Abstract
A panel of eight, second generation triazine dendrimers differing in the number of amines, guanidines, hydroxyls and aliphatic groups on the periphery was synthesized and assayed for gene transfer in an attempt to correlate the effects of surface functionality on transfection efficiency. The physicochemical and biological properties of the dendrimers and dendriplexes, such as condensation of DNA, size, surface charge and morphology of dendriplexes, toxicity and ultimately transfection efficiency in MeWo cells, were analyzed. The results from an ethidium bromide exclusion assay showed that the complexation efficiency of the dendrimers with DNA is moderately affected by surface groups. Increasing the number of surface amines, reducing the number of surface hydroxyl groups, or replacing the amine moiety with guanidines all help strengthen the complex formed. Results from dynamic light scattering and zeta potential analyses indicate that the smallest particles correlate with complexes that exhibit the highest zeta potentials. Cytotoxicity was low for all compounds, particularly for the G2-5 dendrimer containing alkyl groups on the periphery, indicating the benefit of incorporating such neutral functionality onto the surface of the triazine dendrimers. Within this panel, the highest transfection efficiency was observed for the dendrimers that formed the smallest complexes, suggesting that this physicochemical property is an accurate predictor for determining which dendrimers will show high transfection efficiency.
Introduction
While gene therapy has shown promise in clinical trials for the treatment of a variety of diseases, complications, including immunogenicity and the potential for genetic recombination, that are associated with using viral vectors as DNA carriers have motivated numerous chemists to design non-viral carrier systems.1 Non-viral vector systems include cationic polymers,2 peptides,3 liposomes,4 and dendrimers.5 While nearly all of these systems show reduced transfection efficiency compared to viral analogues, their potential for large-scale production and reduced toxicity make these systems a more attractive alternative.6
Among the non-viral vector systems, dendrimers are advantageous because they can be synthesized to afford monodisperse products with multi-functional peripheries. Various dendrimers including polyamidoamine (PAMAM),7 polypropylenimine (PPI),8 dendritic poly-l-lysine (PLL),9 phosphorus-containing dendrimers,10 and carbosilane dendrimers11 have all shown success in a number of transfection studies. However, a thorough analysis of dendrimers based on melamine as candidates for gene therapy is, thus far, unavailable. Cationic triazine dendrimers have shown low cytotoxicity in vitro and in vivo.12 In addition, the synthesis of these compounds allows for the incorporation of multiple functionalities on the surface13 to aid in overcoming the barriers associated with gene therapy. With the recently completed kilogram-scale synthesis of a dendrimer based on melamine,14 commercial availability of these types of compounds could follow.
The obstacles encountered for gene transfer mediated by non-viral vectors can generally be classified into three main groups: (1) toxicity of the vector, (2) affinity of the complex for the cell surface and cell penetration, and (3) stability of the vector–DNA complex. Cytotoxicity of most non-viral carriers stems from their cationic nature, and accordingly, incorporating non-ionic groups to improve biocompatibility is common. Derivatizing cationic dendrimers with neutral groups such as polyethylene glycol (PEG),15 hydroxyl groups,16 or lipids17 has helped reduce cytotoxicity. Our library focuses on the potential of hydroxyl groups as well as an alkyl group to help reduce the toxicity of triazine dendrimers.
To improve the association of non-viral complexes with the cell surface and to increase cell penetration, studies often aim at incorporating functionalities onto the surface of dendrimers that can mimic the effects seen for arginine-rich peptides.18–20 Guanidinylation is an effective and synthetically facile way to probe this effect.21 Our library includes structures to compare amines and guanidinium groups. Studies have also shown that PAMAM dendriplexes with hydrophobic groups on the surface display enhanced affinity for the cell periphery.22 The hydrophobic nature of our dendrimer cores as well as the incorporation of alkyl groups on the surface are hypothesized to promote interactions with cell surface phospholipids. Finally, the number of surface amine groups was varied to gain insight into dendrimer–DNA complex formation.
In this paper we report the synthesis of a library of dendrimers with various surface functional groups to investigate the effect of triazine dendrimer peripheries on transfection efficiency. The physicochemical and biological properties of these structures, including complexation efficiency, zeta potential, hydrodynamic diameter, cytotoxicity, and finally transfection efficiency, were all assayed. Correlations between these properties and the functionality on the peripheries of the triazine dendrimers are reported.
Results and discussion
Synthesis
