Abstract
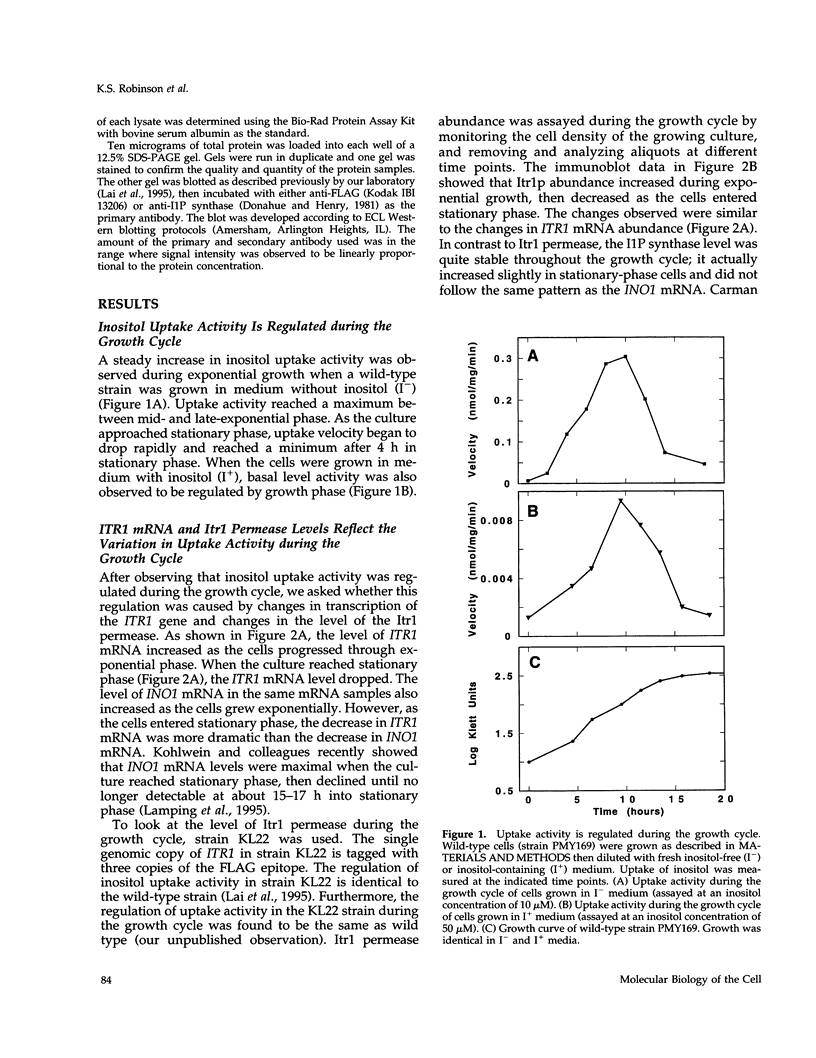
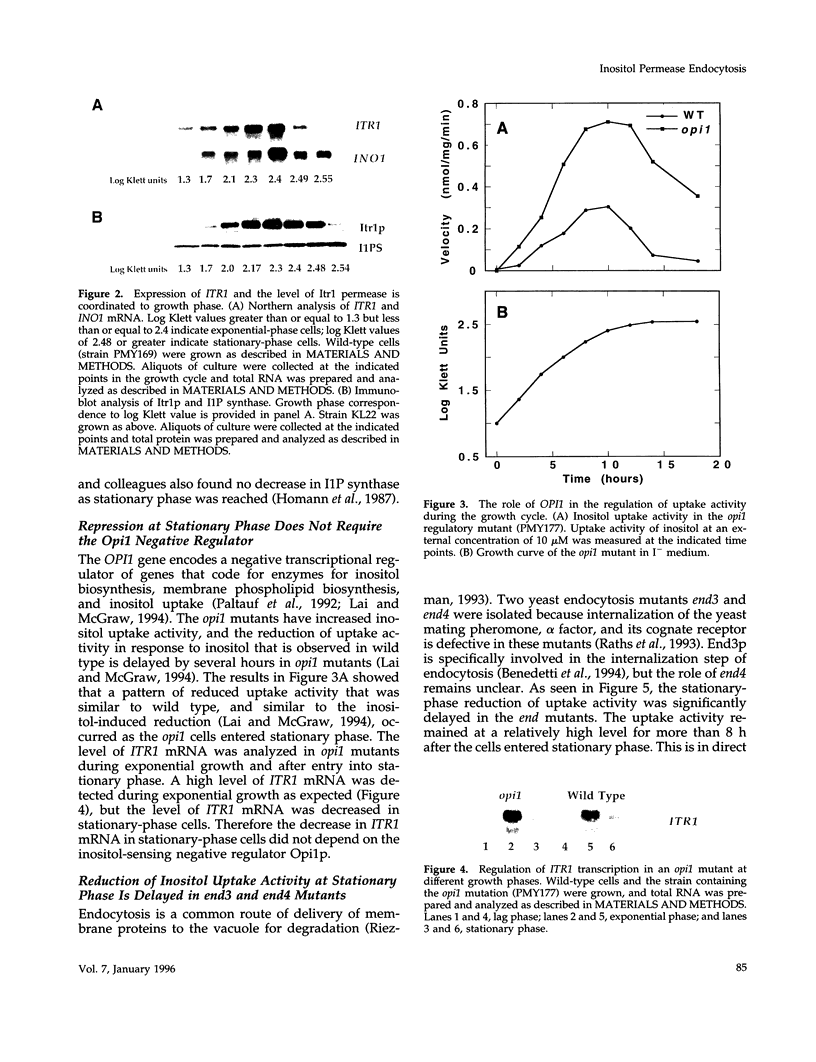
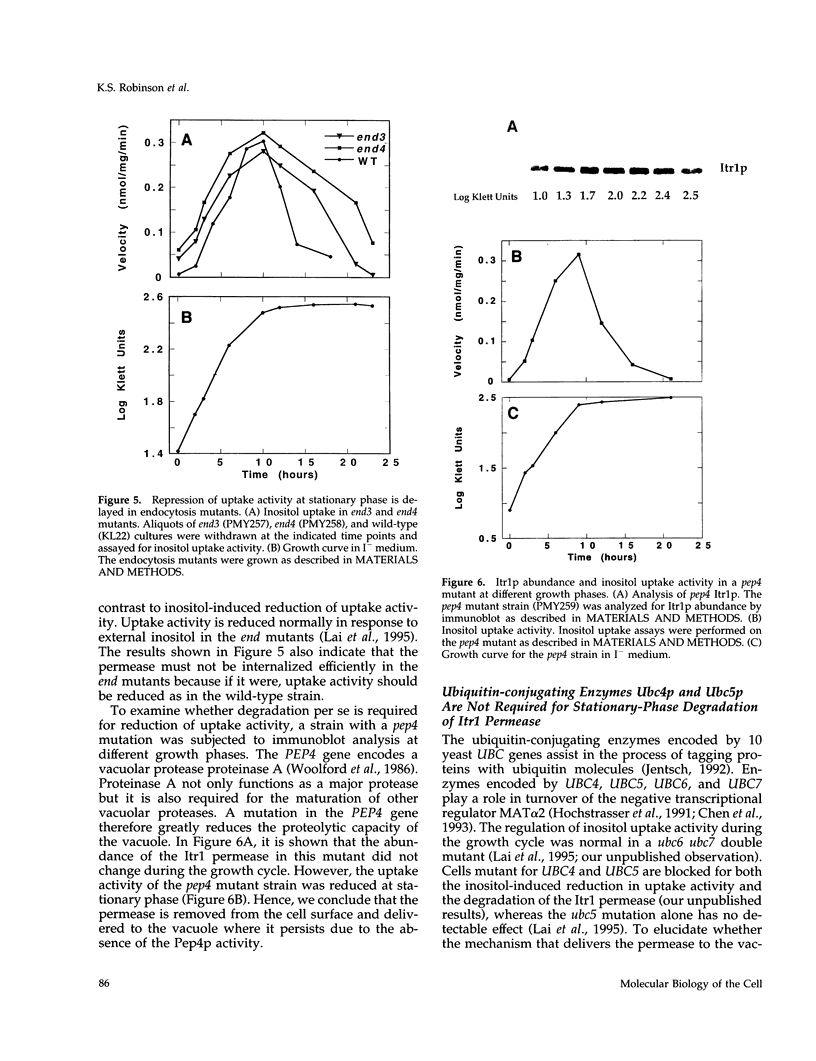
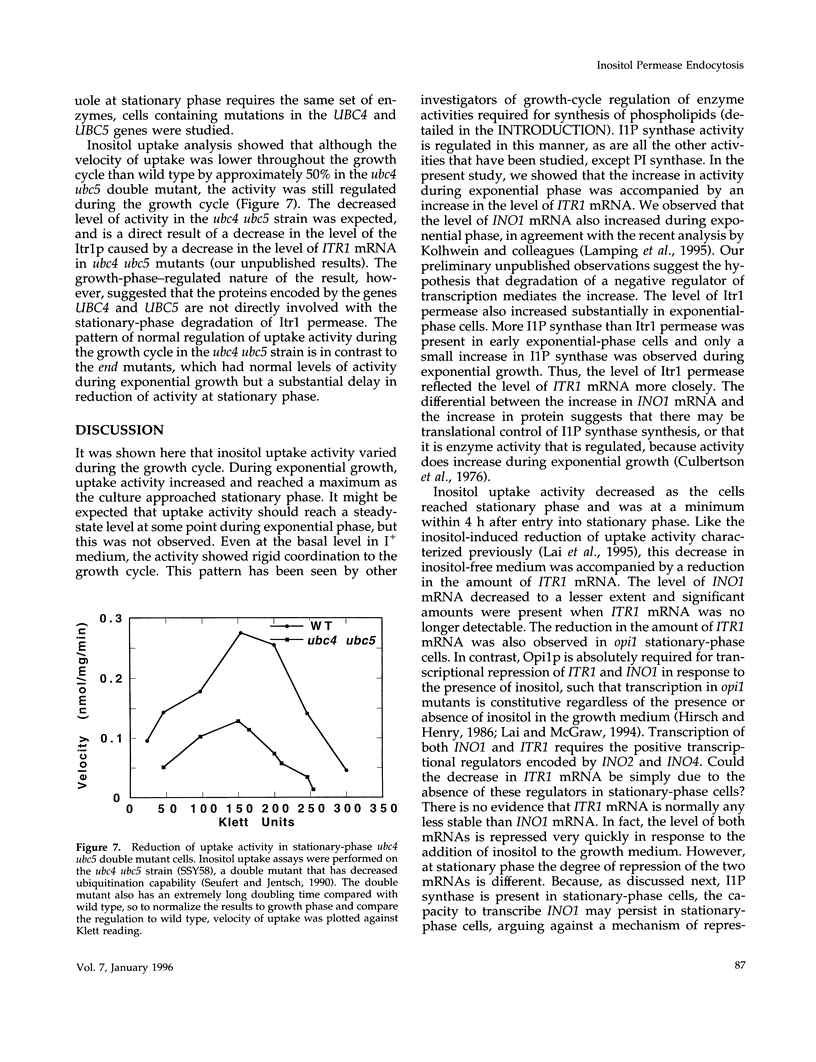
Regulation of inositol uptake activity in Saccharomyces cerevisiae during the growth cycle was examined. Activity increased as the cell population transited from lag phase to exponential growth, and continued to increase until late exponential phase. The increase in activity was due to increased transcription of the ITR1 gene and synthesis of the Itr1 permease. When the culture reached stationary phase, uptake activity decreased and dropped to a minimum within 4 h. The decrease was due to repression of ITR1 transcription, independent of the negative regulator Opi1p, and degradation of the existing permease. Degradation depended on delivery of the permease to the vacuole through the END3/END4 endocytic pathway. During exponential growth in inositol-containing medium the permease is also rapidly degraded, whereas in inositol-free medium the permease is highly stable. Rapid degradation of the permease at stationary phase occurred in inositol-free medium, indicating that there are two distinct mechanisms that trigger endocytosis and degradation in response to different physiological stimuli. In addition, the level of the enzyme required for inositol biosynthesis, inositol-1-phosphate synthase, encoded by INO1, is not reduced in stationary-phase cells, and this contrast in the regulation of inositol supply is discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bénédetti H., Raths S., Crausaz F., Riezman H. