Abstract
A number of hypotheses have been suggested to explain why invasive exotic plants dramatically increase their abundance upon transport to a new range. The novel weapons hypothesis argues that phytotoxins secreted by roots of an exotic plant are more effective against naïve resident competitors in the range being invaded. The common reed Phragmites australis has a diverse population structure including invasive populations that are noxious weeds in North America. P. australis exudes the common phenolic gallic acid, which restricts the growth of native plants. However, the pathway for free gallic acid production in soils colonized by P. australis requires further elucidation. Here, we show that exotic, invasive P. australis contain elevated levels of polymeric gallotannin relative to native, noninvasive P. australis. We hypothesized that polymeric gallotannin can be attacked by tannase, an enzymatic activity produced by native plant and microbial community members, to release gallic acid in the rhizosphere and exacerbate the noxiousness of P. australis. Native plants and microbes were found to produce high levels of tannase while invasive P. australis produced very little tannase. These results suggest that both invasive and native species participate in signaling events that initiate the execution of allelopathy potentially linking native plant and microbial biochemistry to the invasive traits of an exotic species.
Invasive weeds are a major source of agricultural costs due to reduced productivity and the labor expended for weed control. In addition, the extensive use of herbicides to control weed populations has undesirable environmental consequences. Therefore, understanding mechanisms that facilitate exotic plant dispersal and displacement of natives in new ranges is critical to predicting and controlling invasions and may yield insights into the ecological processes that govern homeostasis and perturbation in natural plant communities.
Phragmites australis (Cav.) Trin ex. Steud. (common reed) has been present in the United States for at least 10,000 years as a major component of mixed tidal wetland plant communities (Saltonstall, 2002). However, over the past 200 years its distribution and abundance has expanded rapidly and it is now considered one of the most aggressive invasive species in marsh communities in North America. Chloroplast DNA analysis has shown that 13 native North American Phragmites haplotypes exist, while invasive populations possess a single chloroplast DNA haplotype (M) that is also widespread in Europe and Asia (Saltonstall, 2002). These data are supported by nuclear microsatellite DNA analysis (Saltonstall, 2003) and morphological differences that distinguish native, noninvasive from exotic, invasive Phragmites in North America (Saltonstall et al., 2004). When grown under the same conditions, exotic Phragmites has significantly higher aboveground and belowground biomass than native Phragmites (Vasquez et al., 2005; Saltonstall and Stevenson, 2007), and this pattern is typically observed under field conditions as well although exceptions exist (League et al., 2006; Meadows and Saltonstall, 2007). Unfortunately today, only remnant native P. australis populations remain along the Atlantic Coast of North America, indicating the near total displacement of native populations by exotic P. australis.
Various hypotheses have been forwarded to explain the rapid invasion of P. australis, of which human activities, stress regimes, and hydrologic disturbances have received the greatest attention (Chambers et al., 1999). Compared to invasion in terrestrial ecosystems, invasiveness in marsh communities is less well documented and it is still not clear how environmental factors relate to the establishment of specific dominant marsh species. Although allelopathy has been superficially suggested as the main displacing mechanism in P. australis (Kaneta and Sugiyama, 1972; Drifmeyer and Zieman, 1979), there has been minimal success in characterizing the responsible allelochemical. Interestingly, three triterpenoids (β-amacin, taraxerol, and taraxerone) and a flavone (tricin) have been identified from aerial portions of P. australis (Kaneta and Sugiyama, 1972; Drifmeyer and Zieman, 1979). Regrettably, none of these identified chemicals were tested for possible allelopathic activity.
Previously, we showed that a root exudate component of P. australis roots inhibits seedling growth, and that production of the exudates is higher in the invasive P. australis haplotype (Rudrappa et al., 2007). The active fraction of this exudate was found to be composed of gallic acid (3,4,5-trihydroxybenzoic acid). Gallic acid is toxic to a variety of weeds, crop plant species, and the model plant species Arabidopsis (Arabidopsis thaliana; Rudrappa et al., 2007; Rudrappa and Bais, 2008). Our published results also show the persistence of gallic acid in soil extracts from P. australis-invaded fields, which validates our in vitro results and strongly supports the idea that P. australis' invasive behavior may partly be due to the exudation of gallic acid in the soil/marsh (Rudrappa et al., 2007). Our studies concur with the earlier established reports of phytotoxicity and persistence of gallic acid in soil (Weidenhamer and Romeo, 2004).
