Abstract
BEL1-like transcription factors are ubiquitous in plants and interact with KNOTTED1 types to regulate numerous developmental processes. In potato (Solanum tuberosum subsp. andigena), the BEL1-like transcription factor StBEL5 and its Knox protein partner regulate tuber formation by targeting genes that control growth. RNA detection methods and heterografting experiments demonstrated that StBEL5 transcripts are present in phloem cells and move across a graft union to localize in stolon tips, the site of tuber induction. This movement of RNA originates in leaf veins and petioles and is induced by a short-day photoperiod, regulated by the untranslated regions, and correlated with enhanced tuber production. Assays for RNA mobility suggest that both 5′ and 3′ untranslated regions contribute to the preferential accumulation of the StBEL5 RNA but that the 3′ untranslated region may contribute more to transport from the leaf to the stem and into the stolons. Addition of the StBEL5 untranslated regions to another BEL1-like mRNA resulted in its preferential transport to stolon tips and enhanced tuber production. Transcript stability assays showed that the untranslated regions and a long-day photoperiod enhanced StBEL5 RNA stability in shoot tips. Upon fusion of the untranslated regions of StBEL5 to a β-glucuronidase marker, translation in tobacco (Nicotiana tabacum) protoplasts was repressed by those constructs containing the 3′ untranslated sequence. These results demonstrate that the untranslated regions of the mRNA of StBEL5 are involved in mediating its long-distance transport, in maintaining transcript stability, and in controlling translation.
As part of an elaborate long-distance communication system, plants have evolved a unique signaling pathway that takes advantage of connections in the vascular tissue, predominantly the phloem. This information superhighway has been implicated in regulating development, responding to biotic stress, delivering nutrients, and as a vehicle commandeered by viruses for spreading infections (Lough and Lucas, 2006). Numerous full-length transcripts have been identified in the sieve elements of several plant species (Asano et al., 2002; Vilaine et al., 2003; Omid et al., 2007; Deeken et al., 2008; Gaupels et al., 2008), but only six have been confirmed to be transported (Kehr and Buhtz, 2008). Of these, three are from pumpkin (Cucurbita maxima), CmGAIP, CmNACP, and CmPP16 (Ruiz-Medrano et al., 1999; Xoconostle-Cazares et al., 1999; Haywood et al., 2005); DELLA-GAI is from Arabidopsis (Arabidopsis thaliana; Haywood et al., 2005); PFP-LeT6 is from tomato (Solanum lycopersicum; Kim et al., 2001); and StBEL5, a BEL1-like transcription factor, is expressed in potato (Solanum tuberosum subsp. andigena; Banerjee et al., 2006a). Of this group, information is available for StBEL5 RNA on the role of the untranslated regions (UTRs) and photoperiod in mediating movement and on the function of the mobile RNA (Banerjee et al., 2006a). Gel mobility-shift assays have demonstrated the role of specific polypyrimidine tract-binding motifs in the phloem RNAs of pumpkin, CmGAIP and CmPP16-1, that mediate binding to RNA-binding proteins (Ham et al., 2009). In this system, phloem RNAs and the CmRBP50 protein provide the basis for a mobile ribonucleoprotein complex containing as many as 16 proteins.
In animals, mRNAs are transported within the cell to facilitate their function. With the extensively studied Staufen protein of Drosophila (St. Johnston et al., 1991; Ferrandon et al., 1994), binding to the bicoid and oskar mRNAs occurs to determine their localization in the egg cell, either anterior or posterior. This directed localization involves RNA-protein, RNA-RNA (Ferrandon et al., 1997), and protein-protein interactions (Elvira et al., 2006) during transport and repression of translation of the mRNAs. Repression of translation ensures that the mobile mRNAs are functional only at the target site (King et al., 2005). Commonly, it is the UTRs that function via protein interactions in facilitating movement of a transcript (Jansen, 2001), in mediating RNA stability (Derrigo et al., 2000; Lee and Jeong, 2006), or in regulating the efficiency of translation (Gualerzi et al., 2003; Barreau et al., 2006). Posttranscriptional regulation mediated by protein/mRNA complexes occurs widely among eukaryotes (Keene and Lager, 2005; Lee and Schedl, 2006; Sanchez-Diaz and Penalva, 2006; Keene, 2007; Pullmann et al., 2007; Halbeisen et al., 2008). Without protection, naked cellular RNAs quickly fall prey to degradative processes (Shyu et al., 2008).
While there are several reports on full-length mobile RNAs in plants (for review, see Kehr and Buhtz, 2008), there is a scarcity of information on the RNA sequence that mediates this process. These sequences form recognition stem/loop structures, designated “zip code elements.” These binding motifs have been identified in the RNAs of animals and function in recognizing RNA-binding proteins (for review, see Jansen, 2001). Whereas these zip codes may be located anywhere in the transcript, they are most predominant in the 3′ UTR (Macdonald and Kerr, 1997; Chartrand et al., 1999; Saunders and Cohen, 1999; Corral-Debrinski et al., 2000; Thio et al., 2000). Viroid RNA motifs have also been identified that facilitated selective RNA movement through plant cells (Qi et al., 2004; Zhong et al., 2007, 2008). These short sequences most likely mimic endogenous plant RNA motifs that are recognized by cellular factors for transport.
The mRNA of potato that is the focus of this study, designated StBEL5, is a member of the BEL1-like family of transcription factors. This family of transcription factors is ubiquitous among plant species, and these proteins interact with KNOTTED1 types to regulate numerous developmental processes (Müller et al., 2001; Chen et al., 2003; Smith and Hake, 2003; Kumar et al., 2007; Pagnussat et al., 2007; Kanrar et al., 2008; Ragni et al., 2008). In potato, the BEL1-like transcription factor, StBEL5, and its Knox protein partner, POTH1 (for potato homeobox 1), regulate tuber formation by mediating hormone levels in the stolon tip (Rosin et al., 2003; Chen et al., 2004). This change in hormone levels leads to the creation of a strong sink organ, the tuber, by activating cell division and enlargement of specific cells of the stolon tip (Xu et al., 1998; Hannapel et al., 2004). Upon this activation, abundant amounts of Suc are transported to the stolon tip for the synthesis of starch in the newly formed tuber.
RNA detection methods and heterografting experiments demonstrated that StBEL5 transcripts are present in phloem cells and move across a graft union to localize in stolon tips, the site of tuber induction (Banerjee et al., 2006a; Yu et al., 2007). This movement originates in leaf veins and petioles and is induced by a short-day photoperiod, regulated by the UTRs, and correlated with enhanced tuber production. Whereas photoperiod mediates the movement of StBEL5 RNA, activation of transcription of the StBEL5 gene in leaves is regulated by either red or blue light, regardless of photoperiod or light intensity (Chatterjee et al., 2007). This study demonstrates that one or both of the UTRs of the StBEL5 mRNA are involved in mediating its long-distance transport, in maintaining transcript stability, and in controlling its translation.
RESULTS
Effect of the UTRs of the StBEL5 RNA on Tuber Phenotype and Transcript Mobility
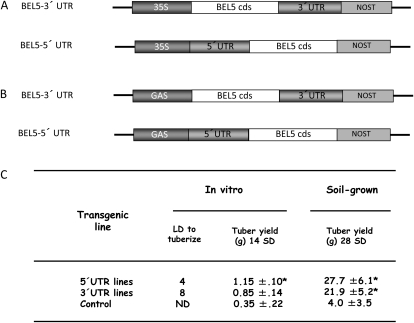
Previous work has shown that both UTRs of StBEL5 together affected RNA mobility (Banerjee et al., 2006a). To determine the relative contributions of these two distinct regions to transcript mobility, constructs were made with the BEL5 coding sequence (cds) with one or the other UTR attached and driven by either the cauliflower mosaic virus (CaMV) 35S (Fig. 1A) or the leaf-abundant galactinol synthase (GAS; Fig. 1B) promoter. As expected, transgenic lines containing each of the constructs driven by the CaMV 35S promoter exhibited enhanced tuber production (Fig. 1C). Soil-grown transgenic lines under short-day conditions produced greater tuber yields after 28 d. Tuber yields were increased by as much as 6-fold in the 5′ UTR lines relative to control lines after 4 weeks. For in vitro-grown plants, the transgenic lines produced tubers under noninductive (long-day) conditions and as much as a 3-fold increase in tuber yields under short-day conditions (Fig. 1C). In addition to the effect on tuberization, these transgenic lines exhibited an overall average increase in plant height of 16% relative to the control lines. Other than these observations, the transgenic lines resembled the wild-type phenotype.