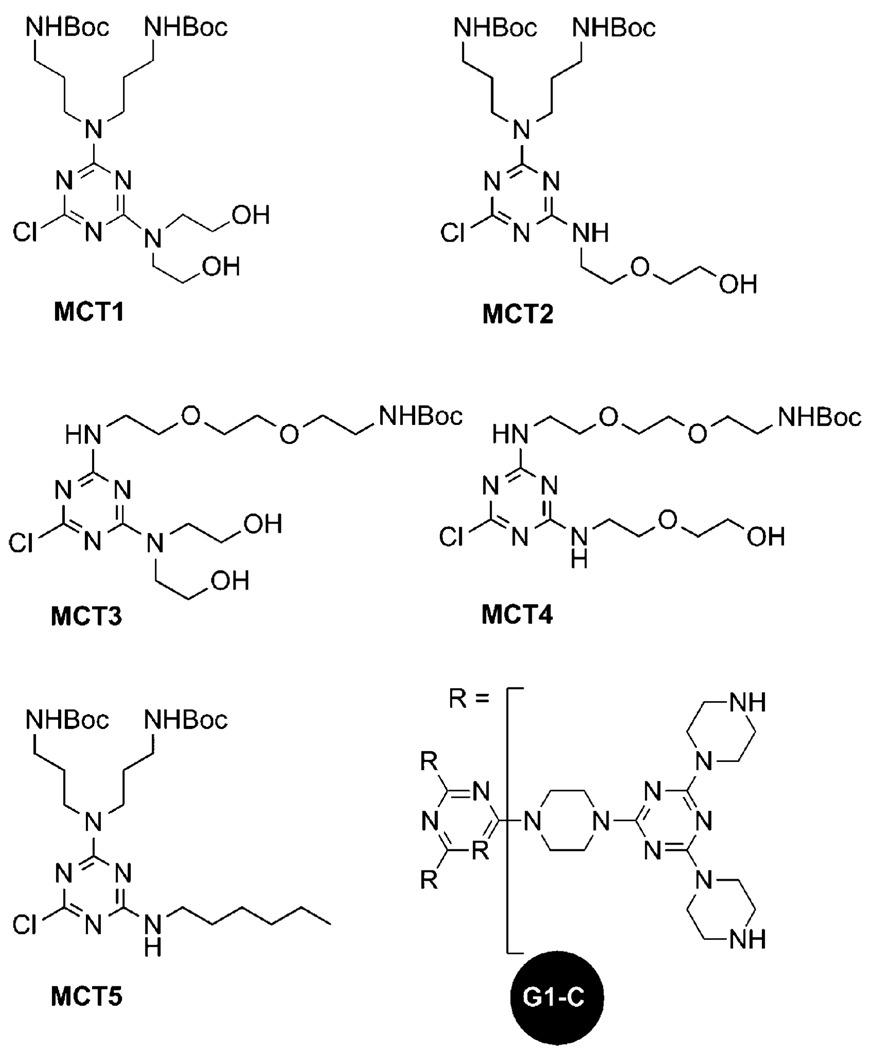
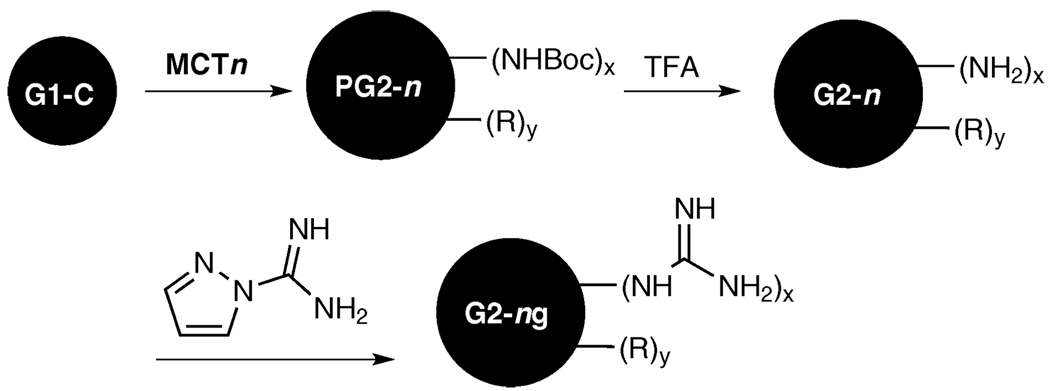
Chart 1 shows the monochlorotriazines and the triazine core used in the synthesis of the panel of dendrimers shown iconically in Chart 2. Scheme 1 shows the general process wherein a first generation triazine core, G1-C,23 is reacted with various monochlorotriazines (MCT1 to MCT5) to form the protected second generation structures (PG2-1 to PG2-5). These Boc-protected intermediates are deprotected to yield dendrimers G2-1 through G2-5. Subsequently, dendrimers G2-1 to G2-3 were guanidinylated following previously described procedures23 to form G2-1g through G2-3g. The monochlorotriazines, illustrated in Chart 1, each contain different functional groups which allow for the creation of a library of dendrimers that are composed of an identical core but which vary in functionality on the periphery. All of the intermediates and products were characterized by 1H and 13C NMR and/or mass spectrometry as described in the experimental section. The results show very high purity for all of the final products except G2-4, for which data are still provided. Schematic representations of the triazine dendrimers are shown in Chart 2.
Chart 1.
Structures of monochlorotriazines and triazine core used in the synthesis of the library of dendrimers.
Chart 2.
Schematic representations of triazine dendrimers showing functional groups used on the periphery.
Scheme 1.
General reaction scheme showing the divergent synthesis of dendrimers used for transfection study.
Ethidium bromide quenching assay
The association of dendrimers and DNA, a so-called dendriplex, can result in the formation of both tightly or loosely associated complexes, with the latter resulting in DNA being more accessible to nucleases and other species. To quantify the amount of unassociated DNA, an ethidium bromide intercalation assay is utilized. In this assay, dendrimer and DNA are complexed and then exposed to ethidium bromide. If unassociated DNA exists, ethidium bromide intercalates and fluoresces. If only tightly bound DNA is present, intercalation is hindered and fluorescence is absent. Therefore, compounds that exhibit low fluorescence typically correlate with highly compacted and protected DNA complexes, which is favorable for in vivo studies. The extent of intercalation quenching by triazine dendrimers is reported in Fig. 1 and various trends that correlate the packaging of DNA with the surface functionality of the dendrimers are evident.
Figure 1.
DNA condensation efficiency was quantified with an ethidium bromide quenching assay.
The choice of the peripheral group of the triazine dendrimers appears to have a notable effect on the packaging of DNA. The dendrimers that contain only six surface amines (G2-3 and G2-4) complexed weakly to DNA as compared to the analogues that contain twelve surface amines (G2-1 and G2-2, respectively). This suggests that condensation efficiency is moderately affected by the number of surface amines, with improved complexation seen for dendrimers with a higher number of surface amines. Guanidinylation appears to significantly increase packaging over primary amine analogues in nearly all cases. This has been seen in earlier studies in which guanidinylated cholesterol derivatives were reported to interact more strongly with DNA than similar amine structures.24 Like the amine analogues, more strongly complexed structures are formed using dendrimers with twelve peripheral guanidines (G2-1g and G2-2g) as compared to dendrimers with only six guanidines (G2-3g).
The formation of strongly complexed structures decreases with increasing number of surface hydroxyl groups, suggesting that this neutral functionality may hinder interactions with DNA. However, for the alkyl group of G2-5, the formation of a strongly complexed dendriplex is observed. In fact the alkylated compound outperforms PEI in complexation efficiency, suggesting that this compound should be competitive in transfection efficiency assays.
Dynamic light scattering and zeta potential measurements
Dynamic light scattering was used to determine the size of the triazine dendrimer–DNA complexes and results are shown in Fig. 2a. In general, the size of the complex can affect both transfection efficiency and toxicity. For in vitro studies, large polyplexes can sediment faster and may result in high transfection efficiency, but can also result in high cytotoxicity.25 However, complexes with diameters of less than 200 nm are endocytosed by most cell lines and are generally believed to be more practical for in vivo and clinical applications.
Figure 2.
Hydrodynamic diameters (a) and zeta potentials (b) of various dendriplexes at an N/P ratio of 5 were determined in a 10 mMHEPES buffer at pH 7.4.
In general, complex size appears to be independent of peripheral functionality. Small complexes were formed using dendrimers G2-1 and G2-5, suggesting these compounds should be the most competitive in transfection efficiency assays. Alternatively, both G2-1g and G2-2g formed complexes with sizes of approximately 1 µm, which is generally a size for which sedimentation occurs, suggested that the compounds may also exhibit notable transfection efficiency in vitro.
