Abstract
Grass species represent a major source of food, feed, and fiber crops and potential feedstocks for biofuel production. Most of the biomass is contributed by cell walls that are distinct in composition from all other flowering plants. Identifying cell wall-related genes and their functions underpins a fundamental understanding of growth and development in these species. Toward this goal, we are building a knowledge base of the maize (Zea mays) genes involved in cell wall biology, their expression profiles, and the phenotypic consequences of mutation. Over 750 maize genes were annotated and assembled into gene families predicted to function in cell wall biogenesis. Comparative genomics of maize, rice (Oryza sativa), and Arabidopsis (Arabidopsis thaliana) sequences reveal differences in gene family structure between grass species and a reference eudicot species. Analysis of transcript profile data for cell wall genes in developing maize ovaries revealed that expression within families differed by up to 100-fold. When transcriptional analyses of developing ovaries before pollination from Arabidopsis, rice, and maize were contrasted, distinct sets of cell wall genes were expressed in grasses. These differences in gene family structure and expression between Arabidopsis and the grasses underscore the requirement for a grass-specific genetic model for functional analyses. A UniformMu population proved to be an important resource in both forward- and reverse-genetics approaches to identify hundreds of mutants in cell wall genes. A forward screen of field-grown lines by near-infrared spectroscopic screen of mature leaves yielded several dozen lines with heritable spectroscopic phenotypes. Pyrolysis-molecular beam mass spectrometry confirmed that several nir mutants had altered carbohydrate-lignin compositions.
The C4 grasses, with their high photosynthetic efficiency compared with C3 plants, provide more than half of the world's calories in human nutrition and for grazing animals (Langenheim and Thimann, 1982). Lignocellulosic biomass of grasses, comprising plant cell walls, is also regarded as a sustainable and renewable feedstock for biofuels (Ragauskas et al., 2006). Although the Arabidopsis (Arabidopsis thaliana) genome sequence has provided entry points for the identification of many wall-related genes, the functions of grass-specific genes need to be elucidated to gain genetic control of biomass yield and quality in food crops and in bioenergy grasses (Carpita and McCann, 2008). The close evolutionary relationship of the C4 grasses of the Panicoid subfamily and the syntenic organization of grass genomes makes possible the rapid translation of genes that impact biomass characteristics identified in maize (Zea mays) into more genetically recalcitrant species. Maize also provides a wealth of tools built on a century of breeding experience and genetic research. In this study, we provide, to our knowledge, the first comprehensive evaluation of the maize genes responsible for the biogenesis of the special type II cell walls of grasses, and we validate a strategy to determine gene function.
The genome sequences of Arabidopsis (Arabidopsis Genome Initiative, 2000), rice (Oryza sativa; International Rice Genome Sequencing Project, 2005), and maize inbred line B73 (Schnable et al., 2009; http://www.maizesequence.org) provide essential inventories for comparative genomic analyses of cell wall-related genes in Arabidopsis and other flowering plants with the grass species. Grasses have cell walls of a distinct composition from all other flowering plants. Arabidopsis has a type I cell wall: a framework of cellulose microfibrils cross-linked by xyloglucans and embedded in a matrix of acid-rich pectic polysaccharides (McCann and Roberts, 1991; Carpita and Gibeaut, 1993). Type II cell walls of the grasses, such as maize and rice, have a framework of cellulose microfibrils cross-linked primarily with glucuronoarabinoxylans (GAXs). Pectins are a small proportion of the matrix polymers, with GAXs providing most of the negatively charged matrix of the type II cell wall (Carpita, 1996). Structural proteins are deposited and cross-linked at the cessation of growth of type I walls, whereas networks of hydroxycinnamic acid-rich phenylpropanoids covalently linked to GAXs are deposited in the type II wall. Furthermore, a mixed-linkage (1→3),(1→4)-β-d-glucan is synthesized during cell expansion of grasses and hydrolyzed when growth ceases (Carpita and Gibeaut, 1993; Carpita, 1996). All angiosperms synthesize certain polymers in common, but these vary in abundance and possess subtle distinctions in fine structure.
With nearly half of the genes in plant genomes yet to be functionally annotated, an estimated 1,000 genes of unknown function may encode cell wall-related proteins (Yong et al., 2005). Analysis of coregulated genes using microarrays in Arabidopsis has implicated unannotated genes as candidates in secondary wall formation (Brown et al., 2005; Kubo et al., 2005; Persson et al., 2005). Microarrays with maize coding sequences are currently available but incomplete (http://www.maizearray.org/), whereas massively parallel sequencing technologies allow relative transcript abundance to be measured on a genome-wide scale (Eveland et al., 2008). From this data set of transcripts expressed in the maize developing ovary before pollination, we profiled the major primary cell wall-related genes. We then compared these profiles with microarray data for Arabidopsis and rice ovaries sampled at the same stage of development. Our results document that orthology often cannot be inferred from homology and that different sets of genes than would be predicted are expressed between a model dicotyledonous species and the model grasses.
Even for Arabidopsis, only a small percentage of genes have a biochemically validated functional annotation. The identification of mutants provides important clues to specific gene function in wall biogenesis. From the T-DNA insertional lines of Arabidopsis (Alonso et al., 2003), we generated over 1,000 homozygous lines mutated in annotated cell wall-related genes (http://cellwall.genomics.purdue.edu/families). To generate a comparable resource of maize mutants, we screened the UniformMu population (McCarty et al., 2005) by both forward- and reverse-genetics approaches. This collection has a uniform, inbred background, and the highly mutagenic lines (transposon-on) can be readily stabilized genetically (transposon-off) to prevent additional insertional events occurring in lines of interest (http://www.plantgdb.org/prj/). We constructed DNA grids from over 15,000 independent, pedigreed lines that enable PCR-based screening of the population for insertional mutants in selected cell wall genes. In addition, we identified Robertson's Mutator (Mu)-tagged maize cell wall mutants by analysis of Mu-flanking DNA sequences derived from the DNA grids and in a forward screen of mutagenized populations by near-infrared (NIR) spectroscopy.
From identification and assembly of the many families of cell wall-related genes in dicots and grasses, we estimate that plants devote 10% of their genomes to cell wall biogenesis (Yong et al., 2005). We have classified cell wall-related genes of Arabidopsis, rice, and maize into gene families whose products function in six stages of wall biogenesis: substrate generation, polysaccharide synthesis, membrane trafficking, assembling and turnover, secondary wall formation, and signaling (http://cellwall.genomics.purdue.edu; McCann and Carpita, 2005; Yong et al., 2005). Here, we present several examples of families from these six functional stages and their organization in species with different cell wall types. A database including other species except maize was developed that adopted our classification scheme (Girke et al., 2004), and an additional resource for a selected number of maize cell wall-related genes that are the most similar sequences to annotated Arabidopsis genes was reported (Guillaumie et al., 2007). However, as we describe here, the structure of the gene families in grasses and their comparative expression have diverged to such an extent that identification of only the closest Arabidopsis homologs will not provide a comprehensive approach to identify and characterize the unique genes that are responsible for the differences in the structures of type I and type II walls. More recently, the members of a great many of the glycosyl transferase (GT) gene families of rice were compared with those of several dicots and inferred to be largely nonorthologous (Cao et al., 2008). We report here a comprehensive inventory of maize cell wall genes, an analysis of comparative expression profiles in a single tissue type between rice, maize, and Arabidopsis, an effective means to generate and identify mutants with defects in cell wall-related genes, and a robust high-throughput screen for cell wall mutants independent of visible phenotypes yet reflective of cell wall composition and architecture.
RESULTS AND DISCUSSION
Building Families of Cell Wall-Related Genes in Arabidopsis, Rice, and Maize
Sequences of annotated cell wall-related genes of Arabidopsis and their paralogs were used in a BLAST search (Altschul et al., 1990) to find the putative orthologs and paralogous sequences in rice and, subsequently, the homologous sequences in maize. Until very recently, the maize sequence was largely unannotated and incomplete; thus, computer algorithms used to find common sequences based on keywords or full-length sequences, as described by Guillaumie et al. (2007), may have missed many relevant genes. We used the newly released maize genome sequence (Schnable et al., 2009) to attain a complete view of many cell wall gene families in maize for comparison with rice and Arabidopsis. Dendrograms of cell wall gene families of Arabidopsis, maize, and rice were developed for each species individually and in combination. In every instance where ambiguity as to classification or large family member expansions occurred, predicted protein sequences were compared and analyzed for motifs that validated the annotation using ProScan (Zdobnov and Apweiler, 2001).
About 60% of the Arabidopsis genome is annotated with respect to predicted function of its protein products (Swarbreck et al., 2008), and several Web sites have assembled gene families based on known functionalities. For example, the Carbohydrate-Active Enzyme database (http://www.cazy.org/) assembles families of GTs, glycosyl hydrolases (GHs), and other carbohydrate-metabolizing enzymes. There are 91 gene families of evolutionarily distinct GTs and 112 GHs, with Arabidopsis and rice genes populating 40 and 34 of them, respectively. Although the total number of GTs is higher in rice than in Arabidopsis, 550 versus 445, the numbers of GHs are about the same, 419 for rice and 403 for Arabidopsis. Only a few groups within families of cell wall-related genes have similar numbers of members for Arabidopsis and grasses (Table I). We observed differences in the number of members of a family, in numbers of members of a single group of a family, and in the presence of new family groups (or loss of family groups). This is likely a consequence of duplication and divergence in the genomes since the last common ancestor, resulting in splitting of a single gene function between paralogs (subfunctionalization), new function in a duplicate gene (neofunctionalization), or a combination of both events (subneofunctionalization). For example, the monosaccharide transporter genes of Arabidopsis and rice have duplicated, adapted, and diverged extensively (Johnson and Thomas, 2007). Differences in the duplication and retention of genes in animals and hominoid lineages are proposed to account for their evolutionary differences (Fortna et al., 2004; Hughes and Friedman, 2004). In 12 genotyped Drosophila species, 41% of the gene families have undergone expansion or contraction in different species, even though the overall gene number in the family remains relatively constant (Hahn et al., 2007). Furthermore, compared with sorghum (Sorghum bicolor; Paterson et al., 2009), maize appears to have retained significantly more duplications resulting from the tetraploidization that occurred before the maize-sorghum divergence rather than from independent tandem duplication (Schnable et al., 2009). As we describe below, EST/cDNA data support differential expression of each of the duplicates, indicating their neofunctionalization and subfunctionalization after the tetraploidization event.
Table I.
Numbers of cell wall genes and numbers expressed in developing ovaries by family and group in Arabidopsis, rice, and maize
4CL, 4-Coumaric acid CoA-ligase; C3'H, p-coumaroyl-shikimate/quinate 3′-hydroxylase; C4H, cinnamate 4-hydroxylase; CCoAOMT, caffeoyl-CoA 3-O-methyl transferase; F5H, ferulate (coniferyl alcohol/aldehyde) 5-hydroxylase.
| Designationa | Class/Family | Family/Group | No. of Genes (Expressants)b |
||
|---|---|---|---|---|---|
| Arabidopsis | Rice | Maize | |||
| 1.1 | Nucleotide-sugar interconversion | AUD/SUD | 6 (2) | 6 (5) | 9 (4) |
| AXS | 2 (1) | 1 (1) | 2 (1) | ||
| GAE | 6 (2) | 5 (4) | 9 (2) | ||
| GER | 2 (0) | 2 (1) | 1 (1) | ||
| GMD | 2 (1) | 1 (1) | 1 (1) | ||
| GME | 1 (1) | 2 (2) | 2 (1) | ||
| RHM/UER | 4 (4) | 3 (2) | 4 (2) | ||
| UGD | 4 (2) | 5 (2) | 3 (2) | ||
| UGE | 5 (1) | 4 (2) | 3 (0) | ||
| UXE | 4 (1) | 3 (3) | 4 (0) | ||
| Total | 36 (15) | 32 (23) | 38 (14) | ||
| 1.3 | Phenylpropanoid biosynthesis | COMT | 16 (0) | 6 (0) | 3 (1) |
| 4CL | 13 (3) | 14 (6) | 11 (5) | ||
| CCoAOMT | 7 (1) | 6 (3) | 6 (4) | ||
| PAL | 4 (0) | 9 (0) | 10 (4) | ||
| CAD | 9 (1) | 14 (1) | 7 (3) | ||
| C3'H, F5H, C4H | 6 (0) | 10 (1) | 8 (3) | ||
| CCR | 7 (2) | 24 (6) | 19 (4) | ||
| HCT | 19 (2) | 5 (3) | 38 (9) | ||
| Total | 81 (9) | 86 (20) | 102 (33) | ||
| 2.1 | CesA/Csl superfamily | CesA | 10 (4) | 10 (4) | 20 (9) |
| 2.2 | CslA | 8 (5) | 11 (5) | 10 (2) | |
| CslB | 6 (2) | 0 (0) | 0 (0) | ||
| CslC | 5 (3) | 6 (4) | 8 (2) | ||
| CslD | 5 (1) | 5 (1) | 5 (1) | ||
| CslE | 1 (0) | 3 (0) | 3 (0) | ||
| CslF | 0 (0) | 8 (2) | 7 (0) | ||
| CslG | 3 (3) | 0 (0) | 0 (0) | ||
| CslH | 0 (0) | 3 (1) | 0 (0) | ||
| Total Csl | 29 (14) | 36 (13) | 33 (5) | ||
| 2.3.1 | Family GT8 | A | 5 (1) | 4 (3) | 7 (4) |
| B | 8 (1) | 2 (2) | 2 (0) | ||
| C | 10 (5) | 8 (3) | 10 (4) | ||
| D | 15 (7) | 22 (14) | 25 (2) | ||
| E | 3 (2) | 5 (3) | 5 (0) | ||
| Total | 41 (16) | 41 (25) | 49 (15) | ||
| 2.3.2 | Family GT47 | A | 11 (3) | 11 (5) | 22 (2) |
| B | 8 (3) | 9 (6) | 9 (2) | ||
| C | 7 (0) | 6 (2) | 3 (0) | ||
| D | 8 (3) | 4 (3) | 5 (3) | ||
| E | 5 (3) | 9 (6) | 11 (9) | ||
| F | 0 (0) | 1 (1) | 4 (1) | ||
| Total | 39 (12) | 40 (23) | 54 (17) | ||
| 2.3.3 | Family GT37 | A | 10 (3) | 1 (0) | 1 (0) |
| B | 0 (0) | 9 (1) | 7 (1) | ||
| C | 0 (0) | 8 (3) | 11 (0) | ||
| Total | 10 (3) | 18 (4) | 19 (1) | ||
| 2.3.4 | Family GT34 | A | 3 (3) | 2 (1) | 6 (0) |
| B | 2 (0) | 2 (0) | 4 (1) | ||
| C | 1 (0) | 3 (1) | 1 (0) | ||
| D | 0 (0) | 2 (0) | 3 (0) | ||
| E | 2 (1) | 1 (1) | 4 (4) | ||
| Total | 8 (4) | 10 (3) | 18 (5) | ||
| 2.3.5 | Family GT31 | A | 12 (5) | 7 (3) | 11 (2) |
| B | 6 (3) | 10 (7) | 12 (5) | ||
| C | 3 (1) | 2 (2) | 3 (0) | ||
| D | 3 (3) | 2 (2) | 3 (2) | ||
| E | 8 (5) | 8 (5) | 7 (3) | ||
| F | 1 (1) | 10 (2) | 5 (0) | ||
| Total | 33 (18) | 39 (21) | 41 (12) | ||
| 4.1.1 | Expansin | α- | 26 (5) | 34 (7) | 36 (5) |
| α-like | 3 (0) | 5 (2) | 4 (2) | ||
| β- | 5 (1) | 18 (4) | 17 (6) | ||
| β-like | 1 (0) | 4 (0) | 0 (0) | ||
| Total | 35 (6) | 61 (13) | 57 (13) | ||
| 4.2 | XTH | A | 11 (3) | 3 (1) | 5 (2) |
| B | 15 (1) | 15 (2) | 17 (7) | ||
| C | 7 (1) | 11 (3) | 10 (5) | ||
| Total | 33 (5) | 29 (6) | 32 (14) | ||
| 4.3.3 | PGase | A | 15 (7) | 14 (9) | 14 (9) |
| B | 10 (3) | 0 (0) | 0 (0) | ||
| C | 8 (3) | 8 (4) | 8 (2) | ||
| D | 7 (0) | 10 (6) | 7 (0) | ||
| E | 8 (0) | 3 (0) | 3 (1) | ||
| F | 10 (2) | 0 (0) | 0 (0) | ||
| G | 8 (1) | 4 (0) | 2 (0) | ||
| H | 0 (0) | 6 (0) | 16 (0) | ||
| Total | 66 (16) | 45 (19) | 50 (12) | ||
| 4.5.1 | PME | A | 21 (5) | 18 (5) | 14 (7) |
| B | 11 (1) | 11 (3) | 11 (1) | ||
| C | 11 (5) | 1 (1) | 1 (1) | ||
| D | 14 (4) | 10 (5) | 7 (4) | ||
| E | 9 (2) | 1 (1) | 0 (0) | ||
| Total | 66 (17) | 42 (15) | 34 (13) | ||
| 6.4.8 | COBRA | 11 (2) | 11 (3) | 9 (4) | |
Our Web site classification number.
