Summary
RNase R readily degrades highly structured RNA, whereas its paralogue, RNase II, is unable to do so. Furthermore, the nuclease domain of RNase R, devoid of all canonical RNA binding domains, is sufficient for this activity. RNase R also binds RNA more tightly within its catalytic channel than does RNase II, which is thought to be important for its unique catalytic properties. To investigate this idea further, certain residues within the nuclease domain channel of RNase R were changed to those found in RNase II. Among the many examined, we identified one amino acid, R572, that plays a significant role in the properties of RNase R. Conversion of this residue to lysine, as found in RNase II, results in weaker substrate binding within the nuclease domain channel, longer limit products, increased activity against a variety of substrates and a faster substrate on-rate. Most importantly, the mutant encounters difficulty in degrading structured RNA, pausing within a double-stranded region. Additional studies show that degradation of structured substrates is dependent upon temperature, suggesting a role for thermal breathing in the mechanism of action of RNase R. Based on these data, we propose a model in which tight binding within the nuclease domain allows RNase R to capitalize on the natural thermal breathing of an RNA duplex to degrade structured RNAs.
Keywords: RNase R, RNase II, RNA degradation, RNA binding, Escherichia coli ribonuclease
Introduction
RNA degradation is an essential component of RNA metabolism. Ribonucleases (RNases) are required for the maturation of stable RNAs and the removal of defective RNAs or RNA species that are no longer required by the cell. In Escherichia coli, mRNA decay is primarily carried out by RNase II and polynucleotide phosphorylase (PNPase)1, although structured mRNAs can only be degraded by PNPase2 or a third enzyme, RNase R2, 3. Similarly, PNPase and/or RNase R are required for the degradation of the highly structured rRNA4 and tRNA5 (S. Chebolu, C. Kim, E. Quesada and M. P. Deutscher, personal communication) molecules.
RNase R and RNase II both belong to the RNR family of processive, hydrolytic 3′ to 5′ exoribonucleases. Nevertheless, consistent with their respective functions in vivo, their catalytic properties differ significantly6, 7. RNase II is specific for single-stranded RNA, generally stalling 6 to 11 nucleotides before a duplex3, 8–10. In contrast, RNase R is able to degrade through an RNA duplex, provided there is a 3′ single-stranded overhang to which it can bind and initiate degradation3, 11. RNase R is the only 3′ to 5′ exoribonuclease able to degrade through extensive secondary structure without the aid of a helicase activity. Consequently, its mode of action is of great interest.
Relatively unstructured substrates such as poly(A) are degraded by both RNase R and RNase II, but even on these substrates differences are discernable. Single-stranded RNA is degraded by RNase II at least 4-fold faster than by RNase R12. Moreover, RNase II becomes distributive as the substrate is degraded to lengths shorter than 10 nucleotides12, 13, typically releasing limit products of tetra and penta-nucleotides3, 12, whereas RNase R remains processive and leaves di- and tri-nucleotides as the limits of digestion3, 12.
Both RNase R and RNase II share a common domain organization with two cold-shock domains (CSDs) near the N-terminus, a central nuclease domain and an S1 domain near the C-terminus. Crystal structures of E. coli RNase II show that the CSDs and the S1 domain come together to form an RNA-binding clamp through which the substrate presumably passes en route to the catalytic center located at the base of a narrow channel within the nuclease domain14, 15.
We have recently presented evidence that, while the cold-shock domains (CSDs) and the S1 domain of RNase R appear to play a role in substrate binding and in ensuring efficient catalysis, especially for longer substrates, they are not essential for RNase R to degrade through double-stranded RNA16. Hence, the nuclease domain alone is sufficient to perform this function16. Furthermore, we found that the nuclease domain of RNase R binds RNA more tightly than the nuclease domain of RNase II16. This tighter binding may help to explain many of the differences between the catalytic properties of RNase R and RNase II, for example, the slower rate of degradation of single-stranded RNA and the greater processivity of RNase R relative to RNase II. It remains to be determined if tighter substrate binding within the catalytic channel can also account for the ability of RNase R to degrade through duplex RNA.