Zeta potential measurements were used to determine the surface charge of the dendriplexes, and results are shown in Fig. 2b. In general, positively charged particles associate well with the cell surface and are internalized by the cell via endocytosis, while negatively charged particles are usually repelled by the negatively charged plasma membrane. Overall, the results of the zeta potential analysis are complementary to the data collected from DLS. The dendrimers that showed the highest surface charge (G2-1, G2-4, G2-3g, and G2-5) also formed the smallest complexes, suggesting that these structures may exhibit notable transfection efficiency. Only dendrimer G2-3 showed a negative zeta potential at an N/P ratio of 5, indicating that this compound will not likely be an effective gene transfer agent due to unfavorable electrostatic interactions.
MTT assays
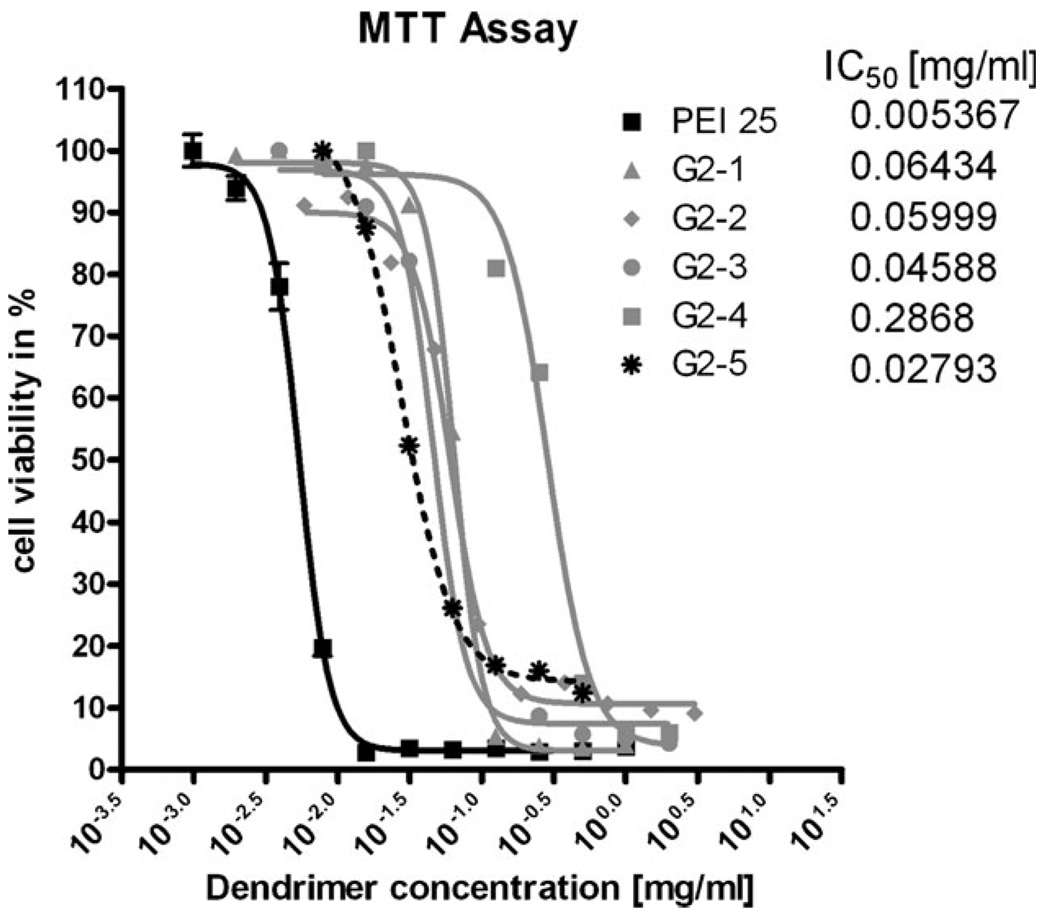
The cytotoxicity observed in this study derives from incubation of various concentrations of the dendrimer (0.001–0.5 mg mL−1) with murine L929 fibroblasts. The cell viability is quantified by mitochondrial enzyme activity, which reduces yellow 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) to its purple formazan. The reduction is quantified spectrophotometrically. The quantified toxicity represents the “worst case scenario”: dendriplexes should display reduced toxicity due to the masking of cationic groups, and this trend has been reported in previous studies investigating the use of PPI for gene delivery.8 The results are shown in Fig. 3.
Figure 3.
Toxicity profiles of all water soluble dendrimers according to the MTT assay were compared to that of PEI 25 kDa.
The cytotoxicity results from this study correlate well with an earlier report that revealed an IC50 value of 0.1 mg mL−1 for a related triazine dendrimer.12 In general there are no clear trends for correlating cytotoxicity with surface functionality. None of the triazine dendrimers used in this study showed substantial cytotoxicity, indicating that this will not be a substantial factor that influences gene transfer activity.
Transfection efficiency
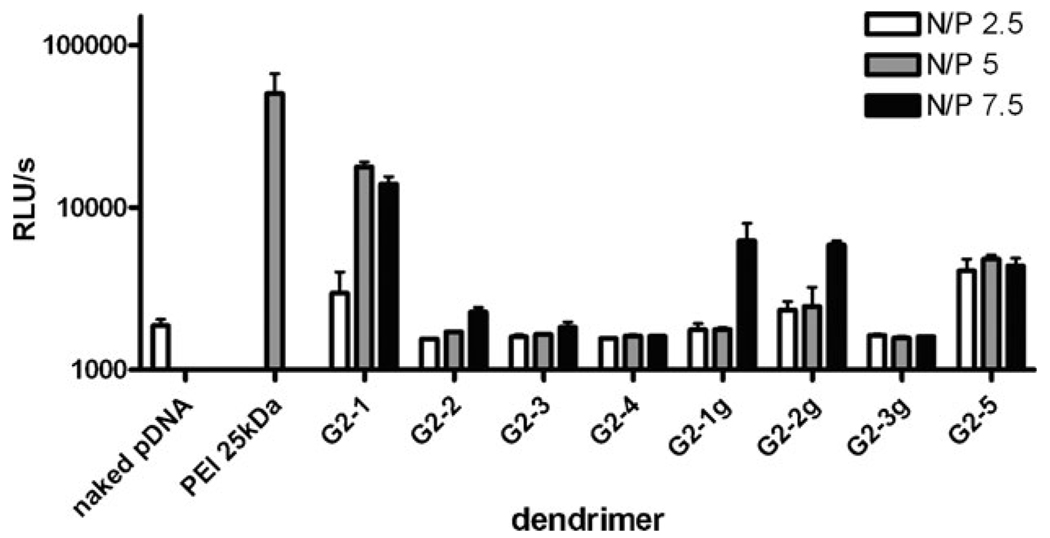
Transfection efficiency of the dendriplexes at three different N/P ratios was quantified using a commercial luciferase assay in a human melanoma cell line (MeWo). The data are reported in relative light units on a log scale. The gene transfer activity was compared to 25 kDa PEI, a gold standard in transfection (Fig. 4).
