Abstract
Loss or inactivation of BLM, a helicase of the RecQ family, causes Bloom syndrome, a genetic disorder with a strong predisposition to cancer. Although the precise function of BLM remains unknown, genetic data has implicated BLM in the process of genetic recombination and DNA repair. Previously, we demonstrated that BLM can disrupt the RAD51-single-stranded DNA filament that promotes the initial steps of homologous recombination. However, this disruption occurs only if RAD51 is present in an inactive ADP-bound form. Here, we investigate interactions of BLM with the active ATP-bound form of the RAD51-single-stranded DNA filament. Surprisingly, we found that BLM stimulates DNA strand exchange activity of RAD51. In contrast to the helicase activity of BLM, this stimulation does not require ATP hydrolysis. These data suggest a novel BLM function that is stimulation of the RAD51 DNA pairing. Our results demonstrate the important role of the RAD51 nucleoprotein filament conformation in stimulation of DNA pairing by BLM.
Mutations of BLM helicase cause Bloom syndrome (BS),2 a rare autosomal disorder, which is associated with stunted growth, facial sun sensitivity, immunodeficiency, fertility defects, and a greatly elevated incidence of many types of cancer occurring at an early age (1). BLM belongs to the highly conserved family of RecQ helicases that are required for the maintenance of genome integrity in all organisms (2, 3). There are five RecQ helicases in humans; mutations in three of them, WRN, RECQ4, and BLM, have been associated with the genetic abnormalities known as Werner, Rothmund-Thomson, and Bloom syndrome, respectively (4, 5). The cells from BS patients display genomic instability; the hallmark of BS is an increase in the frequency of sister chromatid and interhomolog exchanges (1, 6). Because homologous recombination (HR) is responsible for chromosomal exchanges, it is thought that BLM helicase functions in regulating HR (7–9). Also, BLM helicase is required for faithful chromosome segregation (10) and repair of stalled replication forks (11, 12), the processes that are linked to HR (13–15). BLM was found to interact physically with RAD51, a key protein of HR (16) that catalyzes the central steps in HR including the search for homology and the exchange of strands between homologous ssDNA and dsDNA sequences (17). In cells, BLM forms nuclear foci, a subset of which co-localize with RAD51. Interestingly, the extent of RAD51 and BLM co-localization increases in response to ionizing radiation, indicating a possible role of BLM in the repair of DNA double-strand breaks (16).
Biochemical studies suggest that BLM may perform several different functions in HR. BLM was shown to promote the dissociation of HR intermediates (D-loops) (18–20), branch migration of Holliday junctions (21), and dissolution of double Holliday junctions acting in a complex with TopoIIIα and BLAP75 (22–24). BLM may also facilitate DNA synthesis during the repair process by unwinding the DNA template in front of the replication fork (25). In addition, BLM and its yeast homolog Sgs1 may play a role at the initial steps of DNA double-strand break repair by participating in exonucleolitic resection of the DNA ends to generate DNA molecules with the 3′-ssDNA tails, a substrate for RAD51 binding (26–29).
In vivo, the process of HR is tightly regulated by various mechanisms (30). Whereas some proteins promote HR (14, 31), others inhibit this process, thereby preventing its untimely initiation (32, 33). Disruption of the Rad51-ssDNA nucleoprotein filament appears to be an especially important mechanism of controlling HR. This filament disruption activity was demonstrated for the yeast Srs2 helicase (34, 35) and human RECQ5 helicase (36). Recently, we found that BLM can also catalyze disruption of the RAD51-ssDNA filament (25). This disruption only occurs if the filament is present in an inactive ADP-bound form, e.g. in the presence of Mg2+. Conversion of RAD51 into an active ATP-bound form, e.g. in the presence of Ca2+ (37), renders the filament resistant to BLM disruption (25). In this study, we analyze the interactions of BLM with an active ATP-bound RAD51-ssDNA filament. Surprisingly, we found that BLM stimulates the DNA strand exchange activity of RAD51. Thus, depending on the conformational state of the RAD51 nucleoprotein filament, BLM may either inhibit or stimulate the DNA strand exchange activity of RAD51. Our analysis demonstrated that, in contrast to several known stimulatory proteins that act by promoting formation of the RAD51-ssDNA filament, BLM stimulates the DNA strand exchange activity of RAD51 at a later stage, during synapsis. Stimulation appears to be independent of the ATPase activity of BLM. We suggest that this stimulation of RAD51 may represent a novel function of BLM in homologous recombination.
EXPERIMENTAL PROCEDURES
Proteins and DNA
Human RAD51, DMC1, RAD54, RPA, BLM, BLM K695R, murine Hop2/Mnd1, and yeast Rad51 and Rad54 proteins were purified as described (38). The BLM K695A ATPase mutant was kindly provided by P. Sung. Supercoiled pUC19 dsDNA was purified as described (37). All of the oligonucleotides (IDT, Inc.) used in this study (supplemental Table S1) were purified, labeled, and stored as described previously (39). dsDNA substrates were prepared by annealing of oligonucleotides, as described in Ref. 39.
The D-loop Assay
To form the nucleoprotein filament, 32P-labeled ssDNA (ssDNA 90; 3 μm nucleotides) was incubated with human or yeast Rad51 protein (1 μm) in standard buffer containing 25 mm Tris acetate, pH 7.5, 1 mm ATP, 0.5 mm CaCl2, 2 mm DTT, 100 μg/ml BSA, 20 mm phosphocreatine, and 30 units/ml creatine phosphokinase for 15 min (unless indicated otherwise) at 37 °C. When indicated, RPA (0.13 μm) was added to the reaction. BLM (in the indicated concentrations) was added prior to the addition of pUC19 scDNA (50 μm nucleotides), unless indicated otherwise. D-loop formation was terminated after a 15-min incubation. The products were deproteinized, analyzed by electrophoresis in a 1% agarose gel in 1× TAE buffer (40 mm Tris acetate, pH 8.0, and 1 mm EDTA), visualized, and quantified using a Storm 840 PhosphorImager (GE Healthcare). The yield of D-loops was expressed as a percentage of the total plasmid DNA.
dsDNA Capture by the RAD51-ssDNA Presynaptic Filament
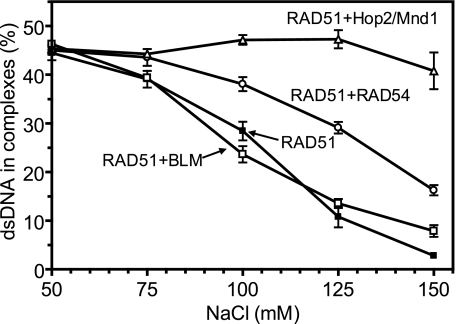
This assay measures homology-independent conjunction of RAD51-ssDNA presynaptic filament with dsDNA in complexes that sediment at more than 10,000 S (Svedberg) (2 min at 15,000 × g) (40). To form the nucleoprotein filament, RAD51 (1 μm) was incubated with ssDNA (ssDNA 71; 3 μm nucleotides) in buffer containing 25 mm Tris acetate, pH 7.5, 1 mm ATP, 0.5 mm CaCl2, 2 mm DTT, 100 μg/ml BSA, 20 mm phosphocreatine, and 30 units/ml creatine phosphokinase for 15 min at 37 °C. Then NaCl was added in the indicated concentration followed by the addition of BLM (100 nm), HOP2/MND1 (100 nm), or RAD54 protein (100 nm). After 10 min of incubation, dsDNA capture was initiated by the addition of 32P-labeled pUC19 (25 μm nucleotides), linearized with BamHI restriction endonuclease, followed by 10 min of incubation at 37 °C. Aliquots (10 μl) were withdrawn from the reaction mixture, and co-aggregates were collected by centrifugation at 15,000 × g for 5 min at 21 °C. The yield of co-aggregates was determined using a Beckman LS 6500 radioactive counter. The residual retention of the radioactive DNA on the tube walls, ∼1–2%, was subtracted from the measurements.
