Abstract
There currently exists a diverse array of molecular probes for the in situ localization of polysaccharides, nucleic acids, and proteins in plant cells, including reporter enzyme strategies (e.g. protein-glucuronidase fusions). In contrast, however, there is a paucity of methods for the direct analysis of endogenous glycoside hydrolases and transglycosidases responsible for cell wall remodeling. To exemplify the potential of fluorogenic resorufin glycosides to address this issue, a resorufin β-glycoside of a xylogluco-oligosaccharide (XXXG-β-Res) was synthesized as a specific substrate for in planta analysis of XEH activity. The resorufin aglycone is particularly distinguished for high sensitivity in muro assays due to a low pKa (5.8) and large extinction coefficient (ε 62,000 m−1cm−1), long-wavelength fluorescence (excitation 571 nm/emission 585 nm), and high quantum yield (0.74) of the corresponding anion. In vitro analyses demonstrated that XXXG-β-Res is hydrolyzed by the archetypal plant XEH, nasturtium (Tropaeolum majus) NXG1, with classical Michaelis-Menten substrate saturation kinetics and a linear dependence on both enzyme concentration and incubation time. Further, XEH activity could be visualized in real time by observing the localized increase in fluorescence in germinating nasturtium seeds and Arabidopsis (Arabidopsis thaliana) inflorescent stems by confocal microscopy. Importantly, this new in situ XEH assay provides an essential complement to the in situ xyloglucan endotransglycosylase assay, thus allowing delineation of the disparate activities encoded by xyloglucan endotransglycosylase/hydrolase genes directly in plant tissues. The observation that XXXG-β-Res is also hydrolyzed by diverse microbial XEHs indicates that this substrate, and resorufin glycosides in general, may find broad applicability for the analysis of wall restructuring by polysaccharide hydrolases during morphogenesis and plant-microbe interactions.
The development and application of molecular probes for the localization of biomolecules in planta continues to have a profound impact on the field of plant physiology. A number of elegant techniques have been devised for the detection of nucleic acids, polypeptides, and polysaccharides in situ, including DNA/RNA hybridization (Jin and Lloyd, 1997), reporter protein fusions (Jefferson et al., 1987; Ehrhardt, 2003; Chapman et al., 2005; Stewart, 2005; Berg and Beachy, 2008; Nelson et al., 2008), immunohistochemical methods (Walker et al., 2001; Chapman et al., 2005), applications of natural carbohydrate-binding proteins (Knox, 2008), and direct spectroscopy (Vicente et al., 2007). While there now exists a considerable toolbox to identify the location to which biomolecules are directed in the cell, elucidation of specific biochemical function at the site of localization often remains challenging.
Presently, there is a growing interest in the roles of glycoside hydrolases (GHs) and transglycosylases in plant cell wall biogenesis, remodeling, and degradation (Minic and Jouanin, 2006; Vicente et al., 2007; Gilbert et al., 2008; Lopez-Casado et al., 2008). A technical limitation of many studies, however, is that enzyme activities can only be measured for crude whole-tissue extracts, or purified or recombinant enzymes, and thus cannot be directly correlated with the high-resolution in situ localization of other biomacromolecules. As such, the in situ analysis of GH activities responsible for the degradation of plant cell wall polysaccharides has received comparatively little attention, primarily due to a paucity of convenient assay methods (Vicente et al., 2007). Some notable exceptions include the use of commercially available X (5-bromo-4-chloro-3-indolyl glycoside) substrates for the detection of exoglycosidase activity (Monroe et al., 1999; Chantarangsee et al., 2007; Macquet et al., 2007; Wen et al., 2008). Likewise, transglycosylase activity has been visualized in higher plant and yeast cell walls using sulforhodamine-oligosaccharide acceptor substrates (Vissenberg et al., 2000; Bourquin et al., 2002; Nishikubo et al., 2007; Cabib et al., 2008). Both types are examples of end point, or stopped, assays, in which precipitated indigoid dyes or incorporated fluorescent oligosaccharide conjugate, respectively, are observed after a terminal incubation time.
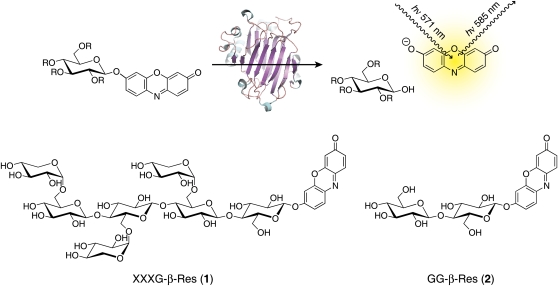
In this study, we have developed the use of resorufin glycosides as substrates for the real-time, continuous observation of GH activity in situ (Fig. 1). Enzymatic hydrolysis of such substrates releases the resorufin aglycone, which is distinguished by a low pKa value (5.8) and a large extinction coefficient (ε 62,000 m−1cm−1), long-wavelength fluorescence (excitation/emission maxima, 571 nm/585 nm), and high quantum yield (0.74) of the resorufinyl anion (Bueno et al., 2002). The pKa value and spectral properties make resorufin glycosides particularly suitable for high sensitivity in muro enzyme activity assays due to significant ionization of resorufin at typical apoplastic pH values (Felle, 2005). To highlight the potential of this class of substrates in cell wall morphological studies, we have chemically synthesized a xylogluco-oligosaccharide (XGO) resorufin β-glycoside (XXXG-β-Res; Fig. 1 [1]; XGO nomenclature according to Fry et al., 1993) and demonstrated its use for the real-time imaging of xyloglucan endohydrolase (XEH) activity in plant tissues from nasturtium (Tropaeolum majus) and Arabidopsis (Arabidopsis thaliana) by confocal fluorescence microscopy.
Figure 1.
Use of resorufin glycosides as fluorogenic substrates for glycosidases. R = saccharide or hydrogen; [1] and [2], substrates for determination of (xylo)glucanase activity. Oligosaccharide nomenclature is according to Fry et al. (1993). [See online article for color version of this figure.]
RESULTS
Synthesis of the Novel XEH Substrate, XXXG-β-Res
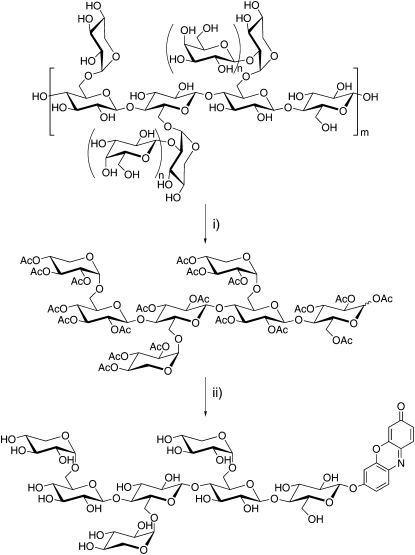
The specific cleavage of tamarind (Tamarindus indica) seed xyloglucan by various endoglucanases to produce a mixture of XXXG, XXLG, XLXG, and XLLG oligosaccharides has been long known (O'Neill and Selvendran, 1985; York et al., 1993). We have previously exploited this technique, together with the ability of β-galactosidase to further reduce xylogluco-oligosaccharide complexity (York et al., 1993), to synthesize phenyl XXXG and XLLG β-glycosides (XXXG-β-Ph) as chromogenic probes for in vitro kinetic analyses of XEHs (alternatively endoxyloglucanases, EC 3.2.1.151; Ibatullin et al., 2008). In this study, we have extended this methodology to the synthesis of XXXG-β-Res as a new, sensitive fluorogenic probe for both in vitro and in situ XEH assays. Thus, XXXG-β-Res was produced from tamarind seed xyloglucan via a straightforward procedure involving endoglucanase hydrolysis, β-galactosidase treatment, per-O-acetylation and separation of XXXG per-O-acetate, α-bromination, phase-transfer glycosylation, and Zemplén deprotection (Fig. 2). This method was readily extended to the synthesis of the unsubstituted glucanase substrate, resorufin β-cellobioside (GG-β-Res; Fig. 1 [2]; Coleman et al., 2007). In a preliminary study, we were likewise able to perform the glycosylation step using variation of the Königs-Knorr method, using Ag+ as a promoter (Baumann, 2004).