Biochemically, the transition from simple galloylglucoses to complex gallotannins is marked by addition of further galloyl moieties to the pentagalloylglucose (Niemetz and Gross, 2005). It is now known that free gallic acid is released from complexed gallotannins by simple hydrolysis reactions, wherein a tannase activity breaks gallate ester to form free gallic acid, ellagic acid, and Glc (Mahoney and Molyneux, 2004). Treatment of fungal tannase from Aspergillus flavus results in hydrolysis of pellicle-localized gallotannin to form gallic acid, and ellagic acid as two phenolic components (Mahoney and Molyneux, 2004). As gallic acid is often complexed as gallotannins (Niemetz and Gross, 2005), we speculated that plant- or microbial-derived tannase may facilitate free gallic acid release in salt marsh soils.
Aside from allelopathy, invasive plants may deleteriously affect interactions between rhizospheric microbial communities and native plant species (Klironomos, 2002; Wardle et al., 2004; Callaway et al., 2008) to promote their expansion in new ranges. One specific example is the disruption of interactions between native species and their arbuscular mycorhizae, upon which the native species rely for nutrient acquisition (Stinson et al., 2006). Another recent study suggests that the recruitment or establishment of an altered soil microbial community may negatively impact the ability of native species to survive in the same soils (Batten et al., 2008). Evidences suggest that soil biota have several effects on the success of invasive plants and the interactions are based in part on the biochemistry, i.e. novel biochemical weapons (Callaway and Ridenour, 2004). However, to our knowledge, no previous studies have directly tested whether P. australis or any other exotic plant may exploit the biochemical potential of native plant and microbial communities to release a phytotoxin (gallic acid) from a relatively benign precursor (gallotannin) in the rhizosphere. This report presents evidence that links native plant and microbial biochemistry to the invasive traits of an exotic species.
RESULTS AND DISCUSSION
Gallotannin Levels in Exotic and Native P. australis Populations
To determine if gallotannin-derived gallic acid may play a significant role in the invasiveness of exotic P. australis, representative samples of exotic (16 populations) and native (eight populations) populations were analyzed for total gallotannin levels. The exotic P. australis showed high endogenous and rhizospheric titers of total gallotannins (194–265 μm g fresh weight [FW]−1; Table I) and consistently elevated levels (11–50 μm g FW−1) of free gallic acid compared to the native populations. Our data supports the fact that the increased gallotannin titers in exotic P. australis populations may enhance the release of free gallic acid in the rhizosphere of exotic strains.
Table I.
Mean total gallotannin (GT) and free gallic acid (GA) contents in the rhizome and the rhizospheric soil samples of exotic and native P. australis collected from different populations in Delmarva Peninsula