Measuring of the Apparent DNA Strand Exchange Promoted by BLM
BLM (in the indicated concentrations) was incubated with ssDNA (ssDNA 71; 32 nm molecules) in buffer containing 25 mm Tris acetate, pH 7.5, 1 mm ATP (or indicated nucleotide co-factor), 0.5 mm CaCl2, 2 mm DTT, 100 μg/ml BSA, 20 mm phosphocreatine, and 30 units/ml creatine phosphokinase for 10 min at 37 °C. To start the reaction 32P-labeled dsDNA (ssDNA 5/6; 32 nm molecules) was added followed by 15 min of incubation. The reactions were terminated and deproteinized by the addition of stop buffer up to 1.5% SDS and proteinase K (800 μg/ml) for 5 min at 25 °C, mixed with a 0.1 volume of loading buffer (70% glycerol, 0.1% bromphenol blue), and analyzed by electrophoresis in 10% native polyacrylamide gels in TBE buffer (90 mm Tris borate, pH 8.3, and 1 mm EDTA) at 135 V for 1.5 h.
RESULTS
BLM Stimulates the DNA Strand Exchange Activity of RAD51
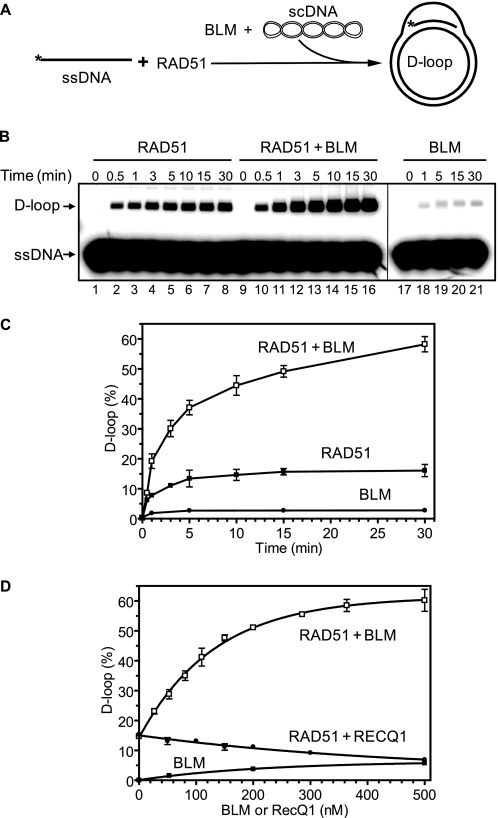
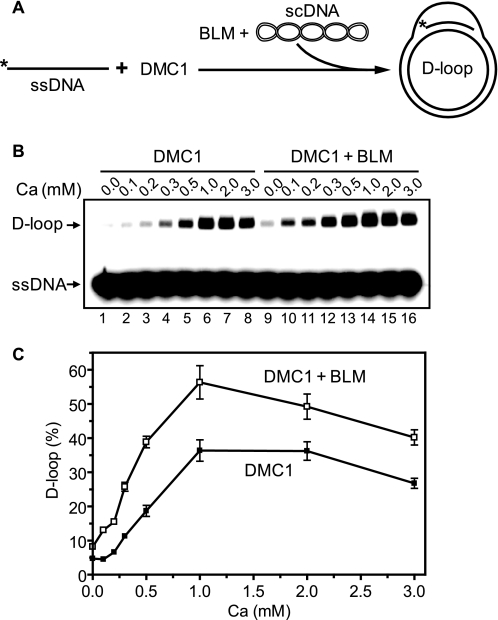
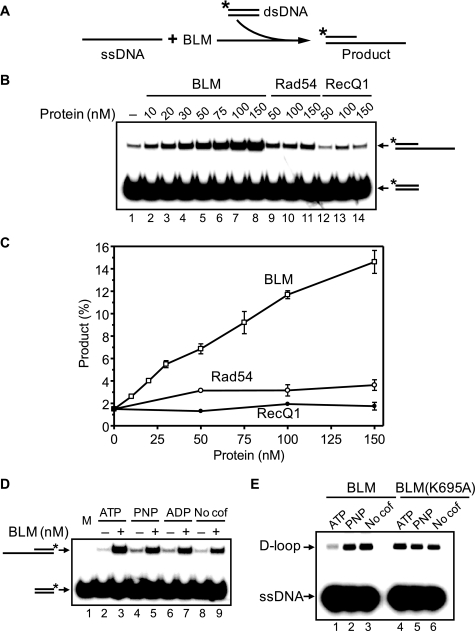
RAD51 shows characteristics of a self-inactivating ATPase; during ATP hydrolysis in the presence of Mg2+, the RAD51-ssDNA filament turns into an inactive ADP-bound form (37). Previously, we showed that BLM can disrupt an inactive ADP-bound RAD51-ssDNA filament by dislodging RAD51 from ssDNA (25). Here we wanted to investigate interactions of BLM with a RAD51-ssDNA filament maintained in an active ATP-bound form in the presence of Ca2+. Using the D-loop assay (Fig. 1A), we found that the addition of BLM to an active RAD51 filament stimulates joint molecule formation 2–4-fold (Fig. 1, B and C). Control experiments showed that BLM alone has some limited ability to promote D-loop formation; however, this activity was too small to account for the observed stimulation of RAD51 DNA pairing (Fig. 1, B, lanes 17–21, and C). With the D-loop assay, we also showed that BLM can stimulate the DNA strand exchange activity of DMC1 protein, a meiosis-specific RAD51 homolog (Fig. 2).
FIGURE 1.
BLM stimulates DNA strand exchange activity of RAD51 protein in the presence of Ca2+. A, the experimental scheme. The asterisk indicates the 32P label. B, lanes 9–16, the kinetics of D-loop formation was initiated by addition of BLM (100 nm) and pUC19 scDNA to RAD51-ssDNA nucleoprotein filaments assembled in the presence of 0.5 mm Ca2+. Lanes 1–8 and 17–21, either BLM or RAD51 were substituted with their storage buffers, respectively. C, the results from B are presented as a graph. D, effect of BLM concentration on the efficiency of D-loop formation promoted by RAD51. BLM or RECQ1 (in the indicated concentrations) were added to the RAD51-ssDNA filaments formed in the presence of 0.5 mm Ca2+. The reactions were carried out for 15 min. The error bars in C and D indicate S.E.
FIGURE 2.
BLM stimulates the DNA strand exchange activity of DMC1. A, the experimental scheme. The asterisk indicates the 32P label. B, nucleoprotein filaments were formed by incubation of DMC1 (1 μm) with 32P-labeled ssDNA (ssDNA 90; 3 μm nucleotides) in buffer containing 25 mm Tris acetate, pH 7.5, 1 mm ATP, 2 mm DTT, 100 μg/ml BSA, 20 mm phosphocreatine, 30 units/ml creatine phosphokinase, 32P-labeled ssDNA (ssDNA 90; 3 μm nucleotides), and the indicated amount of CaCl2 for 15 min at 37 °C. To initiate the reactions, BLM (100 nm) was added followed by the addition of pUC19 scDNA (50 μm nucleotides). Products of DNA strand exchange were analyzed by electrophoresis in a 1% agarose gel in 1× TAE (40 mm Tris acetate, pH 8.0, and 1 mm EDTA). C, the results from B represented as a graph. The error bars indicate S.E.