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994 Sep;5(9):1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Johnson P., Sommer T., Jentsch S., Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993 Jul 30;74(2):357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Culbertson M. R., Donahue T. F., Henry S. A. Control of inositol biosynthesis in Saccharomyces cerevisiae: properties of a repressible enzyme system in extracts of wild-type (Ino+) cells. J Bacteriol. 1976 Apr;126(1):232–242. doi: 10.1128/jb.126.1.232-242.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Henry S. A. myo-Inositol-1-phosphate synthase. Characteristics of the enzyme and identification of its structural gene in yeast. J Biol Chem. 1981 Jul 10;256(13):7077–7085. [PubMed] [Google Scholar]
- Drebot M. A., Johnston G. C., Singer R. A. A yeast mutant conditionally defective only for reentry into the mitotic cell cycle from stationary phase. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7948–7952. doi: 10.1073/pnas.84.22.7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J. P., Henry S. A. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol Cell Biol. 1986 Oct;6(10):3320–3328. doi: 10.1128/mcb.6.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M., Ellison M. J., Chau V., Varshavsky A. The short-lived MAT alpha 2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann M. J., Poole M. A., Gaynor P. M., Ho C. T., Carman G. M. Effect of growth phase on phospholipid biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1987 Feb;169(2):533–539. doi: 10.1128/jb.169.2.533-539.