Figure 2.
Synthesis of XXXG-β-Res. Reagents and conditions are as follows. (i) a, Cellulase from T. reesei, buffer pH 5.0, 40°C 1 h, then β-galactosidase from Aspergillus niger, 20 h. b, Ac2O/pyridine, 60°C, 18 h. (ii) a, HBr/AcOH/CH2Cl2; 0°C, 3 h. b, Resorufin sodium salt, BuBn3N(Cl)/CH2Cl2/H2O, room temperature 20 h. c, NaOMe/MeOH, room temperature 1 h.
In Vitro Kinetic Analysis of Enzyme-Catalyzed XXXG-β-Res Hydrolysis
Visible Light Adsorption Spectrometry
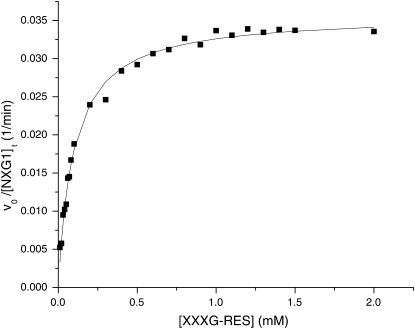
TmNXG1 from nasturtium is a well-characterized, predominant XEH from the xyloglucan endotransglycosylase/hydrolase (XTH) gene subfamily of GH family 16 (Baumann et al., 2007, and refs. therein). This enzyme, which functions in xyloglucan mobilization during seed germination (Edwards et al., 1986), was chosen as the archetype to test the efficacy of XXXG-β-Res as a substrate for plant XEHs. UV spectrophotometric analysis indicated that XXXG-β-Res was indeed a competent substrate for purified, recombinant TmNXG1, resulting in liberation of the resorufin aglycone. The hydrolysis of XXXG-β-Res followed classical Michaelis-Menten kinetics: A plot of vo/[E]t versus [S] over the substrate range 0.01 to 2 mm was hyperbolic (Fig. 3), with kcat = 0.036 ± 0.001 min−1 and Km = 0.098 ± 0.005 mm.
Figure 3.
Dependence of the rate of nasturtium endoxyloglucanase-catalyzed hydrolysis of XXXG-Res on substrate concentration, determined by visible spectrophotometry. The solid line indicates the best fit of the equation vo/[E]t = kcat/(Km + [S]) to the data; kcat = 0.036 ± 0.001 min−1, Km = 0.098 ± 0.005 mm.
XXXG-β-Res was also tested in vitro as a substrate for microbial endoxyloglucanses. Bacillus licheniformis endoxyloglucanase BlXG12 (Gloster et al., 2007) and Paenibacillus pabuli endoxyloglucanase PpXG5 (Gloster et al., 2007) cleaved XXXG-β-Res to release the resorufinyl anion with specific activities of 0.31 mol min−2 mol−1 enzyme and 48 mol min−2 mol−1 enzyme, respectively (0.2 mm XXXG-β-Res in 10 mm NaOAc, pH 5.5, 30°C). The relative activities of these enzymes toward XXXG-β-Res are consistent with their activities toward xyloglucan and other XGO aryl β-glycosides (Gloster et al., 2007; Ibatullin et al., 2008). Preliminary experiments with Trichoderma reesei endoglucanase II (EGII; TrCel5A; Henrissat et al., 1998) indicated that XXXG-β-Res was also a substrate for this fungal enzyme.
Interestingly, XXXG-β-Res was not a substrate for the strict xyloglucan endotransglycosylase (XET) from Populus tremula × tremuloides, PttXET16-34 (Johansson et al., 2004; no detectable activity using 3 μm enzyme and 1 mm XXXG-β-Res in 40 mm NaOAc, pH 5.6, 30°C). PttXET16-34 has previously been shown to be devoid of XEH activity using highly sensitive reducing sugar (Kallas et al., 2005) and HPLC (Baumann et al., 2007) assays. The lack of PttXET16-34 activity toward XXXG-β-Res parallels our earlier studies on XXXG- and XLLG-phenyl glycosides, which indicated that XETs lacking detectable XEH activity do not cleave these artificial substrates (Johansson et al., 2004; Ibatullin et al., 2008).
In Vitro Fluorescence Spectroscopy
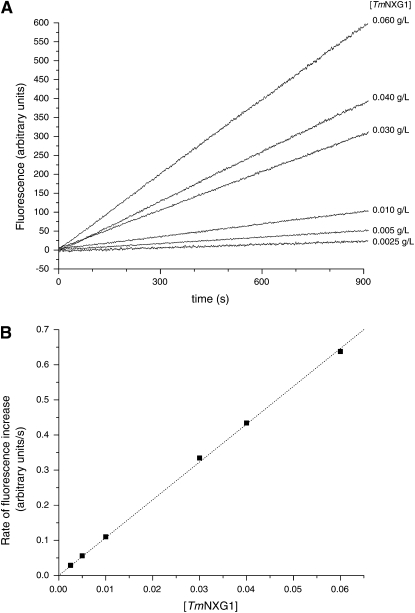
To demonstrate the suitability of resorufin glycosides for the quantitative analysis of enzyme activity by fluorimetry, XXXG-β-Res was incubated with increasing concentrations of TmNXG1 in vitro and substrate hydrolysis was monitored continuously in a spectrofluorimeter. As shown in Figure 4, the rate of release of the resorufinyl anion from XXXG-β-Res (0.05 mm) was linearly dependent on enzyme concentration over the range [TmNXG1] = 0.0025–0.060 g/L (0.079–1.9 μm, Mr 31,548). Furthermore, under these conditions, fluorophore release was linear with respect to time (Fig. 4).
Figure 4.
Time course of release of resorufin from XXXG-Res by TmNXG1 followed by fluorescence spectrometry in vitro.