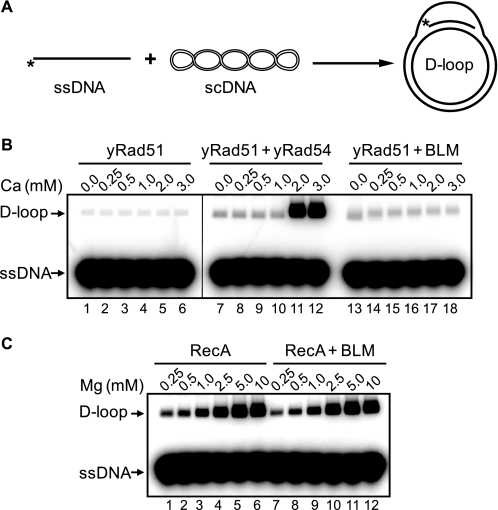
We further found that BLM stimulation was specific for the human recombinases RAD51 and DMC1; no stimulation of Saccharomyces cerevisiae Rad51 (ScRad51) or Escherichia coli RecA was observed (Fig. 3). We did not observe RecA stimulation by BLM also in the presence of Ca2+ and ATP under tested conditions (data not shown). We also tested whether RECQ1 (RECQL1), another member of the human RecQ family, has a stimulatory effect on RAD51. The biochemical properties of RECQ1 and BLM are similar; both proteins physically interact with RAD51 (41–43) and show branch migration activities (21, 44). However, RECQ1 did not stimulate DNA strand exchange promoted by RAD51 under tested conditions; in contrast, at high concentrations RECQ1 inhibited this reaction (Fig. 1D).
FIGURE 3.
BLM helicase does not stimulate DNA strand exchange activity of S. cerevisiae Rad51 or E. coli RecA proteins. A, the experimental scheme. The asterisk indicates the 32P label. B, nucleoprotein filament s were formed by incubation of ScRAD51 (1 μm) with 32P-labeled ssDNA (ssDNA 90; 3 μm nucleotides) in buffer containing 25 mm Tris acetate, pH 7.5, 1 mm ATP, 2 mm DTT, 100 μg/ml BSA, 20 mm phosphocreatine, 30 units/ml creatine phosphokinase, and the indicated concentration of CaCl2. After a 15-min incubation, D-loop formation was initiated by the addition of ScRad54 (100 nm) or BLM (100 nm) and pUC19 scDNA (50 μm nucleotides). C, for RecA, D-loop formation was carried out under similar conditions, except that CaCl2 was replaced by MgCl2 (in indicated concentration), ATP was replaced with ATPγS (1 mm), and the ATP regeneration system was omitted. The reactions promoted by ScRad51 and RecA were stopped after 15 min of incubation. The products were deproteinized, analyzed by electrophoresis in a 1% agarose-TAE (40 mm Tris acetate, pH 8.0, and 1 mm EDTA), visualized, and quantified using a Storm 840 PhosphorImager (GE Healthcare).
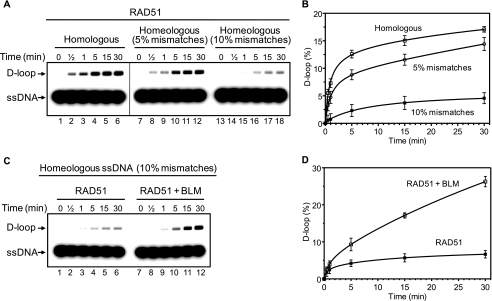
In the accompanying paper, Kikuchi et al. (59) demonstrate that BLM is required for recombination between homeologous DNA sequences (homologous sequences with some mismatches) in the DT-40 cells. Here, we examined the effect of BLM on DNA strand exchange between homeologous ssDNA and pUC19 scDNA promoted by RAD51 (Fig. 4). As expected, mismatches in DNA substrates significantly reduce the efficiency of the RAD51-promoted DNA strand exchange (Fig. 4, A and B). The addition of BLM increases the extent of D-loop formation on DNA substrates containing 10% of mismatches ∼4-fold (Fig. 4, C and D). Thus, BLM stimulates the RAD51 DNA strand exchange activity on both homologous and homeologous DNA substrates. Our current results showed that BLM stimulates DNA strand exchange activity of RAD51 and DMC1. The BLM stimulation appeared to be specific for these two human recombinases.
FIGURE 4.
BLM stimulates RAD51 in DNA strand exchange between homeologous DNA substrates. A, mismatches in the ssDNA reduce the efficiency of DNA strand exchange catalyzed by RAD51 protein. Products of DNA strand exchange were analyzed by electrophoresis in a 1% agarose gel. RAD51-ssDNA nucleoprotein filaments were assembled on ssDNA fully homologous to pUC19 DNA (ssDNA 90, lanes 1–6), on homeologous ssDNA containing 5% of mismatches (ssDNA 423, lanes 7–12), or 10% of mismatches (ssDNA 422, lanes 13–18). The kinetics of D-loop formation was initiated by the addition of pUC19 scDNA. B, the results from A are presented as a graph. C, the kinetics of D-loop formation was initiated by the addition of BLM (100 nm) and pUC19 scDNA to RAD51-ssDNA nucleoprotein filaments assembled on homeologous ssDNA, which contains 10% mismatches. In lanes 1–6, BLM was substituted with its storage buffer (50 mm Tris acetate, pH 7.5, 100 mm KCl, 30% glycerol, and 10 mm β-mercaptoethanol). D, the results from C are presented as a graph.
BLM Stimulates RAD51 at the Synaptic Stage
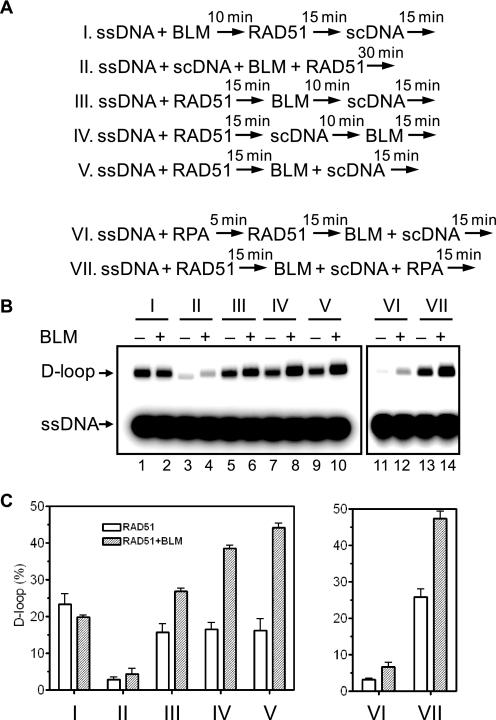
Several known Rad51 stimulatory proteins act by promoting Rad51 filament formation on ssDNA or by facilitating the interaction of the filament with dsDNA (45). To characterize the mechanism of a BLM-dependent RAD51 stimulation, we tested the effect that order of addition of BLM and RAD51 has on this stimulation (Fig. 5A). We found no stimulation of D-loop formation when BLM was incubated with ssDNA before the addition of RAD51 and pUC19 scDNA (Fig. 5, B and C, scheme I). In contrast, stimulation was observed when BLM was added after formation of the RAD51 filament; it reached maximum, ∼3-fold, when BLM was added either after pUC19 DNA or at the same time as pUC19 (Fig. 5, B and C, schemes IV and V). The stimulation was smaller, ∼1.5-fold, when BLM was added to the RAD51-ssDNA prior to pUC19 scDNA (Fig. 5, B and C, scheme III). Thus, these results indicate that BLM exerts its stimulatory effect likely after completion of filament formation, during the search for homology and DNA strand exchange.