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland L. S., Johnston G. C., Drebot M. A., Dhillon N., DeMaggio A. J., Hoekstra M. F., Singer R. A. A member of a novel family of yeast 'zn-finger' proteins mediates the transition from stationary phase to cell proliferation. EMBO J. 1994 Aug 15;13(16):3812–3821. doi: 10.1002/j.1460-2075.1994.tb06692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley M. J., Bailis A. M., Henry S. A., Carman G. M. Regulation of phospholipid biosynthesis in Saccharomyces cerevisiae by inositol. Inositol is an inhibitor of phosphatidylserine synthase activity. J Biol Chem. 1988 Dec 5;263(34):18078–18085. [PubMed] [Google Scholar]
- Lai K., Bolognese C. P., Swift S., McGraw P. Regulation of inositol transport in Saccharomyces cerevisiae involves inositol-induced changes in permease stability and endocytic degradation in the vacuole. J Biol Chem. 1995 Feb 10;270(6):2525–2534. doi: 10.1074/jbc.270.6.2525. [DOI] [PubMed] [Google Scholar]
- Lai K., McGraw P. Dual control of inositol transport in Saccharomyces cerevisiae by irreversible inactivation of permease and regulation of permease synthesis by INO2, INO4, and OPI1. J Biol Chem. 1994 Jan 21;269(3):2245–2251. [PubMed] [Google Scholar]
- Lamping E., Kohlwein S. D., Henry S. A., Paltauf F. Coordinate regulation of phosphatidylserine decarboxylase in Saccharomyces cerevisiae. J Bacteriol. 1991 Oct;173(20):6432–6437. doi: 10.1128/jb.173.20.6432-6437.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamping E., Lückl J., Paltauf F., Henry S. A., Kohlwein S. D. Isolation and characterization of a mutant of Saccharomyces cerevisiae with pleiotropic deficiencies in transcriptional activation and repression. Genetics. 1994 May;137(1):55–65. doi: 10.1093/genetics/137.