Figure 4.
Transfection efficiency of all the water soluble dendrimers at various N/P ratios in MeWo cells are given as relative light unites detected by a luciferase assay.
The highest transfection efficiency was seen for DNA complexed with either G2-1 or G2-5. These complexes have relatively small sizes (<250 nm), which correlates with earlier studies that showed that high transfection efficiency is seen for small complexes.26,27 However, at high N/P ratios (N/P=7.5) high transfection efficiency was also seen for complexes with twelve guanidine surface groups (G2-1g, G2-2g). This high transfection efficiency may be due to a sedimentation effect exerted on these large complexes. However, due to their large size these compounds would not be suitable for in vivo gene delivery studies. For all of the complexes that contain only 6 terminal amines (G2-3, G2-4 and G2-3g), poor transfection efficiency correlates with low cation number.
Conclusions
Second generation triazine dendrimers join the growing list of macromolecules capable of facilitating gene delivery. Even within this small subset of architectures, trends in activity emerge that validate observations made for other classes of molecules. Perhaps surprisingly, small structural perturbations in the periphery of triazine dendrimers influence transfection efficiency by up to 10-fold within the panel probed. The competitive efficacy seen with G2-1 bodes well for future studies, including those aimed at examining the influence of generation and core structure, which we hope to communicate in due course.
Experimental
Materials
All chemicals used for synthesis were of analytical grade. The intermediate structures G1-C and MCT-2 were synthesized as described elsewhere.13,22
Synthesis
Monochlorotriazine 1 (MCT1)
Cyanuric chloride (0.92 g, 5.00 mmol) and Boc-protected 3,3′-bisaminopropylamine (1.66 g, 5.00 mmol) were each dissolved in 60 mL tetrahydrofuran and cooled to 0 °C. The Boc-protected triamine was added dropwise to cyanuric chloride followed by the addition of DIPEA (1.75 mL, 10.05 mmol). After 3 h, diethanolamine (0.53 g, 5.02 mmol) and DIPEA (1.75 mL, 10.05 mmol) were dissolved in 60 mL tetrahydrofuran, cooled to 0 °C, and added dropwise to the reaction mixture. The mixture was warmed to room temperature. After 20 h the solvent was evaporated under reduced pressure. The crude product was dissolved in dichloromethane and washed with distilled water. The organic layer was dried and purified by column chromatography (10 : 1 CH2Cl2–ethyl acetate) to yield MCT1 (2.49 g, 91%). 1H NMR (300 MHz, CDCl3) δ: 5.76 (b, 1H, OH) 5.36 (t, 2H, NHBoc), 3.77 (d, 4H, CH2OH), 3.67 (d, 4H, NCH2CH2OH), 3.51-3.43 (br, 4H, NCH2CH2CH2), 3.01 (m, 4H, CH2NHBoc), 1.79-1.67 (m, 4H, CH2CH2CH2), 1.37 (s, 18H, C(CH3)3). 13C NMR (75 MHz, CDCl3) δ: 168.4 (C3N3), 165.0 (C3N3), 164.1 (C3N3), 156.3 (CO), 79.3 (OC(CH3)3), 61.7 (CH2OH), 51.7 (NCH2CH2OH), 44.6 (NCH2CH2CH2), 38.0 (CH2NHBoc), 28.2 (C(CH3)3), 27.7 (CH2CH2CH2). MS (ESI): calcd 547.29 (M+); found 548.30 (M + H+).
Monochlorotriazine 3 (MCT3)
Mono-Boc-protected 2,2′-(ethylenedioxy)bis(ethylamine) (0.33 g, 1.32 mmol) and cyanuric chloride (0.24 g, 1.33 mmol) were each dissolved in 20 mL tetrahydrofuran, and cooled to 0 °C. The diamine solution was added dropwise to the cyanuric chloride solution followed by the addition of DIPEA (0.75 mL, 4.31 mmol). After 3 h, diethanolamine (0.11 g, 1.326 mmol) and DIPEA (0.75 mL, 4.31 mmol) were dissolved in 20 mL tetrahydrofuran, cooled to 0 °C, and added dropwise to the reaction mixture. The solution was warmed to room temperature. After 18 h the reaction mixture was dried under reduced pressure. The crude product was dissolved in dichloromethane and washed with distilled water. The dichloromethane layer was dried and purified using column chromatography (10 : 1 CH2Cl2–ethyl acetate) to afford MCT3 (0.49 g, 80%). 1H NMR (300 MHz, CDCl3) δ: 6.11 (br, 1H, OH), 5.38 (br, 1H, NHBoc), 3.89 (br, 4H, NCH2CH2OH), 3.77 (br, 2H, NHCH2CH2O) 3.60 (s, 4H, OCH2CH2O), 3.54 (m, 6H, OCH2CH2NHBoc, NCH2CH2OH), 3.29, (q, 2H, CH2NHBoc), 1.96 (br, 2H, OH), 1.42 (s, 9H, C(CH3)3). 13C NMR (75 MHz, CDCl3) δ: 168.7 (C3N3), 165.9 (C3N3), 165.2 (C3N3), 156.4 (CO), 70.6 (CH2OCH2CH2OCH2), 69.5 (OCH2CH2O), 61.8 (CH2OH), 52.3 (NCH2CH2OH), 41.0 (HNCH2), 40.5 (CH2NHBoc), 28.6 (C(CH3)3). MS (ESI): calcd 464.22 (M+); found 465.17 (M + H+).
Monochlorotriazine 4 (MCT4)
Mono-Boc-protected 2,2′-(ethylenedioxy)bis(ethylamine) (0.10 g, 0.40 mmol) and cyanuric chloride (0.07 g, 0.40 mmol) were each dissolved in 7 mL tetrahydrofuran and cooled to 0 °C. The diamine solution was added dropwise to the cyanuric chloride solution followed by the addition of DIPEA (0.21 mL, 1.21 mmol). After 3 h, 2-(2-aminoethoxy)ethanol (0.04 mL, 0.40 mmol) and DIPEA (0.21 mL, 1.21 mmol) were mixed in 7 mL tetrahydrofuran, cooled to 0 °C, and added dropwise to reaction mixture. The solution was allowed to warm to room temperature. After 18 h the reaction mixture was dried under reduced pressure, dissolved in dichloromethane, and washed with distilled water. The dichloromethane layer was dried and the product was purified by column chromatography (1 : 1 CH2Cl2–ethyl acetate → 1 : 1 CH2Cl2–ethyl acetate with 10% CH3OH) to afford MCT4 (0.16 g, 85%). 1H NMR (300 MHz, CDCl3) δ: 3.72 (s, 4H, NCH2), 3.62 (s, 8H, OCH2CH2O), 3.53 (t, 6H, OCH2CH2NH), 3.31 (t, 2H, CH2NHBoc), 2.75 (s, 1H, OH), 1.42 (s, 9H, C(CH3)3). 13C NMR (75 MHz, CDCl3) δ: 166.2 (C3N3), 165.8 (C3N3), 156.3 (CO), 79.3 (OC(CH3)3), 72.6 (OCH2CH2O), 70.6 (OCH2CH2NH), 61.8 (CH2OH), 40.8 (NHCH2), 40.5 (CH2NHBoc), 28.6 (C(CH3)3). MS (ESI): calcd 464.22 (M+); found 465.23 (M + H+).