Values shown are numbers of genes and (in parentheses) numbers of genes expressed in developing ovaries.
Genes of Substrate Generation: Nucleotide-Sugar Interconversion Pathways
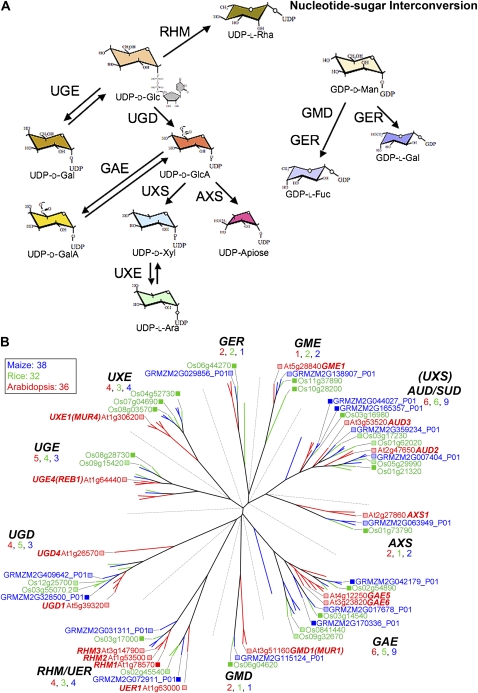
The numbers of genes in the 10 families that encode the enzymes of the nucleotide-sugar interconversion pathways, which are responsible for the formation of the basic sugar building blocks of many cell wall carbohydrates (Fig. 1A; Reiter and Vanzin, 2001), vary little between Arabidopsis and the grasses. These families are combined for convenience into one dendrogram, and evolutionary relationships are relevant within a single family (Fig. 1B). With the exception of maize duplications in several families, genes appear to be in orthologous relationships (Fig. 1B). Deoxysugars Fuc and Rha are characteristic of several polymers in type I walls but are of much lower abundance in type II walls. While Fuc is a key sugar in the structure of rhamnogalacturonan (RG) II and certain type I xyloglucans, it is absent from grass RG II (Thomas et al., 1989) and xyloglucan (Gibeaut et al., 2005). Still, Fuc is a component of N-linked glycoproteins, so retention of GMD and GER genes in rice and maize is expected (Fig. 1B).
Figure 1.
Genes of the nucleotide-sugar interconversion pathways. A, Schematic of pathways for plant nucleotide-sugar interconversion. The committed step to synthesis of uronic acids and pentoses is catalyzed by UDP-Glc dehydrogenase (UGD); isoforms exhibit different catalytic activities that indicate varied functions (Karkonen et al., 2005). The function of the UDP-GlcA decarboxylase (carboxyl-lyase) was established for the UXS family in barley (Zhang et al., 2005), with homology to the SUD/AUD group proposed for Arabidopsis (Reiter and Vanzin, 2001). Apiose, the essential monosaccharide in the boron didiester cross-linking of RG II, is synthesized by enzymes encoded by members of the AXS group, which converts irreversibly UDP-GlcA to a mixture of UDP-apiose and UDP-Xyl. A reduction in the levels of these synthases results in an RG II deficiency and cell wall abnormalities (Ahn et al., 2006). Although pectins are a minor component of the walls of grasses, an apiose-containing RG II with only slightly modified side groups is present (Thomas et al., 1989). B, These evolutionarily distinct families are combined for convenience into one dendrogram; evolutionary relationships are relevant only within a single family. Three groups of C-4 epimerases have been annotated: the UDP-Glc 4-epimerases (UGEs), including REB1, that interconvert UDP-Glc and UDP-Gal (Seifert et al., 2002; Nguema-Ona et al., 2006); the UDP-GlcA 4-epimerases (GAEs) that interconvert UDP-GlcA and UDP-GalA (Mølhøj et al., 2004; Usadel et al., 2004); and the UDP-Xyl 4-epimerases (UXEs), including MUR4, that interconvert UDP-Xyl and UDP-Ara (Burget et al., 2003). GDP-Fuc is synthesized de novo from GDP-Man via two enzymes, a 4,6-dehydratase (GMD), such as MUR1, and a 3,5-epimerase-reductase (GER; Reiter and Vanzin, 2001). For accession numbers of all genes in these families, see http://cellwall.genomics.purdue.edu/families/1-1/. Color coding for all dendrograms of gene families (Figs. 1–7; Supplemental Figs. S1–S3) are Arabidopsis (red), rice (green), and maize (blue), with numbers of genes in each group indicated. Expression levels for maize genes are indicated as the numbers of reads obtained in the sequencing runs, with 10 or more considered highly or moderately expressed (dark blue boxes), whereas one to nine reads are considered low expression (light blue boxes). Maize gene expression was compared with Arabidopsis and rice expression in developing ovary from public sources, primarily NCBI Gene Expression Omnibus (Barrett et al., 2007), as visualized in Genevestigator (https://www.genevestigator.ethz.ch/). Genevestigator was the primary model where expression in ovary is compared with that in other organs and tissues. Genes minimally expressed in Arabidopsis and rice are noted with light red and light green boxes, respectively, whereas genes moderately to highly expressed are noted with dark red and dark green boxes. Whether expressed or not, known mutants are indicated after the gene annotation.
Although UDP-Ara is synthesized in the pyranose form, a substantial portion of the Ara in cell wall polysaccharides is in the furanose form. Konishi et al. (2007) discovered that the “reversibly glycosylated protein” (RGP; Dhugga et al., 1997) can act as an isomerase that interconverts UDP-Arap and UDP-Araf. There are several maize and rice homologs to the five Arabidopsis RGP genes (Supplemental Fig. S1; Drakakaki et al., 2006). There is one known splice variant of the rice gene, Os03g40270; the three maize members closely related to Arabidopsis RGP3 share 93% protein sequence conservation. This could represent a recent duplication event, as only one rice RGP3 homolog (Os07g41360) is found associated with a fourth RGP3-related maize gene in a group separate from the five Arabidopsis RGP genes.
Genes of Substrate Generation: Phenylpropanoid Metabolism
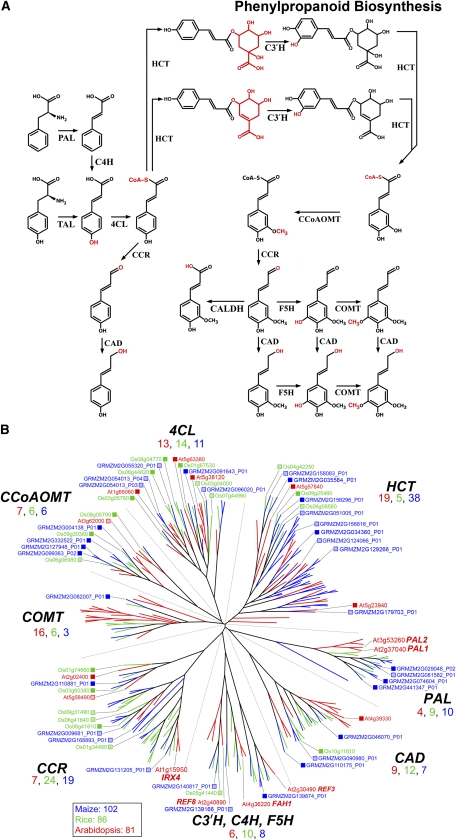
The large and diverse families associated with phenylpropanoid biosynthesis show an expansion of the grass genes relative to those of Arabidopsis, consistent with the relative importance of the phenylpropanoid network in walls of grasses (Carpita, 1996). As with the families of nucleotide-sugar interconversion, these families are combined into a single dendrogram, and evolutionary relationships are relevant within a single family (Fig. 2).
Figure 2.
Genes of phenylpropanoid substrate synthesis. A, The current view of the metabolic pathways from Phe or Tyr to hydroxycinnamic acids and monolignols. The PAL1 and PAL2 genes encoding PAL (Rohde et al., 2004) and CAD-C and CAD-D (Sibout et al., 2005) were identified as genes involved in lignification in the floral stem of Arabidopsis. The fah1 mutant lacking sinapate esters was characterized as ferulate (coniferyl aldehyde/alcohol) 5-hydroxylase (Meyer et al., 1996; Humphreys et al., 1999). An extension of a genetic screen for reduced epidermal fluorescence mutants resulted in the discovery of ref3 and ref8 (Franke et al., 2002a, 2002b), which were found to encode a cinnamate 4-hydroxylase (C4H) and a p-coumaroyl-shikimate/quinate 3′-hydroxylase (C3′H), respectively. The latter enzyme had been identified by Schoch et al. (2001) after a phylogenetic analysis of Arabidopsis cytochrome P450 enzymes. Identification of the substrate of this enzyme was aided by early studies on chlorogenic acid biosynthesis by Heller and Kühnl (1985) and Kühnl et al. (1987). Generation of p-coumaroyl-CoA from p-coumaric acid is catalyzed by HCT. This enzyme is also responsible for the transfer of the caffeoyl moiety of chlorogenic acid (caffeoyl-quinate) and caffeoyl-shikimate to CoA, based on studies by Hoffmann et al. (2003) that built on early studies by Stöckigt and Zenk (1974), Rhodes and Wooltorton (1976), and Ulbrich and Zenk (1980). Selection of irregular xylem mutants also proved helpful in identifying genes of phenylpropanoid metabolism, as the irx4 mutant was determined to result from a defective CCR gene (Jones et al., 2001). A knockout of CAFFEIC ACID O-METHYLTRANSFERASE1 (COMT1) results in lignins with strongly reduced levels of syringyl units in maize (Vignols et al., 1995) and, subsequently, in Arabidopsis (Goujon et al., 2003). 4CL, 4-Coumaric acid CoA-ligase; CALDH, coniferaldehyde dehydrogenase; CCoAOMT, caffeoyl-CoA 3-O-methyl transferase; F5H, ferulate (coniferyl alcohol/aldehyde) 5-hydroxylase; TAL, Tyr ammonia-lyase. B, These evolutionarily distinct families are combined for convenience into one dendrogram; evolutionary relationships are relevant only within a single family. Color scheme and dendrogram labeling are as described in the legend of Figure 1. For accession numbers of all genes in these families, see http://cellwall.genomics.purdue.edu/families/1-3/. Functional classification of phenylpropanoid genes in maize, rice, and Arabidopsis was based on characteristic signal peptides and motifs identified using InterProScan (http://www.ebi.ac.uk/Tools/InterProScan/) but do not imply that enzymatic function has been experimentally verified.
The biosynthetic pathways of monolignols and the hydroxycinnamic acid, ferulic acid, have been largely rewritten following elucidation of the mechanism by which phenylpropanoids are 3-hydroxylated (Schoch et al., 2001; Franke et al., 2002a, 2002b; Boerjan et al., 2003; Hoffmann et al., 2003) and the discovery of the coniferaldehyde dehydrogenase gene (Fig. 2A; Nair et al., 2004). All angiosperms make lignin, and eight large families of genes encode the many enzymes of the pathways to monolignol synthesis. Subgroups in each family have been expanded in the grasses (Fig. 2B). Phe ammonia-lyase (PAL) genes, which encode the enzymes of the first step in phenylpropanoid production (Boerjan et al., 2003), are resolved into three clades, one that is Arabidopsis specific and two that are grass specific, resulting in at least a doubling of the numbers of genes (Fig. 2B). In grasses, PAL also has Tyr ammonia-lyase activity (Roesler et al., 1997). The number of maize hydroxycinnamoyl-CoA transferase (HCT) genes, which encode enzymes necessary to enable 3′ hydroxylation, is remarkably expanded compared with other grasses, with 22 genes homologous with Arabidopsis sequences in one clade with no rice members and additional duplications in another HCT clade dominated by rice and maize (Fig. 2B). Grass-dominated clades are also observed in the gene families encoding cinnamyl alcohol dehydrogenase (CAD) and the cytochrome P450 monooxygenases (trans-cinnamate 4-hydroxylase, p-coumaroyl-shikimate/quinate 3′-hydroxylase, ferulate [coniferyl alcohol/aldehyde] 5-hydroxylase, caffeoyl-CoA 3-O-methyl transferase, and 4-coumaric acid CoA-ligase). The structure of the gene family encoding cinnamoyl-CoA reductases (CCRs) has an overall 3-fold expansion in numbers of grass sequences (Fig. 2B). Broad expansion and divergence among the grass-specific clades of families suggests novel functions associated with the synthesis of the complex phenylpropanoid network that is made in primary as well as secondary walls of grass species.