FIGURE 5.
The order of addition of BLM affects the efficiency of D-loop formation promoted by RAD51. A, the order of addition of individual components of the reaction mixtures. The numbers above the arrow indicate time of incubation. B, the D-loops were analyzed by electrophoresis in a 1% agarose gel. Note, migration of the D-loops was slightly changed in the presence of BLM, which probably indicates some conformational changes in plasmid DNA. C, the results from B presented as a graph. The error bars indicate S.E.
The Rad51 mediator proteins that promote loading of Rad51 on ssDNA are known to alleviate an inhibition of Rad51-ssDNA filament formation by RPA, a single-stranded binding protein, when RPA was bound to ssDNA prior to Rad51 (30). In contrast, we found that BLM does not rescue DNA strand exchange that was inhibited by RPA, when RPA was bound to ssDNA before the addition of RAD51 (Fig. 5, B and C, scheme VI). On the other hand, RPA did not diminish the stimulatory effect of BLM when added to an assembled nucleoprotein filament (Fig. 5, B and C, scheme VII), indicating no specific interactions between BLM and RPA, which could prevent RAD51 stimulation by BLM at the presynaptic stage. Also, consistent with the inability of BLM to promote RAD51 loading on ssDNA, we found that BLM did not rescue DNA strand exchange when RAD51 was added to the mixture of ssDNA and scDNA, instead of being first preincubated with ssDNA to form the filament (Fig. 5, B and C, scheme II). Thus, we concluded that, unlike several known RAD51 mediators that act during RAD51-ssDNA filament formation, BLM stimulates RAD51 at the synaptic or postsynaptic stage.
The ATPase Activity of BLM Is Not Required for RAD51 Stimulation
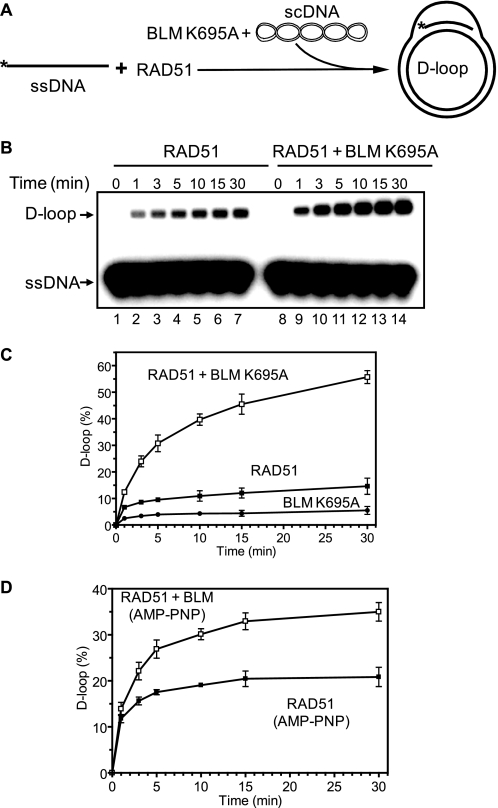
It is known that RAD54, an important HR protein of the Swi2/Snf2 family, also stimulates RAD51-promoted DNA strand exchange at the synaptic or postsynaptic stage. Because RAD54 stimulates RAD51 in an ATPase-dependent manner, we expected that the ATPase activity of BLM, which is required for its translocation on DNA, would also be important for stimulation of the RAD51 DNA strand exchange activity. However, using the D-loop assay, we found that the ATPase-deficient BLM K695A mutant stimulates DNA strand exchange activity of RAD51 similar to wild type BLM (compare Fig. 1C with Fig. 6, B, lanes 8–14, and C). Furthermore, we then found that BLM can stimulate RAD51 in the presence of the nonhydrolyzable ATP analog AMP-PNP (Fig. 6D).
FIGURE 6.
The ATPase activity of BLM is not required for RAD51 stimulation. A, the experimental scheme. The asterisk indicates the 32P label. B, the ATPase-deficient mutant BLM K695A (100 nm) was added to the RAD51-ssDNA filaments formed in the presence of 0.5 mm Ca2+ followed by immediate addition of scDNA. In control, BLM was substituted with storage buffer. At indicated time points aliquots were withdrawn and analyzed on a 1% agarose gel. C, the results as in B represented as a graph. Concentration of BLM K695A was 100 nm. D, BLM (100 nm) stimulates DNA strand exchange activity of RAD51 in the presence of 1 mm AMP-PNP. The reactions were carried out and analyzed as in B, except that the CaCl2 concentration was 0.3 mm. The error bars indicate S.E.
Therefore, the ATPase activity of BLM is not required for stimulation of the DNA strand exchange activity of RAD51. Thus, BLM stimulates the RAD51 strand exchange activity through a novel mechanism that is independent of BLM translocation along DNA.
BLM-dependent Stimulation Requires an Active RAD51-ssDNA Filament
We wanted to explore further the role of an active form of the RAD51 filament in BLM-dependent stimulation of DNA strand exchange. We investigated whether the active form of the filament is merely needed for prevention of the RAD51 filament disruption or whether it plays a significant role in productive interactions of the RAD51 nucleoprotein filament with BLM required for stimulation of DNA strand exchange.
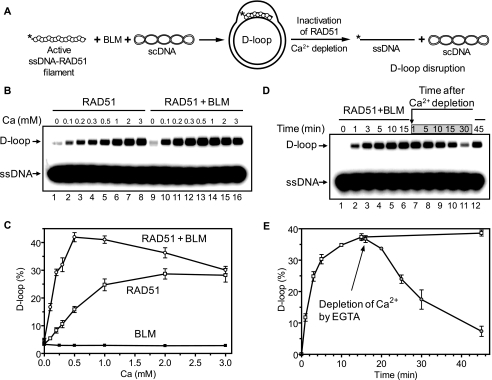
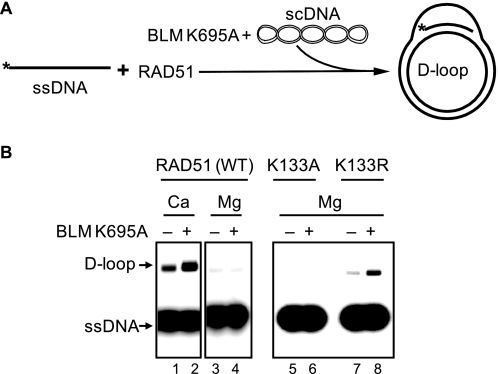
Using wild type BLM we found that the stimulation occurs only in the presence of Ca2+ (Fig. 7, A–C). However, this result did not exclude the possibility that in the absence of Ca2+ BLM efficiently disrupts an inactive RAD51 filament or D-loops containing inactive RAD51 (25), thereby diminishing the effect of stimulation. Indeed, Ca2+ depletion with EGTA during outgoing D-loop formation by Rad51 results in significant dissociation of D-loops by BLM (Fig. 7, D and E). Therefore, we used BLM K695A mutant that cannot disrupt RAD51 filaments (25) but stimulates RAD51 strand exchange activity as efficiently as wild type BLM (Fig. 6). We found that BLM K695A failed to stimulate an inactive RAD51 filament assembled in the absence of Ca2+ (Fig. 8). We then tested the effect of BLM on the RAD51 K133R and K133A ATPase-deficient mutants. Of these two mutants, the RAD51 K133R is known to form an active filament on ssDNA in the presence of ATP, whereas the RAD51 K133A can only form an inactive filament because of poor ATP binding. We found that BLM K695A stimulates DNA pairing activity of the RAD51 K133R but not that of RAD51 K133A mutant (Fig. 8).