Different letters on the columns indicate statistically significant difference between isolates. Means with common letters are not significantly different at P ≤ 0.05 according to Duncan's multiple range test. Pop, Population.
| GA μm/g FW |
GT μm/g FW |
|||
|---|---|---|---|---|
| Rhizome | Rhizospheric Soil | Rhizome | Rhizospheric Soil | |
| Native | ||||
| Pop 1 | 8.70 ± 0.42 c | 5.80 ± 0.28 d | 41.30 ± 0.99 c | 23.80 ± 0.85 b |
| Pop 2 | 11.20 ± 1.41 c | 7.40 ± 0.28 cd | 22.20 ± 7.64 c | 21.56 ± 4.19 b |
| Pop 3 | 9.90 ± 1.56 c | 7.60 ± 1.41 cd | 38.45 ± 17.47 c | 35.95 ± 1.20 b |
| Pop 4 | 10.90 ± 0.42 c | 9.50 ± 0.99 c | 47.60 ± 15.41 c | 38.75 ± 0.21 b |
| Exotic | ||||
| Pop 1 | 15.40 ± 0.28 b | 8.20 ± 0.28 cd | 240.31 ± 40.01 ab | 210.93 ± 0.41 a |
| Pop 2 | 11.30 ± 0.14 c | 14.80 ± 1.13 b | 213.45 ± 2.84 b | 228.70 ± 0.00 a |
| Pop 3 | 12.16 ± 0.62 bc | 22.50 ± 2.97 a | 194.61 ± 0.41 b | 238.80 ± 46.95 a |
| Pop 4 | 48.80 ± 3.39 a | 7.20 ± 0.57 cd | 263.70 ± 37.57 a | 254.90 ± 62.93 a |
Tannase Activity in Native North American Plants
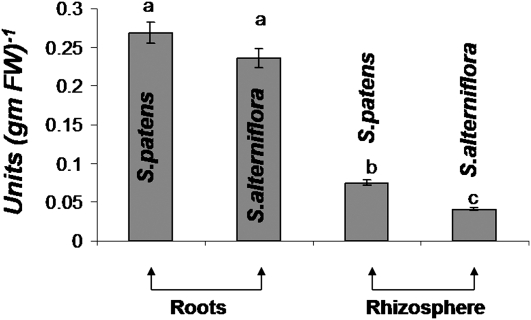
As gallic acid in soils is often complexed as gallotannins (Niemetz and Gross, 2001), we hypothesized that an exogenous source of tannase activity might liberate gallic acid from gallotannins excreted by P. australis into the rhizosphere. This could lead to prolonged toxicity on native plant species even after removal of exotic biomass. Specific tannase activity was measured in the roots and root secretions of native plant species whose ranges overlap with P. australis exotic populations. Tannase activity has been observed in many plant species, but only rarely in aquatic or semiaquatic plants. Spartina alterniflora and Spartina patens are native grasses that are often displaced by exotic P. australis populations in the mid-Atlantic region (Chambers et al., 1999). Interestingly, both native plant species showed high titers of total tannase activity in tissue (Fig. 1) and moderate titers of tannase activity in the rhizosphere (Fig. 1). Exotic P. australis populations only showed trace tannase activity in tissues (data not shown). This evidence suggests that tannase-mediated hydrolysis of polymeric gallotannin produced by exotic P. australis populations triggers the release of free gallic acid that subsequently acts as phytotoxin.
Figure 1.
Specific tannase activity in the root and rhizospheric samples of native plants S. alterniflora and S. patens. Tannase activity is expressed as enzyme units per milligram protein (per gram of FW). One enzyme unit is defined as the release of 1 μmol of gallic acid per minute. Different letters on the bars indicate a statistically significant difference between taxa.
Microbial Biochemistry and Plant Invasion
