μL of methanol:water:acetic acid (90:10:0.1, v/v) and filtered through a 0.45-μm polytetrafluoroethylene filter. Next, a 15-μL aliquot was analyzed using a Thermo Finnigan TSQ Quantum Ultra AM mass spectrometer equipped with an ESI source (Thermo). A reverse-phase Hypersil Gold C18 column (15 cm × 2.1 mm i.d., 5 μm; Thermo) was used in all analyses. For details, see Supplemental Materials and Methods S1.
The standards IAA, IAA-Ala, IAA-Phe, IAA-Asp, D2-IAA, and ABA were purchased from Sigma-Aldrich, while IAA-Leu was obtained from Wako. The optimal parameters for the MRM detection of IAA, D2-IAA, IAA conjugates, and ABA by LC-MS/MS are given in Supplemental Table S1.
To quantify the amount of free IAA by GC-MS, samples were prepared according to the method of Edlund et al. (1995). The analysis was carried out using a gas chromatograph-mass spectrometer (GC-TOF MS; Agilent/LECO) fitted with a capillary column (DA-35 ms; 30 m × 0.25 mm × 0.25 μm; Agilent).
The sequence data from this article can be found in the RiceGE database (http://signal.salk.edu/cgi–bin/RiceGE) under the following accession numbers: OsIAA1 (Os01g08320), OsIAA9 (Os02g56120), OsIAA20 (Os06g07040), OsLEA1 (Os04g49980), OsLEA8a (Os05g50710), OsLEA14a (Os01g50910), OsLEA18 (Os04g52110), OsLEA19a (Os05g46480), OsLEA23 (Os02g44870), OsLEA24 (Os03g45280), OsLEA27 (Os11g26760), OsLEA28 (Os11g26780), OsLEA29 (Os11g26790), OsGH3.1 (Os01g57610), OsGH3.2 (Os01g55940), OsGH3.4 (Os05g42150), and OsGH3.8 (Os07g40290).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Determination of the number of T-DNA insertions in tld1-D by Southern blotting.
Supplemental Figure S2. Productivity of the wild type versus tld1-D.
Supplemental Figure S3. The coding sequences of the TLD1 and TLD2 splice variants.
Supplemental Figure S4. Alignment of the deduced primary sequences of TLD1 and TLD2, together with members of the GH3 families in rice and Arabidopsis.
Supplemental Figure S5. Comparison of culm and root from wild-type and tld1-D plants.
Supplemental Figure S6. Comparison of the lamina joint in cross section between the wild type and tld1-D.
Supplemental Figure S7. Characterization of the TLD1 loss-of-function mutant.
Supplemental Figure S8. No difference between the wild type and tld1 under drought
Supplemental Figure S9. Differential expression of several OsGH3 members under salt, cold, and drought stress.
Supplemental Table S1. Optimal parameters for the MRM detection of plant hormones by LC-MS/MS.
Supplemental Table S2. Primers used for gene cloning.
Supplemental Table S3. Primers used for RT-PCR.
Supplemental Table S4. Primers used for qRT-RCR.
Supplemental Table S5. Primers used for tld1 identification.
Supplemental Materials and Methods S1. Detailed methods used for the quantification of IAA, IAA conjugates, and ABA by LC-MS/MS.
Supplementary Material
Acknowledgments
We are grateful to Lian-Feng Ai at the Hebei Entry-Exit Inspection and Quarantine Bureau of China for quantifying the plant hormones by HPLC-ESI-MS/MS; Zhi-Yong Wang at the Carnegie Institution of Washington for valuable discussions; Hong-Xuan Lin at the Institute of Plant Physiology and Ecology of the Chinese Academy of Sciences for critically reading the manuscript; Kang Chong and Zhen Xue at the Institute of Botany of the Chinese Academy of Sciences for providing the vector PTCK303 and quantifying the level of IAA by GC-MS; and Dr. Gynheung An, at POSTECH Biotech Center, Republic of Korea, for providing OsGH3.13 T-DNA insertion rice seeds.