FIGURE 7.
The effect of Ca2+ concentration on the efficiency of D-loop formation promoted by RAD51 with and without BLM. A, the experimental scheme. The asterisk indicates the 32P label. B, products of D-loop formation were separated in a 1% agarose gel. BLM (100 nm) (lanes 9–16) or storage buffer (lanes 1–8) were mixed with RAD51-ssDNA filament formed in the presence of the indicated concentrations of Ca2+ followed by addition of scDNA to initiate D-loop formation. C, the results from B represented as a graph. D, BLM stimulates D-loop formation by active RAD51-ssDNA filament and disrupts joint molecules after filament inactivation. D-loop formation was initiated by the addition of BLM (100 nm) and scDNA to RAD51-ssDNA filament in the presence of 0.5 mm Ca2+ and 0.3 mm Mg2+. Aliquots (10 μl) were withdrawn at indicated time points (lanes 1–6). After a 15-min incubation EGTA was added to deplete Ca2+ and inactivate RAD51 and followed by additional incubation for the indicated period of time (lanes 7–11). Lane 12 shows the reaction incubated for 45 min (total) without Ca2+ depletion. DNA products were separated in a 1% agarose gel. E, the results from D represented as a graph. The error bars in C and E indicate S.E.
FIGURE 8.
An active ATP-bound state of RAD51 filament is required for BLM stimulation. A, the experimental scheme. The asterisk indicates the 32P label. B, RAD51-ssDNA filaments formed either in the standard buffer containing 0.5 mm CaCl2 (Ca, to produce an active ATP-bound filament) or 0.5 mm MgCl2 (Mg, to produce an inactive ADP-bound filament). In the right panel, the ATPase-deficient mutants RAD51 K133A and K133R were used instead of RAD51 wild type (WT) to form nucleoprotein filaments in standard buffer containing 0.5 mm MgCl2. D-loop formation was initiated by the addition of BLM K695A (100 nm) and pUC19 scDNA (50 μm nucleotides).
Therefore, the conformation of the RAD51-ssDNA filament plays an important role in determining the outcome of interactions with BLM. We found that BLM can only stimulate DNA pairing activity of an active ATP-bound form of the RAD51-ssDNA filament.
BLM Does Not Enhance dsDNA Binding by the RAD51-ssDNA Filament
It was recently shown that Hop2/Mnd1 heterodimer stimulates RAD51 and DMC1 proteins by enhancing dsDNA binding by the RAD51-ssDNA and DMC1-ssDNA filaments (31, 46). Previously, we showed that RAD54 has a similar effect on dsDNA binding by the RAD51 filament (47). Here, we wanted to test whether BLM may stimulate the DNA pairing activity of RAD51 by the same mechanism. As in our previous studies with RAD54 (47), we used a co-aggregation assay that measures formation of low speed sedimenting co-aggregates resulting from the homology-independent uptake of dsDNA by the RAD51-ssDNA filament (48). If BLM were to increase dsDNA binding affinity of the RAD51-ssDNA filament, we would expect to observe an increase in resistance of co-aggregates to solution ion strength.
Using 32P-labeled linearized pUC19 dsDNA as a substrate, we found that Hop2/Mnd1 and, to a smaller degree, RAD54 increases the salt resistance of co-aggregates between RAD51 and dsDNA, as expected (Fig. 9). In contrast, BLM did not significantly increase the resistance of the co-aggregates to the solution ion strength. Thus, the mechanism of stimulation of the RAD51 DNA pairing activity does apparently not include an increase in the dsDNA binding affinity of the RAD51-ssDNA filament.
FIGURE 9.
Effect of BLM, RAD54, and HOP2/MND1 proteins on heterologous dsDNA capture by the RAD51-ssDNA presynaptic filament. The extent of dsDNA binding was determined by measuring of co-aggregation of RAD51-ssDNA-dsDNA tertiary complexes. The RAD51-ssDNA filament was formed by incubation of RAD51 with ssDNA (ssDNA 71, 94-mer) followed by the addition of BLM, HOP2/MND1, or RAD54 protein. In control, BLM was substituted with storage buffer (50 mm Tris acetate, pH 7.5, 100 mm KCl, 30% glycerol, and 10 mm β-mercaptoethanol). Co-aggregation was initiated by the addition of 32P-labeled linear pUC19 dsDNA. The stability of presynaptic filament interaction with dsDNA was challenged by NaCl, which was added prior to dsDNA. The reactions were carried out for 10 min. The results are presented as a graph. Presynaptic filament independent co-aggregation by individual protein under conditions used did not exceed 1–3%. The error bars indicate S.E.
Stimulation of RAD51 Correlates with the BLM Ability to Promote Apparent DNA Strand Exchange between Homologous ss- and dsDNA, but Not with ssDNA Annealing Activity
Previously, it was shown that herpes simplex virus UL9 DNA helicase can promote unwinding of the DNA duplex in an ATPase-independent manner (49). We suggested that BLM binding may also cause a local destabilization of the DNA helix and thereby facilitate the search for homology and DNA strand exchange promoted by RAD51. We expected that if dsDNA were unwound by BLM, it would anneal with complementary ssDNA to form a tailed DNA product (Fig. 10A). Indeed, we observed formation of the tailed DNA during incubation of BLM with dsDNA in the presence of homologous ssDNA (Fig. 10, B, lanes 2–8, and C). In contrast, RAD54 protein and RECQ1 helicase did not efficiently promote formation of the tailed DNA product (Fig. 10, B, lanes 9–14, and C). The observed product could result from resection of the dsDNA by contaminating exonucleases followed by annealing to the homologous ssDNA. However, analysis of the BLM-treated 32P-labeled dsDNA performed by electrophoresis in a 20% polyacrylamide gel containing 8 m urea confirmed full integrity of the dsDNA, ruling out this possibility (data not shown). DNA strand exchange activity of BLM showed no dependence on ATP; omission of ATP or its substitution with either ADP or a nonhydrolyzable analog AMP-PNP did not significantly affect the reaction (Fig. 10D). Thus, the ATP independence of BLM-promoted DNA strand exchange parallels the ATP independence of RAD51 stimulation by BLM (Fig. 8).
FIGURE 10.
BLM promotes apparent DNA strand exchange between homologous DNA sequences. A, the experimental scheme. The asterisk indicates the 32P label. B, the products were analyzed by electrophoresis in a 10% polyacrylamide gel. BLM (lanes 2–8), RAD54 (lanes 9–11), or RECQ1 (lanes 12–14) in the indicated concentrations were incubated with ssDNA. The reactions were started by the addition of 32P-labeled dsDNA. In control, BLM was substituted with storage buffer (lane 1). C, the results from B presented as a graph. The error bars indicate S.E. D, BLM (100 nm) was incubated with ssDNA in the presence of indicated nucleotide co-factors (1 mm) or without nucleotide co-factor (No cof). To start the reaction, 32P-labeled dsDNA was added. The products were separated in 10% PAGE. Lane 1 shows migration of 32P-labeled dsDNA. E, BLM catalyzes D-loop formation in an ATPase- and ATP-independent manner. BLM or BLM(K695A) ATPase mutant (100 nm) were mixed with ssDNA (1 μm nucleotides) in standard buffer for D-loop assay for 10 min in the presence of either ATP (lanes 1 and 4), AMP-PNP (PNP, lanes 2 and 5), or in the absence of nucleotide co-factor (No cof, lanes 3 and 6). D-loop formation was initiated by the addition of pUC19 scDNA (50 μm nucleotides). The products were analyzed in a 1% agarose gel.
