Abstract
Peroxisomes are unique organelles involved in multiple cellular metabolic pathways. Nitric oxide (NO) is a free radical active in many physiological functions under normal and stress conditions. Using Arabidopsis (Arabidopsis thaliana) wild type and mutants expressing green fluorescent protein through the addition of peroxisomal targeting signal 1 (PTS1), which enables peroxisomes to be visualized in vivo, this study analyzes the temporal and cell distribution of NO during the development of 3-, 5-, 8-, and 11-d-old Arabidopsis seedlings and shows that Arabidopsis peroxisomes accumulate NO in vivo. Pharmacological analyses using nitric oxide synthase (NOS) inhibitors detected the presence of putative calcium-dependent NOS activity. Furthermore, peroxins Pex12 and Pex13 appear to be involved in transporting the putative NOS protein to peroxisomes, since pex12 and pex13 mutants, which are defective in PTS1- and PTS2-dependent protein transport to peroxisomes, registered lower NO content. Additionally, we show that under salinity stress (100 mm NaCl), peroxisomes are required for NO accumulation in the cytosol, thereby participating in the generation of peroxynitrite (ONOO−) and in increasing protein tyrosine nitration, which is a marker of nitrosative stress.
Peroxisomes are single membrane-bound organelles whose basic enzymatic constituents are catalase and H2O2-producing flavin oxidases as their basic enzymatic and are found in virtually all eukaryotic cell types (Corpas et al., 2001; Hayashi and Nishimura, 2006; Reumann et al., 2007; Pracharoenwattana and Smith, 2008; Palma et al., 2009). These oxidative organelles are characterized by metabolic plasticity, as their enzymatic content can vary according to the organism, cell/tissue type, and environmental conditions (Mullen et al., 2001; Hayashi and Nishimura, 2003; Corpas et al., 2009a). In higher plants, peroxisomes contain a complex battery of antioxidative enzymes, such as catalase, superoxide dismutase, the components of the ascorbate-glutathione cycle, and the NADP-dehydrogenases of the pentose-P pathway (Corpas et al., 2009a). The generation of superoxide radicals has also been reported in the matrices and membranes of peroxisomes (López-Huertas et al., 1999; del Río et al., 2006). All these findings point to the important role played by peroxisomes in the cellular metabolism of reactive oxygen species (Corpas et al., 2001, 2009a; del Río et al., 2006).
Nitric oxide (NO) is a free radical involved in many physiological functions under normal and stress conditions in both animal and plant cells (Arasimowicz and Floryszak-Wieczorek, 2007; Corpas et al., 2007a, 2008; Neill et al., 2008). Unlike animal systems, knowledge of NO generation and subcellular location in plants remains largely elusive, and the data are sometimes contradictory and ambiguous (Zemojtel et al., 2006; Jasid et al., 2006; Gas et al., 2009). In previous studies, we detected l-Arg-dependent nitric oxide synthase (NOS) activity in isolated pea (Pisum sativum) leaf peroxisomes (Barroso et al., 1999). In a later study, using electron paramagnetic resonance techniques, we demonstrated the presence of NO in these types of peroxisomes (Corpas et al., 2004). However, several issues, such as whether NO is released into the cytosol and the physiological function of this free radical, remain unresolved.
In this study, we provide an in vivo demonstration that Arabidopsis peroxisomes are essential for NO accumulation in the cytosol, thus participating in the generation of nitrosative stress under salinity conditions. In addition, using Arabidopsis mutants pex12 and pex13, we also suggest that these peroxins are involved in importing into peroxisomes the enzyme responsible for NO generation.
RESULTS
Localization of NO during the Development of Arabidopsis Seedlings
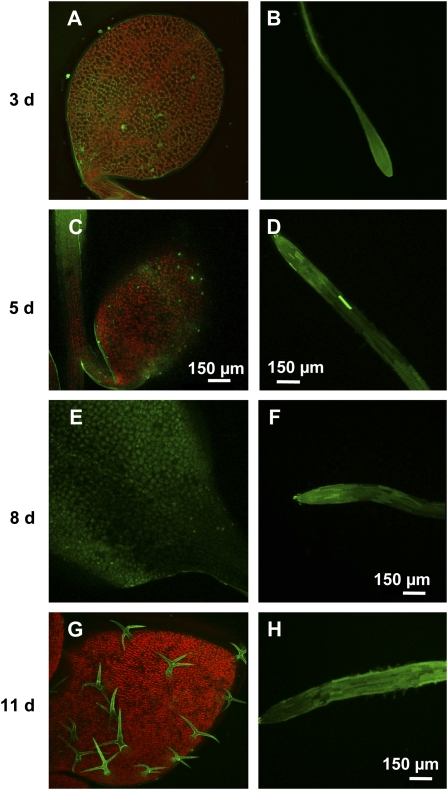
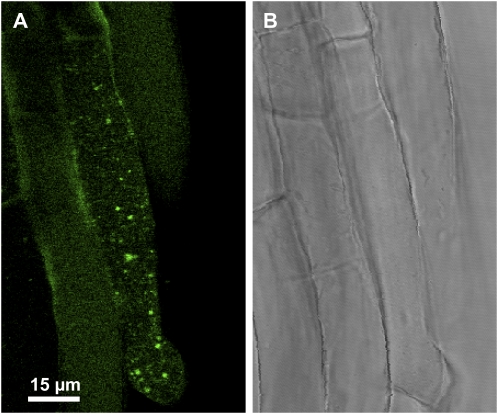
The visualization of endogenous NO in primary roots and cotyledons from 3-, 5-, 8-, and 11-d-old Arabidopsis seedlings was carried out through confocal laser scanning microscopy (CLSM) using 4-aminomethyl-2′,7′-difluorofluorescein diacetate (DAF-FM DA) as a fluorescent probe (Fig. 1, A–H). In cotyledons, an intense green fluorescence caused by NO was observed in the epidermal cells. This fluorescence was more intense in 8-d-old seedlings (Fig. 1E) and 11-d-old trichome seedlings (Fig. 1G). The orange color corresponds to autofluorescence. On the other hand, in primary roots, the green fluorescence was present throughout the entire apical root regardless of length of development (Fig. 1, B, D, F, and H). Figure 2 shows a high magnification image of root cells, where green spherical spots resembling peroxisomes inside the cell can be observed.
Figure 1.
Representative images illustrating the CLSM detection of endogenous NO (green color) in Arabidopsis seedlings at different stages of development. A and B, Cotyledon and primary root of 3-d-old Arabidopsis seedling, respectively. C and D, Cotyledon and primary root of 5-d-old Arabidopsis seedling, respectively. E and F, Cotyledon and primary root of 8-d-old Arabidopsis seedling, respectively. G and H, Cotyledon and primary root of 11-d-old Arabidopsis seedling, respectively. Arabidopsis seedlings were incubated with 10 μm DAF-FM DA as fluorescent probe. The orange-yellow color corresponds to the autofluorescence. [See online article for color version of this figure.]
Figure 2.
High magnification micrograph illustrating the CLSM in vivo detection of NO (green color) in Arabidopsis seedling roots, where punctate spots inside the cells (A) and their corresponding bright-field image (B) can be observed. NO was detected using 10 μm DAF-FM DA (green color; excitation 495 nm and emission 515 nm) as fluorescent probe. [See online article for color version of this figure.]