In Situ Microscopic Analysis
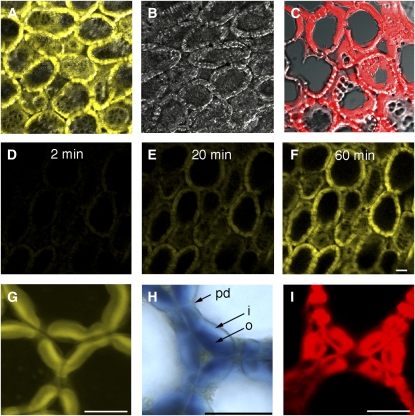
Together, data from in vitro visible absorbance and fluorescence spectrometry indicate that XXXG-β-Res can be used to reliably quantify XEH activity. To test the applicability of this substrate to detect XEH activity in planta, hand-sectioned germinating nasturtium seeds, known to express XEH activity (Edwards et al., 1986) were incubated with XXXG-β-Res and fluorophore liberation was followed by confocal microscopy. Figure 5 shows the yellow-wavelength emission due to the resorufinyl anion accumulation in nasturtium endosperm cells. The signal appeared in the storage cell walls within a few minutes after addition of the XXXG-β-Res substrate and its intensity increased for at least 80 min (Fig. 5, D–F). The control sections that were heated to denature proteins did not show any detectable signal during the incubation with the XXXG-β-Res substrate (Fig. 5B). In the fresh tissue sections, the signal was pH dependent with the maximum at pH 6.5. This value is above the in vitro pH optimum of TmNXG1 of 4.8 (Baumann et al., 2007), but greatly increases the ionization of resorufin to the fluorescent resorufinyl anion (pKa 5.8). No significant signal was observed at pH 5.5 or lower (data not shown). The high-resolution analysis showed that the resorufinyl anion accumulated most prominently in the outer cell wall layers of endosperm cells (Fig. 5G). This localization matched that of polymeric xyloglucan visualized in the same material by iodine staining (Fig. 5H). To compare the distribution of XEH activity with that of XET activity, we followed the incorporation of XXXG-SR to cell walls (Vissenberg et al., 2000; Nishikubo et al., 2007) in the same material (Fig. 5, C and I). XET activity showed a contrasting distribution pattern with the most prominent staining in the inner cell wall layer. The lack of staining of heat-inactivated sections, the pH dependence, the colocalization with the native substrate, and the specific distribution pattern observed in cell walls indicate that the fluorescent signal seen in confocal microscopy corresponds to resorufin released by the native XEH activity.
Figure 5.
Applicability of resorufin glycoside substrates to visualize glycosidase activities in situ: localization of xyloglucanase (XEH, EC 3.2.1.151) activity with XXXG-β-Res in the endosperm of germinating nasturtium seeds. Activity is visualized as yellow fluorescence signal developing in freshly cut tissue sections during the incubation with the XEH substrate XXXG-β-Res (A). The control section (B) was heated at 95°C prior to incubation with the substrate to inactivate enzymatic activities. The fluorescence signal is superimposed on the transmitted light signal to visualize the anatomy of the endosperm in A to C. D to F, Hydrolysis of XXXG-β-Res in cell walls observed in real time by time lapse scanning confocal microscopy. G is a high-magnification fluorescence image of endosperm cell walls shown in A, illustrating the distribution of XEH activity in cell walls. H is a light microscopy image of cell walls showing the distribution of high Mr xyloglucan stained with iodine. Note the colocalization of XEH activity and xyloglucan in the outer cell wall layers. C and I show the distribution of XET activity in the endosperm cell walls visualized by the incorporation of XXXG-SR to the tissue for comparison. Scale bar = 50 μm. o, Outer wall layer; i, inner wall layer; pd, plasmodesmata.
XEH and Cellulase Activity in Arabidopsis Vegetative Tissue
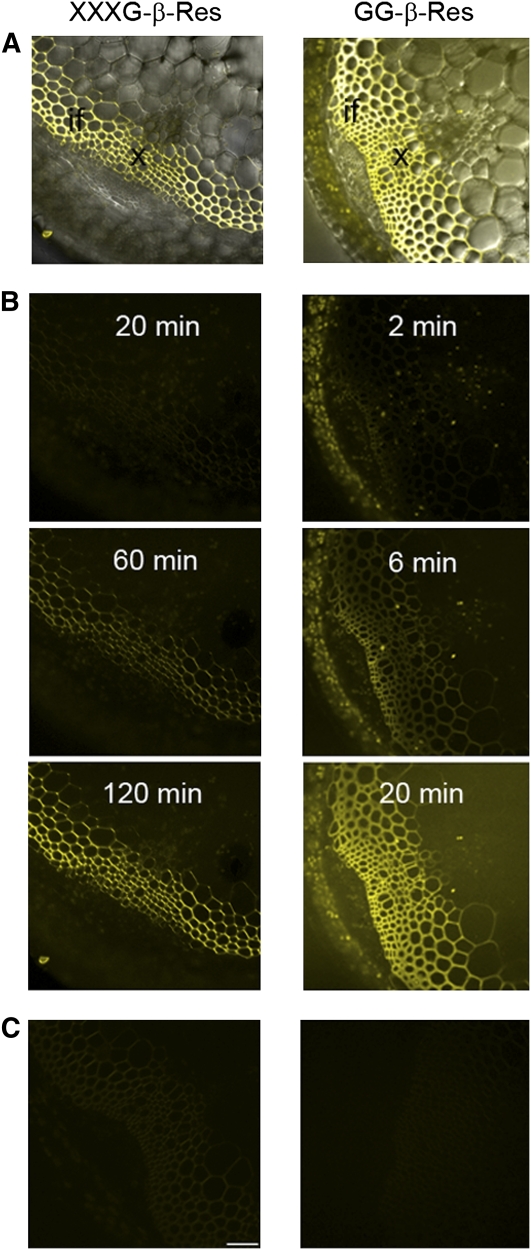
To highlight the generality of resorufin β-glycosides to detect GH activity in diverse tissues, XXXG-β-Res (Fig. 1 [1]) and GG-β-Res (Fig. 1 [2]) were applied to fresh hand-sections of Arabidopsis inflorescent stems. The release of resorufinyl anion from XXXG-β-Res substrate was compared to that from GG-β-Res (a substrate for both cellulases and β-glucosidases), which we have previously employed to demonstrate increased cellulase activity in overexpressing transgenic Arabidopsis lines (Takahashi et al., 2009). Here, incubation of both substrates with Arabidopsis stem sections resulted in a time-dependent increase of the signal from the resorufinyl anion (Fig. 6). Prior heat treatment of the sections prevented liberation of the aglycone due to enzyme denaturation. In the case of GG-β-Res substrate, the accumulation of resorufinyl anion was much more quickly observed, consistent with high expression levels of KORRIGAN1 (Takahashi et al., 2009) and other cellulases from the family GH9 (Park et al., 2003) in the stem tissues, as well as the combined action of endogenous β-glucosidases (Takahashi et al., 2009). The analysis shows that in Arabidopsis stem, the xyloglucanase and cellulase/β-glucosidase activities are most readily observed in secondary cell walls of xylem cells and interfascicular fibers (Fig. 6).
Figure 6.
Comparison of in situ analyses of different enzymatic activities in Arabidopsis inflorescent stems using two different substrates. XXXG-β-Res substrate visualizes the xyloglucanase (XEH, EC 3.2.1.151) activity, whereas GG-β-Res substrate reveals cellulase (endoglucanase, EC 3.2.1.4; or cellobiohydrolase) and β-glucosidase activities (EC 3.2.1.21). A, The fluorescence signal from resorufin (yellow) is superimposed on the transmitted light signal to visualize the anatomy of the stem. The reaction product can be seen in secondary cell walls of xylem (x) and interfacscicular fibers (if). B, A sequence of fluorograms taken at successive time points during the incubation of fresh sections of stems with the substrates. Note that the release of resorufin by the enzymes present in the stem tissues is much slower in the case of xyloglucanases compared to cellulases/glucosidases. C, Control sections that were heated at 95°C to inactivate enzymatic activities prior to incubation with the substrates. Scale bar = 50 μm. It should be noted that XXXG-β-Res is not enzyme isoform specific, and thus may detect enzymes with XEH activity and pH optima distinct from TmNXG1, vide infra.