Genes of Polysaccharide Synthesis: Processive GTs
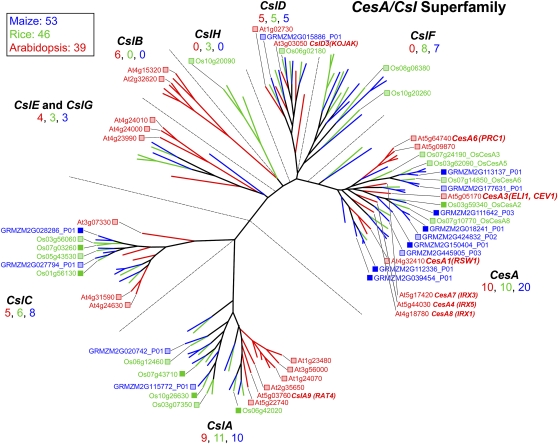
The CesA/Csl (for cellulose synthase-like) superfamily and callose synthases belong to the GT2 family and encode processive synthases of cellulose and other β-linked backbone polymers (Fig. 3; Delmer, 1999; Holland et al., 2000). First discovered in developing cotton (Gossypium hirsutum) fiber cells, plant CesA proteins possess four “U-domains” considered essential for catalysis of (1→4)-β-linked glucans in which one residue is oriented 180° with respect to each neighbor (Pear et al., 1996). The CesA genes of rice and maize appear to form orthologous clusters with Arabidopsis CesAs known to function at specific stages of cell growth and differentiation (Fig. 3; Holland et al., 2000; Vergara and Carpita, 2001; Appenzeller et al., 2004). At least three CesA genes are expressed in growing cells producing primary cell wall cellulose, whereas three different CesA genes are coexpressed in cells engaged in secondary wall cellulose synthesis in Arabidopsis (Taylor et al., 2003), rice (Tanaka et al., 2003), maize (Appenzeller et al., 2004), and barley (Hordeum vulgare; Burton et al., 2004). The clade structure and expression patterns suggest conserved cooperative functions for cellulose synthase catalytic units in primary versus secondary walls. However, six maize CesAs are duplicated relative to their rice and Arabidopsis putative orthologs.
Figure 3.
Genes of the CesA/Csl superfamily. At least three CesA genes are coexpressed during primary wall formation, and mutants in each of them, AtCesA1 (rsw1; Arioli et al., 1998), AtCesA6 (procuste; Fagard et al., 2000), and AtCesA3 (cev1; Ellis et al., 2002; eli1; Caño-Delgado et al., 2003), result in cellulose deficiencies, indicating that all three are essential for cellulose synthesis. The irregular xylem mutants AtCesA8 (irx1), AtCesA7 (irx3), and AtCesA4 (irx5) are deficient in cellulose synthesis specifically in secondary walls (Taylor et al., 2003). The root-hairless mutant kojak was traced to a mutation in the CslD3 gene proposed to be a cellulose synthase in these tip-growing cells (Favery et al., 2001). Rice CslD1 (Kim et al., 2007) and maize CslD5 are apparent orthologs, as mutations in each result in the reduced root hair phenotype. Heterologous expression of the Arabidopsis CslA9 in Drosophila cells in culture confirmed the role of this gene in mannan synthesis (Liepman et al., 2005). Expression of a barley CslF gene in Arabidopsis resulted in the de novo appearance of epitopes of the mixed-linkage (1→3),(1→4)-β-d-glucan (Burton et al., 2006), and characterization of a CslC coexpressed with a xyloglucan-specific xylosyl transferase in Pichia resulted in the synthesis of extended glucan polymers (Cocuron et al., 2007). Color scheme and dendrogram labeling are as described in the legend of Figure 1. See http://cellwall.genomics.purdue.edu/families/2-1/ for accession numbers of all genes in this superfamily.
The Csl genes encode proteins that each contain the four U-motifs but lack several key sequences of CesAs, such as the zinc-finger domains and/or plant-specific sequences (Delmer, 1999). An exception may be the CslD group, some of which encode the zinc-finger domains and may constitute the cellulose synthases of tip-growing cells of root hairs (Favery et al., 2001) and pollen tubes (Doblin et al., 2001), with CSLD1 (Os10g42750) in rice (Kim et al., 2007) having a similar role. Of the eight distinct classes of Csls, several are represented in both grasses and Arabidopsis. From heterologous expression studies, the CslA group encodes (1→4)-β-d-(gluco)mannan synthases (Dhugga et al., 2004; Liepman et al., 2005). While several Arabidopsis CslAs cluster, those of both rice and maize are more diverse (Fig. 3). CslCs encode β-glucans that are possibly the backbones of xyloglucans (Cocuron et al., 2007). The CslF group is unique to the grasses (Hazen et al., 2002) and encodes a synthase of mixed-linkage (1→3),(1→4)-β-d-glucans (Burton et al., 2006), a polysaccharide that among the angiosperms is unique to the grasses (Poales). It is the clearest example of the divergence of a group within an entire family that functions in the synthesis of a grass-specific polysaccharide. The functions of the CslBs, CslEs, and CslGs are unknown, but within the CesA/Csl superfamily CslE genes are the most similar to cyanobacterial CesAs, which are implicated as progenitors of modern plant cellulose synthase genes (Nobles and Brown, 2004). Inclusion of grass sequences alters the clade structures proposed for gene families when Arabidopsis sequences alone are considered (Hazen et al., 2002). For example, CslE and CslG may belong to the same clade; similarly, CslB and CslH are predicted to be derived from a common ancestral gene (Fig. 3).
Genes of Polysaccharide Synthesis: The Nonprocessive GTs
From over 35 nonprocessive GT gene families, we will focus on a subset of five. The families GT8 and GT47 are exceptionally well populated with members that function in substitution of cell wall pectic and cross-linking glycan backbones with several kinds of sugars. Family GT31 of galactosyl transferases may broadly function in the synthesis of the galactan backbones of AGPs. Families GT34 and GT37 encode xylosyl and fucosyl transferases involved in xyloglucan synthesis.
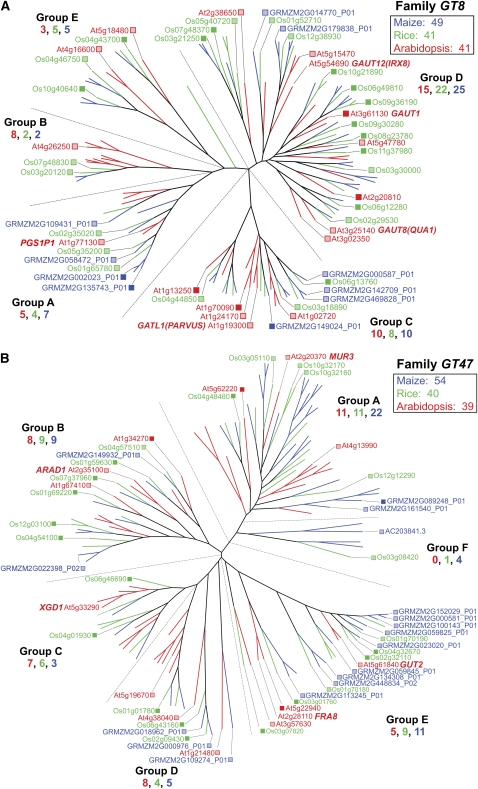
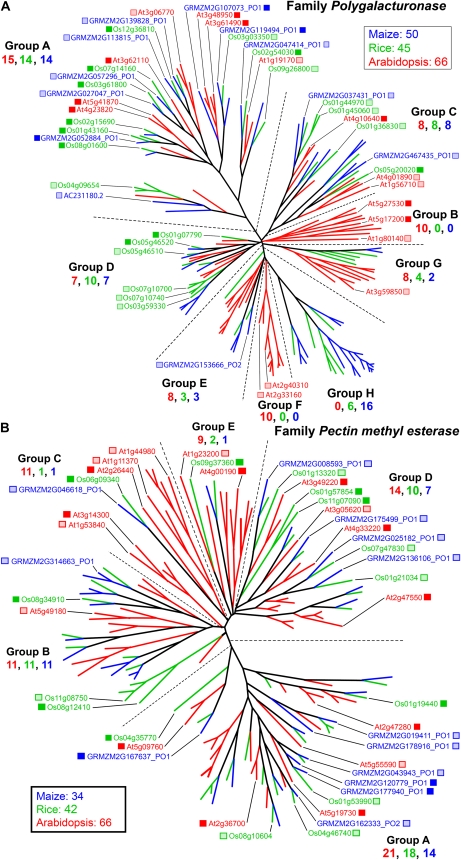
Family GT8 comprises over 40 genes in five groups in both Arabidopsis and the grasses (Fig. 4A). The functions of only a few genes have been determined. Group D, the largest, contains GALACTURONOSYL TRANSFERASE1 (GAUT1 [At3g61130]; Sterling et al., 2006), which encodes what is thought to be a synthase of homogalacturonans. Because GT8 family members encode transferases with retaining mechanisms of glycosyl transfer, resulting in α-linked polysaccharides, they are thought to be primarily involved in pectin synthesis (Scheller et al., 2007). However, α-linked residues are also found in many types of nonpectic polysaccharides, such as the (1→2)-α-GlcA of GAXs (Carpita, 1996), and an enzyme essential for glucuronoxylan synthesis is encoded by the group B gene GAUT-like (GATL1 [PARVUS]; At1g19300; Lee et al., 2007a). Despite roughly equal numbers of total GT8 genes, Arabidopsis group B genes are significantly expanded in number, whereas the grass genes are more abundant in groups D and E (Fig. 4A).
Figure 4.
Genes of major nonprocessive GT families. A, In GT8, the largest clade, group D, encodes GAUT1, the only protein established to iteratively extend GalA units typical of homogalacturonan (Sterling et al., 2006). This large family of retaining transferase genes encodes several putative GAUTs and three distinct subgroups of GATL proteins. GAUT8, or QUASIMODO1 (QUA1), is involved in the synthesis of RG II (Mouille et al., 2007), whereas the group C GATL1 and a secondary wall-associated group A member (Sterling et al., 2006) have no established function. The PARVUS gene (Lao et al., 2004) is involved in the synthesis of the tetrasaccharide primer of xylan synthesis (Lee et al., 2007a). See http://cellwall.genomics.purdue.edu/families/2-3-1/ for accession numbers of all genes in this family. B, Group A of GT47 contains MUR3, which encodes a galactosyl transferase that adds the (1→2)-β-d-Gal residue of the first xylosyl residue from the reducing end of the repeating heptasaccharide unit of xyloglucan (Madson et al., 2003). In group B, ARAD1 functions in (1→5)-α-l-arabinan synthesis (Harholt et al., 2006), and in group C, XYLOGALACTURONAN DEFICIENT1 (XGD1) encodes a xylosyl transferase that adds the (1→3)-β-Xyl units to homogalacturonan (Jensen et al., 2008). Group E contains FRAGILE FIBER8 (FRA8), which functions in the synthesis of the tetrasaccharide primer for xylan synthase (Peña et al., 2007), and putative glucuronosyl transferase genes, GUT1 and GUT2, that add substituents to secondary wall xylans (Zhong et al., 2005). Color scheme and dendrogram labeling are as described in the legend of Figure 1. For accession numbers of all genes in this family, see http://cellwall.genomics.purdue.edu/families/2-3-2/.
The five-group family GT47 in Arabidopsis encodes enzymes with at least four different types of transferase activities, including galactosyl, arabinosyl, xylosyl, and glucuronosyl transferases, all with inverting mechanisms of glycosyl transfer. Numerous duplications of both maize and rice genes are evident in comparison with their closest Arabidopsis homologs in groups A, B, and E. This resulted in a minimum of seven subclades for the grass genomes, one of which is so substantially diverged as to constitute a new grass-specific group F (Fig. 4B). In contrast, Arabidopsis genes form distinct subgroups with numerous duplications in groups A, C, and D compared with their grass homologs (Fig. 4B). With the exception of the group C XYLOGALACTURONAN DEFICIENT1 (At5g33290), which encodes a xylosyl transferase that adds the (1→3)-β-Xyl units to homogalacturonan (Jensen et al., 2008), the three other enzymatic activities are encoded by unclustered Arabidopsis genes. Of these, group A includes MUR3 (At2g20370), which encodes a galactosyl transferase that adds the (1→2)-β-d-Gal residue of the first xylosyl residue from the reducing end of the repeating heptasaccharide unit of xyloglucan (Madson et al., 2003). Group B includes ARABINOSE DEFICIENT1 (ARAD1; At2g35100), which is involved in the (1→5)-α-arabinan synthesis (Harholt et al., 2006), and group E includes AtGUT1 (At1g27440), AtGUT2 (At5g61840), and FRA8 (At2g28110), which encode glucuronosyl transferases (Zhong et al., 2005; Peña et al., 2007). While maize and rice homologs are observed for each of these Arabidopsis transferase genes, a significant challenge remains to establish the function of even the Arabidopsis genes of this large family.
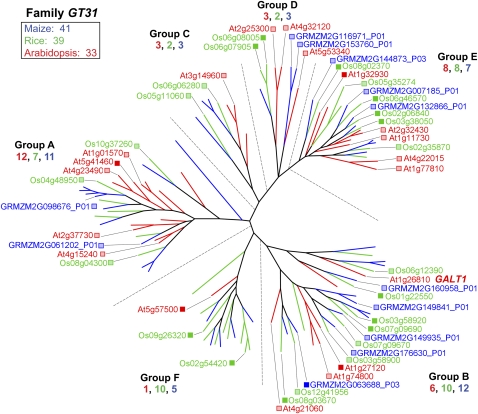
GT31 is a five-group family in Arabidopsis, for which a galactosyl transferase function associated with N-glycan formation is ascribed to a single member in group B (Fig. 5; Strasser et al., 2007). However, Qu et al. (2008) suggest that members of other groups of GT31 function in galactan backbone formation of AGPs. When grass sequences are included, a new group F emerges, which contains a single Arabidopsis gene with weak homology to genes in groups A and B. Five pairs of rice and maize genes in group F appear after the Arabidopsis-grass divergence (Fig. 5).
Figure 5.