Location of NO in Root Peroxisomes (GFP-PTS1)
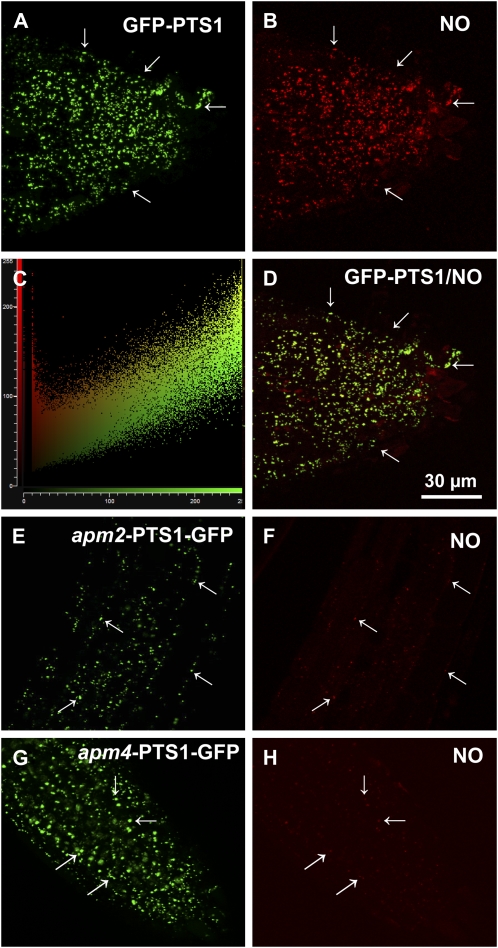
Figure 3A shows in vivo CLSM visualization of peroxisomes in the root tips of transgenic Arabidopsis plants expressing GFP through the addition of peroxisomal targeting signal 1 (PTS1) (Mano et al., 2002). The peroxisomes appeared in the form of spherical spots in all root tip cells. Figure 3B shows the same field analyzed using the diaminorhodamine-4M acetoxymethyl ester (DAR-4M AM) fluorescence probe, which also enabled NO to be detected (Kojima et al., 2001; Tun et al., 2006, 2008). An intense red fluorescence was found in spherical spots with a pattern similar to that of the GFP-PTS1. Figure 3C shows the linear correlation spots obtained using Leica software, indicating that most of these spots coincide. Figure 3D contains a merged image of the overlap of A and B, showing a virtually complete overlap of the two punctate patterns, indicating that NO was present in peroxisomes. The location of NO in two Arabidopsis mutants with an aberrant peroxisome morphology (apm; Mano et al., 2006) was analyzed to gain more knowledge about the transport of the protein assumed to be responsible for NO generation in peroxisomes. Figure 3, E and G, show the green fluorescent patterns corresponding to GFP-PTS1 in apm2/pex13 and apm4/pex12 mutants, respectively. The peroxisomes were detected in the form of spherical spots in all cells. However, GFP fluorescence was also detected in the cytosol, indicating that the targeting of PTS1-containing proteins in apm2/4 mutants was impaired. Figure 3, F and H, show the location of NO in the same fields as those depicted in E and G, respectively. Supplemental Figure S1 shows the overlap of D with the bright-field image of the Arabidopsis root tip.
Figure 3.
Representative images illustrating the CLSM in vivo detection of NO (red color) and peroxisomes (green color) in root tips of 5-d-old Arabidopsis seedlings of parent plants and mutants with aberrant peroxisome morphology (apm2 and apm4) expressing GFP-PTS1. A, Fluorescence punctates (green) attributable to GFP-PTS1, indicating the localization of peroxisomes in parent plants. B, Fluorescence punctates (red) attributable to NO detection in the same root area. C, Plot showing the linear correlation between the punctates corresponding to GFP-PTS1 and NO. D, Merged image of A and B showing colocalized fluorescence punctates (yellow). E, Fluorescence punctates (green) attributable to GFP-PTS1 in apm2/pex13 mutants. F, Fluorescence punctates (red) attributable to NO detection in apm2/pex13 mutants. G, Fluorescence punctates (green) attributable to GFP-PTS1 in apm4/pex12 mutants. H, Fluorescence punctates (red) attributable to NO detection in apm4/pex12 mutants. NO was detected with DAR-4M AM (excitation 543 nm; emission 575 nm) and peroxisomes with GFP (excitation 495 nm; emission 515 nm). Arrows indicate representative punctate spots corresponding to NO and peroxisome localization. [See online article for color version of this figure.]
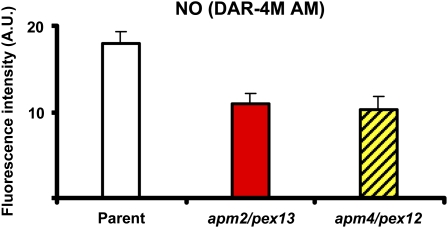
Figure 4 shows the change in the fluorescence intensity of NO in Figure 3, B, F, and H. In the mutants characterized by defective protein transport to peroxisomes, NO production was significantly lower (−39% for apm2 and −43% for apm4), suggesting that the protein responsible for releasing NO into peroxisomes is adversely affected in these mutants.
Figure 4.
Fluorescence intensity of NO in root tips of 5-d-old Arabidopsis seedlings of parent plants and mutants with aberrant peroxisome morphology, apm2/pex13 and apm4/pex12, expressing GFP-PTS1 shown in Figure 3, B, F, and H. The fluorescence produced was expressed as arbitrary units (A.U.) using Leica confocal software. [See online article for color version of this figure.]
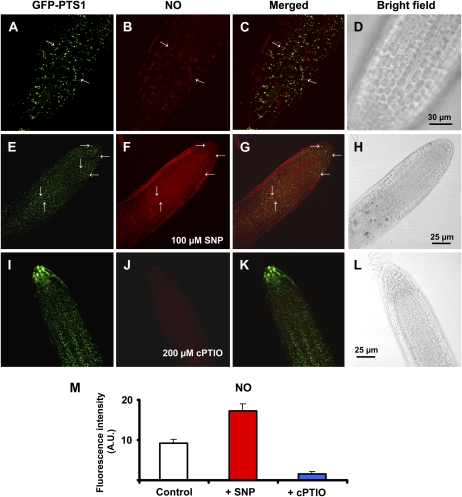
Several controls were used to confirm the specific findings of NO detection using the new fluorescence probe DAR-4M AM in Arabidopsis seedling expressing GFP-PTS1. Figure 5, E to H, show the images of the same tip root of Arabidopsis seedlings preincubated with 100 μm sodium nitroprusside (SNP), a NO donor used as positive control. Under these conditions, a NO-dependent increase in DAR-4M AM fluorescence (red color) was observed compared with the control seedlings (Fig. 5B), showing the specificity of this fluorescence probe in relation to NO, as previously reported for Arabidopsis wild type (Tun et al., 2006, 2008). On the other hand, when the seedlings were preincubated with 200 μm 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO; a NO scavenger), the red fluorescence was greatly reduced (Fig. 5J) without affecting GFP-PTS1 detection (Fig. 5I). The quantification of NO showed that SNP increases fluorescence intensity 1.9-fold and that the cPTIO reduced NO approximately 5.8-fold (Fig. 5M).
Figure 5.
Representative images illustrating the CLSM in vivo detection of NO (red color) and peroxisomes (green color) in primary roots of Arabidopsis seedlings expressing GFP-PTS1 (green color) preincubated with different chemicals as controls. Fourteen-day-old Arabidopsis seedlings expressing GFP-PTS1 incubated only with DAR-4M AM as the fluorescent probe for NO (A–D) preincubated for 2 h 30 min at 25°C with 100 μm SNP as NO donor (E–H) and 200 μm cPTIO as NO scavenger (I and J). A, E, and I show peroxisome detection with GFP (excitation 495 nm; emission 515 nm). B, F, and J show NO detection (red color) with DAR-4M AM (excitation 543 nm; emission 575 nm). C, G, and K show merged images of corresponding treatments. D, H, and L show the bright-field image of the corresponding samples. M, Fluorescence intensity of NO from B, F, and J. Arrows indicate some representative punctate spots corresponding to NO and peroxisome localization. Fluorescence is expressed as arbitrary units (A.U.) using Leica confocal software. [See online article for color version of this figure.]