Figure 1.
Constructs for the functional analysis of the 5′ and 3′ UTRs of StBEL5 (A and B) and tuber yields (C) for transgenic lines expressing these constructs. Constructs were driven by either the CaMV 35S (A) or the leaf-abundant GAS (B) promoter in the binary vector pBI101.2 for movement assays. Transformation, regeneration, and screening of transgenic potato lines were performed as described by Banerjee et al. (2006b). The StBEL5 5′ UTR and 3′ UTRs are 146 and 505 nucleotides in length, respectively. Transgenic lines of potato that overexpress the chimeric constructs of StBEL5 listed in A (driven by the CaMV 35S promoter) were evaluated for their effect on tuberization (C). The rate of tuberization (number of long days [LD] to tuberize) was determined by the first appearance of tubers from among 20 replicates (C). The yield (g fresh weight plant−1) of tubers was scored for four plants after 14 d of short-day (SD) conditions (8 h of light/16 h of dark). In vitro conditions were the same as described previously (Chen et al., 2003). For the soil-grown experiment, plants from one independent transgenic line per construct were grown in 10-cm pots under long days (16 h of light/8 h of dark) until they reached the 16-leaf stage and then transferred to short-day conditions. After 28 d under short days, four plants from a high-expressing independent line for each construct were evaluated for tuber yield (g fresh weight plant−1). Wild-type plants were used as controls. se values are shown, and asterisks indicate significant differences from the control as determined by t test at a confidence interval of 95%. ND, Not detected; NOST, nopaline synthase terminator.
To facilitate the quantification of transcript mobility for numerous transgenic constructs, an assay for movement through whole plants was utilized in this study instead of grafting. This former approach has been developed as an appropriate protocol for accurate and reproducible mobility assays in potato (Banerjee et al., 2006a). To assess the effect on movement, the relative amount of transgenic RNA present in leaf and stolon tips after 12 d of short-day conditions was quantified in the transgenic lines containing the constructs listed in Figure 1, A and B. Reverse transcription (RT)-PCR was performed on three replicates using a transgenic RNA-specific tag and a gene-specific primer from the cds or the 3′ UTR of StBEL5 RNA. Movement was assessed for the BEL5 constructs driven by both the CaMV 35S and GAS promoters. In potato, the GAS promoter is most active in the minor veins of leaves and its activity is generally not detected in roots, stolons, or tubers (Fig. 2; Banerjee et al., 2006a). Predicted representative models for these UTR chimeric RNAs derived from Mfold analyses (Fig. 3A, +5′ UTR and +3′ UTR) indicate that, whereas they both maintained the finger-like structure composed of the UTRs (circles), this latter structure is reduced in complexity in the truncated RNAs relative to the full-length transcript of StBEL5 (compare the +5′ UTR and +3′ UTR models with the full-length model [arrows], Fig. 3A).
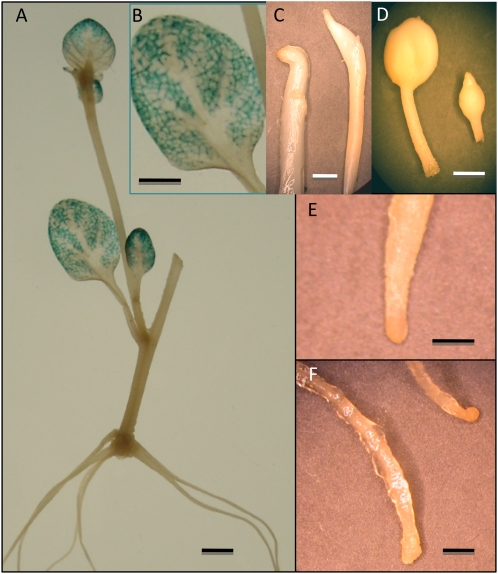
Figure 2.
GUS activity driven by the leaf-abundant promoter (Ayre et al., 2003) for GAS in transgenic potato plants detected by histochemical analysis. With transgenic potato plants containing a GAS::GUS construct, GUS activity was detected in leaves but not in roots or stolons. Activity is generally not detected in tubers, but in a few rare cases, GAS::GUS activity was detected in trace amounts on knobby structures of irregularly formed tubers. A, Transgenic plant grown in vitro under long-day conditions. B, Enlargement of leaf from the plant in A. C, Stolon tips from a short-day plant. D, Newly formed tubers. E, Primary root tip. F, Lateral root tips. C and D are from a soil-grown plant. E and F are from an in vitro-grown long-day plant. These samples are representative of numerous ones that were examined. Bars = 2.0 mm.
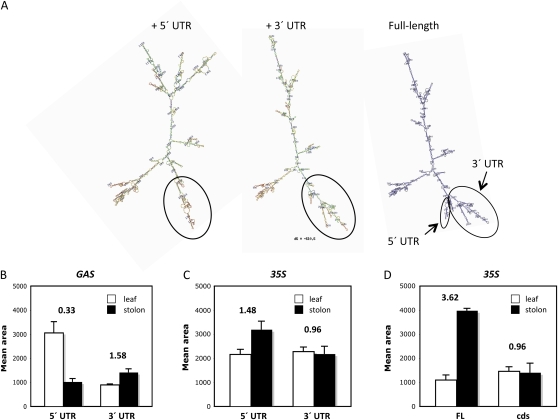
Figure 3.
Effects of UTRs on movement of the StBEL5 mRNA. Representative Mfold models (Zuker, 2003) of two RNA constructs assayed for movement and the full-length StBEL5 RNA (A; from left to right): 5′ UTR + cds, cds + 3′ UTR, and full-length RNA of StBEL5. In nearly all Mfold models, the full-length mRNA has both UTRs in close proximity (arrows, full-length model), forming a multifingered structure. The BEL5 UTRs for each model are highlighted by solid circles. Quantification of movement was performed on transgenic lines with the constructs shown in Figure 1, A and B, and on StBEL5 constructs with or without UTRs (D, FL and cds, respectively), all grown under short-day conditions (8 h of light/16 h of dark). Relative levels of RNA were quantified from total RNA extracted from both new leaves (white bars) and 0.5-cm samples from the tip of the stolon (black bars) from three separate plants for each construct. Homogenous RT-PCR products were quantified using ImageJ software (Abramoff et al., 2004) and normalized using rRNA values. se values of three clones from one independent transgenic line per construct are shown. A movement factor is provided for mobility assays above each set of constructs (B–D). The movement factor is equal to the relative stolon tip RNA quantity divided by the relative leaf RNA quantity. One set of constructs was driven by the GAS promoter of C. melo (B), whereas the other two sets were driven by the CaMV 35S promoter (C and D). The GAS promoter is most active in the minor veins of leaves (Ayre et al., 2003; Fig. 2). [See online article for color version of this figure.]
For the GAS promoter lines, most of the RNA will arise from the leaf (Fig. 2). It is assumed, however, that the CaMV 35S promoter is active equally in leaves, stems, and stolons, so any increase of RNA levels in the stolon tips would indicate enhanced RNA transport. Using the 35S promoter, an assay with a movement factor of approximately 1.0 would indicate no movement (Fig. 3D, cds construct). In one case, the movement factor for a negative control using the GAS promoter was 0.05 (Banerjee et al., 2006a). With the GAS promoter, both constructs exhibited enhanced mobility, but the +3′ UTR transcripts were transported more efficiently than the +5′ UTR transcripts (Fig. 3B). The difference in RNA accumulation in leaves relative to stolon tips between transgenic lines with the 3′ UTR and the 5′ UTR constructs was 4.8-fold (Fig. 3B, 1.58 ÷ 0.33). In other words, with the 3′ UTR, almost 5-fold more BEL5 RNA was being transported from leaf veins to stolon tips than with the 5′ UTR. When considering movement of these same chimeric transcripts expressed by the CaMV 35S promoter, however, mobility for both construct types was less than optimum. With this promoter, the 5′ UTR transcript exhibits a movement factor of 1.48, whereas the 3′ UTR transcript exhibits a movement factor of 0.96 (Fig. 3C). Compare these data with the movement factors of 3.62 and 0.96 for the positive and negative mobility controls, respectively (Fig. 3D). Either only a very slight increase in mobility (5′ UTR construct) or none at all (3′ UTR construct) was observed with these test constructs.