Monochlorotriazine 5 (MCT5)
Boc-protected 3,3′-bisaminopropylamine (0.48 g, 1.45 mmol) and cyanuric chloride (0.27 g, 1.45 mmol) were each dissolved in 4 : 1 CH2Cl2–CH3OH (30 mL) and cooled to 0 °C. The triamine solution was added dropwise to cyanuric chloride followed by the addition of DIPEA (0.25 mL, 1.44 mmol). After 3 h, hexylamine (0.15 g, 1.45 mmol) and DIPEA (0.25 mL, 1.44 mmol) were dissolved in 15 mL of 4 : 1 CH2Cl2–CH3OH, cooled to 0 °C, and added dropwise to the reaction mixture. After 12 h reaction mixture was dried under reduced pressure, dissolved in dichloromethane, and washed with distilled water. The dichloromethane layer was dried and the crude product was purified by column chromatography (10 : 1 CH2Cl2–ethyl acetate) to afford MCT5 (0.72 g, 91%). 1H NMR (300 MHz, CDCl3) δ: 3.59–3.49 (t, 4H, NCH2), 3.37 (q, 2H, NHCH2), 3.09 (m, 4H, CH2NHBoc), 1.78–1.69 (m, 4H, CH2CH2CH2), 1.57 (m, 2H, NHCH2CH2), 1.44 (s, 18H, C(CH3)3), 1.31 (s, 6H, CH2), 0.88 (t, 3H, CH3). 13C NMR (75 MHz, CDCl3) δ: 168.7 (C3N3), 165.6 (C3N3), 165.4 (C3N3), 156 (CO), 79.1 (OC(CH3)3), 44.0 (NCH2CH2CH2), 43.6 (NHCH2), 41.2 (NHCH2CH2), 37.8 (CH2NHBoc), 37.0 (CH2NHBoc), 31.7 (CH2CH2CH3), 28.6 (C(CH3)3), 28.0 (CH2CH2CH2), 26.7 (NHCH2CH2CH2), 22.8 (CH2CH3), 14.2 (CH3). MS (ESI): calcd 543.33 (M+); found 544.33 (M + H+).
Protected G2-1 (P-G2-1)
MCT1 (0.57 g, 1.04 mmol) and G1-C (0.18 g, 0.17 mmol) were each dissolved in 10 mL of 9 : 1 CH2Cl2–CH3OH. The solutions were mixed at room temperature followed by the addition of DIPEA (0.5 mL, 2.9 mmol). The reaction mixture was heated to 70 °C. After 10 days the reaction mixture was dried, re-dissolved in dichloromethane, and washed with distilled water. The organic layer was condensed and purified by column chromatography (4 : 1 CH2Cl2–ethyl acetate with 1 → 3% CH3OH) to yield P-G2-1 (0.43 g, 61%). 1H NMR (300 MHz, CDCl3) δ: 5.64 (s, 12H, OH) 5.09 (s, 12H, NHBoc), 3.87 (br, 24H, CH2OH), 3.81 (br, 72H, piperazine), 3.77 (br, 24H, NCH2CH2OH), 3.61 (br, 24H, NCH2CH2CH2), 3.11 (br, 24H, CH2NHBoc), 1.67 (br, 24H, CH2CH2CH2), 1.44 (s, 108H, C(CH3)3). 13C NMR (75 MHz, CDCl3) δ: 165.6 (C3N3), 165.5 (C3N3), 156.2 (CO), 79.2 (OC(CH3)3), 62.0 (CH2OH), 51.3 (NCH2CH2OH), 44.2 (NCH2CH2CH2), 43.6 (piperazine), 38.2 (CH2NHBoc), 28.7 (C(CH3)3), 27.9 (CH2CH2CH2). MS (MALDI): calcd 4142.57 (M+); found 4142.71 (M + H+).
G2-1
P-G2-1 (0.38 g, 0.09 mmol) was dissolved in 3 M HCl–CH3OH. After 2 days the reaction was condensed and neutralized with NaCO3 to pH = 7. NaCl byproduct was removed from the mixture using an Amicon apparatus and a millipore membrane with NMWL = 1000 under 35 psi N2. The solution remaining inside the Amicon vessel was dried in vacuo to afford G2-1 (0.25 g, 93%). MS (MALDI): calcd 2941.94 (M+); found 4142.90 (M + H+). HPLC (2 : 1 H2O with 0.14% TFA–acetonitrile): 3.196 min.
G2-1g
G2-1 (21.9 mg, 7.4 µmol) was dissolved in 1 mL distilled water. 1H-pyrazole-1-carboxamidine·HCl (0.13 g, 890 µmol) and DIPEA (0.31 mL, 1.78 mmol) were mixed in 1 mL acetonitrile. When the acetonitrile solution became clear, the distilled water solution was added, and the mixture was stirred at room temperature. After 5 days the solvents were removed and the mixture was re-dissolved in diionized water. Excess starting material was removed from the solution using an Amicon apparatus and a millipore membrane with NMWL= 1000 under 35 psi N2. The solution remaining inside the Amicon vessel was dried in vacuo to afford G2-1g dendrimer (18.1 mg, 71%). MS (MALDI): calcd 3446.21 (M+); found 3447.00 (M + H+).
Protected G2-2 (P-G2-2)
MCT2 (0.30 g, 0.55 mmol) and G1-C (0.10 g, 0.09 mmol) were each dissolved in 3 mL 9 : 1 CHCl3–CH3OH. The monochlorotriazine solution was added dropwise to the G1-C solution followed by the addition of DIPEA (0.40 mL, 2.30 mmol). After 5 days the reaction mixture was dried under reduced pressure, re-dissolved in dichloromethane, and washed with distilled water. The organic layer was dried and purified by column chromatography (4 : 1 CH2Cl2–ethyl acetate and 3 → 7% CH3OH) to afford P-G2-2 dendrimer (0.22 g, 58%). 1H NMR (300 MHz, CDCl3) δ: 5.26 (br, 18H, NHBoc), 3.82 (br, 72H, piperazine), 3.75 (br, 36H, CH2OCH2CH2OH), 3.64 (br, 12H, NHCH2), 3.61 (br, 24, NCH2), 3.08 (br, 24H, CH2NHBoc), 1.73 (br, 24H, CH2CH2CH2), 1.44 (s, 108H, C(CH3)3). 13C NMR (75 MHz, CDCl3) δ: 165.6 (C3N3), 165.2 (C3N3), 156.2 (CO), 79.3 (OC(CH3)3), 72.5 (OCH2CH2OH), 70.2 (NHCH2CH2O), 61.8 (CH2OH), 43.3 (piperazine), 42.5 (NCH2CH2CH2), 40.8 (NHCH2CH2O), 37.4 (CH2NHBoc), 28.7 (C(CH3)3), 28.0 (CH2CH2CH2). MS (MALDI): calcd 4142.57 (M+); found 4143.15 (M + H+).