Genes of family GT31. A single member of the GT31, GALT1 from group B, has been shown to be a galactosyl transferase required in the synthesis of N-glycan hybrid structures (Strasser et al., 2007). Recent bioinformatics approaches indicate that many of the members of this family may encode (1→3)-β-galactosyl transferases (Qu et al., 2008). For accession numbers of all genes in this family, see http://cellwall.genomics.purdue.edu/families/2-3-5/.
In Arabidopsis, the eight-membered family GT34 encodes transferases shown to xylosylate the glucan backbone of xyloglucan (Faik et al., 2002) and both (1→2)-α- and (1→6)-α-galactosyl transferases (Edwards et al., 1999). Inclusion of the rice (10) and maize (18) sequences expands the group structure of the gene family from three to five, with several duplications of grass genes homologous to a single Arabidopsis gene (Supplemental Fig. S2A). The XXT1 (At3g62720) and XXT2 (At4g02500) group genes encode the xyloglucan-specific xylosyl transferases (Cavalier et al., 2008). Family GT37 encodes fucosyl transferases, with MUR2 (FUT1; At2g03220) encoding the enzyme that forms the fucosylated trisaccharide side group of xyloglucan in Arabidopsis (Sarria et al., 2001; Vanzin et al., 2002). Consistent with the retention of genes to synthesize Fuc despite the lack of fucosylation of grass RG II (Thomas et al., 1989) and xyloglucan (Gibeaut et al., 2005), the maize and rice sequences fall into two new groups, B and C, distinct from Arabidopsis (Supplemental Fig. S2B).
From informatics-based analyses, GT43 has been predicted to contain genes encoding the (1→4)-β-d-xylan synthase of GAX (Mitchell et al., 2007), despite an expectation that the Csls would encode the processive synthases for (1→4)-β-linked glycans (Hazen et al., 2002). Peña et al. (2007) showed that the irx9 mutant phenotype was a result of the failure to elongate primed xylan chains. Based on the Pfam family PF02458 motif, the designation GT61 could be a misnomer, encoding not a sugar transferase but a GAX-specific feruloyl transferase (Mitchell et al., 2007). The GT77 members, Arabidopsis RGX1 and RGX2, encode (1→3)-α-d-xylosyl transferases thought to be involved in the synthesis of RG II (Egelund et al., 2006). Egelund et al. (2007) showed that the rrd1 and rrd2 mutants of Arabidopsis are Ara deficient in the wall residue after pectin and GAX extraction, consistent with a deficiency in extensin arabinosylation and with the appearance of homologs associated as the major GT family in Chlamydomonas, an organism with Hyp-rich protein walls (Lee et al., 2007b; U. Goodenough, personal communication).
Genes of Wall Assembly and Rearrangement
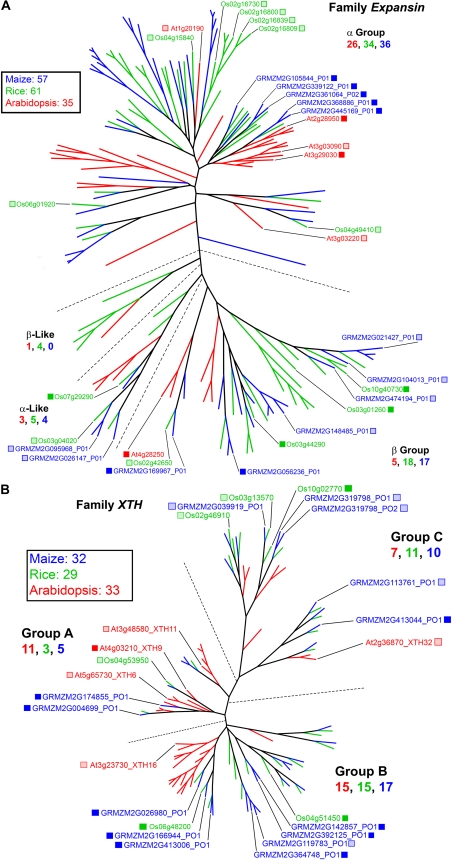
Expansins are involved in wall loosening during growth and do so by disruption of hydrogen bonds (Cosgrove, 2000). Members of the expansin superfamily are related to the GH45 family of glucosidase genes, but the proteins have no measurable glycolytic activity (Sampedro and Cosgrove, 2005). The Arabidopsis expansin family comprises 35 genes in two major groups, those encoding the α-expansins and β-expansins, and two minor groups encoding the α-expansin-like A and β-expansin-like B proteins (Fig. 6A; Sampedro and Cosgrove, 2005). In rice and maize, the expansin family is slightly expanded in the α-expansin group and greatly expanded in the β-expansin group containing the class 1 pollen allergens (Fig. 6A; Sampedro and Cosgrove, 2005; Valdivia et al., 2007). The grasses also have a subgroup of β-expansins, grass group 2 pollen allergens, which lack the GH45-related expansin domain (Sampedro and Cosgrove, 2005). The class B expansin and an original set of class A expansin genes show a conserved microsynteny across many grass species (Valdivia et al., 2007).
Figure 6.
Genes of wall assembly and rearrangement. A, The Expansin gene family. This family comprises two major groups, the α-expansins and β-expansins, and two minor groups, the α-like and β-like expansins (Sampedro and Cosgrove, 2005). The grasses have high numbers of α- and β-expansin genes in all groups, but Arabidopsis has a disproportionately high number of α-expansins. The crystal structure of one expansin, the β-expansin EXPB1 (Zea m1), a maize group 1 pollen allergen, has been solved (Yennawar et al., 2006). Color scheme and dendrogram labeling are as described in the legend of Figure 1. See http://cellwall.genomics.purdue.edu/families/4-1-1/ for accession numbers of all genes in this family. B, The XTH gene family. Three major groups of XTH genes have been identified, but transferase and/or hydrolase activities have not been systematically defined. Mutants with defects in XTH24 (meri5; Verica and Medford, 1997), XTH22 (tch4; Xu et al., 1995), and XTH28 (formerly XTR2; Akamatsu et al., 1999) all result in altered growth responses. Even though xyloglucan is a specific substrate, the xyloglucan-poor grasses have nearly equal representation in all three major groups. For accession numbers of all genes in this family, see http://cellwall.genomics.purdue.edu/families/4-2/.
Two large gene families function in cell wall expansion. The GH16 family contains the xyloglucan endo-β-transglucosylase/hydrolase genes (XTHs), so named because of the two distinct activities of enzymes they encode (Rose et al., 2002). Xyloglucan endo-β-hydrolase cleaves the glucan backbones of xyloglucan, a function essential for mobilization of seed reserve xyloglucan (Buckeridge et al., 2000), and the xyloglucan endo-β-transglucosylase (XET) cleaves xyloglucans but can ligate a donor chain to an acceptor chain, retethering xyloglucans during growth (Rose et al., 2002). The XTH family of Arabidopsis has 33 members assembled in at least three groups, and rice and maize have numerous members in all of them (Fig. 6B). Hrmova et al. (2007) suggested that, in the grasses, XETs may function to couple xyloglucans to cellulosic oligomers or to the mixed-linkage (1→3),(1→4)-β-d-glucan. However, because the rates of reaction with these alternative acceptor substrates are a small fraction of that of galactosylated xyloglucan, the physiological significance in vivo is still in question. In particular, group B XTHs show a tight association of Arabidopsis sequences, with a much looser clustering of grass homologs into three distinct subgroups. Group C is largely populated with grass members, whereas group A includes several Arabidopsis members (Fig. 6B). Xyloglucans can reach an abundance of about 10% of wall mass in growing tissues of grasses (Gibeaut et al., 2005), and XET activity may serve more significant functions in grasses than previously thought (Yokoyama et al., 2004).
Genes of Wall Modification and Disassembly
Most families contain larger numbers of maize and rice genes compared with those of Arabidopsis, but the smaller number of Arabidopsis genes is not strictly a result of its compact genome. This is particularly evident in the genes devoted to pectin metabolism, where the numbers of Arabidopsis genes greatly exceed those of the grasses (Fig. 7). The GH28 family that encodes polygalacturonases (PGases) and carbohydrate esterase family GE8 genes that encode pectin methylesterases (PMEs) constitute two of the larger families found in angiosperms (Kim et al., 2006; González-Carranza et al., 2007). The PGase family consists of seven groups, where multiple duplication events in groups B, E, and G resulted in a much larger number of genes in Arabidopsis than in maize or rice (Fig. 7A). Several multiple duplications are also observed in maize and rice in groups A, C, D, and H, suggesting novel subfunctionalizations in the grasses despite the low amounts of pectins in their cell walls. Group B is Arabidopsis only, and a novel grass-specific group H diverged from an Arabidopsis-only group F. Other subgroups are evident within the group nomenclature established for Arabidopsis sequences (Fig. 7A).
Figure 7.
Genes of pectin modification. A, The PGase gene family. The polygalacturonan hydrolase gene family in Arabidopsis comprises three groups based on protein structure, which can be further divided into subgroups (Kim et al., 2006; González-Carranza et al., 2007). Inclusion of maize and rice sequences defines a new group H with grass-only sequences. A mutation in a member of group A causes failure of separation of pollen tetrads (quartet3 [qrt3]; Rhee et al., 2003). See http://cellwall.genomics.purdue.edu/families/4-3-3/ for accession numbers of all genes in this family. B, The PME gene family. The original Arabidopsis gene family comprises five major groups (Louvet et al., 2006), of which groups C, D, and E are enriched in Arabidopsis sequences. The mutation qrt1 also results in a pollen cell-separation phenotype similar to qrt3 (Francis et al., 2006). For accession numbers of all genes in this family, see http://cellwall.genomics.purdue.edu/families/4-5-1/. Color scheme and dendrogram labeling are as described in the legend of Figure 1.
Similarly, family PME comprises five groups, three of which, C, D, and E, have reduced group members in rice and maize compared with Arabidopsis (Fig. 7B). However, the clustering of these few genes on several chromosomes suggests that they arose by consecutive tandem duplications. The presence of two grass-specific clusters in groups A and C, with one and no close Arabidopsis homolog, respectively, also suggests a novel subfunctionalization in the grasses.
The COBRA Gene Family
The COBRA gene family encodes glycosylphosphatidylinositol-anchored proteins that are associated with cellulose biosynthesis and orientation but for which the biochemical function remains unknown (Supplemental Fig. S3; Schindelman et al., 2001; Roudier et al., 2002, 2005; Brady et al., 2007). A mutation in the founding member of the COBRA family in Arabidopsis results in severe inhibition of root elongation and the progressive radial swelling of the cortical cells away from the root tip (Hauser et al., 1995). Rice Brittle culm1 (Bc1) is expressed primarily in vascular regions of the leaves and culm, and the mutant bc1 phenotype results in organ brittleness (Li et al., 2003). Mutations in Arabidopsis COBRA-like4 (COBL4), the closest homolog of the rice Bc1 gene, result in plants with normal morphology but stems with reduced tensile strength (Brown et al., 2005). The maize ortholog of COBL4 and Bc1 is Brittle stalk2 (Bk2), and there is an additional COBL4 like gene each in rice and maize. Ching et al. (2006) reported that the loss of mechanical strength in bk2 is due to a reduction in the synthesis of secondary wall cellulose, but Sindhu et al. (2007) propose that the protein functions in a patterning of lignin-cellulosic architecture that maintains organ flexibility, as the cellulose deficiency is present in juvenile plants but the brittle phenotype occurs only after the developmental transition to adult plants.
Expression Profiles of Genes for the Primary Cell Wall in Ovary Tissues
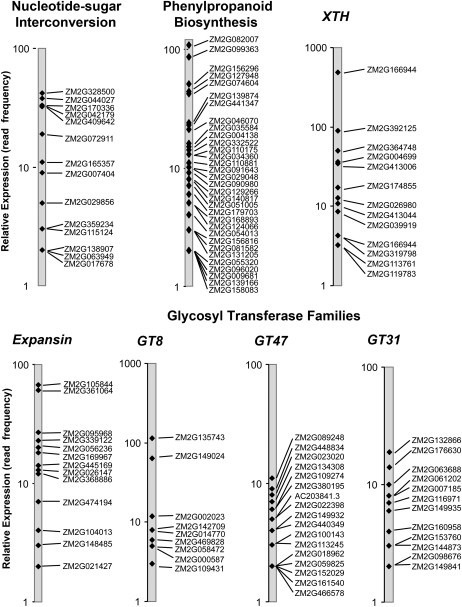
Massively parallel sequencing technologies enhance coverage and quantification of transcripts (Eveland et al., 2008). The long-read, 3′ untranslated region (UTR)-anchored, gene-specific transcript profiling strategy can readily distinguish closely related gene family members (Supplemental Table S1) and quantify their abundance across a wide range of expression (Fig. 8). In a study of developing maize ovaries, Eveland et al. (2008) resolved transcripts for most CesA genes of maize and found that CesA family members exhibited nearly 100-fold differences in transcript abundance.
Figure 8.
Expression profiles of cell wall genes in developing maize ovaries. Maize cell wall genes were classified by function according to pathway and/or gene family (the latter as defined in Figs. 1–7; Supplemental Figs. S1–S3). Relative transcript abundance within each class was quantified by the frequency of 3′ UTR-anchored cDNA sequences (number of reads) from each transcript in the data set of Eveland et al. (2008). The sequence counts for each transcript are plotted on a log scale (note range in abundance). Transcripts are labeled with their ZM2G maize sequence identifiers (maizesequence.org). The groups shown include two major biosynthetic processes, nucleotide sugar interconversion (1.1) and phenylpropanoid biosynthesis (1.3), and five large gene families, xyloglucan endotransglucosylase/hydrolases (4.2), expansins (4.1.1), GT8 (2.3.1), GT47 (2.3.2), and GT31 (2.3.5). See Supplemental Table S1 for mRNAs of cell wall genes not shown here.
As described here, the completion of the maize genome sequence and our annotation of the gene families allowed us to interrogate the database of transcript abundances for many additional, coexpressed members of several cell wall-related gene families. We compared Roche Applied Science 454-based sequencing of 3′ UTR-rich expression profiles generated from developing maize ovaries before pollination with those of the publicly available Arabidopsis and rice microarray data. The Genevestigator anatomy expression tool (Grennan, 2006; https://www.genevestigator.ethz.ch/), combined with the National Center for Biotechnology Information's (NCBI's) Gene Expression Omnibus (Barrett et al., 2007), allowed comparison of homologous sequences expressed in developing ovaries dissected from Arabidopsis and rice before pollination.
From 14,822 unique contigs searched by BLAST analysis, significant matches were made to 30% of the unique maize cell wall genes (167 of 556). In comparison, we identified matches for 28% of expressed cell wall genes in Arabidopsis (141 of 498) and 38% of expressed cell wall genes in rice (192 of 501; https://www.genevestigator.org/). Thus, 454 sequencing compares favorably with microarray analyses for capturing gene expression within a specific tissue when a complete sequence is available. In addition, the specificity of the 3′ UTR sequences often enabled resolution of expression for putative paralogous transcripts that aligned to a single cDNA sequence.