Similarly, Supplemental Figure S2 shows the staining of Arabidopsis apm2/pex13 (A–H) and apm4/pex12 (I–P) mutants with DAR-4M AM preincubated with 100 μm SNP, a NO donor used as positive control. The quantification of the fluorescence intensity of NO showed that SNP increases fluorescence 1.9-fold in apm2/pex13 and 1.6-fold in apm4/pex12 (Supplemental Fig. S2Q). It is therefore possible to conclude that DAR-4M AM uptake and staining in these mutants were not affected by the use of the NO donor.
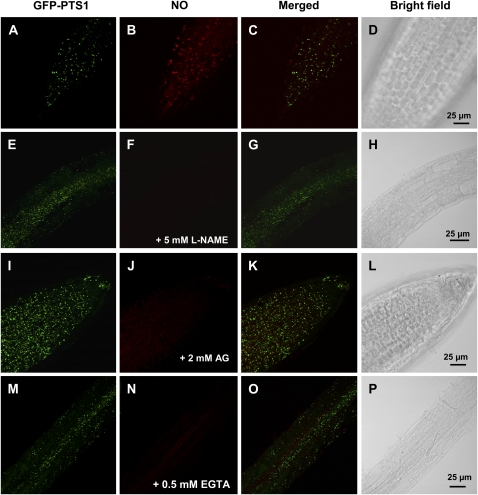
A pharmacological approach was also used to evaluate the putative involvement of a NOS activity in the release of NO into peroxisomes. The preincubation of Arabidopsis seedlings with 5 mm NG-nitro-l-Arg methyl ester (l-NAME; Fig. 6, E–H) and 2 mm aminoguanidine (Fig. 6, I–L), two well-known inhibitors of animal NOS, produced a sharp reduction in NO detected by DAR-4M AM when compared to the control (Fig. 6, A–D), which suggests that a l-Arg-dependent NOS activity is involved in the production of the NO detected. Furthermore, when the Arabidopsis seedlings were preincubated with 0.5 mm EGTA (Fig. 5, M–P) as a calcium chelator, NO content was also significantly reduced, indicating that NO generation was calcium dependent.
Figure 6.
Representative images illustrating the CLSM in vivo detection of NO (red color) and peroxisomes (green color) in primary roots of Arabidopsis seedlings expressing GFP-PTS1 (green color) preincubated with two inhibitors of animal NOS activity (5 mm l-NAME and 2 mm AG) and a calcium chelator (0.5 mm EGTA). Fourteen-day-old Arabidopsis seedlings expressing GFP-PTS1 incubated with 5 μm DAR-4M AM for 1 h at 25°C to detect and visualize NO by CLSM (A–D), preincubated for 2 h 30 min at 25°C with 5 mm l-NAME as competitive inhibitor of animal NOSs (E–H), 2 mm AG as general inhibitor of animal NOSs (I–L), and 0.5 mm EGTA as calcium chelator (M–P) and then incubated with 5 μm DAR-4M AM for 1 h at 25°C to detect and visualize NO by CLSM. A, E, I, and M show peroxisome detection with GFP (excitation 495 nm; emission 515 nm). B, F, J, and N show NO detection (red color) with DAR-4M AM (excitation 543 nm; emission 575 nm). C, G, K, and O show merged images of corresponding treatments. D, H, L, and P show the bright-field image of the corresponding samples. [See online article for color version of this figure.]
NO in Arabidopsis Root Peroxisomes under Salinity Stress
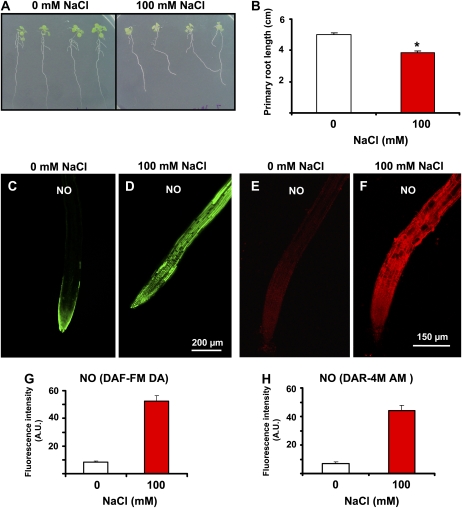
To study the potential physiological function of NO released into peroxisomes, its production was studied under abiotic stress conditions. Previous studies have shown that Arabidopsis seedlings grown with 100 mm NaCl underwent salinity stress (Sun et al., 2007; Nishizawa et al., 2008). Figure 7A shows Arabidopsis seedlings grown with 100 mm NaCl, which significantly reduced root length by 24% (Fig. 7B). When NO generation was analyzed by CLSM, using both DAF-FM DA (Fig. 7, C and D) and DAR-4M AM (Fig. 7, E and F) as fluorescence probes, a significant increase in NO production was observed in roots under salt stress. On the other hand, the fluorescence intensity of NO under salinity conditions, as detected by DAF-FM DA and DAR-4M AM, increased 6.4-fold (Fig. 7G) and 6.3-fold (Fig. 7H), respectively, indicating that both these fluorescence probes are useful tools.
Figure 7.
Effect of salinity on Arabidopsis seedling growth and NO content detected with DAF-FM DA and DAR-4M AM. Appearance (A) and primary root length (B) of 13-d-old Arabidopsis seedling growth in MS medium supplemented and not supplemented with 100 mm NaCl. Results are the mean of three different experiments ± se. Asterisk indicates differences in relation to control values were significant at P < 0.05. Representative images illustrating the CLSM detection of NO (green or red color) in primary roots of 6-d-old Arabidopsis wild-type seedlings exposed and not exposed to 100 mm NaCl using DAF-FM DA (C and D) and DAR-4M AM (E and F) as fluorescent probes. Fluorescence intensity of NO was detected by DAF-FM DA (G) or DAR-4M AM (H). NO was detected using 10 μm DAF-FM DA (green color; excitation 495 nm and emission 515 nm) and DAR-4M AM (red color; excitation 543 nm and emission 575 nm) as fluorescent probes. Fluorescence is expressed as arbitrary units (A.U.) using Leica confocal software. [See online article for color version of this figure.]