This work was supported by the National Key Program on the Development of Basic Research in China (grant no. 2006CB100101), the National Program of High Technology Development of China (grant nos. 2002AA2Z1001–10, 2008ZX08009–003), and the Outstanding Researcher Program of Hebei in China.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ying Sun (yingsun@mail.hebtu.edu.cn).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311 91–94 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602 [DOI] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S (2008) Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo C, Andrés F, Tharreau D, Iglesias D, Talón M (2009) Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol Plant Microbe Interact 22 201–210 [DOI] [PubMed] [Google Scholar]
- Doust A (2007) Grass architecture: genetic and environmental control of branching. Curr Opin Plant Biol 10 21–25 [DOI] [PubMed] [Google Scholar]
- Edlund A, Eklof S, Sundberg B, Moritz T, Sandberg G (1995) A microscale technique for gas chromatography-mass spectrometry measurements of pictogram amounts of indole-3-acetic acid in plant tissues. Plant Physiol 108 1043–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmon CA, Tinsley AG, Ljung K, Sandberg G, Hearne LB, Liscum E (2006) A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc Natl Acad Sci USA 103 236–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jurgens G (2003) Efflux-dependent auxin gradients establish the apical basal axis of Arabidopsis. Nature 426 147–153 [DOI] [PubMed] [Google Scholar]
- Grieneisen V, Xu J, Marée AFM, Hogeweg P, Scheres B (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449 1008–1013 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T (1985) Rapid induction of selective transcription by auxins. Mol Cell Biol 5 1197–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-tanaka M, Matsuoka M (2004) Brassinosteroids and rice architecture. J Pestic Sci 29 184–188 [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Umemura K, Uoza S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Kaur N, Garg R, Thakur J, Tyagi A, Khurana J (2006) Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6 47–59 [DOI] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5 387–405 [Google Scholar]
- Li P, Wang Y, Qian Q, Fu Z, Wang M, Zeng D, Li B, Wang X, Li J (2007) LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res 17 402–410 [DOI] [PubMed] [Google Scholar]
- Liu Y, Chen Y, Zhang Q (2004) Amplification of genomic sequences flanking T-DNA insertions by thermal asymmetric interlaced polymerase chain reaction. In L Peña, ed, Transgenic Plants: Methods and Protocols, Vol 286. Humana Press, Totowa, NJ, pp 341–348 [DOI] [PubMed]
- Matsuda F, Miyazawa H, Wakasa K, Miyagawa H (2005) Quantification of indole-3-acetic acid and amino acid conjugates in rice by liquid chromatography-electrospray ionization-tandem mass spectrometry. Biosci Biotechnol Biochem 69 778–783 [DOI] [PubMed] [Google Scholar]
- McSteen P (2009) Hormonal regulation of branching in grasses. Plant Physiol 149 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M (2001) DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J 25 213–221 [DOI] [PubMed] [Google Scholar]
- Paciorek T, Friml J (2006) Auxin signaling. J Cell Sci 119 1199–1202 [DOI] [PubMed] [Google Scholar]
- Park J, Park J, Kim Y, Staswick P, Jeon J, Yun J, Kim S, Kim J, Lee Y, Park C (2007. a) GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem 282 10036–10046 [DOI] [PubMed] [Google Scholar]
- Park J, Seo P, Lee A, Jung J, Kim Y, Park C (2007. b) An Arabidopsis GH3 gene, encoding an auxin-conjugating enzyme, mediates phytochrome B-regulated light signals in hypocotyl growth. Plant Cell Physiol 48 1236–1241 [DOI] [PubMed] [Google Scholar]
- Qin G, Gu H, Zhao Y, Ma Z, Shi G, Yang Y, Pichersky E, Chen H, Liu M, Chen Z, et al (2005) Regulation of Arabidopsis leaf development by an indole-3-acetic acid carboxyl methyltransferase in Arabidopsis. Plant Cell 17 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C (2000) Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E, Rueb S, Meijer AH (2003) The RADICLELESS1 gene is required for vascular pattern formation in rice. Development 130 645–658 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Xiang F, Qiao M, Park JY, Lee Y, Kim SG, Lee YH, Park WJ, Park CM (2009) The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol 151 275–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58 221–227 [DOI] [PubMed] [Google Scholar]
- Song Y, Wang L, Xiong L (2008) Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta 229 577–591 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase T, Nakazawa M, Ishikawa A, Kawashima M, Ichikawa T, Takahashi N, Shimada H, Manabe K, Matsui M (2004) ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyls and root elongation. Plant J 37 471–483 [DOI] [PubMed] [Google Scholar]
- Takeda K (1977) Internode elongation and dwarfism in some gramineous plants. Gamma Field Symp 16 1–18 [Google Scholar]
- Takeno K, Pharis RP (1982) Brassinosteroid-induced bending of the leaf lamina of dwarf rice seedlings: an auxin mediated phenomenon. Plant Cell Physiol 23 1275–1281 [Google Scholar]
- Terol J, Domingo C, Talón M (2006) The GH3 family in plants: genome wide analysis in rice and evolutionary history based on EST analysis. Gene 371 279–290 [DOI] [PubMed] [Google Scholar]
- Uozu S, Tanaka-Ueguchi M, Kitano H, Hattori K, Matsuoka M (2000) Characterization of XET-related genes of rice. Plant Physiol 122 853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhu H, Jin G, Liu H, Wu W, Zhu J (2007) Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.). Plant Sci 172 414–420 [Google Scholar]
- Wang Z, Chen C, Xu Y, Jiang R, Han Y, Xu Z, Chong K (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22 409–417 [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye H, Xiong L (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148 1938–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Huang Y, Tang N, Xiong L (2007) Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet 115 35–46 [DOI] [PubMed] [Google Scholar]
- Xu D, Duan X, Wang B, Hong B, Ho T, Wu R (1996) Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol 110 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Zhu L, Shou H, Wu P (2005) A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol 46 1674–1681 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T (2007) Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143 1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12 1591–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Peng H, Huang H, Wu J, Jia S, Huang D, Lu T (2004) Large-scale production of enhancer trapping lines for rice functional genomics. Plant Sci 167 281–288 [Google Scholar]
- Zhang Z, Li Q, Li Z, Staswick PE, Wang M, Zhu Y, He Z (2007) Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol 145 450–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.