A recent study suggests that the recruitment or establishment of an altered soil microbial community by invasive plant species may negatively impact native species survival in the same soils either by direct pathogenesis or by enhancing the potency of phytotoxic secretions (Klironomos, 2002; Batten et al., 2006). Alternatively, the invasiveness of an exotic species may result from the fortuitous interaction between the secretions of an exotic species and the native soil microbial community in the new range as recently proposed by van der Putten et al. (2007). If exotic P. australis actively structures the microbial community upon invasion, one might expect that microbial tannase activity and gallotannin-degrading microbes might only be found in exotic P. australis stands. If the alternative is correct, then gallotannin degraders should be present in both native and exotic P. australis stands.
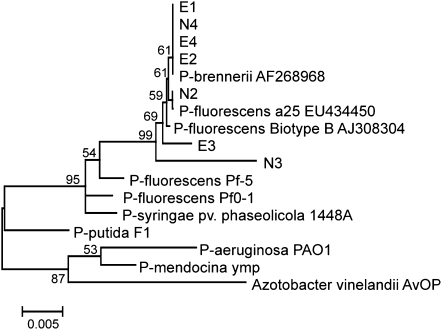
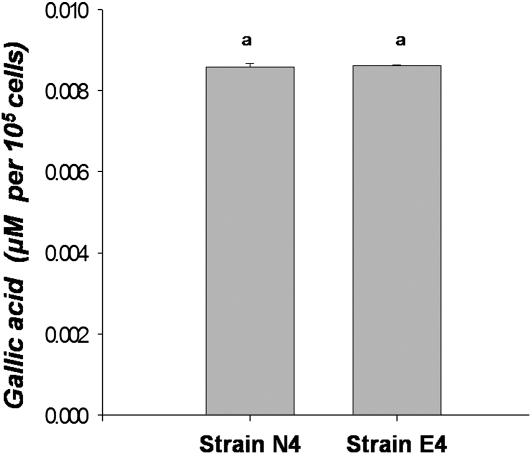
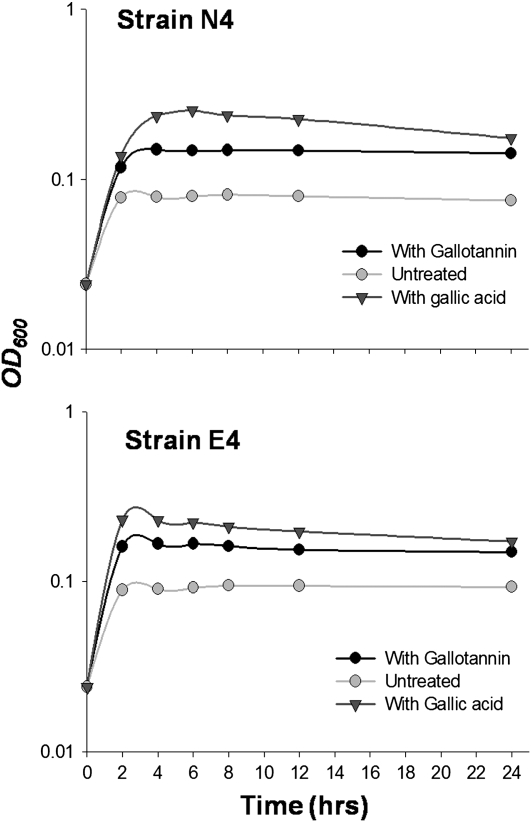
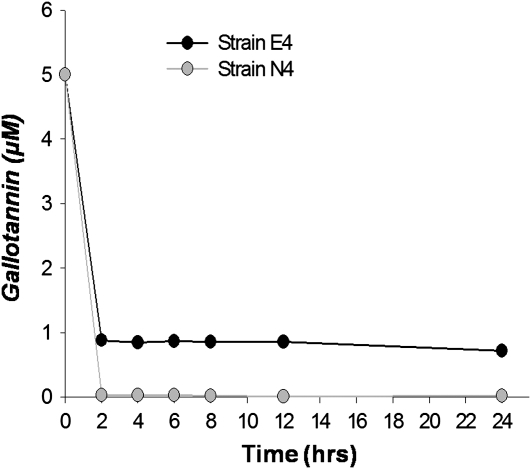
Bacterial strains were isolated from both exotic (16 populations) and native (eight populations) P. australis populations with a rich growth (Luria-Bertani agar) medium. The 16S rRNA gene sequence analysis of randomly selected isolates indicated that culturable microbial populations from both native and exotic P. australis roots are dominated by Pseudomonas spp. closely related to Pseudomonas fluorescens and Pseudomonas brennerii (Fig. 2). Tannin degradation has been observed in other Pseudomonas spp. (Chowdhury et al., 2004). Two of the eight isolates, one each from native (strain N4) and exotic (strain E4) P. australis populations, were assayed for tannase activity and their ability to utilize gallotannin and gallic acid as a carbon source in a minimal medium. Pseudomonas sp. strains from both native and exotic P. australis stands showed moderate tannase activity, though the bacterial tannase-specific activity was less than that obtained from the native plants (Fig. 3). Gallotannin and gallic acid stimulated the growth of both Pseudomonas sp. strains (Fig. 4) and gallotannin was degraded by both strains to similar extents (Fig. 5).
Figure 2.
Neighbor-joining phylogram obtained with 16S rRNA gene sequences of bacterial isolates from exotic (E1–E4) and native (N2–N4) P. australis rhizosphere samples. Numbers at nodes represent bootstrap support.
Figure 3.
Tannase activity in bacterial isolates from exotic (Pseudomonas sp. strain E4) and native (Pseudomonas sp. strain N4) P. australis rhizospheric samples. Specific enzymatic activity is represented as the mean amount of free gallic acid released (μm, ±sd, n = 16) from gallotannin (5 mg L−1) per 105 cells. Same letters on the bars indicate no statistical difference in between treatments.
Figure 4.
Growth curves for Pseudomonas spp. isolated from exotic (strain E4) and native (strain N4). After growth in rich medium, strains were subcultured in minimal medium with either gallotannin (5 mg L−1), gallic acid (5 mg L−1), or no added carbon.
Figure 5.
Degradation of gallotannin by Pseudomonas spp. isolated from exotic (strain E4) and native (strain N4) P. australis stands. Total gallotannin concentration was measured in the medium during growth (Fig. 4) of each strain on gallotannin as the sole carbon source.