G2-2
P-G2-2 (0.22 g, 0.05 mmol) was dissolved in 3 M HCl–CH3OH. After 3 days the reaction mixture was condensed, dissolved in distilled water and neutralized with 1 M NaOH. NaCl byproduct was removed from the solution using an Amicon apparatus and a millipore membrane with NMWL= 1000 under 35 psi N2. The solution remaining inside the Amicon vessel was dried in vacuo to afford G2-2 dendrimer (0.09 g, 59%). MS (MALDI): calcd 2941.94 (M+); found 4142.96 (M + H+). HPLC (2 : 1 H2O with 0.14% TFA–acetonitrile): 3.256 min.
G2-2g
G2-2 (14.6 mg, 5.0 µmol) was dissolved in 1 mL distilled water. 1H-pyrazole-1-carboxamidine·HCl (0.09 g, 590 µmol) and DIPEA (0.21 mL, 1.21 mmol) were mixed in 1 mL acetonitrile. When the acetonitrile solution became clear, the distilled water solution was added, and the mixture was stirred at room temperature. After 5 days the solvents were removed and the mixture was re-dissolved in diionized water. Excess starting material was removed from the solution using an Amicon apparatus and a millipore membrane with NMWL = 1000 under 35 psi N2. The solution remaining inside the Amicon vessel was dried in vacuo to afford G2-2g (12.7 mg, 74%). MS (MALDI): calcd 3446.21 (M+); found 3447.12 (M + H+).
Protected G2-3 (P-G2-3)
MCT3 (0.30 g, 0.65 mmol) and G1-C (0.12 g, 0.11 mmol) were each dissolved in 3 mL 9 : 1 CH2Cl2–CH3OH. The monochlorotriazine solution was added to the G1-C solution followed by the addition of DIPEA (0.34 mL, 1.95 mmol). The reaction mixture was heated to 70 °C. After 4 days the reaction mixture was dried under reduced pressure, dissolved in dichloromethane, and washed with distilled water. The organic layer was dried and purified by column chromatography using 4 : 1 CH2Cl2–ethyl acetate with 7% CH3OH to afford P-G2-3 (0.18 g, 47%). 1H NMR (300 MHz, CDCl3) δ: 5.39 (br, 12H, NH), 3.82 (br, 24 + 12 + 72H, NCH2CH2OH, NHCH2, piperazine), 3.57 (s, 36H, OCH2CH2O, NCH2) 3.53 (t, 24H, OCH2CH2NHBoc), 3.31 (q, 12H, OCH2CH2NHBoc), 1.88 (br, 12H, OH), 1.43 (s, 54H, C(CH3)3). 13C NMR (75 MHz, CDCl3) δ: 167.2 (C3N3), 165.5 (C3N3), 164.6 (C3N3), 156.7 (CO), 79.4 (OC(CH3)3), 70.5 (CH2OCH2CH2OCH2), 70.1 (OCH2CH2O), 63.1 (CH2OH), 51.6 (NCH2CH2OH), 43.4 (piperazine), 40.6 (NHCH2, CH2NHBoc), 28.6 (C(CH3)3). MS (MALDI): calcd 3644.13 (M+); found 3645.86 (M + H+).
G2-3
P-G2-3 (0.18 g, 0.05 mmol) was dissolved in 3 M HCl–CH3OH (10 mL). After 5 days the solvent was evaporated under reduced pressure and the product was re-dissolved in distilled water and neutralized with NaCO3 to pH = 7. NaCl byproduct was removed from the solution using an Amicon apparatus and a millipore membrane with NMWL = 1000 under 35 psi N2. The solution remaining inside the Amicon vessel was dried in vacuo to afford G2-3 (0.12 g, 77%). MS (MALDI): calcd 3043.82 (M+); found 3044.37 (M + H+). HPLC (2 : 1 H2O with 0.14% TFA–acetonitrile): 3.345 min.
G2-3g
G2-3 (19.3 mg, 6.3 µmol) was dissolved in 1 mL distilled water. 1H-pyrazole-1-carboxamidine·HCl (0.06 g, 380 µmol) and DIPEA (0.13 mL, 750 mmol) were mixed in 1 mL acetonitrile. When the acetonitrile solution became clear, the distilled water solution was added and the mixture was stirred at room temperature. After 5 days the solvents were removed and the mixture was re-dissolved in diionized water. Excess starting material was removed from the solution using an Amicon apparatus and a millipore membrane with NMWL= 1000 under 35 psi N2. The solution remaining inside the Amicon vessel was condensed in vacuo to afford G2-3g (18.3 mg, 88%). MS (MALDI): calcd 3295.95 (M+); found 2396.51 (M + H+).
Protected G2-4 (P-G2-4)
MCT4 (0.15 g, 0.32 mmol) and G1-C (0.06 g, 0.05 mmol) were each dissolved in 3 mL 9 : 1 CH2Cl2–CH3OH. The monochlorotriazine solution was added to the G1-C solution at room temperature followed by the addition of DIPEA (0.16 mL, 0.92 mmol). The reaction was heated to 70 °C. After 4 days the reaction mixture was dried in vacuo, dissolved in dichloromethane, and washed with distilled water. The organic layer was dried to afford P-G2-4 (0.18 g, 93%). 1H NMR (300 MHz, CDCl3) δ: 5.39 (br, 18H, NH), 3.80 (br, 72H, piperazine), 3.71 (br, 12, OCH2CH2OH), 3.61 (br, 96H, CH2), 3.31 (br, 12H, CH2NHBoc), 1.43 (s, 54H, C(CH3)3). 13C NMR (75 MHz, CDCl3) δ: 166.2 (C3N3), 165.6 (C3N3), 165.2 (C3N3), 156.3 (CO), 79.3 (OC(CH3)3), 72.7 (CH2OCH2CH2OCH2), 70.4 (OCH2CH2O), 61.7 (CH2OH), 43.3 (piperazine), 40.6 (NHCH2, CH2NHBoc), 28.6 (C(CH3)3). MS (MALDI): calcd 3644.13 (M+); found 3645.49 (M + H+).