DISCUSSION
The favorable spectroscopic properties of resorufin have been previously harnessed for the production of sensitive in vitro substrates for esterases and glycosidases (Tokutake et al., 1990; Beisson et al., 2000; Coleman et al., 2007), including single-molecule detection (English et al., 2006; Gorris et al., 2007). Indeed, earlier work has highlighted the utility of resorufin β-glycosides for imaging the activity of GHs, including exo-β-galactosidase in yeast (Saccharomyces cerevisiae; Wittrup and Bailey, 1988) and exo-β-glucosidase in animal cells (Hays et al., 1998). However, the use of such substrates for visualizing endogenous cell wall polysaccharidases in planta, including endo-acting enzymes, has not been explored in detail thus far. The long excitation wavelength of the resorufinyl anion (571 nm) confers a significant advantage over methylumbelliferyl and fluoromethylumbelliferyl glycosides (Ge et al., 2007; Vrsanska et al., 2008): The excitation maxima of these aglycones are <400 nm, which results in considerable background autofluorescence of plant cell components such as lignin, chlorophyll, and phenolics. To illustrate the wider potential of resorufin glycosides as plant molecular probes, we synthesized the new XEH substrate XXXG-β-Res in a proof-of-principle study including both in vitro and in vivo enzyme activity assays.
The synthesis of XXXG-β-Res (Fig. 2) was facile, essentially following our established method for the production of XXXG phenyl β-glycosides from tamarind seed xyloglucan (Ibatullin et al., 2008). Two enzymatic steps were used to produce the required XXXG heptasaccharide starting material from the polysaccharide. The resorufin aglycone was subsequently installed using established carbohydrate chemistry, which is directly applicable to a wide range of mono- and oligosaccharides. Thus, a library of resorufinyl β-glycosides, each designed to detect a specific glycosidase in planta on the basis of saccharide specificity, can be produced using the method presented herein. Indeed, the simple monosaccharide congeners Glcp-β-Res, Galp-β-Res, and GlcAp-β-Res are known (Tokutake et al., 1990; English et al., 2006), while during the course of this study, the synthesis of resorufinyl β-cellobioside (Fig. 1 [2]) was presented together with its use for cellulase detection in vitro (Coleman et al., 2007).
Our previous data has indicated that hydrolysis of XXXG and XLLG phenyl β-glycosides by TmNXG1 is essentially independent of substrate galactosylation (Ibatullin et al., 2008), consistent with the biological function of this enzyme in the degradation of heterogenously substituted seed galactoxyloglucan (Edwards et al., 1986). In this proof of concept, we elected to produce XXXG-β-Res for this reason, although it should be noted that the synthesis can be readily extended, by analogy (Ibatullin et al., 2008), to mono- and digalactosylated congeners (e.g. XXLG-β-Res, XLXG-β-Res, XLLG-β-Res), the desxylosylated backbone (GGGG-β-Res), and possibly other xyloglucan fragments (Hoffman et al., 2005; Faure et al., 2006). In vivo specificities may therefore be further delineated by the comparative analysis of a range of such substrates, e.g. in reverse genetics or recombinant expression/knock-in studies in planta. Nonetheless, XXXG-β-Res, on its own, is likely to be a useful general diagnostic probe of XEH activity in situ, given the emerging pattern that distal side chain residues in xyloglucans are not major determinants of substrate recognition by plant and microbial XEHs from diverse GH families (for review, see Gilbert et al., 2008).
In addition to exemplifying the general potential of resorufin glycosides for in planta analyses, the novel XXXG-β-Res substrate provides an essential new assay for untangling the distinct hydrolytic and transglycosylating activities of proteins encoded by XTH genes. This group of proteins forms a major clade in family GH16 in the carbohydrate active enzymes classification (Cantarel et al., 2009) and contains members with both strict transglycosylase (XET; EC 2.4.1.207) and predominant hydrolase (XEH; EC 3.2.1.151) activity (Baumann et al., 2007; for review, see Rose et al., 2002; Gilbert et al., 2008). The physiological importance of this catalytic distinction cannot be understated: On one hand, the transient cleavage and religation of xyloglucan polysaccharides catalyzed by XETs is thought to be one of the main contributors to controlled cell wall extension and strengthening, while on the other, irreversible hydrolysis catalyzed by XEHs would lead to wall decomposition (Rose et al., 2002; Cosgrove, 2005; Vicente et al., 2007). Although only few of the vast number of plant XETs and XEHs have been enzymatically characterized thus far, there is a sustained interest in the molecular analysis of these enzymes (Rose et al., 2002; Gilbert et al., 2008), including tissue-specific gene expression patterns and in situ analysis (Becnel et al., 2006).
Direct visualization of XET activity in situ was elegantly achieved nearly 10 years ago using a fluorescent XGO aminoalditol-sulforhodamine conjugate (XGO-SR) as an alternate glycosyl acceptor substrate (Vissenberg et al., 2000). Briefly, colocalization of endogenous XET activity and native xyloglucan, which acts as a glycosyl donor, results in the incorporation of XGO-SR into the wall-bound polysaccharide. After coincubation of XGO-SR with fresh tissue sections or whole mounts, the reaction is stopped by removal of the excess unincorporated XGO-SR by washing, followed by fluorescence microscopy analysis. Since its introduction, this technique has been applied to the analysis of a range of primary and secondary cell wall-forming tissues (Bourquin et al., 2002; Vissenberg et al., 2005; Nishikubo et al., 2007, and refs. therein). Until now, however, there has been no equivalent method for the visualization of XEH activity and thus, half of the picture of xyloglucan restructuring and degradation in the plant cell wall has been missing (Vicente et al., 2007). As shown here, the use of XXXG-β-Res as a fluorogenic XEH substrate compliments the widely used in situ XET activity assay. Together, these methods provide a powerful system to visualize the disparate hydrolytic and transglycosylation activities of xyloglucan-active enzymes in the plant cell. Importantly, the hydrolytic nature of the enzymatic reaction, together with judicious choice of the resorufin aglycone due to its particular spectral properties, allows the continuous visualization of color development in the microscope (thus contrasting the end point XET assay).
One of the biggest advantages of applying in situ glycosidase assays to survey enzymatic activities in cells is that the assays can be conducted under truly native conditions, which is difficult to mimic using in vitro approaches. This is of particular importance for enzymes working in complexes, having a defined orientation, configuration, or attached to a specific membrane. Such in situ assays also allow for a certain degree of control, including examining the effects of factors such as pH or ionic strength. The pH of cell wall is usually around 5.5, whereas the pH of the cytoplasm is neutral, although both are known to vary in response to stimuli (Fasano et al., 2001). Since the pKa of resorufin is 5.8, assay sensitivity in increasingly acidic environments will be affected by resorufin ionization, and this should be considered both when developing the in situ assay and interpreting the results. For example, measurements of local pH could accompany the activity assays and a calibration of the resorufin signal using a control with resorufin alone at a relevant pH (native or buffer controlled) could be included. Such analysis would provide deeper insight into native cell biochemistry.