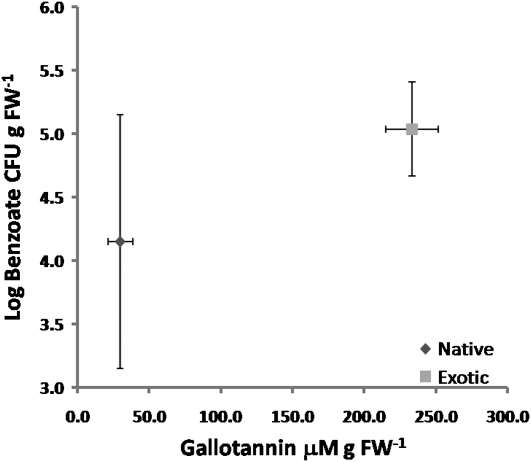
Microbial communities were quantitatively compared between native and exotic P. australis populations via most probable number (MPN) assays with benzoate as the sole carbon source to quantify aromatic acid-degrading microbes in four native and exotic rhizosphere samples with known gallotannin and gallic acid content. Benzoate was utilized as the carbon source because gallic acid is unstable over extended periods in culture media. Furthermore, benzoic acid is thought to be an intermediate in gallic acid degradation and therefore an intermediate in gallotannin degradation. Therefore, this experiment sought to determine if the microbial community was adapted to higher fluxes of aromatic acids from gallotannin in exotic P. australis stands. The native and exotic P. australis populations were found to group together when segregated by gallotannin content and the population size of the benzoate-degrading microbial community (Fig. 6). The differences were significant for both variables with ANOVA P values of 1 × 10−6 for gallotannin content and 0.055 for benzoate degrader community size. The appearance of the bulk rhizosphere samples was quite different among these populations with varying degrees of sand and clay (data not shown). We suspect that this contributed to higher variability in the MPN measurements for the native samples, while the lower degree of variability in the exotic populations for the MPN measurement may indicate a more constant selection for aromatic acid degradation. Gallic acid content was not significantly different in these four native and exotic populations (ANOVA, P = 0.172).
Figure 6.
Separation of native and exotic P. australis populations into coherent groups by benzoate-degrading microbial community size and gallotannin content. Data points are the means for four populations each of native and exotic P. australis. The error bars are the sd for MPN and gallotannin contents.
CONCLUSION
Based on these results, we conclude that the invasiveness of exotic P. australis species results, at least in part, from the increased levels of polymeric gallotannin that serve as a source of gallic acid production via the tannase activities of native microbial and plant species documented here. The limited information covering the two-way chemical interaction between invasive and native plants have largely been on resistance of native plants against invader toxins (Weir et al., 2006). Our work showed that the gallotannin content was higher in exotic P. australis populations relative to natives and that both native plants and microbes produce tannase activity that liberates gallic acid from gallotannins. As gallotannin- and gallic acid-degrading microbes were associated with both native and exotic P. australis populations and the population size of aromatic acid-degrading microbes was significantly higher in exotic rhizosphere samples that contained elevated gallotannin content. This observation suggests that the microbial community is quantitatively altered in the exotic P. australis rhizosphere and supports the hypothesis that the biochemistry of native plants and microbes may enhance the invasive potential of an exotic species.
The data lead us to hypothesize that the high levels of hydrolyzable tannins produced by exotic P. australis roots serve as a passive polymeric precursor that is hydrolyzed by native plant and microbial biochemistry and thereby clears range space for colonization. The data reported here are correlative from an environmental sampling effort. To directly test this hypothesis, experiments are under way to quantitatively examine P. australis-microbe-native plant interactions in microcosms and pure cultures.
To summarize, we propose an intriguing extension of both the novel weapons hypothesis (Callaway and Aschehoug, 2000) and spectrum of potential allelopathic interactions. This model suggests that inhibiting free gallic acid release in the rhizosphere could be targeted to mitigate P. australis phytotoxicity. The model also can explain the phytotoxic persistence of gallic acid even after the removal of invasive P. australis biomass. An open question related to this is how exotic P. australis strains resist gallic acid produced by competing native species and the native microbial community. This resistance is another potential target that may be attacked to mitigate exotic P. australis invasion. Root-derived biochemical changes driving plant invasion are reported in only a few groups of invasive plants. The information gained through this work will provide vital clues regarding how species become invasive and may lead to environmental and economic benefits through better management of invasive species.
MATERIALS AND METHODS
Plant Material
Spartina alterniflora and Spartina patens seeds were procured from Environmental Concern Inc., Native Plant Nursery. Phragmites australis rhizomes and soil core samples were collected from different locations on the Delmarva Peninsula. The populations (16 exotic populations and eight native populations) sampled in this study were all native (haplotype F) and exotic P. australis (haplotype M) strains (Meadows and Saltonstall, 2007). Soil sampling and rhizome analysis for gallic acid was performed as per the description provided in Rudrappa et al. (2007).