Transcript abundances for individual members of several cell wall gene families in maize also revealed wide differences in expression (Fig. 8). Arabidopsis and rice expression from public sources was compared with maize expression patterns in ovary tissue (Fig. 8; Supplemental Table S1) to identify putative orthologous and/or divergent cell wall-related genes. We assume that the functional equivalence of orthologous sequences requires homologous gene expression in the equivalent tissues of different species. For the genes of the nucleotide interconversion pathway, transcripts of at least one family member were detected for all 10 families of rice, nine in Arabidopsis, and eight in maize (Figs. 1B and 8). In families GME, RHM/UER, UGD, and GMD, apparently orthologous genes are expressed in all species, whereas in SUD/AUD (UXS), UGE, and GAE families, more divergent members are expressed in the grasses (Fig. 1B).
Maize and rice showed moderate to high levels of expression for multiple genes in all eight families involved in phenylpropanoid biosynthesis (Figs. 2B and 8). In contrast, very few Arabidopsis genes in these families were expressed in ovary tissue, with isolated instances of coexpression with the closest grass homolog. The much higher number of phenylpropanoid genes expressed in the ovary tissues of the grasses underscores the phenylpropanoid richness of even the primary wall of these species. Guillaumie et al. (2007) reported expression of a limited number of genes in the phenylpropanoid pathway, showing that some were more highly expressed in young stems, whereas expression of others was greater in developing internodes.
All three primary wall CesAs of Arabidopsis (CesA1, CesA3, and CesA6) were expressed in developing ovaries of the grasses, with six in rice and eight in maize (Fig. 3). The CslA and CslC groups show a large departure in sequence between the grasses and Arabidopsis, but few instances of expression of the closest homologs are observed between rice and maize. No expression of the maize CslF genes was detected in the ovary tissue (Fig. 3). The grass-specific (1→3),(1→4)-β-d-glucan synthase encoded by these genes is typically absent from the meristematic tissues, and the β-glucan is synthesized in abundance only when cell expansion ensues (Carpita, 1984; McCann et al., 2007).
For the two major GT families, GT8 and GT47, many Arabidopsis, rice, and maize genes are expressed, but the closest grass homolog to an Arabidopsis gene is rarely expressed (Fig. 4). In GT8, Arabidopsis and rice have at least one member expressed from each of the five groups (Fig. 4A). Three members of maize GT8 are highly expressed in developing maize ovaries (Figs. 4A and 8). Two members in group A are related to Arabidopsis PGS1P1 (At1g77130), and the third is related to group C genes GATL1 (PARVUS; At1g19300; Lee et al., 2007a) and rice Os04g44850.
Family GT47 contains at least one Arabidopsis member expressed in four of its five represented groups (Fig. 4B). Group E is dominated by grass genes, with a total of six rice and nine maize genes, four of the maize genes being coexpressed with rice genes. Two clades of group D show Arabidopsis and maize coexpression, whereas group C shows only rice expression. In group B, grass homologs closest to the arabinosyl transferase gene ARAD1 (At2g35100; Harholt et al., 2006) are expressed, but several other related grass genes are also expressed, including one each of maize and rice in the new group F (Fig. 4B). Collectively, a diverse group of at least 17 different GT47 genes are expressed in developing maize ovaries (Fig. 8). Neofunctionalization of arabinosyl transferases might be expected in the grasses. For example, in grasses, the arabinosyl units are attached mostly on the O-3 positions along the xylan backbone, whereas in type I walls, the arabinosyl units are mostly on the O-2 positions (Carpita and Gibeaut, 1993). Similarly, the expression of the closest homolog between Arabidopsis and the grasses in families GT31 (Fig. 5), GT34, and GT37 (Supplemental Fig. S2) is the exception rather than the rule. Expression within the GT31 family varied over a 10-fold range in developing maize ovaries (Fig. 8).
Thirteen unique maize expansin family members were expressed (Fig. 6A), with mRNAs from all of these abundant in the maize developing ovary (Fig. 8). These transcripts included five α- and six β-group members. Numerous rice expansin genes (13) were also expressed (Fig. 6A), compared with only six in Arabidopsis, with five of the latter limited to the α-group (Fig. 6A). No Arabidopsis genes encoding α-like or β-like expansins were found to be expressed in the developing ovary of that species, whereas two each of the maize and rice genes encoding α-like expansin genes were expressed. The large increase in numbers of expansin genes expressed in grasses compared with Arabidopsis indicates that cell wall loosening by expansins may play a greater role in development of ovary tissue in grasses.
Five members of family XTH showed significant expression in Arabidopsis, compared with 13 in maize and six in rice, with at least one of these genes from each of three XTH subgroups (Fig. 6B). None of these genes was a close homolog between Arabidopsis and either of the grasses (Fig. 6B). At least 11 maize XTHs were among the abundantly expressed genes in developing ovaries (Fig. 8), consistent with their possible contribution to expansion and/or remodeling of the xyloglucan-rich primary cell walls of grasses (Rose et al., 2002, Yokoyama et al., 2004, Gibeaut et al., 2005).
Arabidopsis has markedly expanded PGase and PME gene families compared with the grasses, but multiple members of many groups were highly expressed in Arabidopsis, maize, and rice ovary tissues. For PGases, 16 Arabidopsis, 20 rice, and 12 maize genes show significant expression, with most expressed from group A and one closely homologous expression group for all three species (Fig. 7A). Group B has two highly expressed genes in this Arabidopsis-only grouping. For PMEs, the 17 Arabidopsis, 15 rice, and 13 maize genes showed significant expression in most groups, but the single maize gene in group E was not expressed. In only two instances was expression of the closest homologs between all three species observed (Fig. 7B).
To summarize, the large differences in expression patterns between Arabidopsis and the grasses with respect to the cell wall gene families underscore the vast amount of work needed to establish specific gene function. For small gene families, such as those of nucleotide-sugar interconversion, orthology can be reasonably inferred by expression of closely related homologs within similar cell types. However, our analyses show that this is a rare occurrence rather than a common one. For large families, such as those of GTs, expansion of certain clades through successive duplications in maize and rice indicates a large potential for neofunctionalization and subfunctionalization during evolution. Such relationships are especially prominent in families such as the PGase, PME, and those associated with the phenylpropanoid synthesis pathway, which may relate to the specific architecture of the different cell wall types. While common motif predictions clearly classify genes to a family, protein modeling and BLAST score analyses are currently insufficient to make specific predictions of gene function and orthology. Thus, what is needed is an empirical approach to gene function, such as the characterization of mutants in genes known to be expressed in a cell-specific context. In addition, expression analysis using large-scale techniques, such as 454 sequencing of 3′ UTRs, allows for rapid assignment of expression patterns that, if performed in enough tissue types or time courses, could help to elucidate function in space and time of specific duplicated genes or altered gene patterns in different cell wall types.
Forward and Reverse Genetics to Identify Specific Mutants in Wall-Relevant Genes
For Arabidopsis, the T-DNA insertional mutant lines have provided an invaluable resource for genetic functional analyses (Alonso et al., 2003). The UniformMu population was developed by introgressing Robertson's Mu into genetically uniform W22 and B73 inbreds (McCarty et al., 2005; Settles et al., 2007), and a database was developed as a similar resource to facilitate high-throughput molecular analysis of Mu-tagged mutants and gene knockouts in maize (http://www.maizegdb.org/documentation/uniformmu/). High-throughput sequencing of Mu insertion sites in UniformMu lines has enabled in silico mapping of random Mu insertions in the maize genome (McCarty et al., 2005) and facilitated cloning of transposon-tagged mutants (Suzuki et al., 2005, 2008). Hence, the UniformMu resources enable both forward- and reverse-genetics applications. Over 100 putative transposon knockouts of maize genes have been analyzed by gene-specific PCR and tested to confirm heritability (Settles et al., 2007). We identified 72 insertion sites in 63 maize cell wall genes (Supplemental Table S2). While not meant as an exhaustive search for knockouts of all maize genes, it represents a substantial increase over the handful of previously known cell wall-related mutants in maize. This resource will continue to grow as current and new Mu insertions are placed with the appropriate genes and seed stocks (http://www.maizegdb.org/documentation/uniformmu/).
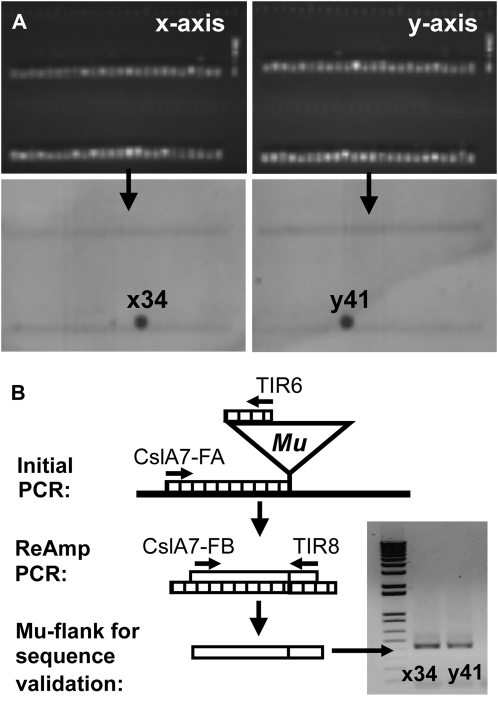
To identify mutants with Mu inserts in specific genes, we have developed DNA grids for high-throughput screening of the UniformMu population. These grids can be screened by PCR with Mu- and gene-specific primer pairs. Resulting candidate mutants are sequence verified and tested for heritability in progeny of the appropriate line. Each UniformMu grid allows concurrent screening of DNA from 2,304 lines in a 48 × 48 arrangement of pooled extracts. Individual lines are traced by the presence of their DNA in given pools from the x and y axes. These materials include a total of eight UniformMu grids, allowing 18,432 lines to be tested. All lines chosen for the grid are transposon-off and genetically stable mutants.
To identify putative Mu inserts in the gene of interest, grids were screened with gene-specific primers in combination with a second primer designed to hybridize with diverse Mu terminal inverted repeats (TIR6; Fig. 9). Two PCR methods were also tested for each gene screened, since standard PCR protocols worked best for some genes and touch-down PCR for others (for the latter, PCR annealing temperatures were decreased in increments so that they could “touch down”). Products were tested by hybridizing Southern blots of resulting PCR products with gene-specific 32P-labeled fragments from wild-type genes. Positive results were followed by dilution of the initial PCR product (from TIR6 + gene-specific primer) and reamplification using nested Mu primers (TIR8 + gene-specific primers where available; Fig. 9). The second product was sequence verified, and plants were grown to test heritability of the mutation. Progressive adaptation of this screening procedure resulted in a many-fold enhancement of specificity for the target genes and gave rise to confirmed gene-specific insertions (Supplemental Table S2).
Figure 9.
Example of results obtained during a reverse-genetics screen for Mu inserts in CslA7 using the UniformMu DNA grids. In this instance, no phenotype was detected, but visible features appear in other mutants (e.g. cslD5 in Fig. 10). A, Top panels show PCR products obtained from a screen of x and y axes from a given DNA grid. A total of 48 lanes appear in double rows on each panel, and each lane represents a pooled fraction of DNA from 48 families (one to two plants each). Collectively, 2,304 families are screened in each grid. Bottom panels localize a gene-specific Mu insert to a grid coordinate of x34, y41 via hybridization to a gene-specific 32P-labeled probe. B, Nested primers are used to amplify a selected Mu flank for sequence validation using a combination of gene-specific and Mu-specific primers. Here, forward primers FA and FB are used for CslA7, in respective combination with the TIR sequences TIR6 and TIR8, characteristic of Mus. The process progresses from the original plant DNA (black line), to the initial PCR product (striped line), and then to the final product (white line) used for sequencing. Confirmation of a Mu insert is followed by field and greenhouse tests for heritability and possible phenotypic features.
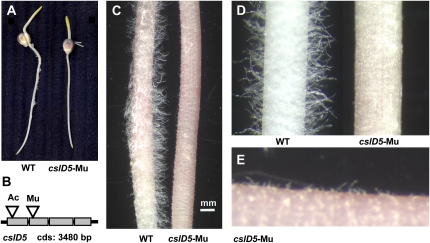
Initial work was directed toward identifying mutants in the Csl gene families. Plants carrying homozygous Mu insertions in CslD5 showed a clear root hair-deficient phenotype (Fig. 10A) and were similar in both Mu- and Ac-induced knockouts of this gene (Fig. 10B) and the Arabidopsis kojak mutant (Favery et al., 2001). In the maize cslD5 mutant, root hairs initiate but fail to elongate to the degree observed for wild-type root hairs (Fig. 10, C–E). Microscopy of live, tip-growing root hairs (data not shown) indicated that those of mutant seedlings were susceptible to bursting at their tips, thus prematurely terminating their elongation. There were no aboveground phenotypic features evident for cslD5 mutant plants in field or greenhouse analyses. Linkage between the root hair-deficient phenotype and the Mu insert in CslD5 was confirmed by testing for a similar phenotype in an Ac insertional allele (accession no. AC027037) obtained from the Ac/Ds resources (Kolkman et al., 2005). These results confirm a conserved role of csld5 orthologs in root hair elongation by Arabidopsis, rice, and maize (Fig. 10; Favery et al., 2001; Kim et al., 2007). Other Mu transposon insertions identified in Csl genes, such as cslA7, have no obvious phenotype.
Figure 10.
Phenotype of a Mu insertion in CslD5. A, Seedlings are root hair deficient. B, Both Mu and Ac inserts near the 5′ end of the CslD5 gene confer a similar phenotype. C to E, Progressive magnification of wild-type (WT) and cslD5 mutant roots reveals the extent of surface differences. Note that although the cslD5 roots appear hairless, they retain a capacity to initiate, but not necessarily elongate, hairs (note hair initials in E.). [See online article for color version of this figure.]