Whereas the proof of concept has been demonstrated with the complex XGO-based substrate XXXG-β-Res, the methodology is nonetheless general and can be applied to a diversity of glycosidases in a range of tissues. Indeed, as part of a functional study of a Populus homolog of KORRIGAN1, PttCel9A1, we were recently able to use comparative time-course analysis of the rates of hydrolysis of resorufinyl β-glucoside and resorufinyl β-cellobioside (Fig. 1 [2]) to evidence functional expression of the endoglucanase PttCel9A1 in secondary cell wall-forming tissues of Arabidopsis (Takahashi et al., 2009). Here, analysis of resorufinyl anion release from XXXG-β-Res (Fig. 1 [1]) and GG-β-Res (Fig. 1 [2]) substrates in Arabidopsis stems (Fig. 6) highlights the different relative amounts of the corresponding glycosidase activities, and underscores the potential for the use of such substrates to map endogenous enzymes throughout plant tissues. It should be noted, however, that these substrates detect activities and are thus not specific for individual gene products [e.g. XEH activity has been found in five GH families thus far (Gilbert et al., 2008), and numerous GH families contain β(1–4)glucanases and glucosidases (Cantarel et al., 2009)]. Thus, unraveling the effects of specific enzyme isoforms on plant morphology will ultimately require a combination of these biochemical probes with genetic tools.
To our knowledge, the identification and special localization of XEH activity in Arabidopsis stems is a novel finding, which requires special consideration in the development of models of cell wall modification. Current models emphasize the importance of xyloglucan endotransglycosylation in the transient scission of the xyloglucan cross-links that bind cellulose microfibrils in the primary wall (Cosgrove, 2005) and that connect different cell wall layers in the secondary walls (Mellerowicz et al., 2008). Indeed, XEH activity has been only been linked to seed storage xyloglucan mobilization thus far (e.g. in nasturtium vide supra). The in situ XEH assay presented here is thus a powerful tool to reveal enzymatic activities in cells, which otherwise could not be intuitively predicted. We suggest that the irreversible hydrolysis of xyloglucan during wall morphological changes, including events such as cell expansion, wall maturation, fruit ripening, and microbial interactions, is a phenomenon worthy of further consideration using these new substrates as molecular probes.
CONCLUSION
Resorufin glycosides of mono- and oligosaccharides are excellent fluorogenic substrates for GHs that are conveniently prepared for the sensitive detection of endogenous enzyme activities in plant tissues. We envision that the straightforward method described here to obtain and utilize these substrates will facilitate their wider use for the screening and in situ kinetic characterization of glycosidases in a variety of fundamental physiological studies of plants.
MATERIALS AND METHODS
General
All chemicals were obtained from Sigma/Aldrich/Fluka and were of reagent grade or better. Ultrapure water (ρ ≥ 18 MΩ cm) was produced on a Milli-Q system (Millipore). Nasturtium (Tropaeolum majus) seed endoxyloglucanase TmNXG1 (GenPept CAA48324; Baumann et al., 2007) and Populus PttXET16-34 (previously PttXET16A, GenPept AAN87142; Kallas et al., 2005) were heterologously expressed in Pichia pastoris and purified from culture media as previously described. Bacillus licheniformis endoxyloglucanase BlXG12 (GenPept AAR65335) and Paenibacillus pabuli endoxyloglucanase PpXG5 (Protein Data Bank identification codes 2JEP and 2JEQ) were kind gifts from Prof. Gideon Davies, University of York, UK (Gloster et al., 2007). Trichoderma reesei EGII (TrCel5A) was a kind gift from Dr. Kathleen Piens, Gent University.
Synthesis of Resorufin β-Glycosides
XXXG-β-Res (resorufinyl α-d-xylopyranosyl-(1→6)-β-d-glucopyranosyl)-(1→4)-[α-d-xylopyranosyl-(1→6)]-β-d-glucopyranosyl-(1→4)-[α-d-xylopyranosyl-(1→6)-]-β-d-glucopyranosyl-(1→4)-β-d-glucopyranoside. Per-O-acetylated XXXG was produced from tamarind (Tamarindus indica) seed xyloglucan as previously described (Greffe et al., 2005; Ibatullin et al., 2008). Following conversion to the corresponding α-glycosyl bromide with 33% solution of HBr in glacial HOAc (Ibatullin et al., 2008), aglycone coupling was performed under phase-transfer conditions (Dess et al., 1981; Kleine et al., 1985): A solution of resorufin sodium salt (Aldrich; 1.5–2 mmol) and benzyltributylammonium chloride (Aldrich; 1 mmol) in water (10–15 mL) was added to a stirred solution of the α-glycosyl bromide (1 mmol) in CH2Cl2 (15–20 mL), followed by vigorous stirring overnight at room temperature. The resulting suspension was filtered through Celite pad, the organic phase was washed with water, dried (Mg2SO4), and evaporated. The residue was purified by flash chromatography to give per-O-acetylated β-resorufinyl XXXG in 63% yield.
13C NMR (CDCl3, 125 MHz): δ (ppm) 186.16 (C=O Res), 170.39, 170.18, 170.15, 170.09, 169.99, 169.95, 169.91, 169.895, 169.89, 169.82, 169.77 (2C), 169.54, 169.50, 169.46, 169.25, 169.20, 168.77, 168.48 (19C, C=O Ac), 159.65, 149.42, 146.84, 145.01, 134.71, 134.63, 131.49, 129.46, 114.84, 106.87, 103.38 (11C, Resorufin), 100.46, 100.30, 100.17, 97.68, 97.01, 96.95, 95.82 (7 × C-1), 76.42, 74.97, 74.94, 74.64, 74.61, 73.22, 73.17, 72.82, 72.55, 72.33, 71.94, 71.75, 71.68, 71.51, 71.42, 70.61, 70.57, 70.34, 69.27, 69.13 (2C), 69.09, 69.02, 68.91, 68.88, 67.23, 65.41, 64.87 (7 × C-2, 7 × C-3, 7 × C-4, 7 × C-5), 62.06, 59.00, 58.86, 58.66 (4 × C-6), 20.84, 20.69, 20.67, 20.63, 20.62 (3C), 20.58 (2C), 20.57(4C), 20.55, 20.53, 20.52, 20.47(2C), 20.42 (19C, CH3 Ac). Electrospray ionization quadrupole-time-of-flight high-resolution mass spectrometry (ESI Q-TOF HRMS; mass-to-charge ratio [m/z]): 1,050.7836 (1,050.7805 calculated for C89H109NNa2O54 [M+Na]2+).
Zemplén de-O-acetylation (catalytic NaOMe in anhydrous CH2Cl2/MeOH) of the per-O-acetate and subsequent purification by reversed-phase chromatography using a 20-mL RP-18 cartridge (Supelco) using stepwise elution with a gradient of acetonitrile in water afforded resorufinyl α-d-xylopyranosyl-(1→6)-β-d-glucopyranosyl)-(1→4)-α-d-xylopyranosyl-(1→6)-β-d-glucopyranosyl-(1→4)-α-d-xylopyranosyl-(1→6)-β-d-glucopyranosyl-(1→4)-β-d-glucopyranoside (XXXG-β-Res [1]) in 76% yield.
13C-NMR (DMSO-d6, 125 MHz): δ (ppm) 185.36, 160.67, 149.63, 145.81, 144.89, 134.93, 133.89, 131.22, 128.51, 114.69, 105.70, 103.17 (12C, Resorufin), 102.55, 102.47, 102.46, 99.25, 99.18, 99.05, 98.66 (7 × C-1), 80.33, 80.29, 79.68, 76.26, 74.91, 74.63, 74.51 (2C), 74.38, 72.98, 72.96, 72.92, 72.90, 72.83, 72.75, 72.55 (2C), 72.48, 72.08, 72.06, 72.02, 70.09, 70.01 (2C), 69.97, 66.65, 66.64, 66.19, 66.16 (7 × C-2, 7 × C-3, 7 × C-4, 7 × C-5), 61.87, 61.86, 61.81, 59.74 (4 × C-6). ESI Q-TOF HRMS (m/z): 1,280.3695 (1,280.3704 calculated for C51H71NNaO35 [M+Na]+).