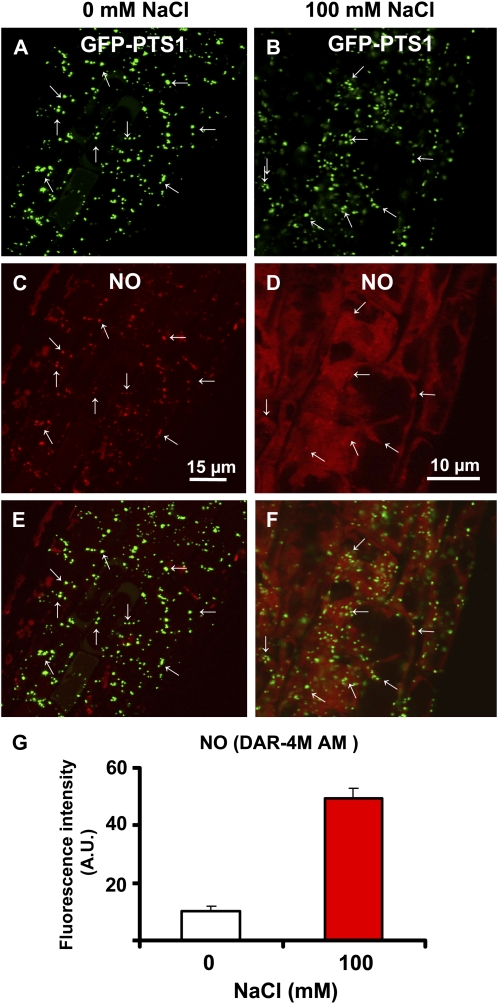
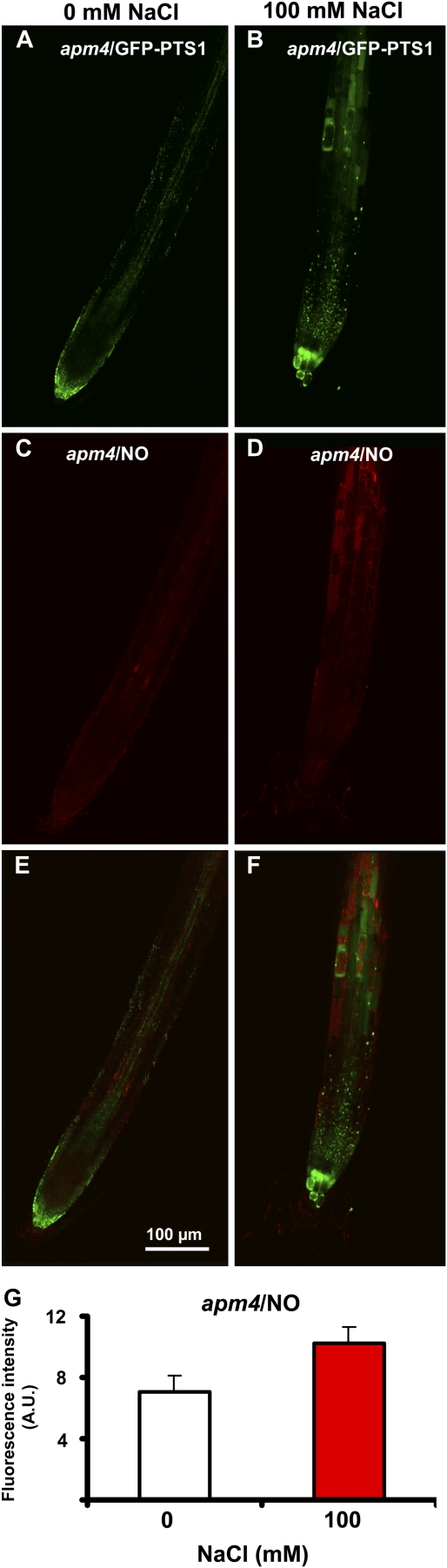
Mutants expressing GFP-PTS1 were used to evaluate if peroxisomes are a potential source of NO in Arabidopsis under salinity conditions. Figure 8 shows NO location and production in Arabidopsis roots expressing GFP-PTS1 under salinity stress (100 mm NaCl). Figure 8, A and C, show that peroxisomes are subcellular compartments where NO is produced mainly in the roots of control (0 mm NaCl) plants, as both punctate patterns are seen to overlap in the merged image (Fig. 8E). Under salinity conditions (100 mm NaCl), the pattern of peroxisomes did not appreciably change compared with control (0 mm NaCl) plants; however, NO production increased significantly (4.8-fold) under salinity conditions (Fig. 8G). The distribution of fluorescence detected in both peroxisomes and cytosol also varied considerably (Fig. 8, D and F). This suggests that NO is released from the peroxisomes into the cytosol under salinity stress. To corroborate this hypothesis, the effect of salinity stress was studied in apm4/pex12 mutants, where the import of proteins into the peroxisomal matrix is affected, including probably the protein that generates NO and showed a low NO content (see Fig. 3H). In Figure 9, A and B, the pattern of peroxisomes appears in the form of green spots in the roots of control and stressed plants, with a slight increase in the number of peroxisomes being observed under salinity stress. Figure 9C shows the location of NO (red color) in the same root area of Figure 9A (0 mm NaCl) where NO was almost totally absent. Under salinity stress (Fig. 9D), NO slightly increased. Figure 9, E and F, show the merged images of control and stressed roots, respectively, where it can be clearly observed that NO is present in cytosol, indicating that the peroxisomal protein responsible for NO generation was not imported into the peroxisomes. Figure 9J shows the relative quantities of NO production in apm4/pex12 mutants under control and salinity conditions, where NO increased 1.4-fold, a lower increase than that the observed in parent plants expressing GFP-PTS1 (Fig. 8G). This suggests that peroxisomes are the main source of NO under normal and stress conditions in Arabidopsis roots.
Figure 8.
Representative images illustrating the CLSM in vivo detection of NO (red color) in root tips of 6-d-old Arabidopsis seedlings expressing GFP-PTS1 (green color) exposed to 100 mm NaCl for 7 d. A, Fluorescence punctates (green) attributable to GFP-PTS1, indicating the localization of peroxisomes in control (0 mm NaCl) plants. B, Fluorescence punctates (green) attributable to GFP-PTS1, indicating the location of peroxisomes in Arabidopsis plants exposed to 100 mm NaCl. C, Fluorescence punctates (red) attributable to NO detection in the same root area of A. D, NO detection (red) in the same root area of B. E, Merged image of A and C showing colocalized fluorescence punctates (yellow). F, Merged image of B and D showing colocalized fluorescence punctates (yellow). G, Fluorescence intensity of NO from C and D. NO was detected with DAR-4M AM (excitation 543 nm; emission 575 nm) and peroxisomes with GFP (excitation 495 nm; emission 515 nm). Fluorescence is expressed as arbitrary units (A.U.) using Leica confocal software. [See online article for color version of this figure.]
Figure 9.
Representative images illustrating the CLSM in vivo detection of NO (red color) in root tips of 6-d-old Arabidopsis apm4/pex12 mutant seedlings expressing GFP-PTS1 (green color) exposed to 100 mm NaCl for 7 d. A, Fluorescence punctates (green) attributable to GFP-PTS1, indicating the location of peroxisomes in control (0 mm NaCl) plants. B, Fluorescence punctates (green) attributable to GFP-PTS1, indicating the location of peroxisomes in Arabidopsis apm4/pex12 mutants exposed to 100 mm NaCl. C, Fluorescence punctates (red) attributable to NO detection in the same root area of A. D, NO detection (red) in the same root area of B. E, Merged image of A and C showing colocalized fluorescence punctates (yellow). F, Merged image of B and D showing colocalized fluorescence punctates (yellow). G, Fluorescence intensity of NO from C and D. NO was detected with DAR-4M AM (excitation 543 nm; emission 575 nm) and peroxisomes with GFP (excitation 495 nm; emission 515 nm). Fluorescence is expressed as arbitrary units (A.U.) using Leica confocal software. [See online article for color version of this figure.]
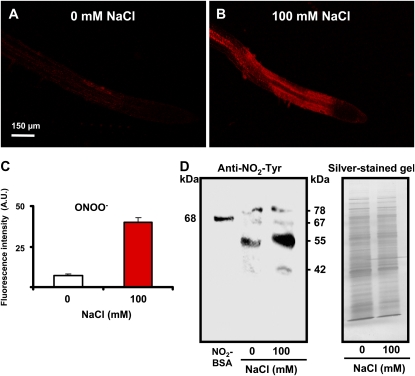
The reaction of NO with the superoxide radical (O2·−) generates peroxynitrite (ONOO−), which has been shown to mediate an increase in the Tyr nitration of proteins under stress conditions in animal cells (Radi, 2004; Szabó et al., 2007). To evaluate this possible correlation between ONOO− and protein nitration in Arabidopsis under salinity conditions, ONOO− was analyzed in roots by CLSM using the fluorescence probe 3′-(p-aminophenyl) fluorescein (APF; Chaki et al., 2009a, 2009b), while the presence of Tyr nitration was studied by immunoblot analysis using a well-characterized antibody against 3-nitrotyrosine (Valderrama et al., 2007; Corpas et al., 2008; Chaki et al., 2009a, 2009b). Figure 10A shows the location of ONOO− in the control roots of Arabidopsis wild-type seedlings and ONOO− significantly increased in roots under salinity stress (Fig. 10B). Figure 10C shows that the fluorescence intensity of ONOO− increased 5.6-fold under salinity conditions. On the other hand, Figure 10D depicts the immunoblot analysis of protein Tyr nitration in Arabidopsis roots using an antibody against 3-nitrotyrosine (NO2-Tyr) and the corresponding silver-stained SDS (10%) gel. Thus, in control roots, a three-immunoreactive-band pattern with molecular masses of 78, 67, and 55 kD, respectively, was observed. This resembled the pattern observed in plant roots subjected to salinity conditions but with an additional band of 42 kD and an intensification of the 55-kD immunoreactive band.