Enhancing the Mobility of Another BEL1-Like RNA
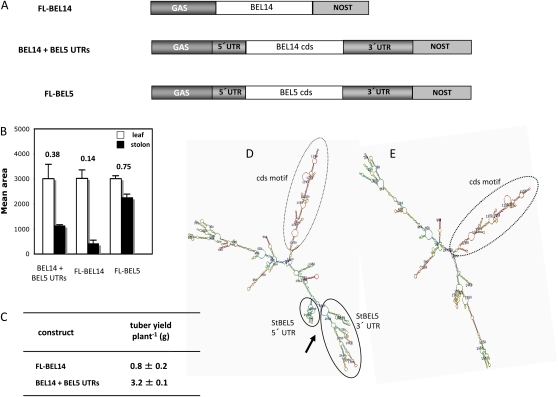
To determine if StBEL5 UTRs are sufficient to mediate the transport of a mRNA from the leaf through the phloem to the stolon tip, a chimeric construct containing both StBEL5 UTRs fused to the cds of another potato BEL1-like RNA was created (Fig. 4A, BEL14 + BEL5 UTRs). The sequence of StBEL14 (accession no. AF406700) was selected because the RNA for this BEL1-like gene of potato was detected only at very low levels in stolons and tubers (Chen et al., 2003) and was not detected in stem phloem cells using laser-capture microdissection (Yu et al., 2007). As a baseline control, a construct consisting of the full-length StBEL14 sequence without StBEL5 UTRs was evaluated for movement (Fig. 4A, FL-BEL14). As a positive control, movement was assayed for a transgenic line characterized previously (Banerjee et al., 2006a) expressing the full-length StBEL5 transcript (Fig. 4A, FL-BEL5). To ensure that the source of most of these transgenic RNAs was the leaf (similar to the StBEL5 promoter), the leaf-abundant GAS promoter was used for all three constructs (Fig. 4A). The RT-PCR quantification procedure was the same as described for Figure 3. As expected, the most efficient movement of transgenic RNA was observed with the full-length StBEL5 RNA, with a movement factor of 0.75 (Fig. 4B, FL-BEL5). The difference in RNA accumulation in leaves relative to stolon tips between transgenic lines with BEL14 plus the StBEL5 UTRs or BEL14 without these UTRs is 2.7-fold (Fig. 4B, 0.38 ÷ 0.14). With the UTRs, almost 3-fold more BEL14 transcript is being transported to stolon tips than without UTRs. In correlation with this enhanced movement, tuber yields for the BEL14 + BEL5 UTR lines were 4-fold greater than those lines with the FL-BEL14 sequence (Fig. 4C). Mfold analysis of the BEL14 constructs used in this experiment showed that fusion of the BEL5 UTRs resulted in a representative model structure that exhibited the multifingered structure (Fig. 4D, arrow) characteristic of the full-length StBEL5 RNA (Fig. 3A, full-length) but absent in the model for FL-BEL14 (Fig. 4E). In addition, 100% of the StBEL14 RNA models derived from the Mfold analysis exhibited a stable stem/loop motif arising from the cds (Fig. 4, D and E, cds motif).
Figure 4.
A, Three RNA constructs driven by the leaf-abundant GAS promoter were tested for their capacity to move from leaf veins to stolon tips: FL-BEL14 (contains BEL14 cds plus both native BEL14 UTRs), BEL14 + both StBEL5 UTRs replacing the StBEL14 UTRs, and full-length StBEL5. StBEL14 was chosen as a test RNA because it is not abundant in stems or stolons (Chen et al., 2003). B, Relative RNA accumulation in new leaves and 0.5-cm samples from the tip of the stolon was quantified in transgenic lines expressing these chimeric transcripts. Transgenic plants were grown under short days (8 h of light/16 h of dark) for 12 d, and the RNA was extracted from new leaves (white bars) and from 0.5-cm stolon tips (black bars) from three separate plants for each construct. C, At the same time, tuber yields were scored for BEL14 lines with and without UTR fusions. Homogenous RT-PCR products were quantified using ImageJ software (Abramoff et al., 2004) and normalized using rRNA values. se values of three clones from one independent transgenic line per construct are shown. A movement factor is provided for mobility assays above each set of constructs (B). The movement factor is equal to the relative stolon tip RNA quantity divided by the relative leaf RNA quantity. D and E, Representative Mfold models of the two BEL14 RNA constructs assayed for movement. Models for the RNA structure of BEL14 + the BEL5 UTRs exhibited the same multifingered structure (D, arrow) formed by the UTRs (D, solid circles) of the full-length StBEL5 transcript (Fig. 3A, full-length model, solid circles). All predicted Mfold models for StBEL14 exhibited a stable, conserved motif arising from its cds (D and E, dotted circles). NOST, Nopaline synthase terminator. [See online article for color version of this figure.]
Factors That Affect the Stability of StBEL5 Transcripts
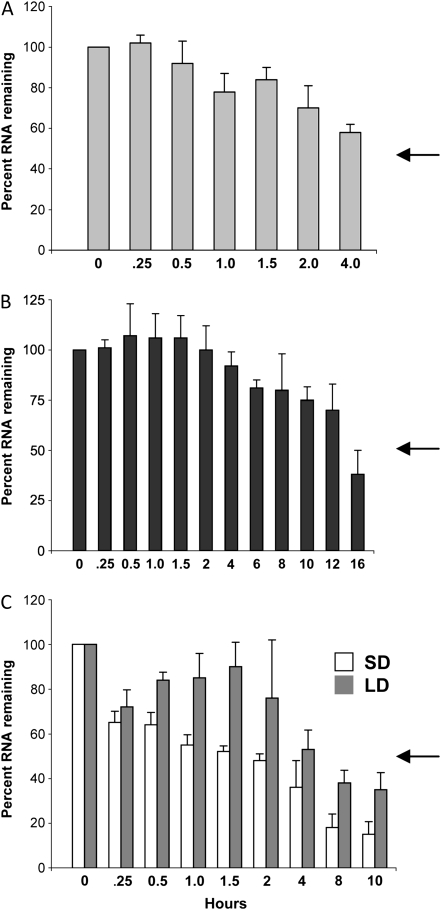
To determine if the UTRs of StBEL5 have an effect on transcript turnover, a stability assay was employed using in vitro-harvested shoot tips from transgenic lines expressing either the full-length StBEL5 transcript or the cds alone. After 7 d of incubation in vitro under short days, shoot tips were harvested, total RNA was extracted, and the StBEL5 RNA was assayed for RNA stability. Quantitative RT-PCR was used for measuring mRNA half-lives because it is more sensitive than northern-blot hybridization. As described before, transgenic RNA- and gene-specific primers were used in the RT-PCR. Not surprisingly, the presence of the StBEL5 UTRs enhanced the stability of its mRNA. Whereas approximately 60% of the transcript without any UTRs remained after 4 h (Fig. 5A), those transcripts that included both UTRs were approximately 90% intact after 4 h and did not drop below 60% until after 12 h (Fig. 5B). The half-life of the full-length transcript was approximately 14 h, whereas for the cds alone it was extrapolated to be 5 h.
Figure 5.
Effects of the UTRs (A and B) and photoperiod (C) on the stability of StBEL5 RNA. Transcript stability time lines for three different RNA molecules, transgenic cds of StBEL5 (A), transgenic full-length StBEL5 (B), and native StBEL5 (C), were determined in potato plants. One-centimeter shoot tips from in vitro-grown plants were harvested, RNA was extracted, and quantitative RT-PCR with gene-specific primers was performed as described previously (Banerjee et al., 2006a). The percentage of RNA remaining for each sample is relative to the amount of RNA present when the experiment was started (0 h). Samples were harvested from plants grown under short-day (SD) conditions (A and B) or both long-day (LD) and short-day conditions (C). The arrows indicate the approximate half-life point for each experimental time line. The UTRs of StBEL5 (accession no. AF406697) include 146 nucleotides from the 5′ end and 505 nucleotides from the 3′ end. The CaMV 35S promoter was used for the transgenic lines in A and B.