Plant Culture Conditions
Seeds were washed with double-distilled water three times, dehusked, and surface sterilized with 50% bleach (sodium hypochlorite) for 7 to 10 min followed by three to four washes with sterile double-distilled water. Seeds were cultured on Murashige and Skoog (1962) solid medium with 3% Suc and allowed to germinate for 5 to 10 d. Subsequently, seeds were incubated at 25°C under 16:8 h light/dark regime. For total tannase activity from Spartina spp., the seedlings were added to 100 mL Murashige and Skoog 3% Suc liquid medium in 250 mL Erlenmeyer flasks and incubated on a rotary shaker.
Gallotannin, Gallic Acid, and Tannase Assay
Quantitative analysis of gallic acid in the hydrolysate extract was used for the determination of total gallotannins (Ossipov et al., 1997). Between 100 and 200 mg of lyophilized rhizomes were extracted for 30 min with 1 mL of 70% (v/v) acetone/water per l00 mg sample at 4°C. The extract was vacuum filtered through a coarse-sintered glass filter into a vial. The filter was washed with 5 mL of 2 n H2SO4 that was added to the vial. The remaining solid portion was discarded. The vial was vacuum sealed, and the sample was heated for 4 h at 100°C. A similar hydrolysis procedure was used for samples of commercial tannic acid, using 1 mL of 2 n H2SO4/mg of tannic acid. The hydrolysate was diluted 10-fold with double-distilled water. In a tube 1.5 mL of 0.667% methanolic rhodanine was added to 1 mL of sample. After exactly 5 min, 1 mL of 0.5 n aqueous KOH solution was added. After 5 min the absorbance was recorded at 520 nm. Gallic acid accumulation and tannase activity was assayed spectrophotometrically by the methanolic-rhodanine method (Sharma et al., 2000) based on chromogen formation between gallic acid (released by the action of tannase on methyl gallate) and rhodanine (2-thio-4-ketothiazolidine). A 2 g plant sample was extracted in 0.05 m citrate buffer, pH 5.0, and frozen overnight at −20°C. The plant material was finely ground in a chilled pestle mortar kept in an ice bath. The homogenate was centrifuged at 12,000g for 30 min at 4°C. The supernatant was used for the tannase assay. The substrate solution (0.01 m methyl gallate prepared in 0.05 m citrate buffer, pH 5.0), enzyme sample, and buffer (0.05 m citrate buffer, pH 5.0) were preincubated at 30°C for 5 to 10 min before the enzyme reaction was started. The reaction mixture in the blank, test, and control tubes contained 0.25 mL of substrate solution to which 0.15 mL of buffer and 0.25 mL of enzyme sample were added to the blank and test, respectively. The tubes were incubated at 30°C for 5 min, and 0.3 mL of methanolic rhodanine (0.667%; w/v) was added to all the tubes that were then placed in 30°C for 5 min. Next, 0.2 mL of 0.5 n potassium hydroxide was added to each tube, and these were incubated at 30°C for 5 min. The absorbance was recorded at 520 nm using a spectrophotometer. Gallic acid was obtained from Sigma-Aldrich, gallotannin from VWR, rhodanine from MP Biomedicals, methyl gallate from Pfaltz & Bauer, and tannase was obtained from Fluka Biomedica.
16S rRNA Gene Sequencing and Analysis
Cells collected from well-isolated colonies of each isolate with a sterile pipette tip were used directly for the PCR amplification of 16S rRNA using the universal primers 519F and 1406R as previously described (Suzuki and Giovannoni, 1996). Amplification products were purified by a column-based kit (Qiagen) and directly sequenced with the amplification primers using standard automated sequencing protocols at the University of Delaware Sequencing and Genotyping Center. Sequences were combined into contigs and edited for quality in the VectorNTI software suite (Invitrogen) and then aligned to reference bacterial 16S rRNA gene sequences in MEGA4 (Tamura et al., 2007). The phylogenetic tree was determined by the neighbor-joining method with pairwise gap deletion over 1,000 bootstrap replicates.