G2-4
P-G2-4 (0.16 g, 0.04 mmol) was dissolved in 3 M HCl : CH3OH. After 5 days the solvent was evaporated under reduced pressure and the product re-dissolved in distilled water and neutralized with NaCO3 to pH = 7. NaCl byproduct was removed from the solution using an Amicon apparatus and a millipore membrane with NMWL = 1000 under 35 psi N2. The solution remaining inside the Amicon vessel was condensed in vacuo to afford G2-4 (0.05 g, 40%). MS (MALDI): calcd 3043.82 (M+); found 3043.62 (M + H+).
Protected G2-5 (P-G2-5)
MCT5 (0.36 g, 0.67 mmol) and G1-C (0.06 g, 0.06 mmol) were each dissolved in 5 mL of 10 : 1 CH2Cl2–CH3OH. The monochlorotriazine solution was added to the G1-C solution at room temperature followed by the addition of DIPEA (0.25 mL, 1.44 mmol). The reaction was heated to 70 °C. After 5 days the reaction mixture was dried in vacuo, dissolved in dichloromethane, and washed three times with distilled water. The organic layer was dried to afford P-G2-5 (0.20 g, 86%). 1H NMR (300 MHz, CDCl3) δ: 5.25 (br, 5H, NH), 4.91 (br, 8H, NH), 3.82 (s, 72H, piperazine), 3.58 (br, 24H, NCH2), 3.39 (q, 12H, NHCH2), 3.08 (br, 24H, CH2NHBoc), 1.77 (br, 24H, CH2CH2CH2), 1.59 (p, 12H, NHCH2CH2), 1.44 (s, 108H, C(CH3)3), 1.32 (br, 36H, CH2), 0.89 (t, 18H, CH3). 13C NMR (75 MHz, CDCl3) δ: 165.6 (C3N3), 165.5 (C3N3), 165.3 (C3N3), 156.2 (CO), 79.2 (OC(CH3)3), 43.3 (NCH2CH2CH2, NHCH2, piperazine), 41.0 (NHCH2CH2), 31.8 (CH2CH2CH3), 30.1 (CH2), 28.7 (C(CH3)3, CH2 CH2CH2), 26.8 (NHCH2CH2CH2), 22.8 (CH2CH3), 14.3 (CH3). MS (MALDI): calcd 4118.82 (M+); found 4121.46 (M + H+).
G2-5
P-G2-5 (0.18 g, 0.04 mmol) was dissolved in 5 mL CH3OH. Concentrated HCl (5 mL) was added to the methanol solution. After 18 h the reaction mixture was condensed in vacuo and neutralized with 5 M NaOH. At basic pH, product precipitated to yield G2-5 (0.12 g, 98%). MS (MALDI): calcd 2918.19 (M+); found 2919.32 (M + H+).
Cell culture
L929 and MeWo cells were purchased from LG Promochem, Wesel, Germany, and were maintained in Dulbecco’s Modified Eagle Medium (DMEM) at low or high glucose (PAA Laboratories, Cölbe, Germany), respectively, supplemented with 10% fetal calf serum (Cytogen, Sinn, Germany) in humidified atmosphere with 5% CO2 at 37 °C.
Preparation of dendriplexes
Dendriplexes were formed by adding a solution of calculated concentration of dendrimer to an equal volume of DNA solution (pCMV-Luc, Plasmid Factory, Bielefeld, Germany). The two components were mixed and left for 20 min to allow for complex formation. Both pDNA and dendrimer dilutions were prepared in 10 mM HEPES buffer, pH 7.4, and the amount of dendrimer needed to afford a certain N/P ratio was calculated by considering the “protonable unit” of each dendrimer which represents the mass of dendrimer per protonable nitrogen atom. For comparison, bulk polyethylenimine (Polymin, 25 kDa), a gift from BASF (Ludwigshafen, Germany), was used.
Ethidium bromide quenching assay
In order to investigate condensation efficiency of the dendrimers, 4 µg of herring testes DNA (Sigma-Aldrich Chemie GmbH, Schnelldorf, Germany) were mixed with increasing amounts of dendrimers in a final volume of 280 µL 10 mM HEPES buffer and pipetted into opaque FluoroNunc™ 96-well plates (Nunc, Thermo Fisher Scientific, Langenselbold, Germany). After incubation for 20 min at room temperature, 20 µL of a 0.1 mg mL−1 ethidium bromide solution (Carl Roth GmbH, Karlsruhe, Germany) were added to each well and briefly incubated in the dark under constant shaking. Intercalation-induced fluorescence was quantified using a fluorescence plate reader (LS 50 B, Perkin-Elmer, Rodgau-Jü gesheim, Germany) at 518 nm excitation and 605 nm emission wavelengths. Results are given as mean relative fluorescence intensity values with intercalation of free DNA representing 100% fluorescence and non-intercalating ethidium bromide in buffer representing 0% remaining fluorescence. Results are given as means of triplicate measurements ± standard deviation (SD).
Dynamic light scattering and zeta potential analysis
For measurements of size and zeta potential by dynamic light scattering (DLS) and laser Doppler anemometry (LDA), respectively, dendriplexes of 0.6 µg pCMV-Luc and the according amount of dendrimer were formed as described above. Complexes were incubated for 20 min and diluted to a total volume of 800 µL with 10 mM HEPES buffer, pH 7.4, to be measured into a folded capillary cell (Malvern Instruments, Herrenberg, Germany) using a Zetasizer Nano ZS, Malvern, Herrenberg, Germany. Measurements were conducted in triplicates in the “General Purpose” mode, where position and attenuator were optimized by the device.
MTT assay
The influence of dendrimers on metabolic cell activity was investigated using the MTT assay. For this proliferation assay, murine L929 fibroblasts were seeded on 96-well plates (Nunc, Wiesbaden, Germany) at a density of 8000 cells per well 24 h before they were treated with dendrimer solutions of increasing concentrations in the range of 0.001 to 1 mg mL−1. After another 24 h, the medium was changed, and the MTT solution was added into fresh serum-free medium and incubated for 4 h. Cells were lysed with 200 µL DMSO per well, and remaining mitochondrial enzyme activity compared to untreated cells was determined by measuring the absorbance of enzymatically formed formazan at 580 nm with 690 nm background correction.