Because movement of the StBEL5 RNA is regulated by photoperiod (Banerjee et al., 2006a), the effect of photoperiod on transcript stability was assessed. Wild-type plants of the photoperiod-responsive potato were grown under either long- or short-day conditions. Quantitative RT-PCR was performed as described previously except that a pair of gene-specific primers for wild-type StBEL5 was used. The stability assays revealed that StBEL5 RNA was more stable after long-day than after short-day conditions (Fig. 5C). From the onset of the experiment, after short-day conditions the StBEL5 transcript exhibited a steady decrease in RNA integrity. A half-life of approximately 1.7 h was observed for StBEL5 RNA from short-day-grown plants. After 10 h, the RNA remaining had decreased to less than 20% (Fig. 5C). Conversely, the StBEL5 RNA extracted from long-day-grown plants remained relatively stable through 2 h (approximately 75% remaining) and exhibited a half-life of approximately 5.0 h (Fig. 5C).
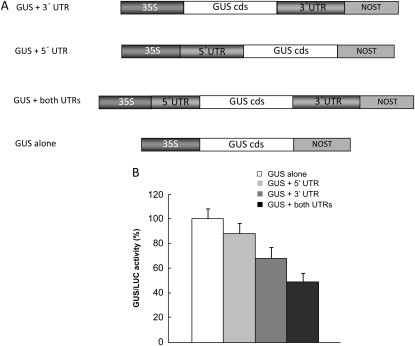
The Effect of the UTRs of StBEL5 RNA on Translation
To assess the effect of the UTRs of StBEL5 mRNA on translation, the cds of GUS with and without the UTRs was cloned into pBI221 and assayed in a transient translation system in tobacco (Nicotiana tabacum) protoplasts. This in vitro procedure allows for the rapid and efficient analysis of numerous constructs. Four constructs were analyzed: GUS + the 3′ UTR of StBEL5, the 5′ UTR of StBEL5 + GUS, GUS plus both UTRs attached, and the GUS cds alone (Fig. 6A). Translation of GUS was reduced in the construct containing the 5′ UTR of StBEL5 by 12% relative to GUS alone (Fig. 6B). With the GUS + 3′ UTR and the 5′ UTR + GUS + 3′ UTR constructs, translation was repressed by 32% and 50%, respectively (Fig. 6B). As a comparison, using in vitro systems with marker genes, regulatory sequence from the transcript, and coexpression of established RNA-binding proteins, levels of translational repression for four mRNAs, GluR2, Vg1, Ringo/SPY, and ASH1, were 30% to 40%, 40% to 60%, 70% to 90%, and 80% to 90%, respectively (Colegrove-Otero et al., 2005; Huang et al., 2006; Padmanabhan and Richter, 2006; Deng et al., 2008). Using RT-PCR, transcript levels for GUS in transfected cells were verified to be essentially equivalent for all four constructs except GUS plus both UTRs (data not shown). These transcripts were approximately one-third more abundant than the GUS RNA without UTRs. This was consistent with the stability assays for full-length StBEL5 RNA (Fig. 5, A and B). In Figure 5, BEL5 RNA containing both UTRs was approximately 30% more stable than cds transcripts (no UTRs) over the 1.0- to 4.0-h harvests.
Figure 6.
The effects of the UTRs of StBEL5 on the translation efficiency of GUS in tobacco protoplasts. A, Four constructs driven by the 35S promoter in pBI221 were analyzed: GUS + the 3′ UTR of StBEL5, the 5′ UTR of StBEL5 + GUS, GUS plus both UTRs attached, and the GUS cds alone. B, The LUC gene in pBI221 under the control of the CaMV 35S promoter was included as an internal control. A relative value for GUS expression was obtained by dividing the GUS activity by the specific LUC activity. Each transfection was performed three times. Data are means ± se. NOST, Nopaline synthase terminator. The levels of GUS translation have been adjusted to reflect differences in RNA accumulation for each of the constructs.
DISCUSSION
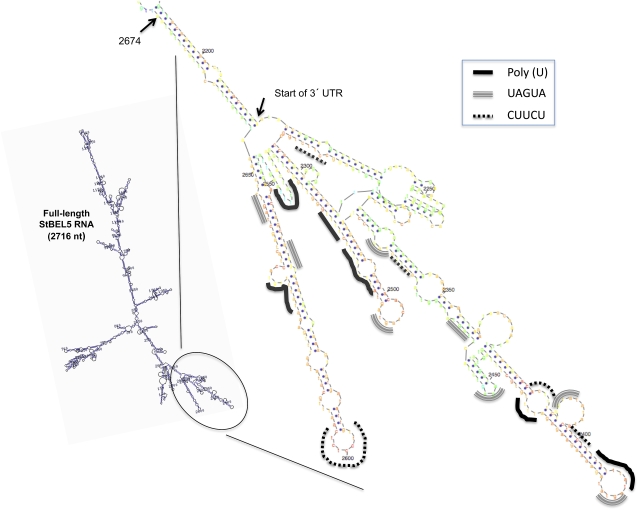
The mRNA of StBEL5 is one of the few full-length mRNAs in plants that have been confirmed to move a long distance through the phloem from leaf veins to stolon tips underground (Banerjee et al., 2006a; Kehr and Buhtz, 2008). This movement has been consistently associated with enhanced tuber production (Banerjee et al., 2006a). Our results here confirm that the UTRs of the StBEL5 RNA are involved in mediating this transport. Whereas either UTR alone may enhance mobility (Fig. 3B), our results suggest that both UTRs together contribute to optimum mobility of the StBEL5 RNA. The contributing effects of the respective UTRs are not clear, but there are numerous examples demonstrating the importance of the 3′ UTRs in recognizing RNA-binding proteins that regulate metabolism and movement (Ferrandon et al., 1994; Corral-Debrinski et al., 2000; Padmanabhan and Richter, 2006; Irion and St. Johnston, 2007). A careful analysis of the 3′ UTR of StBEL5 reveals the presence of three clusters of conserved motifs (Fig. 7). These include five polypyrimidine tracts, six poly-U motifs, and eight novel UAGUA motifs. It is conceivable that these motifs function in interactions with RNA-binding proteins to regulate StBEL5 RNA metabolism. For example, PTB proteins are RNA-binding proteins that bind to clusters of polypyrimidine tracts (Auweter and Allain, 2008).
Figure 7.
Enlarged computer-generated version (Zuker, 2003) of the 3′ UTR of StBEL5 from nucleotide (nt) 2,215 to 2,674 (arrows). Three types of conserved motifs are highlighted: poly-U (solid line), a unique UAGUA motif (double-striped line), and the polypyrimidine tract, CUUCU (dotted line). The enlarged figure (at right) is derived from the Mfold model of full-length StBEL5 RNA (at left and Fig. 3A). Full-length StBEL5 RNA is 2,716 nucleotides in length. The 3′ UTR begins at nucleotide 2,215 (designated by the lower arrow). It should be noted, however, that the existence of these conserved motifs does not validate their function. [See online article for color version of this figure.]
Adding both UTRs to another BEL RNA of potato made it more mobile, targeted it to stolon tips, and enhanced tuber formation (Fig. 4). Although very little is known about this delivery system in plants, the temporal and spatial coordination of the transport of functional mRNAs is a well-documented phenomenon in biology (Kloc et al., 2002; King et al., 2005). For example, in complexes with other proteins, the Staufen family of proteins plays a key role in transporting specific RNAs to the anterior or posterior end of the egg cell in Drosophila. In this system, delivery of the mRNAs for oskar and bicoid to one end or the other defines the primary axis of the developing embryo (St. Johnston et al., 1991). In Saccharomyces cerevisiae, ASH1 mRNA is localized to the bud tip of daughter cells, where it plays key roles in development (Olivier et al., 2005). The localization of these transcripts depends on interactions between cis-acting localization elements in ASH1 that recognize the RNA-binding protein, She2p. The ASH1 mRNA contains four elements that are essential for the proper localization of this transcript (Chartrand et al., 1999). Three of these elements (E1, E2A, and E2B) are located within the coding region of the ASH1 mRNA, whereas the remaining element (E3) includes the termination codon and is located primarily within the ASH1 3′ UTR. These localization elements were predicted to form RNA secondary structures containing stem loops (Gonzalez et al., 1999) that facilitate binding to She2p and are conserved in other bud-localized mRNAs. A cis-acting element in the cds of the Arabidopsis FLOWERING LOCUS T (FT) transcript facilitated movement of the FT mRNA in Nicotiana benthamiana. A fusion of the FT RNA with RNA of Potato virus X enhanced spread and systemic infection of the virus relative to the control (Li et al., 2009). For the GAI RNA of Arabidopsis, motifs in the cds and the 3′ UTR display functional roles in regulating RNA movement (Huang and Yu, 2009).