Screening for “Invisible” Phenotypes
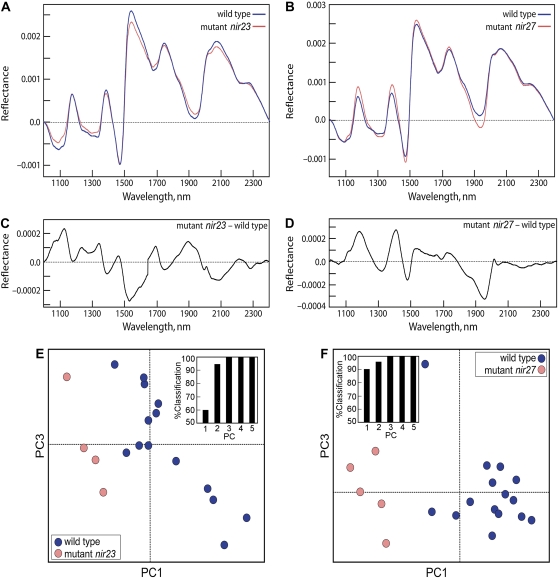
The forward-genetics screen of the UniformMu population identified independently 29 maize mutants with distinct visible phenotypes, including dwarfs, mutants with altered morphology or altered leaf texture, mutants affecting cell-cell adhesion, and a novel brown midrib mutant (Supplemental Fig. S4). We also used NIR spectroscopy in a high-throughput screen for alterations in wall composition in leaves for which no visible phenotype was obvious (Vermerris et al., 2007). We acquired NIR spectra from mature dried leaves representing 2,200 segregating F2 families of the UniformMu population. Spectral data were subjected to a multivariate statistical analysis designed to identify unusual spectra indicative of putative mutants. Only those Mu insertion lines where 15% to 30%, or three to six per 20 plants of the F2 siblings, gave similar “spectrotypes” that were distinct from the wild type were counted in this total. Of several hundred lines that were tentatively selected as potential mutants, 39 NIR spectrotype (nir) mutants were identified (1.8% of the lines screened) and found to be heritable. Six of these nir mutants displayed subtle changes in leaf texture, color, architecture, or disease susceptibility/disease lesion mimic, but the other 33 were indistinguishable from the W22 wild-type controls under normal field conditions. The NIR spectra, principal components analysis (PCA), and a characterization of probable chemical phenotypes for all of these are provided at our Web site (http://cellwall.genomics.purdue.edu/families/7/). Examples of two of these mutants with distinct spectral differences are shown in Figure 11.
Figure 11.
Classification of maize mutants nir23 (A, C, and E) and nir27 (B, D, and F) based on NIR spectra. A and B, Baseline-corrected and area-normalized NIR spectra for the mutant (red) and wild-type control (W22; blue). C and D, Digital subtractions (mutant – wild type) of the spectra shown in A and B, respectively. E and F, PCA score plots showing how the mutant and wild-type leaf samples can be distinguished from each other based on their spectral characteristics. The insets show the percentage of correctly classified samples using a multivariate model with increasing numbers of PCs. In both mutants, a model based on the first three PCs results in 100% correct classification. NIR spectra and their digital subtractions from W22 for all 39 nir mutants can be found at http://cellwall.genomics.purdue.edu/families/7/. [See online article for color version of this figure.]
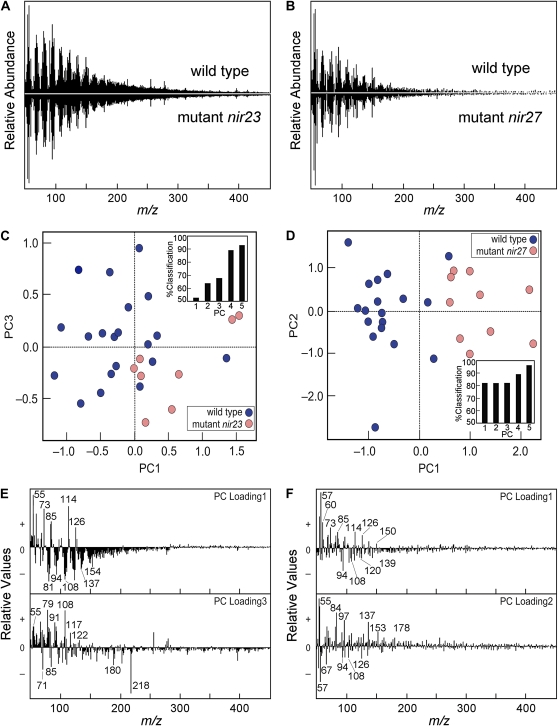
The cell wall compositions of the nir mutants were analyzed using pyrolysis-molecular beam mass spectrometry (PyMBMS) as described (Evans and Milne, 1987; Tuskan et al., 1999). This method relies on thermal degradation of the cell wall constituents under anoxic conditions to provide information about hexose and pentose content and the content and composition of phenolic compounds derived from lignin and hydroxycinnamic acids. PCA of the PyMBMS spectra from isolated cell walls showed that at least six of the 39 nir mutants had altered carbohydrate-lignin interactions of different kinds. When applied to PyMBMS profiles, PCA resolves the nir23 and nir27 mutants based on differences in the relative abundance of ions diagnostic for carbohydrate and aromatics (Supplemental Table S3), indicating a low-lignin phenotype (Fig. 12). Seeds from the 39 nir mutants are available through the Maize Genetics Stock Center (http://maizecoop.cropsci.uiuc.edu/).
Figure 12.
Characterization of mutants nir23 (A, C, and E) and nir27 (B, D, and F) based on PyMBMS analysis. A and B, Mass spectra of the wild type (top) and mutant (bottom; mirror image) in the m/z 50 to 450 range. The spectra were normalized to total ion current. C and D, PCA score plots showing how the mutant and wild-type leaf samples can be distinguished from each other based on their mass spectra. The insets show the percentage of correctly classified samples with increasing numbers of PCs. E and F, Loadings of the two PCs represented in score plots C and D, respectively, with a set of diagnostic ions marked. Diagnostic masses for known carbohydrate and aromatic fragments are provided in Supplemental Table S3. Populations of samples of mutant and W22 (wild type) are classified correctly on the basis of five PCs, accounting for over 90% of the correct classification for both mutants. Loadings of PC1 to PC3 indicate that the cell walls of the mutant have a higher carbohydrate-to-lignin ratio in the mutant, as evidenced by the following ions in PC1 of m/z 55, 73, 85, and 98 diagnostic of carbohydrate, m/z 114 specifically from pentoses, and m/z 126 specifically from hexoses, whereas the other peaks represent either hexoses or pentoses. Based on the presence of m/z 150 in PC1, the cell walls of mutant nir27 contain more ferulic acid. High PC1 translates to less phenolic compounds, including negative values for mass fragments from phenol (m/z 94) and G- and S-lignin derivatives from lignin, such as guaiacyl (m/z 124), 4-vinylphenol (m/z 120), ethylguaiacyl (m/z 137), syringol (m/z 154), isomers of eugenol (m/z 164), and coniferaldehyde (m/z 178). [See online article for color version of this figure.]
CONCLUSIONS AND FUTURE PROSPECTS
Genes that encode the enzymes for cell wall biogenesis are assembled in families common to all plants. However, publication of the maize genome sequence (Schnable et al., 2009) has allowed a comparative genomics analysis with rice and Arabidopsis genome sequences that highlights distinctions in the family substructure for cell wall-related genes of the grasses. In a few instances, homologous sequences indicate potential orthologous functions, whereas in others, a clear expansion of groups is observed that suggests the evolution of novel functions specific to the type II-walled grasses or the type I-walled Arabidopsis. Expression profiling by massively parallel cDNA sequencing indicates that many of these novel genes are expressed in addition to the much smaller proportion of the apparent orthologs. We conclude that potential orthologs cannot be identified solely by identifying sequences with the highest sequence similarities.
As the C4 grasses are key bioenergy species (Ragauskas et al., 2006; Carpita and McCann, 2008), knowledge of cell wall structure and biosynthesis will be pivotal to improve biomass yield and quality. Gene discovery in maize can be translated to increased yields, improved digestibility, and saccharification potential by the microsynteny among bioenergy grass genomes. As described here, forward- and reverse-genetics screens of a genetically stabilized insertional mutant collection like UniformMu (http://maizeGDB.org/documentation/UniformMu) offer a powerful means to establish gene function comparable to the resources available for Arabidopsis. Activator (Bai et al., 2007) and RescueMu (Fernandes et al., 2004) are additional resources of insertional lines for forward- or reverse-genetics screens, and TILLING (for Targeting-Induced Local Lesions in Genomes) in maize (Weil and Monde, 2007) provides a means of generating rich allelic series in essentially any gene. The natural diversity of a population can be exploited in conventional breeding, but having well-mapped populations of recombinant inbred lines, such as the intermated B73 × Mo17 (IBM) population, allows rapid identification of quantitative trait loci of the genes relevant for biomass improvement (Lee et al., 2002). In fact, Hazen and colleagues (2003) discovered several quantitative trait loci affecting cell wall sugar composition in the caryopsis pericarp using the IBM lines. The Nested Association Mapping lines, 200 recombinant inbred lines each derived from crosses of B73 with 25 diverse inbreds, includes many high-biomass tropical maize inbreds that capture a substantial amount of the existing genetic diversity of the species (Yu et al., 2008; http://www.panzea.org). These rich genetic resources greatly facilitate the discovery of biomass-relevant genes, especially when coupled with the rapid screens of carbohydrate and aromatic composition by NIR spectroscopy and PyMBMS that we describe here.
Not only for biofuels production but also for food and feed derived from grass species, maize offers an ideal model system because of a sequenced genome, an accounting of a great many cell wall-related genes, a single-gene knockout strategy, high-throughput screens for nonvisible phenotypes, and a vast genetic diversity that can be exploited in testing individual and multiple gene effects (Carpita and McCann, 2008). The next challenge will be to define the novel functions of grass cell wall-related genes at the biochemical level and their roles in plant growth and development.
MATERIALS AND METHODS
Arabidopsis Family Construction
Construction of most Arabidopsis (Arabidopsis thaliana) families was described previously (Yong et al., 2005). Newly added or expanded gene families are as follows: GT8 from Sterling et al. (2006); XTHs from Yokoyama et al. (2004); Expansins from Sampedro and Cosgrove (2005); and the COBRA glycosylphosphatidylinositol-anchored proteins from Roudier et al. (2002).
Rice and Maize Sequences
Rice (Oryza sativa) gene families were constructed by querying the most current rice peptide sequence database from the J. Craig Venter Institute (http://www.jcvi.org; formerly www.tigr.org) with Arabidopsis protein sequences by downloading the flat file and converting it to a usable database for NCBI's BLAST with formatdb (Altschul et al., 1990). A custom DOSshell script was used to direct the BLAST through multiple sequence files using the following parameters: protein-protein BLAST search, expect value of 10−20, and no alignment output. The BLAST results were parsed using a custom C++ script to scan and place the queried Arabidopsis gene name, associated rice gene names, and match score values for any score greater than 200 into a Microsoft Excel file. Duplicate matches due to multiple hits to the same rice sequence from closely related Arabidopsis sequences were eliminated to generate a unique rice gene list to extract sequences from the rice database using the fastacmd program from NCBI (Altschul et al., 1990) in a custom DOSshell script.
Maize (Zea mays) gene families were identified as described for rice genes using the newly completed gene annotations from the maize sequencing project (Schnable et al., 2009; http://www.maizesequence.org). All protein sequences used in the construction of the dendrograms are available at our Web site (http://cellwall.genomics.purdue.edu).
Sequence Alignments and Dendrogram Development
Dendrograms were assembled from protein-coding sequences by the neighbor-joining method in ClustalW (Saitou and Nei, 1987; Chenna et al., 2003). The parameters used were for a slow, accurate tree with gap open penalty of 10, gap extension penalty of 0.05, and a Gonnet weight matrix for proteins for multiple alignments; a gap open penalty of 10, gap extension penalty of 0.1, and a Gonnet weight matrix for proteins for pairwise alignments; and bootstrapped 1,000 times. After the initial multiple alignment, individual clade alignments were checked using Multalin (Corpet, 1988; http://www-archbac.u-psud.fr/genomics/multalin.html). Matches to conserved regions within groups of family clades with suspect alignments were manually checked using InterProScan (Zdobnov and Apweiler, 2001; http://www.ebi.ac.uk/Tools/InterProScan/), and nonmatching members of the families were removed. Dendrograms were drawn using TreeDyn (Chevenet et al., 2006; http://www.treedyn.org/). The dendrograms exist as FLASH files with interactive links on our Web site (http://cellwall.genomics.purdue.edu).
To evaluate relative transcript abundances of phylogenetically classified maize cell wall genes in an expanding tissue (Figs. 1–8), maize cell wall genes were matched (greater than 85% identity) to 3′ UTR-anchored cDNA consensus sequences in the developing ovary dataset of Eveland et al. (2008) using BLASTN. Transcript abundance was quantified as the number of 454-sequence reads assigned to each consensus sequence (Eveland et al., 2008).
Rice and Arabidopsis Sequence Location and Protein Motif Analyses
Sequence locations and distances were verified using the Gbrowse designed for rice (http://www.jcvi.org) and Arabidopsis (http://www.arabidopsis.org). Proteins were checked for conserved motifs using ProScan (Zdobnov and Apweiler, 2001) from the European Bioinformatics Institute Web site (http://www.ebi.ac.uk/interpro/).
Mu Flank Matches to Maize Genes
A data set of transposon-flanking sequences was constructed from the B73 draft genome by extracting 1-kb segments flanking 2,725 independent, germinal insertion sites identified in UniformMu (MaizeGDB.org/documentation/UniformMu/). The flanking sequences were annotated by BLASTX searches of rice (Michigan State University Osa1; http://rice.plantbiology.msu.edu/) and Arabidopsis (Arabidopsis Genome Initiative) protein sequence databases using a cutoff expect value of 10−7.
Sequences related to individual maize genes for each family were found using BLASTN using an expect value of 10−10 and a score of greater than 100. The results were parsed with a custom C++ script and unique Mu flanks matched with their best corresponding maize gene sequence. Some variation from exact matches was allowed due to comparing sequences of W22 with B73 inbreds; however, most expect values were well above 10−100.
Screening of the UniformMu Population by NIR
Over 2,200 F2 families of a segregating UniformMu population were sown in 16 to 20 seeds per 17-foot (5.2 m) row, with 30-inch-wide (76.2 cm) spacing between rows (over 40,000 plants) during the 2003 and 2004 seasons on irrigated fields, rotated with soybeans (Glycine max), at Purdue University's Agronomy Center for Research and Education in West Lafayette, Indiana. Plants were bar-code tagged with unique identifiers at the five-leaf stage, and visible segregant phenotypes were documented photographically. In a single day of collection, a crew of 16 workers excised 7.5-cm sections at mid-leaf blade from each plant at mid height (adult leaf 5 or leaf 6), mounted them flat with bar-code tags in glassine envelopes, and air dried them at 50°C. The majority of the viable plants were self-pollinated to preserve potential homozygous mutant lines.