Figure 10.
Representative images illustrating the CLSM in vivo detection of ONOO− (red color) and immunoblot showing the protein Tyr nitration in root tips of 6-d-old Arabidopsis wild-type seedlings exposed to 100 mm NaCl for 7 d. A and B, Root tips of control (0 mm NaCl) plants. ONOO− was detected using the fluorescent reagent APF (excitation 495 nm; emission 515 nm). C, Fluorescence intensity of ONOO− observed in A and B. D, Representative immunoblot showing protein Tyr nitration in the roots of 6-d-old Arabidopsis wild-type seedlings exposed to 100 mm NaCl for 7 d and the corresponding silver-stained gel. Root samples (5 μg of protein per lane) were subjected to SDS-PAGE (10%) and western analysis using an antibody against 3-nitrotyrosine (NO2-Tyr; dilution 1:8,000). NO2-BSA, Commercial nitrated bovine serum albumin. Fluorescence is expressed as arbitrary units (A.U.) using Leica confocal software. [See online article for color version of this figure.]
DISCUSSION
Peroxisomes are cell compartments involved in many physiological functions, such as lipid mobilization, photorespiration, and hormone biosynthesis under normal and stress conditions (del Río et al., 1992; Hayashi and Nishimura, 2003; Reumann, 2004; Pracharoenwattana and Smith, 2008; Corpas et al., 2009a). NO, a free radical generated in animal and plant cells, has attracted the attention of many researchers due to its involvement in various physiological processes, such as seed germination, plant development, and senescence (Leshem, 1996; Corpas et al., 2004, 2006) as well as abiotic and biotic stress (Corpas et al., 2007a, 2008; Besson-Bard et al., 2008; Neill et al., 2008; Chaki et al., 2009a). However, several key questions, such as how NO is produced and its subcellular location in plants, are still a subject of debate (Corpas et al., 2009b). On the basis of previous experimental data, we had hypothesized that pea leaf peroxisomes could be a potential source of NO in plant cells (Corpas et al., 2001). This study therefore aims to demonstrate the presence of NO in the peroxisomes of other plant species (using Arabidopsis as a model for plant analysis), to determine whether peroxisomal NO is released into the cytosol and to analyze its physiological function. To achieve these objectives, we used a combination of biochemical, genetic, and cell biology tools.
GFP from the jellyfish Aequorea victoria and its variants has become a highly effective tool to study the subcellular location of many proteins and specifically plant peroxisomes (Hayashi et al., 2000; Mano et al., 2002, 2006; Leterrier et al., 2005; Reumann et al., 2007). Thus, the use of Arabidopsis mutants expressing GFP fused with PTS1 has enabled us to visualize the peroxisomes in vivo. Simultaneous visualization of this GFP-PTS1 using the available fluorescent probes for NO, such as DAF-2 DA and DAF-FM DA (Corpas et al., 2006), was not possible due to the overlap of the excitation and emission wavelengths. However, relatively new fluorescent probes for detecting NO, such as DAR-4M AM (Kojima et al., 2001), which have been successfully tested in plant cells (Tun et al., 2006, 2008), have enabled us to resolve this technical problem, as the excitation and emission wavelengths of neither GFP nor DAR-4M AM overlap. The only drawback is that DAR-4M AM provides a less-intense NO signal than DAF-FM DA.
NO Is Present in Cotyledons and Roots during the Development of Arabidopsis Seedlings
Arabidopsis, a small flowering plant commonly used as a model in plant biology, offers important advantages for basic research in genetics and molecular biology. This plant has been used in many plant studies to determine the involvement of NO in seed germination (Beligni and Lamattina, 2000; Batak et al., 2002; Bethke et al., 2006), biotic stress (Delledonne et al., 1998, 2001), abiotic stress (Mackerness et al., 2001; Huang et al., 2004), programmed cell death (Clarke et al., 2000; Zhang et al., 2003), stomatal closure (Garcia-Mata et al., 2003; Desikan et al., 2004), iron metabolism (Murgia et al., 2004; Perazzolli et al., 2004), flowering (He et al., 2004; Simpson, 2005), and the protein S-nitrosylation (Feechan et al., 2005; Lindermayr et al., 2005), among others. Using the DAF-FM DA as a fluorescence probe, our results provide a detailed picture of the overall distribution of NO during the early development of 3- to 11-d-old Arabidopsis seedlings. Thus, primary roots were observed to have high NO content as compared with cotyledons, which resembles the level detected in Arabidopsis seedlings with a different development period and using different fluorescence probes for NO (DAF-2 DA or DAR-4M AM; Tun et al., 2006, 2008; Kolbert et al., 2008).
NO Is Generated in Arabidopsis Root Peroxisomes
CLSM analysis of Arabidopsis mutants expressing the GFP-PTS1 demonstrated that NO is present in Arabidopsis peroxisomes in vivo (Fig. 3, A–D). The presence of NO has been shown only in isolated pea leaf peroxisomes by using the electron paramagnetic resonance technique (Corpas et al., 2004). It has also been suggested that the small spots, similar to peroxisomes, detected using CLSM, could be a source of NO involved in growth regulation and reorientation of Lilium longiflorum pollen tubes (Prado et al., 2004). These new findings based on a different approach confirm that NO is present in the root peroxisomes of Arabidopsis, indicating that the presence of NO in peroxisomes could be a general feature of plant cells.
Pex12 and Pex13 Appear to Be Involved in the Peroxisomal Import of the NO-Generating Protein
Peroxisomal proteins are selectively targeted for import from the cytosol posttranslationally by either a peroxisomal targeting sequence or protein-protein associations. So-called PEX genes are also involved in regulating peroxisomal biogenesis. At least 22 PEX genes have been identified in Arabidopsis (Nito et al., 2007), and it has been demonstrated that APM2 and APM4 encode proteins homologous to peroxins Pex13 and Pex12, respectively (Mano et al., 2006). Pex12 is an integral membrane protein containing a RING-finger domain that functions as an ubiquitin ligase, an essential component of a multiprotein complex for peroxisomal matrix protein import (Albertini et al., 2001; Mano et al., 2006). It has also been suggested that Pex13 as well as Pex14 and Pex17 are membrane-bound peroxins that act as a docking complex to import proteins into the peroxisomal matrix (Mullen et al., 2001). Thus, Arabidopsis mutant defects in the PEX13 gene cause loss of peroxisomal function due to misdistribution of peroxisomal matrix proteins in the cytosol (Mano et al., 2006). The Pex13 and Pex14 proteins also appear to operate stoichiometrically in vivo, acting as the ideal docking proteins for the receptor-cargo complexes (Azevedo and Schliebs, 2006). Furthermore, it has been demonstrated that apm2/4 mutants have also disturbed the PTS2-dependent protein transport mechanism (Mano et al., 2006), indicating that the apm2/4 mutants are characterized by defective targeting of both PTS1- and PTS2-containing proteins. Under our experimental conditions, we observed that NO production was lower in both apm2/pex13 and apm4/pex12 Arabidopsis mutants (Fig. 3), indicating that the peroxisomal transport of a protein responsible for NO generation in peroxisomes was affected. Additionally, it was demonstrated that peroxisomal NO generation was sensitive to animal NOS inhibitors and was calcium dependent. This data is in line with the biochemical evidence on the presence of NOS activity reported in pea leaf peroxisomes (Barroso et al., 1999; Corpas et al., 2004), olive (Olea europaea) leaf (Valderrama et al., 2007), and sunflower (Helianthus annuus) hypocotyls (Chaki et al., 2009a). It is worth noting that some of the cofactors involved in the l-Arg-dependent NOS activity in plant peroxisomes have been described in previous studies (Corpas et al., 2009b). Several NADP-dehydrogenases can produce NADPH (Corpas et al., 1998, 1999), while the presence of calmodulin in plant peroxisomes has been also demonstrated (Yang and Poovaiah, 2002) being both essential elements for this activity. These results could be important, particularly given that inducible NOS has also been reported in peroxisomes of rat hepatocytes (Stolz et al., 2002; Loughran et al., 2005).