GG-β-Res (resorufinyl β-d-glucopyranosyl-(1→4)-β-d-glucopyranoside [2]). GG-β-Res was synthesized from per-O-acetyl cellobiose following the procedure described above for the synthesis of XXXG-β-Res. The intermediate resorufinyl hepta-O-acetyl-β-cellobioside and the final product had 1H-NMR spectra in accordance with those reported previously (Coleman et al., 2007). Additional spectral data were as follows.
Resorufinyl 4-O-(2,3,4,6-Tetra-O-Acetyl-β-d-Glucopyranosyl)-2,3,4,6-Tetra-O-Acetyl-β-d-Glucopyranoside (Resorufinyl Hepta-O-Acetyl-β-Cellobioside)
1H-NMR (CDCl3, 500 MHz): δ (ppm) 7.71 (d, 1H, J = 8.8 Hz), 7.41 (d, 1H, J = 9.8 Hz), 6.97 (dd, 1H, J1 = 8.7 Hz, J2 = 2.4 Hz), 6.92 (d, 1H, J = 2.5 Hz), 6.84 (dd, 1H, J1 = 9.9 Hz, J2 = 2.0 Hz), 6.29 (m, 1H) − 6H of resorufin, 5.29 (dd, 1H, J3,4 = 8.9 Hz, H-3), 5.22 (dd, 1H, J2,3 = 9.25 Hz, H-2), 5.16 (dd, 1H, J3′,4′ = 9.3 Hz, H-3′), 5.15 (d, 1H, J1,2 = 7.0 Hz, H-1), 5.07 (dd, 1H, J4′,5′ = 9.6 Hz, H-4′), 4.94 (dd, 1H, J2′,3′ = 9.3 Hz, H-2′), 4.57 (dd, 1H, J6a,6b = 11.8 Hz, H-6a), 4.54 (d, 1H, J1′,2′ = 8.0 Hz, H-1′), 4.37 (dd, 1H, J6′a,6′b = 12.5 Hz, H-6′a), 4.13 (dd, 1H. H-6b), 4.06 (dd, 1H, H-6′b), 3.86 (m, 2H, H-4, H-5), 3.68 (ddd, 1H, J5′,6′a = 4.3 Hz, J5′,6′b = 2.3 Hz, H-5′), 2.12, 2.09, 2.06, 2.05, 2.048, 2.01, 1.98 (7 s, 7 × 3H, CH3CO).
13C-NMR (CDCl3, 125 MHz): δ (ppm) 170.47, 170.19, 170.11, 169.70, 169.45, 169.31, 169.07 (7 × CO-Ac), 186.20, 159.86, 149.52, 146.88, 145.11, 134.81, 134.67, 131.60, 129.59, 115.03, 106.92, 103.41 (12 × C-Ar), 100.86, 98.22 (C-1′, C-1), 76.27, 73.37, 72.83, 72.31, 72.09, 71.60, 71.08, 67.75 (C-2, C-2′, C-3, C-3′, C-4, C-4′, C-5, C-5′); 61.88, 61.56 (C-6, C-6′), 20.70, 20.67, 20.64, 20.55, 20.54 (2C), 20.51 (7 × CH3-Ac).
ESI Q-TOF HRMS (m/z): 854.2131 (854.2120 calculated for C38H41NNaO20 [M+Na]+).
Resorufinyl 4-O-(β-d-Glucopyranosyl)-β-d-Glucopyranoside (GG-β-Res, Resorufinyl β-Cellobioside [2])
Resorufinyl β-cellobioside 13C-NMR (DMSO-d6, 125 MHz): δ (ppm) 185.37, 160.74, 149.62, 145.78, 144.89, 134.91, 133.87, 131.20, 128.48, 114.75, 105.71, 102.57 (12C of resorufin), 103.06 (C-1′), 99.41 (C-1), 79.61, 76.77, 76.42, 75.02, 74.67, 73.24, 72.77, 70.00 (C-2, C-2′, C-3, C-3′, C-4, C-4′, C-5, C-5′), 61.00, 59.89 (C-6, C-6′).
ESI Q-TOF HRMS (m/z): 560.1369 (560.1380 calculated for C24H27NNaO13 [M+Na]+).
Michaelis-Menten Enzyme Kinetic Analysis
The enzymatic hydrolysis of XXXG-β-Res was followed by continuously monitoring the release of resorufinyl anion (λmax 571 nm, ε 46,500 m−1cm−1, 50 mm MES buffer, pH 6.0) using a Cary 300 Bio UV/visible spectrophotometer (Varian). A total assay volume of 100 μL was used in 1-cm path length quartz cells equilibrated and maintained at 30°C ± 0.1°C in a Peltier-controlled cell block. Initial rates were determined from the slope of the linear region of the reaction time course corresponding to no more than 10% substrate conversion (often <1%). In these assays, the total concentration of TmNXG1 was 0.2 mg/mL (6.3 μm). Kinetic constants were obtained from nonlinear curve fits of the Michaelis-Menten equation to plots of vo/[E]t versus [S] using Microcal Origin v6.0; [E]t was assumed to be equivalent to the total protein concentration in all cases, i.e. 100% active enzyme.
In Vitro Fluorimetry
The time- and enzyme concentration-dependent release of resorufinyl anion was monitored by fluorimetry using a Perkin-Elmer LS 50B luminescence spectrometer, in 10/2 mm luminescence spectroscopy quartz microcells (Perkin-Elmer), in 80 mm MES buffer pH 6.0.
The volume of the analyzed sample was 500 μL. The excitation and the emission wavelength were correspondingly 571 and 584 nm, the emission filter was open, and the excitation and emission slits were set at 10.
In Situ Confocal Fluorescence Microscopy
Nasturtium seeds were imbibed in water for 5 d to trigger the reserve mobilization. Free-hand sections of the seeds were placed in the assay buffer containing 25 mm MES at pH 6.5. XXXG-β-Res was dissolved in this buffer at concentration 9.3 × 10−4 m, selected after optimization test using 4.7 × 10−4 m, 9.3 × 10−4 m, 4.7 × 10−4 m, 9.7 × 10−5 m, and 4.7 × 10−5 m concentrations. The optimal pH was selected after experiments with buffers having pH of 5.0, 5.5, 6.0, 6.5, and 7.0. The sections were transferred to the substrate (50 μL per section) on a glass slide, covered with coverslips, and sealed with the nail polish, and the evolution of the fluorescent signal from resorufin was monitored by time-lapse function of LSM 510 confocal microscope (Carl Zeiss) during 80 min with scans gathered every 5 min. The argon-krypton laser line at 568 nm was used for the excitation and the emission over 570 nm was recorded. The transmitted light signal was used to visualize anatomical details of the tissue. Control sections were heated at 95°C in the assay buffer for 30 min prior to the incubation with the substrate. Identical scanning conditions were used for the experimental sections and control sections.