NIR spectra (average of 30 spectral acquisitions) were obtained from dried leaf samples using a hand-held probe connected to a FieldSpec Pro NIR spectrometer (Analytical Spectral Devices). The reflectance of light from the leaf segment in the range between 350 and 2,500 nm was recorded relative to a Gore-Tex disc as white reference. Baseline-corrected and area-normalized data sets of the spectra were then used in the chemometric analyses. Most of the PCA was carried out using WIN-DAS software (Kemsley, 1998). Multivariate partial least squares and some of the PCAs were also carried out using Matlab 6.5.1 (The MathWorks). Linear discriminant analysis was used to develop a discriminative calibration model to classify spectra into groups. Mahalanobis distance was used as the distance metric to measure the distance of each observation (spectrum) from each group center. Linear discriminant analysis using squared Mahalanobis distance metrics was applied to the PCA scores of original data (Kemsley, 1998).
PyMBMS
A custom-built molecular beam mass spectrometer using an Extrel model TQMS C50 mass spectrometer was used for pyrolysis vapor analysis (Evans and Milne, 1987; Tuskan et al., 1999). The quartz pyrolysis reactor described previously was replaced with a commercially available pyrolysis unit and autosampler (Frontier model no. PY-2020 iD). Minor modifications were made to incorporate the autosampler inlet pyrolysis system onto the molecular beam mass spectrometer. Maize samples were ground to pass through a 2-mm screen and were placed into 80-μL stainless-steel sample cups. The samples were inserted into the pyrolysis oven with helium flowing through at 2 L min−1 (at standard pressure and temperature). The autosampler furnace was electronically maintained at 500°C, and the interface was set to 350°C. A quarter-inch transfer line used to interface the autosampler pyrolysis unit to the molecular beam mass spectrometer was electronically heated to approximately 350°C. The total pyrolysis time was 2 min, although the pyrolysis reaction was completed in less than 20 s. The mass-to-charge ratio was set between m/z 30 and m/z 450.
The Unscrambler software program (CAMO; version 9.7) was used to normalize background-subtracted data based on total ion content and to perform PCA. The normalized triplicate MBMS spectra for each sample were averaged before PCA. Peak assignments (Supplemental Table S3) were made according to Evans and Milne (1987) and Boon (1989).
Reverse-Genetics Screens of DNA Grids
Eight grids were constructed for reverse genetics from the UniformMu population at the University of Florida. All seeds used for grids were Mu-off individuals, each carrying an estimated 10 unique, stable inserts, plus 50 other known inserts from W22 and parental sources. Each seed was also pedigreed, with ancestral information for seven generations. The screens focused on 50 cell wall-related genes in families hypothesized to have specific roles in grass-type cell walls.
We developed a two-tiered approach to these screens. The first was a “standard PCR screen” used for all 50 target genes, and the second was an “advanced screen” used where more intensive effort was warranted to identify a mutant. Standard screens were done with a Mu-specific primer plus a forward-directed, gene-specific primer to target approximately 1.5 kb of the upper coding sequence of each gene in x axis samples. Advanced screens expanded the target sequence to 2.5 kb, incorporated additional PCR protocols, and tested samples from both axes of the grids. This approach more than doubled the return of Mu inserts sought and was used to supplement the standard screen wherever possible (42% of grid screenings).
Sequences of the reference chromosomes have been deposited in GenBank as accession numbers CM000777 to CM000786. Sequences are also available at http://www.maizesequence.org.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The RGP gene family.
Supplemental Figure S2. Genes of nonprocessive xylosyl and fucosyl transferase families.
Supplemental Figure S3. The Cobra gene family.
Supplemental Figure S4. Examples of visible mutants selected from the UniformMu population.
Supplemental Table S1. Genes expressed in maize ovaries before pollination.
Supplemental Table S2. Currently available cell wall-related Mu knockout insertion lines.
Supplemental Table S3. Origins of m/z fragments from PyMBMS.
Supplementary Material
Acknowledgments
We thank Bill Foster, Phil Devillez, and Javier Campos for technical expertise in the field operations and NIR screens at Purdue, and Wayne Avigne and Susan Latshaw for technical expertise and reverse-genetics PCR at the University of Florida. We thank Robert Sykes for technical assistance with the PyMBMS. We also thank Cristal Musser for her dedicated service as project coordinator and the many National Science Foundation Research Experience for Undergraduates and undergraduate research interns who contributed throughout many phases of this work.
This work was supported by the National Science Foundation Plant Genome Research Program (grant no. DBI–0217552 to W.V., K.E.K., D.R.M., S.R.T., M.C.M., and N.C.C.) and the Office of Science, U.S. Department of Energy (grant no. DE–FG02–08ER64702 to N.C.C. and M.C.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Nicholas C. Carpita (carpita@purdue.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ahn JW, Verma R, Kim M, Lee JY, Kim YK, Bang JW, Reiter WD, Pal HS (2006) Depletion of UDP-D-apiose/UDP-D-xylose synthases results in rhamnogalacturonan II deficiency, cell wall thickening, and cell death in higher plants. J Biol Chem 281 13708–13716 [DOI] [PubMed] [Google Scholar]
- Akamatsu T, Hanzawa Y, Ohtake Y, Takahashi T, Nishitani K, Komeda Y (1999) Expression of endoxyloglucan transferase genes in acualis mutants of Arabidopsis. Plant Physiol 121 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215 403–410 [DOI] [PubMed] [Google Scholar]
- Appenzeller L, Doblin M, Barreiro R, Wang HY, Niu XM, Kollipara K, Carrigan L, Tomes D, Chapman M, Dhugga KS (2004) Cellulose synthesis in maize: isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose 11 287–299 [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Arioli T, Peng LC, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279 717–720 [DOI] [PubMed] [Google Scholar]
- Bai L, Singh M, Pitt L, Sweeney M, Brutnell TP (2007) Generating novel allelic variation through Activator insertional mutagenesis in maize. Genetics 175 981–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R (2007) NCBI GEO: mining tens of millions of expression profiles. Database and tools update. Nucleic Acids Res 35 D760–D765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54 519–546 [DOI] [PubMed] [Google Scholar]
- Boon JJ (1989) An introduction to pyrolysis mass spectrometry of lignocellulosic material: case studies of barley straw, corn stem and Agropyron. In A Chesson, ER Ørskov, eds, Physico-Chemical Characterization of Plant Residues for Industrial and Feed Use. Elsevier Applied Science, London, pp 25–49
- Brady SM, Song S, Dhugga KS, Rafalski JA, Benfey PN (2007) Combining expression and comparative evolutionary analysis: the COBRA gene family. Plant Physiol 143 172–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckeridge MS, dos Santos HP, Tiné MAS (2000) Mobilisation of storage cell wall polysaccharides in seeds. Plant Physiol Biochem 38 141–156 [Google Scholar]
- Burget EG, Verma R, Mølhøj M, Reiter WD (2003) The biosynthesis of L-arabinose in plants: molecular cloning and characterization of a Golgi-localized UDP-D-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell 15 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Shirley NJ, King BJ, Harvey AJ, Fincher GB (2004) The CesA gene family of barley: quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol 134 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Stone BA, Newbigin EJ, Bacic A, Fincher GB (2006) Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-D-glucans. Science 311 1940–1942 [DOI] [PubMed] [Google Scholar]
- Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34 351–362 [DOI] [PubMed] [Google Scholar]
- Cao PJ, Bartley LE, Jung KH, Ronald PC (2008) Construction of a rice glycosyltransferase phylogenomic database and identification of rice-diverged glycosyltransferases. Mol. Plant 1 858–877 [DOI] [PubMed] [Google Scholar]
- Carpita NC (1984) Cell wall development in maize coleoptiles. Plant Physiol 76 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC (1996) Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol 47 445–476 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3 1–30 [DOI] [PubMed] [Google Scholar]
- Carpita NC, McCann MC (2008) Maize and sorghum: genetic resources for the bioenergy grasses. Trends Plant Sci 13 415–420 [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Bergert I, Pauly M, et al (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevenet F, Brun C, Banuls AL, Jacq B, Christen R (2006) TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching A, Dhugga KS, Appenzeller L, Meeley R, Bourett TM, Howard RJ, Rafalski A (2006) Brittle stalk2 encodes a putative glycosylphosphatidylinositol-anchored protein that affects mechanical strength of maize tissues by altering the composition and structure of secondary cell walls. Planta 224 1174–1184 [DOI] [PubMed] [Google Scholar]
- Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG (2007) A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc Natl Acad Sci USA 104 8550–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ (2000) Loosening of plant cell walls by expansins. Nature 407 321–326 [DOI] [PubMed] [Google Scholar]
- Delmer DP (1999) Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol 50 245–276 [DOI] [PubMed] [Google Scholar]
- Dhugga KS, Barreiro R, Whitten B, Stecca K, Hazebroek J, Randhawa GS, Dolan M, Kinney AJ, Tomes D, Nichols S, et al (2004) Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science 303 363–366 [DOI] [PubMed] [Google Scholar]
- Dhugga KS, Tiwari SC, Ray PM (1997) A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: purification, gene cloning, and trans-Golgi localization. Proc Natl Acad Sci USA 94 7679–7684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin MS, De Melis L, Newbigin E, Bacic A, Read SM (2001) Pollen tubes of Nicotiana alata express two genes from different β-glucan synthase families. Plant Physiol 125 2040–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G, Zabotina O, Delgado I, Robert S, Keegstra K, Raikhel N (2006) Arabidopsis reversibly glycosylated polypeptides 1 and 2 are essential for pollen development. Plant Physiol 142 1480–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards ME, Dickson CA, Chengappa S, Sidebottom C, Gidley MJ, Reid JSG (1999) Molecular characterisation of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J 19 691–697 [DOI] [PubMed] [Google Scholar]
- Egelund J, Obel N, Ulvskov P, Geshi N, Pauly M, Bacic A, Petersen BL (2007) Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol Biol 64 439–451 [DOI] [PubMed] [Google Scholar]
- Egelund J, Peterson BL, Motawia MS, Damager I, Faik A, Olsen CE, Ishii T, Clausen H, Ulvskov P, Geshi N (2006) Arabidopsis thaliana RGX1 and RGX2 encode Golgi-localized (1,3)-α-D-xylosyltransferases involved in the synthesis of pectic rhamnogalacturonan II. Plant Cell 18 2593–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RJ, Milne TA (1987) Molecular characterization of the pyrolysis of biomass. 1. Fundamentals. Energy Fuels 1 123–137 [Google Scholar]
- Eveland AL, McCarty DR, Koch KE (2008) Transcript profiling by 3′-untranslated region sequencing resolves expression of gene families. Plant Physiol 146 32–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12 2409–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Price NJ, Raikhel NV, Keegstra K (2002) An Arabidopsis gene encoding an α-xylosyltransferase involved in xyloglucan biosynthesis. Proc Natl Acad Sci USA 99 7797–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L (2001) KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev 15 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J, Dong QF, Schneider B, Morrow DJ, Nan GL, Brendel V, Walbot V (2004) Genome-wide mutagenesis of Zea mays L. using RescueMu transposons. Genome Biol 5 R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T, et al (2004) Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol 2 937–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis KE, Lam SY, Copenhaver GP (2006) Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol 142 1004–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Hemm MR, Denault JW, Ruegger MO, Humphreys JM, Chapple C (2002. a) Changes in the secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J 30 47–59 [DOI] [PubMed] [Google Scholar]
- Franke R, Humphreys JM, Hemm MR, Denault JW, Ruegger MO, Cusumano JC, Chapple C (2002. b) The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J 30 33–45 [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Pauly M, Bacic A, Fincher GB (2005) Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta 221 729–738 [DOI] [PubMed] [Google Scholar]
- Girke T, Lauricha J, Tran H, Keegstra K, Raikhel N (2004) The Cell Wall Navigator database: a systems-based approach to organism-unrestricted mining of protein families involved in cell wall metabolism. Plant Physiol 136 3003–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Carranza ZH, Elliott KA, Roberts JA (2007) Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J Exp Bot 58 3719–3730 [DOI] [PubMed] [Google Scholar]
- Goujon T, Sibout R, Pollet B, Maba B, Nussaume L, Bechtold N, Lu F, Ralph J, Mila I, Barrière Y, et al (2003) A new Arabidopsis thaliana mutant deficient in the expression of O-methyltransferase impacts lignins and sinapoyl esters. Plant Mol Biol 51 973–989 [DOI] [PubMed] [Google Scholar]
- Grennan AK (2006) Genevestigator: facilitating Web-based gene-expression analysis. Plant Physiol 141 1164–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumie S, San-Clemente H, Deswarte C, Martinez Y, Lapierre C, Murigneux A, Berrière Y, Pichon M, Goffner D (2007) MAIZEWALL: database and developmental gene expression profiling of cell wall biogenesis and assembly in maize. Plant Physiol 143 339–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Han MV, Han SG (2007) Gene family evolution across 12 Drosophila genomes. PLoS Genet 3 e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Sørensen SO, Orfila C, Pauly M, Scheller HV (2006) ARABINAN DEFICIENT1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol 140 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser MT, Morikami A, Benfey PN (1995) Conditional root expansion mutants of Arabidopsis. Development 121 1237–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Hawley RM, Davis GL, Henrissat B, Walton J (2003) Quantitative trait loci and comparative genomics of cereal cell wall composition. Plant Physiol 132 263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Scott-Craig JS, Walton JD (2002) Cellulose synthase-like genes of rice. Plant Physiol 128 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller W, Kühnl T (1985) Elicitor induction of a microsomal 5-O-(4-coumaroyl) shikimate 3′-hydroxylase in parsley cell suspension cultures. Arch Biochem Biophys 241 453–460 [DOI] [PubMed] [Google Scholar]
- Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M (2003) Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J Biol Chem 278 95–103 [DOI] [PubMed] [Google Scholar]
- Holland N, Holland D, Helentjaris T, Dhugga K, Xoconostle-Cazares B, Delmer DP (2000) A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiol 123 1313–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrmova M, Farkas V, Lahnstein J, Fincher GB (2007) A barley xyloglucan xyloglucosyl transferase covalently links xyloglucan, cellulosic substrates, and (1-3;1-4)-β-D-glucans. J Biol Chem 282 12951–12962 [DOI] [PubMed] [Google Scholar]