On the other hand, it has been established that Pex12 is not only required for peroxisome biogenesis but is also essential for plant development (Fan et al., 2005). This could plausibly be explained by Pex12's effect on the import of the peroxisomal NO-generating protein given that NO is also involved in plant germination and development processes (Lamattina et al., 2003; Corpas et al., 2006).
Peroxisomes Release NO under Salinity Stress That Is Involved in the Generation of ONOO−
To investigate the possibility of peroxisomal NO release into cytosol and its physiological function, we analyzed NO production under salinity conditions in GFP-PTS1 parent plants (Fig. 8) and apm4/Pex12 mutants (Fig. 9). It was demonstrated that under these stress conditions the generation of NO increased significantly both in peroxisomes and in the cytosol. However, this increase in NO was not found in apm4 mutants exposed to salinity stress, suggesting that the peroxisomal NO-generating protein is involved in the process. These findings also suggest that NO could be released into the cytosol under salinity stress, which is a prerequisite for ONOO− generation, resulting in nitrosative stress, as indicated by the increase in protein Tyr nitration (Fig. 10D). In a previous work involving olive plant growth under salinity (200 mm NaCl) stress conditions, similar behavior was observed, with an increase in the number and intensity of leaf proteins undergoing Tyr nitration (Valderrama et al., 2007). In this context, it must be mentioned that a rise in protein Tyr nitration is considered an indicator of ONOO− activity and a marker of pathological diseases and oxidative stress in animals (Ischiropoulos, 1998; Radi 2004; Szabó et al., 2007) and plants (Corpas et al., 2007b, 2008, 2009c; Chaki et al., 2009a). Under salinity stress conditions, peroxisomes, as a source of NO, can therefore be expected to play a significant role in increasing nitrated proteins in either the peroxisomes or the cytosol. On the other hand, the release of NO into the cytosol could be explained by a protection mechanism triggered by overproduction due to peroxisomal NOS activity. These findings suggest that peroxisomes are required for NO accumulation into the cytosol, which is a prerequisite for the nitrosative stress process mediated by ONOO− observed in Arabidopsis under salinity conditions.
In summary, the data reported in this study provide experimental evidence that Arabidopsis NO production under normal and salinity stress depends on peroxisomal protein import genes PEX12 and PEX13 and generates nitrosative stress mediated by ONOO− overproduction. All these data constitute a significant advance in our knowledge of the metabolism of NO in plant peroxisomes and their involvement in the response to abiotic stress. Furthermore, the importance of human peroxisomes in biomedicine as these organelles are associated with several important genetic diseases caused by peroxisomal dysfunction (Steinberg et al., 2006) suggests that significant advances in human health research could be made on the basis of these results. Although the identification of the peroxisomal protein responsible for NO production remains a major challenge, the involvement of specific peroxins (Pex12 and Pex13) could be a useful tool in its identification.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia and mutant (apm2 and apm4) seeds expressing GFP-PTS1 (Mano et al., 2006) were surface sterilized for 5 min in 70% ethanol containing 0.1% SDS and then placed for 20 min in sterile water containing 20% bleach and 0.1% SDS and washed four times in sterile water. The seeds were sown for 2 d at 4°C in the dark for vernalization on the basal growth medium composed of 4.32 g/L commercial Murashige and Skoog (MS) medium (Sigma-Aldrich) with a pH of 5.5, containing 1% Suc and 0.8% phyto-agar. The petri plates containing the Arabidopsis seeds were then grown at 16 h light, 22°C/8 h dark, at 18°C (long-day conditions) under a light intensity of 100 μE m−2 s−1. For the experiments with NaCl stress, 6-d-old seedlings, the wild type, and apm4 mutants were transferred to MS medium plates both with and without 100 mm NaCl for another 7 d under long-day conditions (Sun et al., 2007).
Crude Extracts of Plant Tissues
Arabidopsis roots were frozen in liquid N2 and ground in a mortar with a pestle. The powder was suspended in a homogenizing medium containing 100 mm Tris-HCl, pH 8.0, 1 mm EDTA, and 10% (v/v) glycerol. Homogenates were centrifuged at 17,000g for 30 min, and the supernatants were used for the assays.
SDS-PAGE and Immunoblot Analysis
SDS-PAGE was carried out according to the Laemmli method (Laemmli, 1970) in 10% acrylamide-slab gels. Gels were stained with silver according to the modifications described in detail by Jiang et al. (1994) for the Heukeshoven and Dernick method (Heukeshoven and Dernick, 1985). For western-blot analysis, proteins were electroblotted to polyvinylidene difluoride membranes with a semidry Trans-Blot cell (Bio-Rad). For immunodetection of nitrotyrosine, a rabbit polyclonal antibody against 3-nitrotyrosine (NO2-Tyr) (Valderrama et al., 2007) diluted 1:3,000 was used with an enhanced chemiluminescence kit (ECL-PLUS; Amersham Pharmacia Biotech), and immunoreactive bands were detected with a photographic film (Hyperfilm; Amersham Pharmacia Biotech).
Detection of NO in Mutants Expressing GFP-PTS1 Using CLSM
NO was detected using the fluorescent reagent DAR-4M AM (Calbiochem). Arabidopsis seedlings were incubated at 25°C for 1 h in darkness with 5 μm DAR-4M AM prepared in 100 mm potassium phosphate buffer, pH 7.4 (Tun et al., 2006, 2008). The samples were then washed twice in the same buffer for 15 min each and mounted on a microscope slide for examination with a confocal laser scanning microscope (Leica TCS SL) using standard filters and collection modalities for DAR-4M AM (excitation 543 nm; emission 575 nm) and GFP (excitation 495 nm; emission 515 nm). As controls, 14-d-old Arabidopsis seedlings expressing GFP-PTS1 were preincubated for 2 h 30 min at 25°C with 100 μm SNP as NO donor, 200 μm cPTIO as NO scavenger, 5 mm l-NAME as competitive inhibitor of animal NOSs, and 2 mm aminoguanidine (AG) as general inhibitor of animal NOSs and then incubated with 5 μm DAR-4M AM for 1 h at 25°C to detect and visualize NO using CLSM.
Alternatively, NO was detected using the fluorescent reagent 10 μm DAF-FM DA (Calbiochem) prepared in 10 mm Tris-HCl (pH 7.4). These probes are highly specific for NO (Nakatsubo et al., 1998; Zhang et al., 2003; Corpas et al., 2006), which is detected using standard filters and collection modalities for DAF-2 green fluorescence (excitation 495 nm; emission 515 nm).
In all cases, the images obtained by CLSM from control and treated Arabidopsis seedlings were held constant during the course of the experiment in order to produce comparable data. The images were processed and analyzed using statistical Leica confocal software.
Detection of ONOO− by CLSM
ONOO− was detected using the fluorescent reagent APF (Invitrogen). Arabidopsis seedlings were incubated at 25°C for 1 h in darkness with 10 μm APF prepared in 10 mm Tris-HCl (pH 7.4; Chaki et al., 2009a). Then, the samples were washed twice in the same buffer for 15 min each and mounted on a microscope slide for examination with CLSM using standard filters and collection modalities for APF fluorescence (excitation 495 nm; emission 515 nm).
Other Assays
Protein concentration was determined with the Bio-Rad protein assay using bovine serum albumin as standard. To estimate the statistical significance between means, the data were analyzed by Student's t test.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Image showing overlap between Figure 3D with the corresponding bright-field image of the Arabidopsis root tip.
Supplemental Figure S2. Representative images illustrating the CLSM in vivo detection of NO (red color) in the root tips of 14-d-old Arabidopsis apm2/pex13 (–H) and apm4/pex12 (I–P) mutant seedlings expressing GFP-PTS1 (green color) preincubated for 2 h and 30 min at 25°C with 100 μm SNP as NO donor.