Bacterial Growth Assay
Pseudomonas fluorescens (strain E4) and Pseudomonas brennerii (strain N4) isolated from exotic and native P. australis were grown on Luria-Bertani broth agar plates. A single colony from a freshly streaked plate was used to grow overnight cultures from which approximately 0.02 OD600 culture was prepared and used in all the experiments. In 96-well plates, 20 μL of the 0.02 OD600 culture was added to 200 μL of minimal growth medium (Wahlund and Madigan, 1995) for control wells. Gallic acid (5 mg L−1) and tannic acid (5 mg L−1) were added to individual wells of separate plates along with the previously mentioned bacteria titer and growth media.
MPN Analysis
Rhizosphere samples were resuspended in carbon-free minimal medium (Wahlund and Madigan, 1995) to a concentration of 0.1 g FW mL−1. This suspension was used to inoculate serial 10-fold dilution series in 96-well plates that contained minimal medium with sodium benzoate (1 g L−1) as the sole carbon source. Plates were incubated with humidified room air at room temperature for 5 d prior to scoring for growth and interpretation by standard MPN tables (de Man, 1983). Three independent-sample replicates were performed for each native and exotic population rhizosphere.
Statistical Analysis
All the data were averaged from two separate experiments and the data have been presented as means with ses of the means. The data were analyzed by one-way ANOVA using Microsoft Excel 2007 and post-hoc mean separation was performed by Duncan's multiple range test at P ≤ 0.05 (Harter, 1960) by using the software SPSS version 12.0.
Acknowledgments
The authors thank Drs. Janine Sherrier, Nicole Donofrio, Jed Fahey, Wim van der Putten, Judith Hough-Goldstein, and Kristin Saltonstall for critical reading of the manuscript.
This work was supported by the University of Delaware (to T.E.H.), the University of Delaware Research Foundation (to H.P.B.), and the Department of Science and Technology (India) for BOYSCAST fellowship (to G.B.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Harsh P. Bais (hbais@udel.edu).
Open Access articles can be viewed online without a subscription.
References
- Batten KM, Scow KM, Davies KF, Harrison SP (2006) Two invasive plants alter soil microbial community composition in serpentine grasslands. Biol Invasions 8 217–230 [Google Scholar]
- Batten KM, Scow KM, Espeland EK (2008) Soil microbial community associated with an invasive grass differentially impacts native plant performance. Microb Ecol 55 220–228 [DOI] [PubMed] [Google Scholar]
- Callaway RM, Aschehoug ET (2000) Invasive plants versus their new and old neighbors: a mechanism for exotic invasion. Science 290 521–523 [DOI] [PubMed] [Google Scholar]
- Callaway RM, Cipollini D, Barto K, Thelen GC, Hallett SG, Prati D, Stinson K, Klironomos J (2008) Novel weapons: invasive plant suppresses fungal mutualists in America but not in its native Europe. Ecology 89 1043–1055 [DOI] [PubMed] [Google Scholar]
- Chambers RM, Meyerson LA, Saltonstall K (1999) Expansion of Phragmites australis in to tidal wetlands of North America. Aquat Bot 4 261–273 [Google Scholar]
- Callaway RM, Ridenour WM (2004) Novel weapons: invasive success and the evolution of increased competitive ability. Front Ecol Environ 2 436–443 [Google Scholar]
- Chowdhury SP, Khanna S, Verma SC, Tripathi AK (2004) Molecular diversity of tannic acid degrading bacteria isolated from tannery soil. J Appl Microbiol 97 1210–1219 [DOI] [PubMed] [Google Scholar]
- de Man JC (1983) MPN tables, corrected. Appl Microbiol Biotechnol 17 301–305 [Google Scholar]
- Drifmeyer JE, Zieman JC (1979) Germination enhancement and inhibition of Distichlis spicata and Scirpus robustus seeds from Virginia. Estuaries 2 16–21 [Google Scholar]
- Harter LN (1960) Critical values for Duncan's new multiple range test. Biometrics 16 671–685 [Google Scholar]
- Kaneta M, Sugiyama N (1972) The constituents of Arthraxon hispidus, Miscanthus tinctorius and Phragmites australis. Bull Chem Soc Jpn 45 528–531 [Google Scholar]
- Klironomos JN (2002) Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 217 67–70 [DOI] [PubMed] [Google Scholar]