Transfection experiments
Human melanoma cells (MeWo) were seeded in 96-well plates (Nunc, Wiesbaden, Germany) at a seeding density of 8000 cells per well. Twenty four hours later, cells were transfected with dendriplexes made of 0.25 µg pCMV-Luc per well and the according amount of dendrimer calculated to be N/P ratios of 2.5, 5, or 7.5. Branched 25 kDa PEI was used as a positive control and free plasmid as a negative control. Dendriplexes were added to full serum-containing medium and cells were incubated for 4 h before medium was changed. After additional 44 h, cells were washed with PBS, lysed will Cell Culture Lysis Reagent, CCLR (Promega, Mannheim, Germany), and assayed for luciferase expression with a commercial luciferase assay kit (Promega, Mannheim, Germany) on a LUMIstar OPTIMA plate luminometer (BMG Labtech, Offenburg, Germany).
Supplementary Material
Acknowledgements
We thank Eva Mohr and Sandra Engel (DPB, Marburg, Germany) for their support in the cell culture lab. EES acknowledges support of the NIH (NIGMS R01 64560). MM acknowledges a predoctoral fellowship from NSF (GK12-DGE 0538547).
Footnotes
Electronic supplementary information (ESI) available: NMR and mass spectra. See DOI: 10.1039/b908735d
Notes and references
- 1.Mintzer MA, Simanek EE. Chem. Rev. 2009;109:259. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 2.Merdan T, Kopecek J, Kissel T. Adv. Drug Delivery Rev. 2002;54:715. doi: 10.1016/s0169-409x(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 3.Niidome T, Katayama Y. In: Non-viral Gene Therapy: Gene Design and Delivery. Taira K, Kataoka K, Niidome T, editors. Tokyo: Springer-Verlag; 2005. pp. 87–102. [Google Scholar]
- 4.Li W, Szoka FC., Jr. Pharm. Res. 2007;24:438. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- 5.Guillot-Nieckowski M, Eisler S, Diederich F. New J. Chem. 2007;31:1111. [Google Scholar]
- 6.Behr J-P. Acc. Chem. Res. 1993;26:274. [Google Scholar]
- 7.Haensler J, Szoka FC., Jr. Bioconjugate Chem. 1993;4:372. doi: 10.1021/bc00023a012. [DOI] [PubMed] [Google Scholar]
- 8.Zinselmeyer BH, Mackay SP, Schatzlein AG, Uchegbu IF. Pharm. Res. 2002;19:960. doi: 10.1023/a:1016458104359. [DOI] [PubMed] [Google Scholar]
- 9.Shah DS, Sakthivel T, Toth I, Florence AT, Wilderspin AF. Int. J. Pharm. 2000;208:41. doi: 10.1016/s0378-5173(00)00534-2. [DOI] [PubMed] [Google Scholar]
- 10.(a) Loup C, Zanta M-A, Caminade A-M, Majoral J-P, Meunier B. Chem.–Eur. J. 1999;5:3644. [Google Scholar]; (b) Padié C, Maszewska M, Majchrzak K, Nawrot B, Caminade A-M, Majoral J-P. New J. Chem. 2009;33:318. [Google Scholar]
- 11.Weber N, Ortega P, Clemente MI, Shsharbin D, Bryszewska M, Javier de la Mata F, Gómez R, Muóoz-Fernández MA. J. Controlled Release. 2008;132:55. doi: 10.1016/j.jconrel.2008.07.035. [DOI] [PubMed] [Google Scholar]
- 12.Neerman MF, Zhang W, Parrish AR, Simanek EE. Int. J. Pharm. 2004;281:129. doi: 10.1016/j.ijpharm.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 13.Steffensen MB, Simanek EE. Angew. Chem., Int. Ed. 2004;43:5178. doi: 10.1002/anie.200460031. [DOI] [PubMed] [Google Scholar]
- 14.Chouai A, Simanek EE. J. Org. Chem. 2008;73:2357. doi: 10.1021/jo702462t. [DOI] [PubMed] [Google Scholar]
- 15.Gajbhiye V, Kumar PV, Tekade RK, Jain NK. Curr. Pharm. Des. 2007;13:415. [Google Scholar]
- 16.Lee JH, Lim Y-B, Choi JS, Lee Y, Kim T-I, Kim HJ, Yoon JK, Kim K, Park J-S. Bioconjugate Chem. 2003;14:1214. doi: 10.1021/bc034095g. [DOI] [PubMed] [Google Scholar]
- 17.Toth I, Sakthivel T, Wilderspin AF, Bayele H, O’Donnell M, Perry DJ, Pasi KJ, Lee CA, Florence AT. STP Pharm. Sci. 1999;9:93. doi: 10.1002/jps.20230. [DOI] [PubMed] [Google Scholar]
- 18.Choi JS, Nam K, Park J-Y, Kim J-B, Lee J-K, Park J-S. J. Controlled Release. 2004;99:445. doi: 10.1016/j.jconrel.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 19.Kim T-I, Baek J-U, Bai CZ, Park J-S. Biomaterials. 2007;28:2061. doi: 10.1016/j.biomaterials.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 20.Okuda T, Sugiyama A, Niidome T, Aoyagi H. Biomaterials. 2004;25:537. doi: 10.1016/s0142-9612(03)00542-8. [DOI] [PubMed] [Google Scholar]
- 21.Theodossiou TA, Pantos A, Tsogas I, Paleos CM. ChemMedChem. 2008;3:1635. doi: 10.1002/cmdc.200800190. [DOI] [PubMed] [Google Scholar]
- 22.Kono K, Akiyama H, Takahashi T, Takagishi T, Harada A. Bioconjugate Chem. 2005;16:208. doi: 10.1021/bc049785e. [DOI] [PubMed] [Google Scholar]
- 23.Chen H-T, Neerman MF, Parrish AR, Simanek EE. J. Am. Chem. Soc. 2004;126:10044. doi: 10.1021/ja048548j. [DOI] [PubMed] [Google Scholar]
- 24.Aissaoui A, Oudrhiri N, Petit L, Hauchecorne M, Kan E, Sainlos M, Julia S, Navarro J, Vigneron JP, Lehn JM, Lehn P. Curr. Drug Targets. 2002;3:1. doi: 10.2174/1389450023348082. [DOI] [PubMed] [Google Scholar]
- 25.Mahato RI, Rolland A, Tomlinson E. Pharm. Res. 1997;14:853. doi: 10.1023/a:1012187414126. [DOI] [PubMed] [Google Scholar]
- 26.Prabha S, Zhou W-Z, Panyam J, Labhasetwar V. Int. J. Pharm. 2002;244:105. doi: 10.1016/s0378-5173(02)00315-0. [DOI] [PubMed] [Google Scholar]
- 27.Xu D-M, Yao S-D, Liu Y-B, Sheng K-L, Hong J, Gong P-J, Dong L. Int. J. Pharm. 2007;338:291. doi: 10.1016/j.ijpharm.2007.01.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.