In the oocyte of Xenopus laevis, numerous RNAs are localized to generate developmental polarity along the animal or vegetal axis (for review, see King et al., 2005). Fifteen RNAs have been identified that are localized to the animal hemisphere and 20 that localize to distinct domains within the cortex. Vg1 RNA is transported to the vegetal cytoplasm, where localized expression of the encoded protein is critical for embryonic polarity. Interaction between specific repeated motifs within the 3′ UTR and RNA-binding proteins mediates the localization of Vg1 RNA to the vegetal pole of Xenopus oocytes (Lewis et al., 2004). The Vg1 localization pathway is directed by key protein factors, such as the RNA-binding proteins Vg1RBP/vera and PTB/hnRNP I, which recognize and bind to these localization elements (Lewis et al., 2008). As many as 16 phloem proteins have been identified that are associated with mobile RNAs in pumpkin (Ham et al., 2009). This ribonucleoprotein complex moves through a graft union between pumpkin and cucumber (Cucumis sativus), but the precise roles of the proteins in this complex in transport and RNA metabolism are still not clear.
Using an RNA stability assay, our results indicate that the stability of StBEL5 RNA is photoperiod dependent (Fig. 5C) and mediated by its UTRs. That UTRs enhanced RNA integrity is not an unusual observation. UTRs are known to mediate transcript degradation rates of numerous RNAs. A genome-wide analysis of mRNA decay rates in Arabidopsis showed that sequence elements in the 3′ UTRs were enriched in short- and long-lived transcripts (Narsai et al., 2007). Light regulation of RNA stability has also been documented. Transcripts of ferredoxin are stabilized in light and destabilized in dark by their association with polyribosomes and their accessibility to ribonucleases. The element responsible for this light regulation is the internal light-regulatory element located within the 5′ UTR of the ferredoxin mRNA (Hansen et al., 2001; Bhat et al., 2004). The stability of the mRNA of the fatty acid desaturase, FAD7, is reduced in response to light, whereas the transcript is relatively stable under dark conditions (Collados et al., 2006). Stability of the mRNA of one of the Arabidopsis oscillator genes for the circadian clock, CIRCADIAN CLOCK-ASSOCIATED1 (CCA1), is stable in the dark and in far-red light but has a shorter half-life in red and blue light (Yakir et al., 2007). Light-mediated stability of CCA1 mRNA is an important mechanism for ensuring that the circadian oscillator is accurately entrained by environmental changes. StBEL5 mRNA accumulates in leaf veins and petioles under the long days of summer, along with sugars from photosynthesis, and is induced to move during the onset of short days. In this system, it is logical that signal mRNA accumulating under long days has some degree of enhanced protection from degradation until transport is activated by short days late in the summer.
As previously discussed, the Staufen protein of Drosophila binds to bicoid and oskar mRNAs to determine their localization in the egg cell and to repress translation until this occurs (St. Johnston et al., 1991; Ferrandon et al., 1994). Results from the translation assay indicate that the 3′ UTR of StBEL5 is involved in the repression of translation (Fig. 6B). Whereas there are numerous examples of the 3′ UTR playing a role in the repression of translation of animal mRNAs (Gavis and Lehmann, 1994; Kim-Ha et al., 1995; Huang et al., 2006; Padmanabhan and Richter, 2006), there is very little information on such a role for 3′ UTRs in plants. In contrast, the role of the 5′ UTR in enhancing translation has been well established (Dansako et al., 2003; Patel et al., 2004; Dugré -Brisson et al., 2005; Sugio et al., 2008). Because of its prominent role in regulating tuber development and its capacity for long-distance transport to stolon tips (Chen et al., 2003; Banerjee et al., 2006a), control of translation of the mRNA encoding the BEL5 transcription factor in the correct place and at the correct time in potato is imperative. Normally, the BEL5 promoter is very active in leaf vasculature in response to light but is inactive in stems (Chatterjee et al., 2007). As an example of what happens when misexpression occurs in nontarget organs, consider the phenotype exhibited by transgenic lines that constitutively overexpress the cds (no UTRs) of StBEL5. Extreme phenotypes of these lines are characterized by the formation of aerial tubers from stems and by a reduction in the yield of underground tubers (Fig. 8). Such extreme phenotypes are not normally observed in transformants expressing the full-length StBEL5 construct (Banerjee et al., 2006a).
Figure 8.
The formation of aerial tubers (black arrows) from stem axillary buds of a transgenic line of potato that constitutively overexpresses the cds (no UTRs) of StBEL5. Expression of the transgene was driven by the CaMV 35S promoter. This BEL5 transgenic line, designated no. 20, exhibited an extreme phenotype characterized by the formation of aerial stem tubers, stunted growth, and a reduction in the yield of underground tubers (Chen et al., 2003). A leaf has been excised (white arrow) to enhance visibility.
Remarkably, translation repression occurs for all four mobile nonplant mRNAs previously discussed, oskar and bicoid mediated by Staufen, ASH1 mediated by Kdh1p and Puf6p (Gu et al., 2004; Paquin et al., 2007), and Vg1 mediated by ELAV (for embryonic lethal and abnormal vision) proteins (Colegrove-Otero et al., 2005). Vg1 RNA is abundant during the earliest stages of oogenesis in Xenopus (Melton, 1987) but is not translated until the later stages of development during localization to the vegetal cortex (Wilhelm et al., 2000). In the cases of oskar and ASH1, repression is spatially regulated (Micklem et al., 2000; Paquin and Chartrand, 2008). As has been postulated for StBEL5 RNA, repression of translation ensures that the mobile mRNAs mentioned above, most of which encode developmentally important transcription factors, are functional only at their target sites. Consistent with the aberrant phenotype for the BEL5 overexpression line shown in Figure 8, misexpression of Vg1 in the animal hemisphere results in severe patterning defects (Dale et al., 1993).
The long-distance transport of StBEL5 RNA from leaves to stolon tips to activate tuber formation in potato represents a complex and dynamic signaling system. In this model system of plants for non-cell-autonomous movement of full-length mRNAs, it is readily apparent that escort factors that facilitate transport are not the only interactions putatively mediated by UTRs that affect RNA metabolism and target efficient expression of the mobile RNA. That so many proteins have been identified in the ribonucleoprotein complex of a phloem-mobile RNA of pumpkin (Ham et al., 2009) is consistent with this multifaceted role of RNA-binding proteins in RNA metabolism.
MATERIALS AND METHODS
Construct Design
For the transgenic lines in Figure 1A, both constructs, cds + 5 UTR and cds + 3 UTR, were PCR cloned downstream from the CaMV 35S promoter into the XbaI/SacI site of the binary vector pCB201 (Xiang et al., 1999). For the GAS promoter lines in Figure 1B, both UTR constructs were cloned into the XmaI/SacI site downstream from the GAS promoter that had been cloned previously into pBI101.2 (Banerjee et al., 2006a). For the StBEL14 constructs in Figure 4A, full-length StBEL14 sequence (1,690 nucleotides including its native UTRs [GenBank accession no. AF406700.1]; designated FL-StBEL14) was cloned into the XbaI/SmaI site of pBI101.2 vector with the GAS promoter inserted previously. For the BEL14 plus both BEL5 UTRs construct, cloning was performed sequentially. First, the cds of a PCR-generated fragment of StBEL14 with the KpnI/BamHI site and the 500-nucleotide 3′ UTR of StBEL5 with BamHI and SacI sites were subcloned into pBluescript SK+. The KpnI/SacI fragment of the BEL14 cds + the 3′ BEL5 UTR fusion and a XmaI/KpnI fragment of the 5′ UTR of BEL5 were purified via restriction digestion and then cloned into the XmaI/SacI site of pBI101.2 containing the GAS promoter in a one-step ligation. Cloning of the full-length and cds constructs of StBEL5 driven by the CaMV 35S and GAS promoters, shown in Figures 3D and 4A, respectively, was described previously (Banerjee et al., 2006a, 2006b).