- League MT, Colbert EP, Seliskar DM, Gallagher JL (2006) Rhizome growth dynamics of native and exotic haplotypes of Phragmites australis (common reed). Estuaries 29 269–276 [Google Scholar]
- Mahoney N, Molyneux RJ (2004) Phytochemical inhibition of aflatoxigenicity in Aspergillus flavus by constituents of walnut (Juglans regia). J Agric Food Chem 52 882–889 [DOI] [PubMed] [Google Scholar]
- Meadows RE, Saltonstall K (2007) Distribution of native and non-native populations of Phragmites australis in oligohaline marshes of the Delmarva Peninsula and southern New Jersey. J Torrey Bot Soc 134 99–107 [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Niemetz R, Gross GG (2001) Gallotannin biosynthesis: β-glucogallin:hexagallolyl 3-O-galloyltransferase from Rhus typhina leaves. Phytochemistry 58 657–661 [DOI] [PubMed] [Google Scholar]
- Niemetz R, Gross GG (2005) Enzymology of gallotannin and ellagitannin biosynthesis. Phymtochemistry 66 2001–2011 [DOI] [PubMed] [Google Scholar]
- Ossipov V, Loponen J, Ossipova S, Haukioja E, Pihlaja K (1997) Gallotannins of birch Betula pubescens leaves: HPLC separation and quantification. Biochem Syst Ecol 25 493–504 [Google Scholar]
- Rudrappa T, Bais H (2008) Genetics, novel weapons and rhizospheric microcosmal signaling in the invasion of Phragmites australis. Plant Signal Behav 3 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrappa T, Bonsall J, Gallagar J, Seliskar DM, Bais HP (2007) Root secreted allelochemical in noxious weed Phragmites australis deploys a reactive oxygen species response and microtubule assembly disruption to execute rhizotoxicity. J Chem Ecol 33 1898–1918 [DOI] [PubMed] [Google Scholar]
- Saltonstall K (2002) Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proc Natl Acad Sci USA 99 2445–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltonstall K (2003) Microsatellite variation within and among North American lineages of Phragmites australis. Mol Ecol 12 1689–1702 [DOI] [PubMed] [Google Scholar]
- Saltonstall K, Peterson P, Soreng R (2004) Recognition of Phragmites australis subsp. americanus (Poaceae: Arundinoideae) in North America: evidence from morphological and genetic analyses. SIDA Contrib Bot 21 683–692 [Google Scholar]
- Saltonstall K, Stevenson JC (2007) The effect of nutrients on seedling growth of native and introduced Phragmites australis. Aquat Bot 86 331–336 [Google Scholar]
- Sharma S, Bhat TK, Dawra RK (2000) A spectrophotometric method for assay of tannase using rhodanine. Anal Biochem 279 85–89 [DOI] [PubMed] [Google Scholar]
- Stinson KA, Campbell SA, Powell JR, Wolfe BE, Callaway RM, Thelen GC, Hallett SG, Prati D, Klironomos JN (2006) Invasive plant suppresses the growth of native tree seedlings by disrupting belowground mutualisms. PLoS Biol 4 e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MT, Giovannoni SJ (1996) Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol 62 625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4. Mol Biol Evol 24 1596–1599 [DOI] [PubMed] [Google Scholar]
- van der Putten WH, Klironomos JN, Wardle DA (2007) Microbial ecology of biological invasions. ISME J 1 28–37 [DOI] [PubMed] [Google Scholar]
- Vasquez EA, Glenn EP, Brown JJ, Guntenspergen GR, Nelson SC (2005) Salt tolerance underlies the cryptic invasion of North American salt marshes by an introduced haplotype of the common reed Phragmites australis (Poaceae). Mar Ecol Prog Ser 298 1–8 [Google Scholar]
- Wahlund TM, Madigan MT (1995) Genetic transfer by conjugation in the thermophilic green sulfur bacterium Chlorobium tepidum. J Bacteriol 177 2583–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardle DA, Bardgett RD, Klironomos JN, Setala H, van der Putten WH, Wall DH (2004) Ecological linkages between aboveground and belowground biota. Science 304 1629–1633 [DOI] [PubMed] [Google Scholar]
- Weidenhamer JD, Romeo JT (2004) Allelochemicals of Polygonella myriophylla: chemistry and soil degradation. J Chem Ecol 30 1067–1082 [DOI] [PubMed] [Google Scholar]
- Weir TL, Bais HP, Stull VJ, Callaway RM, Thelen GC, Ridenour WM,Bhamidi S, Stermitz FR, Vivanco JM (2006) Oxalate contributes to the resistance of Gaillardia grandiflora and Lupinus sericeus to a phytotoxin produced by Centaurea maculosa. Planta 223 785–795 [DOI] [PubMed] [Google Scholar]