Arabidopsis (Arabidopsis thaliana; Columbia) plants were grown for 6 weeks in long-day conditions, and the basal portion of inflorescent stems were used to study the distribution of xyloglucanase activity. The same labeling procedures were used as in the case of nasturtium seeds, except that the incubation time with the substrate was increased (up to 120 min). For the cellulase/glucosidase activities the procedures described in Takahashi et al. (2009) were used.
The procedures for in situ XET activity detection were based on Vissenberg et al. (2000) with modifications (Nishikubo et al., 2007). Tissue sections were incubated in 6.5 μm XXXG-sulforodamine (XXXG-SR, prepared as described by Nishikubo et al., 2007), dissolved in 25 mm MES buffer at pH 5.5 for 15 min in the dark at room temperature. The reaction was stopped by a 10 min wash in ethanol/formic acid/water (15:1:4), followed by an incubation in 5% formic acid for 1 h. Sections were mounted in Citifluor antifading medium (Citifluor Ltd.) and observed by confocal laser-scanning microscopy using an excitation wavelength of 568 nm and detection wavelengths over 585 nm.
Xyloglucan was detected by staining the fresh sections of seeds with iodine-potassium iodide and observed using the light microscope Axioplan 2 equipped with AxioVision camera (Carl Zeiss).
Acknowledgments
Prof. Gideon Davies (York Structural Biology Lab, University of York) is thanked for providing the PpXG5 and BlXG12 xyloglucanases. Dr. Kathleen Piens (Gent University) is thanked for a sample of T. reesei EGII (TrCel5A). Fredrika Gullfot (KTH Biotechnology) is gratefully acknowledged for producing TmNXG1. Gustav Sundqvist (KTH Biotechnology) is thanked for performing mass spectrometry analysis.
This work was supported by grants from the Swedish Foundation for Strategic Research, the Swedish Research Council, and the Knut and Alice Wallenberg Foundation. A.B. was the recipient of COST 50 STSM. M.B. was the recipient of predoctoral funding from the KTH Biofiber Material Centre. H.B. is a Special Research Fellow (Rådsforskare) of the Swedish Research Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Harry Brumer (harry@biotech.kth.se).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Baumann MJ (2004) Design and synthesis of xyloglucan oligosaccharides: structure-function studies and application of xyloglucan endotransglycosylase PttXET16A. Licentiate thesis. Royal Institute of Technology, Stockholm
- Baumann MJ, Eklöf JM, Michel G, Kallas ÅM, Teeri TT, Czjzek M, Brumer H (2007) Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: biological implications for cell wall metabolism. Plant Cell 19 1947–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becnel J, Natarajan M, Kipp A, Braam J (2006) Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol Biol 61 451–467 [DOI] [PubMed] [Google Scholar]
- Beisson F, Arondel V, Verger R (2000) Assaying Arabidopsis lipase activity. Biochem Soc Trans 28 773–775 [DOI] [PubMed] [Google Scholar]
- Berg RH, Beachy RN (2008) Fluorescent protein applications in plants. In KF Sullivan, ed, Fluorescent Proteins, Ed 2. Methods in Cell Biology, Vol 85. Elsevier Academic Press, San Diego, 153–179 [DOI] [PubMed]
- Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M, Teeri TT, Sundberg B, Mellerowicz EJ (2002) Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell 14 3073–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno C, Villegas ML, Bertolotti SG, Previtali CM, Neumann MG, Encinas MV (2002) The excited-state interaction of resazurin and resorufin with amines in aqueous solutions: photophysics and photochemical reaction. Photochem Photobiol 76 385–390 [DOI] [PubMed] [Google Scholar]
- Cabib E, Farkas V, Kosik O, Blanco N, Arroyo J, McPhie P (2008) Assembly of the yeast cell wall: Crh1p and Crh2p act as transglycosylases in vivo and in vitro. J Biol Chem 283 29859–29872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37 D233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantarangsee M, Tanthanuch W, Fujimura T, Fry SC, Cairns JK (2007) Molecular characterization of beta-galactosidases from germinating rice (Oryza sativa). Plant Sci 173 118–134 [Google Scholar]
- Chapman S, Oparka KJ, Roberts AG (2005) New tools for in vivo fluorescence tagging. Curr Opin Plant Biol 8 565–573 [DOI] [PubMed] [Google Scholar]
- Coleman DJ, Studler MJ, Naleway JJ (2007) A long-wavelength fluorescent substrate for continuous fluorometric determination of cellulase activity: resorufin-beta-D-cellobioside. Anal Biochem 371 146–153 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6 850–861 [DOI] [PubMed] [Google Scholar]
- Dess D, Kleine HP, Weinberg DV, Kaufman RJ, Sidhu RS (1981) Phase-transfer catalyzed synthesis of acetylated aryl beta-D-glucopyranosides and aryl beta-D-galactopyranosides. Synthesis 883–885
- Edwards M, Dea ICM, Bulpin PV, Reid JSG (1986) Purification and properties of a novel xyloglucan-specific endo-(1→4)-beta-D-glucanase from germinated nasturtium seeds (Tropaeolum majus L). J Biol Chem 261 9489–9494 [PubMed] [Google Scholar]
- Ehrhardt D (2003) GFP technology for live cell imaging. Curr Opin Plant Biol 6 622–628 [DOI] [PubMed] [Google Scholar]
- English BP, Min W, van Oijen AM, Lee KT, Luo GB, Sun HY, Cherayil BJ, Kou SC, Xie XS (2006) Ever-fluctuating single enzyme molecules: Michaelis-Menten equation revisited. Nat Chem Biol 2 87–94 [DOI] [PubMed] [Google Scholar]
- Fasano JM, Swanson SJ, Blancaflor EB, Dowd PE, Kao TH, Gilroy S (2001) Changes in root cap pH are required for the gravity response of the Arabidopsis root. Plant Cell 13 907–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure R, Saura-Valls M, Brumer H, Planas A, Cottaz S, Driguez H (2006) Synthesis of a library of xylogluco-oligosaccharides for active-site mapping of xyloglucan endo-transglycosylase. J Org Chem 71 5151–5161 [DOI] [PubMed] [Google Scholar]
- Felle HH (2005) pH regulation in anoxic plants. Ann Bot (Lond) 96 519–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau JP, Kato Y, Lorences EP, Maclachlan GA, McNeil M, et al (1993) An unambiguous nomenclature for xyloglucan-serived oligosaccharides. Physiol Plant 89 1–3 [Google Scholar]
- Ge Y, Antoulinakis EG, Gee KR, Johnson L (2007) An ultrasensitive, continuous assay for xylanase using the fluorogenic substrate 6,8-difluoro-4-methylumbelliferyl beta-D-xylobloside. Anal Biochem 362 63–68 [DOI] [PubMed] [Google Scholar]
- Gilbert HJ, Stålbrand H, Brumer H (2008) How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Curr Opin Plant Biol 11 338–348 [DOI] [PubMed] [Google Scholar]
- Gloster TM, Ibatullin FM, Macauley K, Eklöf JM, Roberts S, Turkenburg JP, Bjørnvad ME, Linå Jørgensen P, Danielsen S, Johansen KS, et al (2007) Characterization and three-dimensional structures of two distinct bacterial xyloglucanases from families GH5 and GH12. J Biol Chem 282 19177–19189 [DOI] [PubMed] [Google Scholar]