We also tested the ability of BLM to promote D-loop formation (Fig. 10E). In the presence of ATP, BLM did not efficiently promote formation of D-loops (Fig. 10E, lane 1). However, it was previously demonstrated that in the presence of ATP BLM can disassemble D-loops through its helicase or branch migration activities (19, 25) (see also Fig. 7). Indeed, D-loop formation noticeably increased when we inhibited these BLM activities by omitting ATP or substituting it with the nonhydrolyzable analog AMP-PNP or when we replaced BLM with the BLM K695A ATPase-deficient mutant that lacks D-loop dissociation activity (Fig. 10E). Note that this D-loop formation activity of BLM alone was still not large enough to account for the additive increase in the reaction with RAD51 (Figs. 1C and 6C).
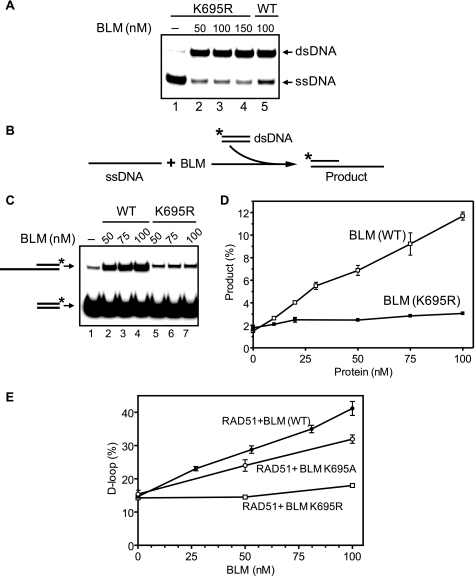
We also tested the possibility that BLM may stimulate RAD51 DNA strand exchange because of its known ssDNA annealing activity (50). In preliminary experiments, we found that one of the BLM mutants, K695R, was efficient in ssDNA annealing but showed a very low ability to promote DNA strand exchange (Fig. 11, A–D). We tested this mutant for the ability to stimulate RAD51 DNA strand exchange and found that it was significantly less efficient than BLM wild type and the BLM K695A mutant (Fig. 11E).
FIGURE 11.
The BLM K695R ATPase-deficient mutant promotes ssDNA annealing but does not promote apparent DNA strand exchange or stimulate the RAD51 strand exchange. A, DNA annealing. Lanes 2–5, BLM wild type (WT) or K695R protein was added in the indicated concentrations to the reaction mixture containing 25 mm Tris acetate, pH 7.5, 1 mm ATP, 1 mm MgCl2, 2 mm DTT, 100 μg/ml BSA, 20 mm phosphocreatine, and 30 units/ml creatine phosphokinase, 32P-labeled ssDNA (ssDNA 90; 1 nm molecules), and complementary ssDNA (ssDNA 91; 1 nm molecules) for 30 min at 22 °C. Lane 1, in control BLM storage buffer (50 mm Tris acetate, pH 7.5, 100 mm KCl, 30% glycerol, and 10 mm β-mercaptoethanol) was added instead of BLM. The reactions were terminated, and DNA samples were deproteinized by adding stop buffer to 1.5% SDS and proteinase K (800 μg/ml) followed by incubation for 5 min at 22 °C. The samples were mixed with a 0.1 volume of loading buffer (70% glycerol, 0.1% bromphenol blue) and analyzed by electrophoresis in 8% native polyacrylamide gels in TBE buffer (90 mm Tris borate, pH 8.3, and 1 mm EDTA). B, the experimental scheme. The BLM K695R mutant poorly promotes apparent DNA strand exchange between homologous DNA sequences. The asterisk indicates the 32P label. C, DNA products were separated in a 10% native polyacrylamide gel in TBE buffer. BLM (lanes 2–4) and BLM K695R (lanes 5–7) in the indicated concentrations were incubated with ssDNA (ssDNA 71; 32 nm molecules) in buffer containing 25 mm Tris acetate, pH 7.5, 1 mm ATP, 0.5 mm CaCl2, 2 mm DTT, 100 μg/ml BSA, 20 mm phosphocreatine, and 30 units/ml creatine phosphokinase for 10 min at 37 °C. The reactions were started by the addition of 32P-labeled dsDNA (ssDNA 5/6; 32 nm molecules) followed by 15 min of incubation. In control (lane 1), BLM was substituted with the storage buffer (50 mm Tris acetate, pH 7.5, 100 mm KCl, 30% glycerol, and 10 mm β-mercaptoethanol). D, the results from C represented as a graph. E, the BLM K695R mutant poorly stimulates DNA strand exchange promoted by RAD51 protein. DNA strand exchange was examined using the D-loop assay. To form the nucleoprotein filament, 32P-labeled ssDNA oligonucleotide (ssDNA 90; 3 μm nucleotides) was incubated with RAD51 protein (1 μm) in standard buffer containing 25 mm Tris acetate, pH 7.5, 1 mm ATP, 0.5 mm CaCl2, 2 mm DTT, 100 μg/ml BSA, 20 mm phosphocreatine, and 30 units/ml creatine phosphokinase for 15 min at 37 °C. The BLM wild type or K695R and K695A mutants (in the indicated concentrations) were added to the reaction mixture prior to addition of pUC19 scDNA (50 mm, nucleotides). D-loop formation was terminated after a 15-min incubation at 37 °C. The products were deproteinized, analyzed by electrophoresis in a 1% agarose gel, visualized, and quantified using a Storm 840 PhosphorImager (GE Healthcare). The results of the quantification are represented as a graph. The error bars indicate S.E.
Thus, stimulation of RAD51 by BLM correlates with the ability of BLM to promote apparent exchange of DNA strands between homologous ss- and dsDNA substrates (dsDNA melting followed by annealing with ssDNA) not with the BLM annealing activity. We suggest that, similar to UL9 DNA helicase, BLM may destabilize dsDNA and thereby facilitate the search for homology and DNA strand exchange promoted by RAD51.
DISCUSSION
Mutations in the BLM helicase cause BS, which is associated with profound genomic instability. Because the frequency of certain recombination events, especially sister chromatid exchanges, is elevated in BS cells, it was suggested that the function of BLM is to down-regulate HR. Several biochemical activities of BLM have been identified in previous studies that are consistent with this view; BLM may dissolve double Holliday junctions (22), dissociate joint molecules (19, 20), and disrupt the RAD51 nucleoprotein filament (25).
However, genetic studies indicate that BLM may also play a role in promoting HR (51, 52). In this study, we present data indicating a new possible function of BLM in HR: the stimulation of the DNA strand exchange activity of RAD51. BLM shows specificity in stimulation of DNA strand exchange promoted by human RAD51 and DMC1. We show that BLM does not stimulate RAD51 orthologs, either E. coli RecA or S. cerevisiae Rad51. Stimulation of RAD51 is not a common property of human RecQ helicases; RECQ1 does not stimulate RAD51 activity under our tested conditions, despite the fact that RECQ1 physically interacts with RAD51 (43). The interaction between RAD51 and BLM may also be important in recruiting BLM to the sites of recombinational repair for serving downstream functions, e.g. stimulation of DNA repair synthesis after strand invasion and dissociation of D-loops after their extension by DNA polymerase and RAD51 inactivation (25, 51, 52).