For translation studies in the tobacco (Nicotiana tabacum) protoplasts, GUS constructs were PCR cloned into the transient expression vector pBI221 (Clontech). First, the full-length StBEL5 3′ UTR was cloned into the SacI site of pBI221 to create the GUS + 3′ UTR construct. The SacI primers used were 5′-AGTCCTGAGCTCGAAAGTCTCGTATTGATAGC-3′ and 5′-GTCACTGAGCTCCTAATCTAATAATGATAGCAC-3′. The full-length 5′ UTR of StBEL5 was then cloned into the XbaI/BamHI site of the GUS + 3′ UTR construct to create the GUS + both UTRs construct. The primers used for this cloning step were 5′-CTACTCTAGAGATAGTACTTATCTGCAA-3′ with XbaI at the 5′ end and 5′-CATCGAGGATCCTTCTTTCTTCTTCGGATTTCTCCTTTTATCT-3′ with BamHI at the 3′ end. Finally, the 5′ UTR + GUS construct was created by cutting the full-length (both UTRs) vector with SacI to release only the BEL5 3′ UTR. All constructs were confirmed via sequencing at the DNA Facility at Iowa State University.
Plant Material and Generation of Transgenic Lines
Transformation was implemented on the photoperiod-responsive potato (Solanum tuberosum subsp. andigena; Banerjee et al., 2006b). In photoperiod-adapted genotypes like andigena, short-day photoperiods (less than 12 h of light) are required for tuber formation, whereas under long-day conditions, no tubers are produced. The in vitro transgenic potato plants were maintained in a growth chamber (Percival Scientific) at 27°C with a photoperiod of 16 h of light/8 h of dark and a fluence rate of 40 μmol m−2 s−1. Soil-grown plants were maintained at 22°C in a growth chamber under either a long-day (16 h of light/8 h of dark) or short-day (8 h of light/16 h of dark) photoperiod with a fluence rate of 100 μmol m−2 s−1. Expression of the GUS reporter gene driven by the GAS promoter (Fig. 2) was analyzed by incubating the samples overnight at 37°C in GUS buffer containing 1.0 m NaHPO4, pH 7.0, 0.25 m EDTA, pH 8.0, 0.5 mm potassium ferrocyanide, 0.5 mm potassium ferricyanide, 10% Triton X-100, 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-GlcUA. Samples were cleared with 100% ethanol and photographed employing a Nikon COOLPXX995 digital camera.
Movement Assays
Transgenic lines expressing the various transcript constructs were driven by either the CaMV 35S promoter or the leaf-abundant GAS promoter from Cucumis melo (Ayre et al., 2003). Transgenic plants were grown under short or long days for 12 d, and the RNA was extracted from 0.5-cm stolon tips and new leaves from three separate plants for each construct. One-step RT-PCR was performed using 20 ng of total RNA, a non-plant-sequence primer fused to all transgenic RNAs, and a gene-specific primer from the test mRNA. Use of the non-plant-sequence primer, NT-2, corresponding to sequence from the NOS terminator (Banerjee et al., 2006a), makes it possible to discriminate transgenic RNA from native BEL5 RNAs. The internal control for PCR was18S rRNA. All PCRs were standardized and optimized to yield product in the linear range, and conditions were adjusted based on the primers used. Gene-specific primers for the BEL5 cds + 3′ UTR and 5′ UTR + BEL5 cds constructs were 5′-TTTCTTTTGGGTTGGCTTGGAGTA-3′ and 5′-GGGAGATTTTGGAAGGTTTG-3′, respectively. The conditions for RT-PCR were set as follows: 50°C for 30 min, 94°C for 2 min, 94°C for 15 s, 56°C for 30 s, 68°C for 30 s, followed by one cycle of incubation at 68°C for 5 min. After determining the linear range for all of the PCR products, 38 and 18 cycles were used for BEL5 and 18S rRNA amplifications, respectively.
Primers for the 18S rRNA were provided in the manufacturer's kit (Ambion Quantum RNA Universal 18S, catalog no. 1718). For constructs driven by the GAS promoter, the PCR cycle numbers were adjusted to 40 for BEL5 and to 17 for the 18S rRNA to be in the linear range. For analysis of the full-length StBEL5 RNA, the conditions and primers were described previously (Banerjee et al., 2006a). For the GAS:StBEL14 quantitative RT-PCR analysis, the StBEL14 gene-specific primer, 5′-CATCAATGGGGTTAACTACA-3′, and NT-2 were used. For quantitative analysis of BEL14 + UTRs (Fig. 4B), the StBEL5 3′ UTR primer, 5′-TTTCTTTTGGGTTGGCTTGGAGTA-3′, and the NT-2 primer were used. The same RT-PCR conditions were used as described previously, with the cycles adjusted to 32 for gene-specific RNA and 17 for the 18S rRNA. Homogenous PCR products were quantified using ImageJ software (Abramoff et al., 2004) and normalized with the rRNA values. se values of three clones from one independent transgenic line per construct are shown. A movement factor is provided for mobility assays in Figures 3 and 4. The movement factor is equal to the relative stolon tip RNA quantity divided by the relative leaf RNA quantity. Structural modeling of mRNAs was performed using Mfold software (Zuker, 2003). Predicted models were evaluated for relative stability and frequency of conserved motifs. Based on these criteria, representative models were presented for each mRNA sequence (Figs. 3A and 4, D and E). Mfold models are based on statistical analysis and do not represent the actual native structure of folded RNAs.
Stability Assays
Shoot tips of potato, approximately 2.0 cm in length, were excised and harvested from aseptically grown 4-week-old wild-type or transgenic lines. Excised shoot tips were then cultured in semisolid (0.2% Gelrite) growth medium supplemented with Murashige and Skoog (1962) basal salt mixtures and 2.0% Suc. About 150 shoot tips were cultured for each experimental design with two replicates each. Shoot tips were cultured at 22°C under either a long-day (16 h of light/8 h of dark) or a short-day (8 h of light/16 h of dark) photoperiod. After 7 d in culture, 1.0-cm shoot tips were excised and transferred to a 250-mL flask containing 80 mL of incubation buffer (Seeley et al., 1992) and shaken at 75 rpm for 30 min at room temperature. At the end of this period, cordycepin (Sigma) was added to a final concentration of 0.6 mm and the samples were vacuum infiltrated for 45 s at 18 in Hg. Samples were then harvested from the 250-mL flask at regular time intervals starting from time zero up to 16 h, blotted, and immediately frozen in liquid nitrogen. Total RNA isolation and quantitative RT-PCR with gene-specific primers were performed as described previously (Banerjee et al., 2006a).
For StBEL5 native transcript analysis, the BEL5 gene-specific primers 5′-GGGAGATTTTGGAAGGTTTG-3′ and 5′-TCAAATTGGGTCCTCCGACT-3′ were used. For quantitative analysis of StBEL5 cds and StBEL5 full-length transcripts (Fig. 5, A and B), the primers and RT-PCR conditions described previously were used (Banerjee et al., 2006a).
Translation Assay
The CaMV 35S promoter in pBI221 (Clontech) was used for translation assays with the three combinations of UTR fusions to the GUS reporter construct (Fig. 6A). The reporter alone in pBI221 was used as a translation control. A CaMV 35S promoter-driven luciferase (LUC) construct, 35S-LUC (obtained from Dr. Y. Takahashi, Department of Biological Sciences, Graduate School of Science, University of Tokyo), was used as an internal control. Fully expanded leaves from 3- to 4-week-old tobacco plants were excised, placed in K3 basal medium (Kao and Michayluk, 1975) supplemented with 0.4 m Suc, 0.25% (w/v) cellulases (Karlan Research Products), and 0.05% (w/v) macerases (Calbiochem), and incubated overnight at 28°C. The protoplasts were then filtered through sterile cheesecloth into a Babcock bottle and centrifuged for 10 min at 1,000 rpm. Protoplasts were collected from the bottleneck area and washed once in K3 medium with 0.4 m Suc and resuspended in K3 medium containing 0.4 m Glc to a final concentration of 4 × 106 protoplasts mL−1.