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw P., Henry S. A. Mutations in the Saccharomyces cerevisiae opi3 gene: effects on phospholipid methylation, growth and cross-pathway regulation of inositol synthesis. Genetics. 1989 Jun;122(2):317–330. doi: 10.1093/genetics/122.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raths S., Rohrer J., Crausaz F., Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993 Jan;120(1):55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman H. Yeast endocytosis. Trends Cell Biol. 1993 Aug;3(8):273–277. doi: 10.1016/0962-8924(93)90056-7. [DOI] [PubMed] [Google Scholar]
- Seufert W., Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990 Feb;9(2):543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift S., McGraw P. INO1-100: an allele of the Saccharomyces cerevisiae INO1 gene that is transcribed without the action of the positive factors encoded by the INO2, INO4, SWI1, SWI2 and SWI3 genes. Nucleic Acids Res. 1995 Apr 25;23(8):1426–1433. doi: 10.1093/nar/23.8.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volland C., Urban-Grimal D., Géraud G., Haguenauer-Tsapis R. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J Biol Chem. 1994 Apr 1;269(13):9833–9841. [PubMed] [Google Scholar]
- Werner-Washburne M., Braun E., Johnston G. C., Singer R. A. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1993 Jun;57(2):383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford C. A., Daniels L. B., Park F. J., Jones E. W., Van Arsdell J. N., Innis M. A. The PEP4 gene encodes an aspartyl protease implicated in the posttranslational regulation of Saccharomyces cerevisiae vacuolar hydrolases. Mol Cell Biol. 1986 Jul;6(7):2500–2510. doi: 10.1128/mcb.6.7.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]