We demonstrate here that BLM can also stimulate the DNA pairing activity of DMC1, a meiosis-specific homolog of RAD51 (Fig. 2). It is known that BLM plays an important role in meiosis. The expression of BLM is greatly elevated in meiotic prophase, and the protein co-localizes with RAD51/DMC1 foci (53). In humans, males homozygous for BLM mutations are infertile, and heterozygous individuals display increased frequencies of chromosome abnormalities in their spermatozoa (1). In Drosophila melanogaster, mutations in the BLM gene reduce the frequency of meiotic recombination and alter cross-over distribution (54). Stimulation of the DNA strand exchange activity of DMC1 may be related to a meiotic function of BLM.
We showed that BLM stimulates the RAD51 DNA strand exchange on homeologous DNA substrates containing 10% of heterology. This result is consistent with the accompanying report by Kikuchi et al. (59) demonstrating that BLM is required for Ig gene conversion, in which recombination occurs between homeologous DNA sequences containing approximately the same degree of heterology. The apparent BLM independence of recombination between fully homologous substrates observed in this work may indicate that the efficiency of DNA strand exchange promoted by RAD51 on such substrates ceases to be the limited step during HR. Thus, RAD51 stimulation by BLM may be especially important on “difficult” DNA substrates containing mismatches.
BLM shows significant similarity in its biochemical properties with RAD54. Like RAD54, BLM can translocate on DNA in an ATPase-dependent manner, bind specifically to Holliday junctions, and promote their migration, physically interact with RAD51, and disrupt joint molecules formed by RAD51 protein (2, 16). Here we show that, like RAD54, BLM can stimulate RAD51 strand exchange activity. However, the mechanisms of BLM and RAD54 stimulation of the RAD51 DNA strand exchange activity appeared to be different. In contrast with BLM, the ATPase activity of RAD54 protein is required for stimulation of RAD51 (47, 55). It was suggested that RAD54 protein stimulates RAD51 by promoting binding and ATPase-dependent translocation of dsDNA during the search for homologous sequence. An ATP independence of RAD51 stimulation by BLM observed in this study likely excludes a role of DNA translocation in RAD51 stimulation by BLM. Also, in contrast to RAD54 and HOP2/MND1, BLM apparently does not stimulate dsDNA binding by the RAD51-ssDNA filament. However, the ATP independence of RAD51 stimulation does not necessarily exclude a role of the BLM helicase activity in this stimulation. It is known that ATP hydrolysis is required for DNA translocation of helicases rather than for duplex destabilization (49). Consequently, we suggest that BLM may stimulate RAD51 by promoting an ATPase-independent destabilization of DNA duplex during the search for homology. Consistent with this hypothesis, we show that BLM catalyzes an ATPase-independent DNA strand exchange, albeit with a relatively low efficiency.
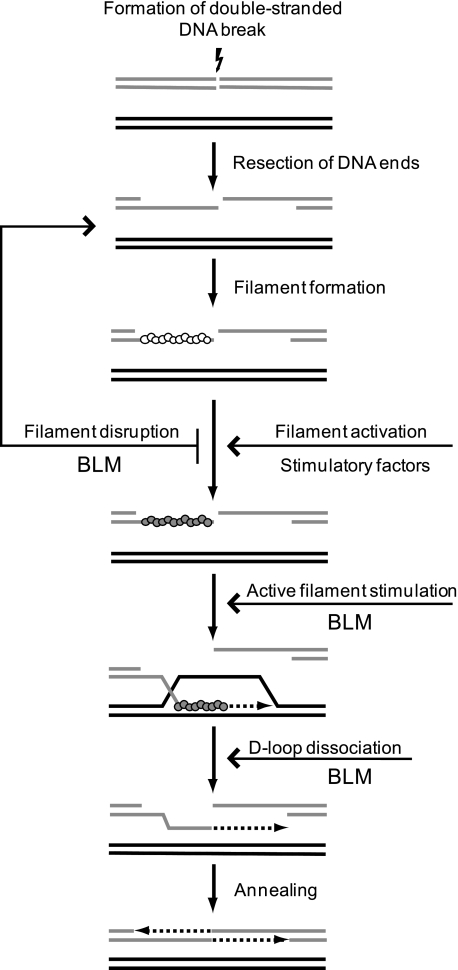
The RAD51-ssDNA filament, an active species that promotes HR, exists in two major conformations, ADP-bound or inactive and ATP-bound or active (56). We found that the conformational state of the RAD51-ssDNA filament plays an important role in interactions with BLM. We previously showed that BLM efficiently disrupts the RAD51-ssDNA filament when RAD51 is present in an inactive ADP-bound form (25). In this study, we show that stimulation of RAD51 by BLM requires the active ATP-bound conformation of the RAD51-ssDNA filament. We suggest that some stimulatory factors, such as accessory proteins (57), specific co-factors, or proper ionic environment may stabilize the presynaptic RAD51-ssDNA filament, making it not only resistant for BLM disruption but also susceptible for BLM stimulation (Fig. 12). Thus, the conformational state of the RAD51-ssDNA filament may play a pivotal role in determining the outcome of the filament interaction with BLM and, as we have showed previously, with RAD54 (18, 58). The observed dependence of the BLM function on the filament conformation suggests that BLM may play a quality control function in HR. It may prevent premature initiation of recombination events by partially active RAD51 filament, but after filament activation BLM may stimulate DNA strand exchange, ensuring an efficient continuation of the proper HR. Further work is needed to determine the role of the observed stimulation of the RAD51 and DMC1 DNA strand exchange activity by BLM in vivo.
FIGURE 12.
Functions of BLM in homologous recombination. BLM may disrupt an inactive ADP-bound RAD51 filament preventing premature initiation of HR. After formation of an active ATP-bound RAD51-ssDNA filament in the presence of mediator proteins or other stimulatory factors, BLM may stimulate DNA pairing of RAD51 and promote late steps of HR by dissociating D-loop after completion of DNA repair synthesis.
Supplementary Material
Acknowledgments
We thank P. Sung, M. Wold, E. Golub, W. Holloman, and D. Camerini-Otero for human RAD51, RPA, DMC1, BLM, and murine Hop2/Mnd1 expression vectors; R. Brosh, Jr., and S. Kowalczykowski for RecQ1 and yeast Rad51 and Rad54; P. Sung for the BLM K695A mutant; and M. Rossi and F. Huang for the comments and discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants CA100839 and MH084119. This work was also supported by Leukemia and Lymphoma Society Scholar Award 1054-09 (to A. V. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
- BS
- Bloom syndrome
- dsDNA
- double-stranded DNA
- ssDNA
- single-stranded DNA
- scDNA
- supercoiled DNA
- BSA
- bovine serum albumin
- DTT
- dithiothreitol
- HR
- homologous recombination
- RPA
- replication protein A
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate.