Supplementary Material
Acknowledgments
Dr. M. Chaki is acknowledged for her help in the immunoblot analyses. CLSM analyses were carried out at the Technical Services of the University of Jaén, and special thanks are given to Miss Nieves de la Casa-Adán for her technical assistance. Mr. Carmelo Ruíz-Torres is also acknowledged for his excellent technical support.
This work was supported by the Ministry of Education and Science (grant nos. BIO2006–14949–C02–01 and BIO2006–14949–C02–02) and Junta de Andalucía (project P06–CVI–1820).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Francisco J. Corpas (javier.corpas@eez.csic.es).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Albertini M, Girzalsky W, Veenhuis M, Kunau WH (2001) Pex12p of Saccharomyces cerevisiae is a component of a multi-protein complex essential for peroxisomal matrix protein import. Eur J Cell Biol 80 257–270 [DOI] [PubMed] [Google Scholar]
- Arasimowicz M, Floryszak-Wieczorek J (2007) Nitric oxide as a bioactive signalling molecule in plant stress responses. Plant Sci 172 876–887 [Google Scholar]
- Azevedo JE, Schliebs W (2006) Pex14p, more than just a docking protein. Biochim Biophys Acta 1763 1574–1584 [DOI] [PubMed] [Google Scholar]
- Barroso JB, Corpas FJ, Carreras A, Sandalio LM, Valderrama R, Palma JM, Lupiáñez JA, del Río LA (1999) Localization of nitric oxide synthase in plant peroxisomes. J Biol Chem 274 36729–36733 [DOI] [PubMed] [Google Scholar]
- Batak I, Devic M, Giba Z, Grubisic D, Poff KL, Konjevic R (2002) The effects of potassium nitrate and NO-donors on phytochrome A- and phytochrome B-specific induced germination of Arabidopsis thaliana seeds. Seed Sci Res 12 253–259 [Google Scholar]
- Beligni MV, Lamattina L (2000) Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 210 215–221 [DOI] [PubMed] [Google Scholar]
- Besson-Bard A, Pugin A, Wendehenne D (2008) New insights into nitric oxide signaling in plants. Annu Rev Plant Biol 59 21–39 [DOI] [PubMed] [Google Scholar]
- Bethke PC, Libourel IG, Jones RL (2006) Nitric oxide reduces seed dormancy in Arabidopsis. J Exp Bot 57 517–526 [DOI] [PubMed] [Google Scholar]
- Chaki M, Fernández-Ocaña A, Valderrama R, Carreras A, Esteban F, Luque F, Gómez-Rodríguez M, Begara-Morales JC, Corpas FJ, Barroso JB (2009. a) Involvement of reactive nitrogen and oxygen species (RNS and ROS) in sunflower-mildew interaction. Plant Cell Physiol 50 265–279 [DOI] [PubMed] [Google Scholar]
- Chaki M, Valderrama R, Fernández-Ocaña AM, Carreras A, López-Jaramillo J, Luque F, Palma JM, Pedradas JR, Begara-Morales JC, Sánchez-Calvo B, et al (2009. b) Protein targets of tyrosine nitration in sunflower (Helianthus annuus L) hypocotyls. J Exp Bot (in press) [DOI] [PubMed]
- Clarke A, Desikan R, Hurst RD, Hancock JT, Neill SJ (2000) NO way back: nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J 24 667–677 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, Carreras A, Quirós M, León AM, Romero-Puertas MC, Esteban FJ, Valderrama R, Palma JM, Sandalio LM, et al (2004) Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant Physiol 136 2722–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, Carreras A, Valderrama R, Palma JM, León AM, Sandalio LM, del Río LA (2006) Constitutive arginine-dependent nitric oxide synthase activity in different organs of pea seedlings during plant development. Planta 224 246–254 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, del Río LA (2001) Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci 6 145–150 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, Palma JM, del Río LA (2009. a) Peroxisomes as key organelles in the metabolism of reactive oxygen species, reactive nitrogen species and reactive sulfur species. In S Terlecky, V Titorenko, eds, Emergent Functions of the Peroxisome. Research Signpost, Kerala, India, pp 97–124
- Corpas FJ, Barroso JB, Sandalio LM, Distefano S, Palma JM, Lupiáñez JA, del Río LA (1998) A dehydrogenase-mediated recycling system of NADPH in plant peroxisomes.. Biochem J 330 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, Sandalio LM, Palma JM, Lupiáñez JA, del Río LA (1999) Peroxisomal NADP-dependent isocitrate dehydrogenase. Characterization and activity regulation during natural senescence. Plant Physiol 121 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Carreras A, Valderrama R, Chaki M, Palma JM, del Río LA, Barroso JB (2007. a) Reactive nitrogen species and nitrosative stress in plants. Plant Stress 1 37–41 [Google Scholar]
- Corpas FJ, Chaki M, Fernández-Ocaña A, Valderrama R, Palma JM, Carreras A, Begara-Morales JC, Airaki M, del Río LA, Barroso JB (2008) Metabolism of reactive nitrogen species in pea plants under abiotic stress conditions. Plant Cell Physiol 49 1711–1722 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Chaki M, Leterrier M, Barroso JB (2009. c) Protein tyrosine nitration: a new challenge in plants. Plant Signal Behav 4 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Palma JM, del Río LA, Barroso JB (2009. b) Evidence supporting the existence of L-arginine-dependent nitric oxide synthase (NOS) activity in plants. New Phytol 184 9–14 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, del Río LA, Barroso JB (2007. b) Need of biomarkers of nitrosative stress in plants. Trends Plant Sci 12 436–438 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394 585–588 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Zeier J, Marocco A, Lamb C (2001) Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc Natl Acad Sci USA 98 13454–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río LA, Sandalio LM, Palma JM, Bueno P, Corpas FJ (1992) Metabolism of oxygen radicals in peroxisomes and cellular implications. Free Radic Biol Med 13 557–580 [DOI] [PubMed] [Google Scholar]
- del Río LA, Sandalio LM, Corpas FJ, Palma JM, Barroso JB (2006) Reactive oxygen species and reactive nitrogen species in peroxisomes. Production, scavenging, and role in cell signaling. Plant Physiol 141 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Cheung MK, Bright J, Henson D, Hancock JT, Neill SJ (2004) ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J Exp Bot 55 205–212 [DOI] [PubMed] [Google Scholar]
- Fan J, Quan S, Orth T, Awai C, Chory J, Hu J (2005) The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol 139 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan A, Kwon E, Yun B-W, Wang Y, Pallas JA, Loake G (2005) A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci USA 102 8054–8059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata C, Gay R, Sokolovski S, Hills A, Lamattina L, Blatt MR (2003) Nitric oxide regulates K+ and Cl− channels in guard cells through a subset of abscisic acid-evoked signaling pathways. Proc Natl Acad Sci USA 100 11116–11121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gas E, Flores-Pérez U, Sauret-Güeto S, Rodríguez-Concepción M (2009) Hunting for plant nitric oxide synthase provides new evidence of a central role for plastids in nitric oxide metabolism. Plant Cell 21 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Nishimura M (2003) Entering a new era of research on plant peroxisomes. Curr Opin Plant Biol 6 577–582 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Nishimura M (2006) Arabidopsis thaliana—a model organism to study plant peroxisomes. Biochim Biophys Acta 1763 1382–1391 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Nito K, Toriyama-Kato K, Kondo M, Yamaya T, Nishimura M (2000) AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J 19 5701–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, et al (2004) Nitric oxide represses the Arabidopsis fl oral transition. Science 305 1968–1971 [DOI] [PubMed] [Google Scholar]