- Hughes AL, Friedman R (2004) Shedding genomic ballast: extensive parallel loss of ancestral gene families in animals. J Mol Evol 59 827–833 [DOI] [PubMed] [Google Scholar]
- Humphreys JM, Hemm MR, Chapple C (1999) New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci USA 96 10045–10050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436 793–800 [DOI] [PubMed] [Google Scholar]
- Jensen JK, Sørensen SO, Harholt J, Geshi N, Sakuragi Y, Møller I, Zandleven J, Bernal AJ, Jensen NB, Sørensen C, et al (2008) Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. Plant Cell 20 1289–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DA, Thomas MA (2007) The monosaccharide transporter gene family in Arabidopsis and rice: a history of duplications, adaptive evolution, and functional divergence. Mol Biol Evol 24 2412–2423 [DOI] [PubMed] [Google Scholar]
- Jones L, Ennos AR, Turner SR (2001) Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J 26 205–216 [DOI] [PubMed] [Google Scholar]
- Karkonen A, Murigneux A, Martinant JP, Pepey E, Tatout C, Dudley BJ, Fry SC (2005) UDP-glucose dehydrogenases of maize: a role in cell wall pentose biosynthesis. Biochem J 391 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemsley EK (1998) Discriminant Analysis of Spectroscopic Data. John Wiley & Sons, Chichester, UK
- Kim CM, Park SH, Il Je B, Park SH, Park SJ, Piao HL, Eun MY, Dolan L, Han CD (2007) OsCSLD1, a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice. Plant Physiol 143 1220–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Shiu SH, Thoma S, Li WH, Patterson SE (2006) Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol 7 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolkman JM, Conrad LJ, Farmer PR, Hardeman K, Ahern KR, Lewis PE, Sawers RJH, Lebejko S, Chomet P, Brutnell TP (2005) Distribution of Activator (Ac) throughout the maize genome for use in regional mutagenesis. Genetics 169 981–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T, Takeda T, Miyazaki Y, Ohnishi-Kameyama M, Hayashi T, O'Neill MA, Ishii T (2007) A plant mutase that interconverts UDP-arabinofuranose and UDP-arabinopyranose. Glycobiology 17 345–354 [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nichikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühnl T, Koch U, Heller W, Wellmann E (1987) Chlorogenic acid biosynthesis: characterization of a light-induced microsomal 5-O-(4-coumaroyl)-D-quinate/shikimate 3′-hydroxylase from carrot (Daucus carota L.) cell suspension cultures. Arch Biochem Biophys 258 226–232 [DOI] [PubMed] [Google Scholar]
- Langenheim JH, Thimann KV (1982) Botany: Plant Biology and Its Relation to Human Affairs. John Wiley & Sons, New York
- Lao NT, Long D, Kiang S, Coupland G, Shoue DA, Carpita NC, Kavanagh TA (2004) Mutation of a family 8 glycosyltransferase gene alters cell wall carbohydrate composition and causes a humidity-sensitive semi-sterile dwarf phenotype in Arabidopsis. Plant Mol Biol 53 647–663 [DOI] [PubMed] [Google Scholar]
- Lee C, Zhong R, Richardson EA, Himmelsbach DS, McPhail BT, Ye ZH (2007. a) The PARVUS gene is expressed in cells undergoing secondary wall thickening and is essential for glucuronoxylan biosynthesis. Plant Cell Physiol 48 1659–1672 [DOI] [PubMed] [Google Scholar]
- Lee JH, Waffenschmidt S, Small L, Goodenough U (2007. b) Between-species analysis of short-repeat modules in cell wall and sex-related hydroxyproline-rich glycoproteins of Chlamydomonas. Plant Physiol 144 1813–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Sharopova N, Beavis WD, Grant D, Katt M, Blair DL, Hallauer AR (2002) Expanding the genetic map of maize with the intermated B73 x Mo17 (IBM) population. Plant Mol Biol 48 453–461 [DOI] [PubMed] [Google Scholar]
- Li YH, Qian O, Zhou YH, Yan MX, Sun L, Zhang M, Fu ZM, Wang YH, Han B, Pang XM, et al (2003) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15 2020–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K (2005) Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA 102 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvet R, Cavel E, Gutierrez L, Guénin S, Roger D, Gillet F, Guerineau F, Pelloux J (2006) Comprehensive expression profiling of the pectin methylesterase gene family during silique development in Arabidopsis thaliana. Planta 224 782–791 [DOI] [PubMed] [Google Scholar]
- Madson M, Dunand C, Li XM, Verma R, Vanzin GF, Calplan J, Shoue DA, Carpita NC, Reiter WD (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MC, Carpita NC (2005) Looking for invisible phenotypes in cell wall mutants of Arabidopsis thaliana. Plant Biosyst 139 80–83 [Google Scholar]
- McCann MC, Defernez M, Urbanowicz BR, Tewari JC, Langewisch T, Olek A, Wells B, Wilson RH, Carpita NC (2007) Neural network analyses of infrared spectra for classifying cell wall architectures. Plant Physiol 143 1314–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MC, Roberts K (1991) Architecture of the primary cell wall. In CW Lloyd, ed, Cytoskeletal Basis of Plant Growth and Form. Academic Press, New York, pp 109–129
- McCarty DR, Settles AM, Suzuki M, Tan BC, Latshaw S, Porch T, Robin K, Baier J, Avigne W, Lai JS, et al (2005) Steady-state transposon mutagenesis in inbred maize. Plant J 44 52–61 [DOI] [PubMed] [Google Scholar]
- Meyer K, Cusumano JC, Somerville C, Chapple CCS (1996) Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monooxygenases. Proc Natl Acad Sci USA 93 6869–6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RAC, Dupree P, Shewry PR (2007) A novel bioinformatics approach identifies candidate genes for the synthesis and feruloylation of arabinoxylan. Plant Physiol 144 43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mølhøj M, Verma R, Reiter WD (2004) The biosynthesis of D-galacturonate in plants: functional cloning and characterization of a membrane-anchored UDP-D-glucuronate 4-epimerase from Arabidopsis. Plant Physiol 135 1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille G, Ralet MC, Cavelier C, Eland C, Effroy D, Hematy K, McCartney L, Truong HN, Gaudon V, Thibault JF, et al (2007) Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J 50 605–614 [DOI] [PubMed] [Google Scholar]
- Nair RB, Bastress KL, Ruegger MO, Denault JW, Chapple C (2004) The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguema-Ona E, Andeme-Onzighi C, Aboughe-Angone S, Bardor M, Ishii T, Lerouge P, Driouich A (2006) The reb1-1 mutation of Arabidopsis: effect on the structure and localization of galactose-containing cell wall polysaccharides. Plant Physiol 140 1406–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobles DR, Brown RM Jr (2004) The pivotal role of cyanobacteria in the evolution of cellulose synthases and cellulose synthase-like proteins. Cellulose 11 437–448 [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457 551–556 [DOI] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93 12637–12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña MJ, Zhong RQ, Zhou GK, Richardson EA, O'Neill MA, Darvill AG, York WS, Ye ZH (2007) Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei HR, Milne J, Page GP, Somerville CR (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Egelund J, Gilson PR, Houghton F, Gleeson PA, Schultz CJ, Bacic A (2008) Identification of a novel group of putative Arabidopsis thaliana β-(1→3)-galactosyltransferases. Plant Mol Biol 68 43–59 [DOI] [PubMed] [Google Scholar]
- Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA, Frederick WJ, Hallet JP, Leak DJ, Liotta CL, et al (2006) The path forward for biofuels and biomaterials. Science 311 484–489 [DOI] [PubMed] [Google Scholar]
- Reiter WD, Vanzin GF (2001) Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant Mol Biol 47 95–113 [PubMed] [Google Scholar]
- Rhee SY, Osborne E, Poindexter PD, Somerville CR (2003) Microspore separation in the quartet3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol 133 1170–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes MJC, Wooltorton LSC (1976) The enzymatic conversion of hydroxycinnamic acids to p-coumarylquinic and chlorogenic acids in tomato fruits. Phytochemistry 15 947–951 [Google Scholar]
- Roesler J, Krekel F, Amrhein N, Schmid J (1997) Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol 113 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Doorsselaere JV, Messens E, Boerjan W (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43 1421–1435 [DOI] [PubMed] [Google Scholar]
- Roudier F, Fernandez AG, Fujita M, Himmelspach R, Borner GHH, Schindelman G, Song S, Baskin TI, Dupree P, Wasteneys GO, et al (2005) COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17 1749–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Schindelman G, DeSalle R, Benfey PN (2002) The COBRA family of putative GPI-anchored proteins in Arabidopsis: a new fellowship in expansion. Plant Physiol 130 538–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4 406–425 [DOI] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ (2005) The expansin superfamily. Genome Biol 6 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarria R, Wagner TA, O'Neill MA, Faik A, Wilkerson CG, Keegstra NV, Raikhel NV (2001) Characterization of a family of Arabidopsis genes related to xyloglucan fucosyltransferase1. Plant Physiol 127 1595–1606 [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Jensen JK, Sørensen SO, Harholt J, Geshi N (2007) Biosynthesis of pectin. Physiol Plant 129 283–295 [Google Scholar]
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN (2001) COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 15 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schoch G, Goepfert S, Morant M, Hehn A, Meyer D, Ullmann P, Werck-Reichhart D (2001) CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J Biol Chem 276 36566–36574 [DOI] [PubMed] [Google Scholar]
- Seifert GJ, Barbar C, Wells B, Dolan L, Roberts K (2002) Galactose biosynthesis in Arabidopsis: genetic evidence for substrate channeling from UDP-D-galactose into cell wall polymers. Curr Biol 12 1840–1845 [DOI] [PubMed] [Google Scholar]
- Settles AM, Holding DR, Tan BC, Latshaw S, Liu J, Suzuki M, Li L, O'Brien BA, Fajardo DS, Wroclawska E, et al (2007) Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics 8 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Séguin A (2005) CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary gene involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhu A, Langewisch T, Olek A, Multani DS, McCann MC, Vermerris W, Carpita NC, Johal G (2007) Maize Brittle stalk2 encodes a COBRA-like protein expressed in early organ development but required for tissue flexibility at maturity. Plant Physiol 145 1444–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling JD, Atmodjo MA, Inwood SE, Kolli VSK, Quigley HF, Hahn MG, Mohnen D (2006) Functional identification of an Arabidopsis pectin biosynthesis homogalacturonan galacturonosyltransferase. Proc Natl Acad Sci USA 103 5236–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöckigt J, Zenk MH (1974) Enzymic synthesis of chlorogenic acid from caffeoyl coenzyme A and quinic acid. FEBS Lett 42 131–134 [DOI] [PubMed] [Google Scholar]
- Strasser R, Bondili JS, Vavra U, Schoberer J, Svoboda B, Glössl J, Léonard R, Stadlmann J, Altmann F, Steinkellner H, et al (2007) A unique β-1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing Lewis a structures in Arabidopsis thaliana. Plant Cell 19 2278–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Latshaw S, Sato Y, Settles AM, Koch KE, Hannah LC, Kojima M, Sakakibara H, McCarty DR (2008) The maize Viviparous8 locus, encoding a putative ALTERED MERISTEM PROGRAM1-like peptidase, regulates abscisic acid accumulation and coordinates embryo and endosperm development. Plant Physiol 146 1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Settles AM, Tseung CW, Li QB, Latshaw S, Wu S, Porch TG, Schmelz EA, James MG, McCarty DR (2005) The maize viviparous15 locus encodes the molybdopterin synthase small subunit. Plant J 45 264–274 [DOI] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36 D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Murata K, Yamazaki M, Onosato K, Miyao A, Hirochika H (2003) Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol 133 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JR, Darvill AG, Albersheim P (1989) Structure of plant cell walls. 24. Isolation and structural characterization of the pectic polysaccharide rhamnogalacturonan II from walls of suspension-cultured rice cells. Carbohydr Res 185 261–277 [Google Scholar]
- Tuskan G, West D, Bradshaw HD, Neale D, Sewell M, Wheeler N, Megraw R, Jeck K, Wiselogel A, Evans R, et al (1999) Two high-throughput techniques for determining wood properties as part of a molecular genetics analysis of hybrid poplar and loblolly pine. Appl Biochem Biotechnol 77–79 55–65 [Google Scholar]
- Ulbrich B, Zenk MH (1980) Partial purification and properties of p-hydroxycinnamoyl-CoA:shikimate-p-hydroxycinnamoyl transferase from higher plants. Phytochemistry 19 1625–1629 [Google Scholar]
- Usadel B, Schluter U, Mølhøj M, Gipmans M, Verma R, Kossman J, Reiter WD, Pauly M (2004) Identification and characterization of a UDP-D-glucuronate 4-epimerase in Arabidopsis. FEBS Lett 569 327–331 [DOI] [PubMed] [Google Scholar]
- Valdivia ER, Sampedro J, Lamb JC, Chopra S, Cosgrove DJ (2007) Recent proliferation and translocation of pollen group allergen genes in the maize genome. Plant Physiol 143 1269–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD (2002) The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci USA 99 3340–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara CE, Carpita NC (2001) β-D-Glucan synthases and the CesA gene family: lessons to be learned from the mixed-linkage (1→3),(1→4)-β-D-glucan synthase. Plant Mol Biol 47 145–160 [PubMed] [Google Scholar]
- Verica JA, Medford JI (1997) Modified MERI5 expression alters cell expansion in transgenic Arabidopsis plants. Plant Sci 125 201–210 [Google Scholar]
- Vermerris W, Saballos A, Ejeta G, Mosier NS, Ladisch MR, Carpita NC (2007) Molecular breeding to enhance ethanol production from maize and sorghum stover. Crop Sci 47 S142–S153 [Google Scholar]
- Vignols F, Rigau J, Torres M, Capellades M, Puigdomènech P (1995) The brown-midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil CF, Monde RA (2007) Getting the point: mutations in maize. Crop Sci 47 S60–S67 [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J (1995) Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yennawar NH, Li LC, Dudzinski DM, Tabuchi A, Cosgrove DJ (2006) Crystal structure and activities of EXPB1 (Zea m 1), a β-expansin and group-1 pollen allergen from maize. Proc Natl Acad Sci USA 103 14664–14671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Rose JKC, Nishitani K (2004) A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice: classification and expression analysis. Plant Physiol 134 1088–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong W, O'Malley R, Link B, Binder B, Bleecker A, Koch KE, McCann MC, McCarty DR, Patterson S, Reiter WD, et al (2005) Plant cell wall genomics. Planta 221 747–751 [DOI] [PubMed] [Google Scholar]
- Yu JM, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R (2001) InterProScan: an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17 847–848 [DOI] [PubMed] [Google Scholar]
- Zhang QS, Shirley N, Lahnstein J, Fincher GB (2005) Characterization and expression patterns of UDP-D-glucuronate decarboxylase genes in barley. Plant Physiol 138 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong RQ, Peña MJ, Zhou GK, Nairn CJ, Wood-Jones A, Richardson EA, Morrison WH, Darvill AG, York WS, Ye ZH (2005) Arabidopsis fragile fiber8, which encodes a putative glucuronosyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17 3390–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.