- Gorris HH, Rissin DM, Walt DR (2007) Stochastic inhibitor release and binding from single-enzyme molecules. Proc Natl Acad Sci USA 104 17680–17685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greffe L, Bessueille L, Bulone V, Brumer H (2005) Synthesis, preliminary characterization, and application of novel surfactants from highly branched xyloglucan oligosaccharides. Glycobiology 15 437–445 [DOI] [PubMed] [Google Scholar]
- Hays WS, Wheeler DE, Eghtesad B, Glew RH, Johnston DE (1998) Expression of cytosolic beta-glucosidase in guinea pig liver cells. Hepatology 28 156–163 [DOI] [PubMed] [Google Scholar]
- Henrissat B, Teeri TT, Warren RAJ (1998) A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. FEBS Lett 425 352–354 [DOI] [PubMed] [Google Scholar]
- Hoffman M, Jia ZH, Peña MJ, Cash M, Harper A, Blackburn AR, Darvill A, York WS (2005) Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr Res 340 1826–1840 [DOI] [PubMed] [Google Scholar]
- Ibatullin FM, Baumann MJ, Greffe L, Brumer H (2008) Kinetic analyses of retaining endo-(xylo)glucanases from plant and microbial sources using new chromogenic xylogluco-oligosaccharide aryl glycosides. Biochemistry 47 7762–7769 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions—beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Lloyd RV (1997) In situ hybridization: methods and applications. J Clin Lab Anal 11 2–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson P, Brumer H, Baumann MJ, Kallas ÅM, Henriksson H, Denman SE, Teeri TT, Jones TA (2004) Crystal structures of a poplar xyloglucan endotransglycosylase reveal details of transglycosylation acceptor binding. Plant Cell 16 874–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallas ÅM, Piens K, Denman SE, Henriksson H, Fäldt J, Johansson P, Brumer H, Teeri TT (2005) Enzymatic properties of native and deglycosylated hybrid aspen (Populus tremula×tremuloides) xyloglucan endotransglycosylase 16A expressed in Pichia pastoris. Biochem J 390 105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine HP, Weinberg DV, Kaufman RJ, Sidhu RS (1985) Phase-transfer-catalyzed synthesis of 2,3,4,6-tetra-O-acetyl-beta-D-galacto-pyranosides. Carbohydr Res 142 333–337 [Google Scholar]
- Knox JP (2008) Revealing the structural and functional diversity of plant cell walls. Curr Opin Plant Biol 11 308–313 [DOI] [PubMed] [Google Scholar]
- Lopez-Casado G, Urbanowicz BR, Damasceno CMB, Rose JKC (2008) Plant glycosyl hydrolases and biofuels: a natural marriage. Curr Opin Plant Biol 11 329–337 [DOI] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Loudet O, Kronenberger J, Mouille G, Marion-Poll A, North HM (2007) A naturally occurring mutation in an Arabidopsis accession affects a beta-D-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19 3990–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerowicz EJ, Immerzeel P, Hayashi T (2008) Xyloglucan: the molecular muscle of trees. Ann Bot (Lond) 102 659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minic Z, Jouanin L (2006) Plant glycoside hydrolases involved in cell wall polysaccharide degradation. Plant Physiol Biochem 44 435–449 [DOI] [PubMed] [Google Scholar]
- Monroe JD, Gough CM, Chandler LE, Loch CM, Ferrante JE, Wright PW (1999) Structure, properties, and tissue localization of apoplastic alpha-glucosidase in crucifers. Plant Physiol 119 385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T, Gandotra N, Tausta SL (2008) Plant cell types: reporting and sampling with new technologies. Curr Opin Plant Biol 11 567–573 [DOI] [PubMed] [Google Scholar]
- Nishikubo N, Awano T, Banasiak A, Bourquin V, Ibatullin F, Funada R, Brumer H, Teeri TT, Hayashi T, Sundberg B, et al (2007) Xyloglucan endo-transglycosylase (XET) functions in gelatinous layers of tension wood fibers in poplar—a glimpse into the mechanism of the balancing act of trees. Plant Cell Physiol 48 843–855 [DOI] [PubMed] [Google Scholar]
- O'Neill MA, Selvendran RR (1985) Structural analysis of the xyloglucan from Phaseolus coccineus cell walls using cellulase-derived oligosaccharides. Carbohydr Res 145 45–58 [Google Scholar]
- Park YW, Tominaga R, Sugiyama J, Furuta Y, Tanimoto E, Samejima M, Sakai F, Hayashi T (2003) Enhancement of growth by expression of poplar cellulase in Arabidopsis thaliana. Plant J 33 1099–1106 [DOI] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43 1421–1435 [DOI] [PubMed] [Google Scholar]
- Stewart CN (2005) Monitoring the presence and expression of transgenes in living plants. Trends Plant Sci 10 390–396 [DOI] [PubMed] [Google Scholar]
- Takahashi J, Rudsander UJ, Hedenström M, Banasiak A, Harholt J, Amelot N, Immerzeel P, Ryden P, Endo S, Ibatullin FM, et al (2009) KORRIGAN1 and its aspen homolog PttCel9A1 decrease cellulose crystallinity in Arabidopsis stems. Plant Cell Physiol 50 1099–1115 [DOI] [PubMed] [Google Scholar]
- Tokutake S, Kasai K, Tomikura T, Yamaji N, Kato M (1990) Glycosides having chromophores as substrates for sensitive enzyme analysis. 2. Synthesis of phenolindophenyl-beta-D-glucopyranosides having an electron-withdrawing substituent as substrates for beta-glucosidase. Chem Pharm Bull (Tokyo) 38 3466–3470 [Google Scholar]
- Vicente AR, Saladie M, Rose JKC, Labavitch JM (2007) The linkage between cell wall metabolism and fruit softening: looking to the future. J Sci Food Agric 87 1435–1448 [Google Scholar]
- Vissenberg K, Fry SC, Pauly M, Hofte H, Verbelen JP (2005) XTH acts at the microfibril-matrix interface during cell elongation. J Exp Bot 56 673–683 [DOI] [PubMed] [Google Scholar]
- Vissenberg K, Martinez-Vilchez IM, Verbelen JP, Miller JG, Fry SC (2000) In vivo colocalization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of Arabidopsis roots. Plant Cell 12 1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrsanska M, Nerinckx W, Claeyssens M, Biely P (2008) An alternative approach for the synthesis of fluorogenic substrates of endo-beta-(1→4)-xylanases and some applications. Carbohydr Res 343 541–548 [DOI] [PubMed] [Google Scholar]
- Walker RP, Chen ZH, Johnson KE, Famiani F, Tecsi L, Leegood RC (2001) Using immunohistochemistry to study plant metabolism: the examples of its use in the localization of amino acids in plant tissues, and of phosphoenolpyruvate carboxykinase and its possible role in pH regulation. J Exp Bot 52 565–576 [PubMed] [Google Scholar]
- Wen F, Celoy R, Price I, Ebolo JJ, Hawes MC (2008) Identification and characterization of a rhizosphere beta-galactosidase from Pisum sativum L. Plant Soil 304 133–144 [Google Scholar]
- Wittrup KD, Bailey JE (1988) A single-cell assay of beta-galactosidase activity in Saccharomyces cerevisiae. Cytometry 9 394–404 [DOI] [PubMed] [Google Scholar]
- York WS, Harvey LK, Guillen R, Albersheim P, Darvill AG (1993) The structure of plant cell walls. 36. Structural-analysis of tamarind seed xyloglucan oligosaccharides using beta-galactosidase digestion and spectroscopic methods. Carbohydr Res 248 285–301 [DOI] [PubMed] [Google Scholar]