- Heukeshoven J, Dernick R (1985) Simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. Electrophoresis 6 103–112 [Google Scholar]
- Huang X, Stettmaier K, Michel C, Hutzler P, Mueller MJ, Durner J (2004) Nitric oxide is induced by wounding and influences jasmonic acid signaling in Arabidopsis thaliana. Planta 218 938–946 [DOI] [PubMed] [Google Scholar]
- Ischiropoulos H (1998) Biological tyrosine nitration: a pathophysiological function of nitric oxide and reactive oxygen species. Arch Biochem Biophys 356 1–11 [DOI] [PubMed] [Google Scholar]
- Jasid S, Simontacchi M, Bartoli CG, Puntarulo S (2006) Chloroplasts as a nitric oxide cellular source. Effect of reactive nitrogen species on chloroplastic lipids and proteins. Plant Physiol 142 1246–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang LW, Bunkelmann J, Towill L, Kleff S, Trelease RN (1994) Identification of peroxisome membrane proteins (PMPs) in sunflower (Helianthus annuus L.) cotyledons and influence of light on the PMP developmental pattern. Plant Physiol 106 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Hirotani M, Nakatsubo N, Kikuchi K, Urano Y, Higuchi T, Hirata Y, Nagano T (2001) Bioimaging of nitric oxide with fluorescent indicators based on the rhodamine chromophore. Anal Chem 73 1967–1973 [DOI] [PubMed] [Google Scholar]
- Kolbert Z, Bartha B, Erdei L (2008) Exogenous auxin-induced NO synthesis is nitrate reductase-associated in Arabidopsis thaliana root primordia. J Plant Physiol 165 967–975 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lamattina L, García-Mata C, Graziano M, Pagnussat G (2003) Nitric oxide: the versatility of an extensive signal molecule. Annu Rev Plant Biol 54 109–136 [DOI] [PubMed] [Google Scholar]
- Leshem YY (1996) Nitric oxide in biological systems. Plant Growth Regul 18 155–159 [Google Scholar]
- Leterrier M, Corpas FJ, Barroso JB, Sandalio LM, del Río LA (2005) Peroxisomal monodehydroascorbate reductase. Genomic clone characterization and functional analysis under environmental stress conditions. Plant Physiol 138 2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Huertas E, Corpas FJ, Sandalio LM, del Río LA (1999) Characterization of membrane polypeptides from pea leaf peroxisomes involved in superoxide radical generation. Biochem J 337 531–536 [PMC free article] [PubMed] [Google Scholar]
- Loughran PA, Stolz DB, Vodovotz Y, Watkins SC, Simmons RL, Billiar TR (2005) Monomeric in ducible nitric oxide synthase localizes to peroxisomes in hepatocytes. Proc Natl Acad Sci USA 102 13837–13842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackerness SAH, John CF, Jordan B, Thomas B (2001) Early signaling components in ultraviolet-B responses: distinct roles for different reactive oxygen species and nitric oxide. FEBS Lett 489 237–242 [DOI] [PubMed] [Google Scholar]
- Mano S, Nakamori C, Hayashi M, Kato A, Kondo M, Nishimura M (2002) Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: dynamic morphology and actin-dependent movement. Plant Cell Physiol 43 331–341 [DOI] [PubMed] [Google Scholar]
- Mano S, Nakamori C, Nito K, Kondo M, Nishimura M (2006) The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J 47 604–618 [DOI] [PubMed] [Google Scholar]
- Mullen RT, Flynn CR, Trelease RN (2001) How are peroxisomes formed? The role of the endoplasmic reticulum and peroxins. Trends Plant Sci 6 256–261 [DOI] [PubMed] [Google Scholar]
- Murgia I, de Pinto MC, Delledonne M, Soave C, de Gara L (2004) Comparative effects of various nitric oxide donors on ferritin regulation, programmed cell death, and cell redox state in plant cells. J Plant Physiol 161 777–783 [DOI] [PubMed] [Google Scholar]
- Nakatsubo N, Kojima H, Kikuchi K, Nagoshi H, Hirata Y, Maeda D, Imai Y, Irimura T, Nagano T (1998) Direct evidence of nitric oxide production from bovine aortic endothelial cells using new fluorescence indicators: diaminofluoresceins. FEBS Lett 427 263–266 [DOI] [PubMed] [Google Scholar]
- Neill S, Bright J, Desikan R, Hancock J, Harrison J, Wilson I (2008) Nitric oxide evolution and perception. J Exp Bot 59 25–35 [DOI] [PubMed] [Google Scholar]
- Nito K, Kamigaki A, Kondo M, Hayashi M, Nishimura M (2007) Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol 48 763–774 [DOI] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma JM, Corpas FJ, del Río LA (2009) Proteome of plant peroxisomes: new perspectives on the role of these organelles in cell biology. Proteomics 9 2301–2312 [DOI] [PubMed] [Google Scholar]
- Perazzolli M, Dominici P, Romero-Puertas MC, Zago E, Zeier J, Sonoda M, Lamb C, Delledonne M (2004) Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. Plant Cell 16 2785–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I, Smith SM (2008) When is a peroxisome not a peroxisome? Trends Plant Sci 13 522–525 [DOI] [PubMed] [Google Scholar]
- Prado AM, Porterfield DM, Feijó JA (2004) Nitric oxide is involved in growth regulation and re-orientation of pollen tubes. Development 131 2707–2714 [DOI] [PubMed] [Google Scholar]
- Radi R (2004) Nitric oxide, oxidants, and protein tyrosine nitration. Proc Natl Acad Sci USA 101 4003–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S (2004) Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol 135 783–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Lüder F, Weckwerth W, Jahn O (2007) Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG (2005) NO flowering. Bioessays 27 239–241 [DOI] [PubMed] [Google Scholar]
- Steinberg SJ, Dodt G, Raymond GV, Braverman NE, Moser AB, Moser HW (2006) Peroxisome biogenesis disorders. Biochim Biophys Acta 1763 1733–1748 [DOI] [PubMed] [Google Scholar]
- Stolz DB, Zamora R, Vodovotz Y, Loughran PA, Billiar TR, Kim YM, Simmons RL, Watkins SC (2002) Peroxisomal localization of inducible nitric oxide synthase in hepatocytes. Hepatology 36 81–93 [DOI] [PubMed] [Google Scholar]
- Sun J, Jiang H, Xu Y, Li H, Wu X, Xie Q, Li C (2007) The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol 48 1148–1158 [DOI] [PubMed] [Google Scholar]
- Szabó C, Ischiropoulos H, Radi R (2007) Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov 6 662–680 [DOI] [PubMed] [Google Scholar]
- Tun NN, Livaja M, Kieber JJ, Scherer GF (2008) Zeatin-induced nitric oxide (NO) biosynthesis in Arabidopsis thaliana mutants of NO biosynthesis and of two-component signaling genes. New Phytol 178 515–531 [DOI] [PubMed] [Google Scholar]
- Tun NN, Santa-Catarina C, Begum T, Silveira V, Handro W, Floh EI, Scherer GF (2006) Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant Cell Physiol 47 346–354 [DOI] [PubMed] [Google Scholar]
- Valderrama R, Corpas FJ, Carreras A, Fernández-Ocaña A, Chaki M, Luque F, Gómez-Rodríguez MV, Colmenero-Varea P, del Río LA, Barroso JB (2007) Nitrosative stress in plants. FEBS Lett 581 453–461 [DOI] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW (2002) Hydrogen peroxide homeostasis: activation of plant catalase by calcium/calmodulin. Proc Natl Acad Sci USA 99 4097–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemojtel T, Fröhlich A, Palmieri MC, Kolanczyk M, Mikula I, Wyrwicz LS, Wanker EE, Mundlos S, Vingron M, Martasek P, et al (2006) Plant nitric oxide synthase: a never-ending story? Trends Plant Sci 11 524–525 [DOI] [PubMed] [Google Scholar]
- Zhang C, Czymmek KJ, Shapiro AD (2003) Nitric oxide does not trigger early programmed cell death events but may contribute to cell-to-cell signaling governing progression of the Arabidopsis hypersensitive response. Mol Plant Microbe Interact 16 962–972 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.