For each transfection analysis, 700 μL of tobacco protoplasts (prepared as described above) was mixed with 30 μL of 2.0 m KCl and plasmid DNA in an electroporation cuvette with a 0.4-cm electrode gap. The plasmid DNA was a mixture of 5.0 μg of the GUS reporter construct and 0.5 μg of the 35S-LUC construct as an internal control. After electroporation (voltage = 170 V, capacitance = 125 μF; Gene Pulser Transfection Apparatus [Bio-Rad]), 4.0 mL of Murashige and Skoog (1962) basal medium was added, and the protoplasts were incubated in the dark at room temperature for 48 h before conducting GUS and LUC activity assays. Transfections were performed three times for each construct.
LUC assays were performed by injecting 100 μL of LUC substrate (Promega) into 20 μL of extract and measuring the emitted photons for 15 s in a TD-20 luminometer (Turner Designs). Fluorometric GUS assays were performed as described (Jefferson, 1987). A fluorescence multiwell plate reader (Fluoroskan II; MTX Labs) was used to measure GUS activity at 365 nm (excitation) and 455 nm (emission). Each sample was measured three times for both LUC and GUS activity. A relative value for GUS expression was obtained by dividing the GUS activity by the specific LUC activity. Relative activities were calculated from three transfection replications and are presented as means ± se.
Acknowledgments
Thanks to Brian A. Campbell for his valuable technical assistance.
This work was supported by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (grant no. 2008–02806) and the National Science Foundation Plant Genome Research Program (grant no. 0820659).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: David J. Hannapel (djh@iastate.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophot Int 11 36–42 [Google Scholar]
- Asano T, Masumura T, Kusano H, Kikuchi S, Kurita A, Shimada H, Kadowaki K (2002) Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: toward comprehensive analysis of the genes expressed in the rice phloem. Plant J 32 401–408 [DOI] [PubMed] [Google Scholar]
- Auweter SD, Allain FHT (2008) Structure-function relationships of the polypyrimidine tract binding protein. Cell Mol Life Sci 65 516–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayre BG, Blair JE, Turgeon R (2003) Functional and phylogenetic analyses of a conserved regulatory program in the phloem of minor veins. Plant Physiol 133 1229–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Chatterjee M, Yu Y, Suh SG, Miller WA, Hannapel DJ (2006. a) Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18 3443–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Prat S, Hannapel DJ (2006. b) Efficient production of transgenic potato (S. tuberosum L. ssp. andigena) plants via Agrobacterium tumefaciens-mediated transformation. Plant Sci 170 732–738 [Google Scholar]
- Barreau C, Paillard L, Osborne HB (2006) AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res 33 7138–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat S, Tang L, Krueger AD, Smith CL, Ford SR, Dickey LF, Petracek ME (2004) The Fed-1 (CAUU)4 element is a 5′ UTR dark-responsive mRNA instability element that functions independently of dark-induced polyribosome dissociation. Plant Mol Biol 56 761–773 [DOI] [PubMed] [Google Scholar]
- Chartrand P, Meng XH, Singer RH, Long RM (1999) Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol 9 333–336 [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Banerjee AK, Hannapel DJ (2007) A BELL1-like gene of potato is light activated and wound inducible. Plant Physiol 145 1435–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Banerjee AK, Hannapel DJ (2004) The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J 38 276–284 [DOI] [PubMed] [Google Scholar]
- Chen H, Rosin FM, Prat S, Hannapel DJ (2003) Interacting transcription factors from the TALE superclass regulate tuber formation. Plant Physiol 132 1391–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colegrove-Otero LJ, Devaux A, Standart N (2005) The Xenopus ELAV protein ElrB represses Vg1 mRNA translation during oogenesis. Mol Cell Biol 25 9028–9039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collados R, Andreu V, Picorel R, Alfonso M (2006) A light-sensitive mechanism differently regulates transcription and transcript stability of omega3 fatty-acid desaturases (FAD3, FAD7 and FAD8) in soybean photosynthetic cell suspensions. FEBS Lett 58 4934–4940 [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M, Blugeon C, Jacq C (2000) In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol Cell Biol 20 7881–7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale L, Matthews G, Colman A (1993) Secretion and mesoderm-inducing activity of the TGF-β-related domain of Xenopus Vg1. EMBO J 12 4471–4480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansako T, Kato K, Satoh J, Sekine M, Yoshida K, Shinmyo A (2003) 5′ untranslated region of the HSP18.2 gene contributes to efficient translation in plant cells. J Biosci Bioeng 95 52–58 [DOI] [PubMed] [Google Scholar]
- Deeken R, Ache P, Kajahn I, Klinkenberg J, Bringmann G, Hedrich R (2008) Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex data sets of microarray experiments. Plant J 55 746–759 [DOI] [PubMed] [Google Scholar]
- Deng Y, Singer RH, Gu W (2008) Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev 22 1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrigo M, Cestelli A, Savettieri G, Di Liegro I (2000) RNA-protein interactions in the control of stability and localization of messenger RNA. Int J Mol Med 5 111–123 [PubMed] [Google Scholar]
- Dugré-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L (2005) Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res 33 4797–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira G, Massie B, DesGroseillers L (2006) The zinc-finger protein ZFR is critical for Staufen 2 isoform specific nucleocytoplasmic shuttling in neurons. J Neurochem 96 105–117 [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Elphick L, Nusslein-Volhard C, St Johnston D (1994) Staufen protein associates with the 3′ UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 79 1221–1223 [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Koch I, Westhof E, Nusslein-Volhard C (1997) RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J 16 1751–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaupels F, Buhtz A, Knauer T, Deshmukh S, Waller F, van Bel AJE, Kogel KH, Kehr J (2008) Adaptation of aphid stylectomy for analyses of proteins and mRNAs in barley phloem sap. J Exp Bot 59 3297–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavis ER, Lehmann R (1994) Translational regulation of nanos by RNA localization. Nature 369 315–318 [DOI] [PubMed] [Google Scholar]
- Gonzalez I, Buonomo SBC, Nasmyth K, von Ahsen U (1999) ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr Biol 9 337–340 [DOI] [PubMed] [Google Scholar]
- Gu W, Deng Y, Zenklusen D, Singer RH (2004) A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev 18 1452–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi CO, Giuliodori AM, Pon CL (2003) Transcriptional and post-transcriptional control of cold-shock genes. J Mol Biol 331 527–539 [DOI] [PubMed] [Google Scholar]
- Halbeisen RE, Galgano A, Scherrer T, Gerber AP (2008) Post-transcriptional gene regulation: from genome-wide studies to principles. Cell Mol Life Sci 65 798–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham BK, Brandom JL, Xoconostle-Cazares B, Ringgold V, Lough TL, Lucas WJ (2009) A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 21 197–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannapel DJ, Chen H, Rosin FM, Banerjee AK, Davies PJ (2004) Molecular controls of tuberization. Am J Potato Res 81 5–16 [Google Scholar]
- Hansen ER, Petracek ME, Dickey LF, Thompson WF (2001) The 5′ end of the pea ferredoxin-1 mRNA mediates rapid and reversible light-directed changes in translation in tobacco. Plant Physiol 125 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood V, Yu TS, Huang NC, Lucas WJ (2005) Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J 42 49–68 [DOI] [PubMed] [Google Scholar]
- Huang NC, Yu TS (2009) The sequences of Arabidopsis GA-INSENSITIVE RNA constitute the motifs that are necessary and sufficient for RNA long-distance trafficking. Plant J 59 921– 929 [DOI] [PubMed] [Google Scholar]
- Huang YS, Kan MC, Lin CL, Richter JD (2006) CPEB3 and CPEB4 in neurons: analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J 25 4865–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U, St Johnston D (2007) bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature 445 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RP (2001) mRNA localization: message on the move. Nat Rev Mol Cell Biol 2 247–256 [DOI] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5 387–405 [Google Scholar]