REFERENCES
- 1.German J. (1993) Medicine 72, 393–406 [PubMed] [Google Scholar]
- 2.Hickson I. D. (2003) Nat. Rev. Cancer 3, 169–178 [DOI] [PubMed] [Google Scholar]
- 3.Wu L., Hickson I. D. (2006) Annu. Rev. Genet. 40, 279–306 [DOI] [PubMed] [Google Scholar]
- 4.van Brabant A. J., Stan R., Ellis N. A. (2000) Annu. Rev. Genomics Hum. Genet. 1, 409–459 [DOI] [PubMed] [Google Scholar]
- 5.Sharma S., Doherty K. M., Brosh R. M., Jr. (2006) Biochem. J. 398, 319–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaganti R. S., Schonberg S., German J. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 4508–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.González-Barrera S., Cortés-Ledesma F., Wellinger R. E., Aguilera A. (2003) Mol. Cell 11, 1661–1671 [DOI] [PubMed] [Google Scholar]
- 8.Steiner W. W., Kuempel P. L. (1998) J. Bacteriol. 180, 6269–6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonoda E., Sasaki M. S., Morrison C., Yamaguchi-Iwai Y., Takata M., Takeda S. (1999) Mol. Cell. Biol. 19, 5166–5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan K. L., North P. S., Hickson I. D. (2007) EMBO J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ralf C., Hickson I. D., Wu L. (2006) J. Biol. Chem. 281, 22839–22846 [DOI] [PubMed] [Google Scholar]
- 12.Davies S. L., North P. S., Hickson I. D. (2007) Nat. Struct. Mol. Biol. 14, 677–679 [DOI] [PubMed] [Google Scholar]
- 13.Wyman C., Kanaar R. (2006) Annu. Rev. Genet. 40, 363–383 [DOI] [PubMed] [Google Scholar]
- 14.Sung P., Klein H. (2006) Nat. Rev. Mol. Cell Biol. 7, 739–750 [DOI] [PubMed] [Google Scholar]
- 15.Whitby M. C. (2005) Biochem. Soc. Trans. 33, 1451–1455 [DOI] [PubMed] [Google Scholar]
- 16.Wu L., Davies S. L., Levitt N. C., Hickson I. D. (2001) J. Biol. Chem. 276, 19375–19381 [DOI] [PubMed] [Google Scholar]
- 17.Bianco P. R., Tracy R. B., Kowalczykowski S. C. (1998) Front. Biosci. 3, D570–D603 [DOI] [PubMed] [Google Scholar]
- 18.Bugreev D. V., Hanaoka F., Mazin A. V. (2007) Nat. Struct. Mol. Biol. 14, 746–753 [DOI] [PubMed] [Google Scholar]
- 19.Bachrati C. Z., Borts R. H., Hickson I. D. (2006) Nucleic Acids Res. 34, 2269–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Brabant A. J., Ye T., Sanz M., German J. L., 3rd, Ellis N. A., Holloman W. K. (2000) Biochemistry 39, 14617–14625 [DOI] [PubMed] [Google Scholar]
- 21.Karow J. K., Constantinou A., Li J. L., West S. C., Hickson I. D. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6504–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu L., Hickson I. D. (2003) Nature 426, 870–874 [DOI] [PubMed] [Google Scholar]
- 23.Wu L., Bachrati C. Z., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G. W., Hickson I. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4068–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raynard S., Bussen W., Sung P. (2006) J. Biol. Chem. 281, 13861–13864 [DOI] [PubMed] [Google Scholar]
- 25.Bugreev D. V., Yu X., Egelman E. H., Mazin A. V. (2007) Genes Dev. 21, 3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mimitou E. P., Symington L. S. (2008) Nature 455, 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gravel S., Chapman J. R., Magill C., Jackson S. P. (2008) Genes Dev. 22, 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G. (2008) Cell 134, 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nimonkar A. V., Ozsoy A. Z., Genschel J., Modrich P., Kowalczykowski S. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16906–16911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sung P., Krejci L., Van Komen S., Sehorn M. G. (2003) J. Biol. Chem. 278, 42729–42732 [DOI] [PubMed] [Google Scholar]
- 31.Chi P., San Filippo J., Sehorn M. G., Petukhova G. V., Sung P. (2007) Genes Dev. 21, 1747–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Branzei D., Foiani M. (2007) Genes Dev. 21, 3019–3026 [DOI] [PubMed] [Google Scholar]
- 33.Foiani M. (2003) Nature 423, 234–235 [DOI] [PubMed] [Google Scholar]
- 34.Krejci L., Van Komen S., Li Y., Villemain J., Reddy M. S., Klein H., Ellenberger T., Sung P. (2003) Nature 423, 305–309 [DOI] [PubMed] [Google Scholar]
- 35.Veaute X., Jeusset J., Soustelle C., Kowalczykowski S. C., Le Cam E., Fabre F. (2003) Nature 423, 309–312 [DOI] [PubMed] [Google Scholar]
- 36.Hu Y., Raynard S., Sehorn M. G., Lu X., Bussen W., Zheng L., Stark J. M., Barnes E. L., Chi P., Janscak P., Jasin M., Vogel H., Sung P., Luo G. (2007) Genes Dev. 21, 3073–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bugreev D. V., Mazin A. V. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9988–9993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petukhova G. V., Pezza R. J., Vanevski F., Ploquin M., Masson J. Y., Camerini-Otero R. D. (2005) Nat. Struct. Mol. Biol. 12, 449–453 [DOI] [PubMed] [Google Scholar]
- 39.Bugreev D. V., Mazina O. M., Mazin A. V. (2006) Nat. Prot., DOI:2010.1038/nprot.2006.2217 [DOI] [PubMed] [Google Scholar]
- 40.Tsang S. S., Chow S. A., Radding C. M. (1985) Biochemistry 24, 3226–3232 [DOI] [PubMed] [Google Scholar]
- 41.Cui S., Klima R., Ochem A., Arosio D., Falaschi A., Vindigni A. (2003) J. Biol. Chem. 278, 1424–1432 [DOI] [PubMed] [Google Scholar]
- 42.Sharma S., Sommers J. A., Choudhary S., Faulkner J. K., Cui S., Andreoli L., Muzzolini L., Vindigni A., Brosh R. M., Jr. (2005) J. Biol. Chem. 280, 28072–28084 [DOI] [PubMed] [Google Scholar]
- 43.Sharma S., Brosh R. M., Jr. (2007) PLoS ONE 2, e1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bugreev D. V., Brosh R. M., Jr., Mazin A. V. (2008) J. Biol. Chem. 283, 20231–20242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.San Filippo J., Sung P., Klein H. (2008) Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 46.Pezza R. J., Voloshin O. N., Vanevski F., Camerini-Otero R. D. (2007) Genes Dev. 21, 1758–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mazina O. M., Mazin A. V. (2004) J. Biol. Chem. 279, 52042–52051 [DOI] [PubMed] [Google Scholar]
- 48.Chow S. A., Radding C. M. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 5646–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He X., Lehman I. R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3024–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheok C. F., Wu L., Garcia P. L., Janscak P., Hickson I. D. (2005) Nucleic Acids Res. 33, 3932–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McVey M., Larocque J. R., Adams M. D., Sekelsky J. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15694–15699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams M. D., McVey M., Sekelsky J. J. (2003) Science 299, 265–267 [DOI] [PubMed] [Google Scholar]
- 53.Moens P. B., Freire R., Tarsounas M., Spyropoulos B., Jackson S. P. (2000) J. Cell Sci. 113, 663–672 [DOI] [PubMed] [Google Scholar]
- 54.McVey M., Andersen S. L., Broze Y., Sekelsky J. (2007) Genetics 176, 1979–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sigurdsson S., Van Komen S., Petukhova G., Sung P. (2002) J. Biol. Chem. 277, 42790–42794 [DOI] [PubMed] [Google Scholar]
- 56.Yu X., Jacobs S. A., West S. C., Ogawa T., Egelman E. H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8419–8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shim K. S., Schmutte C., Tombline G., Heinen C. D., Fishel R. (2004) J. Biol. Chem. 279, 30385–30394 [DOI] [PubMed] [Google Scholar]
- 58.Rossi M. J., Mazin A. V. (2008) J. Biol. Chem. 283, 24698–24706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kikuchi K., Abdel-Aziz H. I., Taniguchi Y., Yamazoe M., Takeda S., Hirota K. (2009) J. Biol. Chem. 284, 26360–26367 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.