- Kanrar S, Bhattacharya M, Arthur B, Courtier J, Smith HMS (2008) Regulatory networks that function to specify flower meristems require the function of homeobox genes PENNYWISE and POUND-FOOLISH in Arabidopsis. Plant J 54 924–937 [DOI] [PubMed] [Google Scholar]
- Kao KN, Michayluk MR (1975) Nutritional requirements for growth of Vicia hajastana cells and protoplasts at a very low density in liquid media. Planta 126 105–110 [DOI] [PubMed] [Google Scholar]
- Keene JD (2007) RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8 533–543 [DOI] [PubMed] [Google Scholar]
- Keene JD, Lager PJ (2005) Post-transcriptional operons and regulons coordinating gene expression. Chrom Res 13 327–337 [DOI] [PubMed] [Google Scholar]
- Kehr J, Buhtz A (2008) Long distance transport and movement of RNA through the phloem. J Exp Bot 59 85–92 [DOI] [PubMed] [Google Scholar]
- Kim M, Canio W, Kessler S, Sinha N (2001) Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293 287–289 [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, Macdonald PM (1995) Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell 81 403–412 [DOI] [PubMed] [Google Scholar]
- King ML, Messitt TJ, Mowry KL (2005) Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell 97 19–33 [DOI] [PubMed] [Google Scholar]
- Kloc M, Zearfoss NR, Etkin LD (2002) Mechanisms of subcellular mRNA localization. Cell 108 533–544 [DOI] [PubMed] [Google Scholar]
- Kumar R, Kushalappa K, Godt D, Pidkowich MS, Pastorelli S, Hepworth SR, Haughn GW (2007) The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell 19 2719–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Jeong S (2006) Beta-catenin stabilizes cyclooxygenase-2 mRNA by interacting with AU-rich elements of 3′-UTR. Nucleic Acids Res 34 5705–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Schedl T (2006) RNA-binding proteins. WormBook 18 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RA, Gagnon JA, Mowry KL (2008) PTB/hnRNP I is required for RNP remodeling during RNA localization in Xenopus oocytes. Mol Cell Biol 28 678–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RA, Kress TL, Cote CA, Gautreau D, Rokop ME, Mowry KL (2004) Conserved and clustered RNA recognition sequences are a critical feature of signals directing RNA localization in Xenopus oocytes. Mech Dev 121 101–109 [DOI] [PubMed] [Google Scholar]
- Li C, Zhang K, Zeng X, Jackson S, Zhou Y, Hong Y (2009) A cis element within Flowering Locus T mRNA determines its mobility and facilitates trafficking of heterologous viral RNA. J Virol 83 3540–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lough TJ, Lucas WJ (2006) Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu Rev Plant Biol 57 203–232 [DOI] [PubMed] [Google Scholar]
- Macdonald PM, Kerr K (1997) Redundant RNA recognition events in bicoid mRNA localization. RNA 3 1413–1420 [PMC free article] [PubMed] [Google Scholar]
- Melton DA (1987) Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature 328 80–82 [DOI] [PubMed] [Google Scholar]
- Micklem DR, Adams J, Grunert S, St Johnston D (2000) Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J 19 1366–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Wang Y, Franzen R, Santi L, Salamini F, Rohde W (2001) In vitro interactions between barley TALE proteins suggest a role for protein-protein associations in the regulation of Knox gene function. Plant J 27 13–23 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Narsai R, Howell KA, Millar AH, O'Toole N, Small I, Whelana J (2007) Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19 3418–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier C, Poirier G, Gendron P, Boisgontier A, Major F, Chartrand P (2005) Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol Cell Biol 25 4752–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omid A, Keilin T, Glass A, Leshkowitz D, Wolf S (2007) Characterization of phloem-sap transcription profile in melon plants. J Exp Bot 58 3645–3656 [DOI] [PubMed] [Google Scholar]
- Padmanabhan K, Richter JD (2006) Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes Dev 20 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Sundaresana V (2007) Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19 3578–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin N, Chartrand P (2008) Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol 18 105–111 [DOI] [PubMed] [Google Scholar]
- Paquin N, Menade M, Poirier G, Donato D, Drouet E, Chartrand P (2007) Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol Cell 26 795–809 [DOI] [PubMed] [Google Scholar]
- Patel M, Corey AC, Yin LP, Ali S, Taylor WC, Berry JO (2004) Untranslated regions from C4 amaranth AhRbcS1 mRNAs confer translational enhancement and preferential bundle sheath cell expression in transgenic C4 Flaveria bidentis. Plant Physiol 136 3550–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullmann R Jr, Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, Gorospe M (2007) Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol Cell Biol 27 6265–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Pelissier T, Itaya A, Hunt E, Wassenegger M, Ding B (2004) Direct role of a viroid RNA motif in mediating directional RNA trafficking across a specific cellular boundary. Plant Cell 16 1741–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni L, Belles-Boix E, Günl M, Pautot V (2008) Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell 20 888–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin FM, Hart JK, Horner HT, Davies PJ, Hannapel DJ (2003) Overexpression of a knox gene of potato alters vegetative development by decreasing gibberellin accumulation. Plant Physiol 132 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano R, Xoconostle-Cazares B, Lucas WJ (1999) Phloem long-distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Development 126 4405–4419 [DOI] [PubMed] [Google Scholar]
- Sanchez-Diaz P, Penalva LO (2006) Post-transcription meets post-genomic: the saga of RNA binding proteins in a new era. RNA Biol 3 101–109 [DOI] [PubMed] [Google Scholar]
- Saunders C, Cohen RS (1999) The role of oocyte transcription, the 5′ UTR, and translation repression and derepression in Drosophila gurken mRNA and protein localization. Mol Cell 3 43–45 [DOI] [PubMed] [Google Scholar]
- Seeley KA, Byrne DH, Colbert JT (1992) Red light-independent instability of oat phytochrome mRNA in vivo. Plant Cell 4 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF, van Hoof A (2008) Messenger RNA regulation: to translate or to degrade. EMBO J 27 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HMS, Hake S (2003) The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nusslein-Volhard C (1991) Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 66 51–63 [DOI] [PubMed] [Google Scholar]
- Sugio T, Satoh J, Matsuura H, Shinmyo A, Kato K (2008) The 5′-untranslated region of the Oryza sativa alcohol dehydrogenase gene functions as a translational enhancer in monocotyledonous plant cells. J Biosci Bioeng 105 300–302 [DOI] [PubMed] [Google Scholar]
- Thio GL, Ray RP, Barcelo G, Schupbach T (2000) Localization of gurken RNA in Drosophila oogenesis requires elements in the 5′ and 3′ regions of the transcript. Dev Biol 15 435–446 [DOI] [PubMed] [Google Scholar]
- Vilaine F, Palauqui JC, Amselem J, Kusiak C, Lemoine R, Dinant S (2003) Towards deciphering phloem: a transcriptome analysis of the phloem of Apium graveolens. Plant J 36 67–81 [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Vale RD, Hegde RS (2000) Coordinate control of translation and localization of Vg1 mRNA in Xenopus oocytes. Proc Natl Acad Sci USA 97 13132–13137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ (1999) A mini-binary vector series for plant transformation. Plant Mol Biol 40 711–717 [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cazares B, Xiang Y, Ruiz-Medrano R, Wang HL, Monzer J, Yoo BC, McFarland KC, Franceschi VR, Lucas WJ (1999) Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283 94–98 [DOI] [PubMed] [Google Scholar]
- Xu X, Vreugdenhil D, van Lammeren AAM (1998) Cell division and cell enlargement during potato tuber formation. J Exp Bot 49 573–582 [Google Scholar]
- Yakir E, Dror Hilman D, Hassidim M, Green RM (2007) CIRCADIAN CLOCK ASSOCIATED1 transcript stability and the entrainment of the circadian clock in Arabidopsis. Plant Physiol 145 925–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YY, Lashbrook CC, Hannapel DJ (2007) Tissue integrity and RNA quality of laser microdissected phloem of potato. Planta 226 797–803 [DOI] [PubMed] [Google Scholar]
- Zhong X, Archual AJ, Amin AA, Ding B (2008) A genomic map of viroid RNA motifs critical for replication and systemic trafficking. Plant Cell 20 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Tao X, Stombaugh J, Leontis N, Ding B (2007) Tertiary structure and function of an RNA motif required for plant vascular entry to initiate systemic trafficking. EMBO J 26 3836–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M (2003) Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]