Based on sequence alignments and the available structural information for RNR family proteins, we now identify residues within the nuclease domain of RNase R that contribute to tight substrate binding and the related RNase R-specific catalytic properties. We focus on residues that differ between RNase R and RNase II. Candidate residues in RNase R are mutated to the amino acid present in RNase II and the resulting mutant proteins are evaluated for their ability to digest double-stranded RNA. We examine in detail one mutant protein, RNase R R572K, that releases longer limit products than wild-type RNase R, and binds A4 with a higher dissociation constant (Kd) value, indicating that this residue contributes to substrate binding within the nuclease domain channel. Furthermore, this mutant protein is more active, more distributive as substrates get shorter and stalls more frequently than wild-type RNase R when encountering double-stranded RNA, all properties consistent with RNase II. We discuss the role that RNA binding may play in the degradation of structured RNA by RNase R and show that the ability of RNase R to degrade through structured RNA is strongly dependent on temperature, suggesting that thermal breathing of the RNA may play an important role in the mechanism. From this and other available information, we propose a model to explain the ability of RNase R to degrade structured RNAs.
Results
Identification of Residues Within the Nuclease Domain Channel of RNase R That Determine the Catalytic Properties of RNase R
The nuclease domain of RNase R is sufficient for RNase R to degrade through double-stranded RNA16. Based upon the crystal structures of the homologous E. coli RNase II14, 15 and Saccharomyces cerevisiae Rrp4417 proteins, this domain contains a narrow channel which can accommodate 5 single-stranded nucleotides. The catalytic center lies at the base of this channel14, 15, 17. It seemed most likely that the residues responsible for the ability of RNase R to degrade through duplex RNA would be those in the nuclease domain that interacted with the RNA substrate. Such residues could be located within the nuclease domain channel itself, where they could directly contribute to tighter RNA-binding, or they could be at the entrance to the channel, where they could be actively involved in separation of the RNA strands.
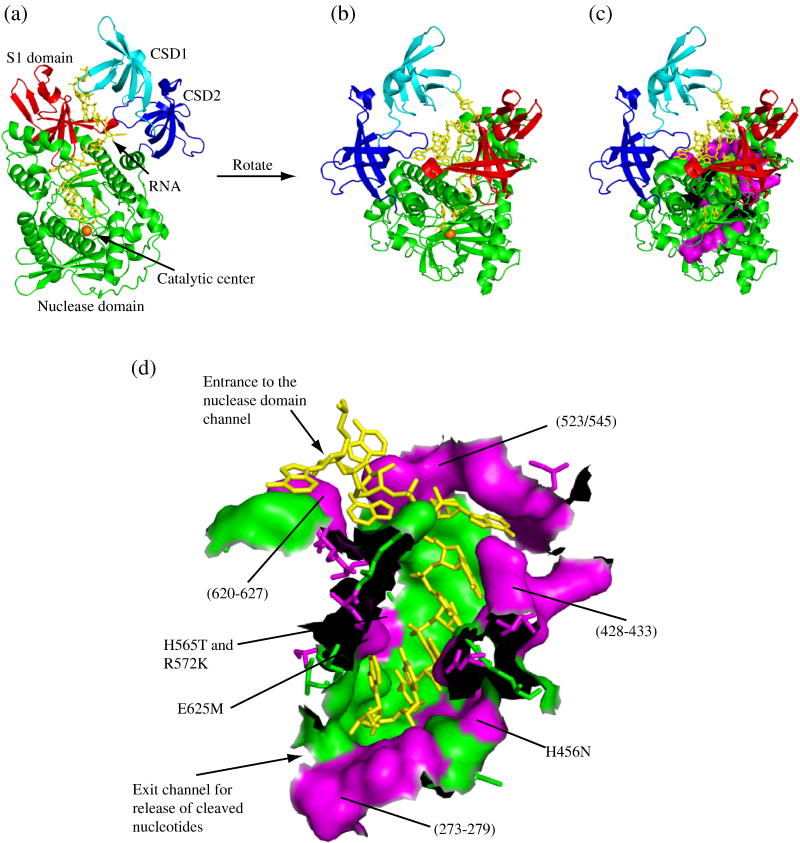
For the purpose of this study, we defined the residues of the nuclease domain channel as all of those from the nuclease domain that lie within 5 Å of the RNA substrate. We then identified such residues in RNase II from the crystal structure of a catalytic site mutant in complex with single-stranded RNA14 using PyMOL (DeLano, W. L. The PyMOL Molecular Graphics System (2002). DeLano Scientific, San Carlos, CA. CA.[Online.] http://www.pymol.org). The equivalent residues were then identified in RNase R based upon sequence alignment of the two proteins using Clustal X18. Of the 45 residues which met this criteria, 21 were unchanged in RNase R compared to RNase II. The majority of these invariant residues corresponded to those that contact the single-stranded RNA in the RNase II crystal structure14, suggesting that similar contacts are made to the RNA by RNase R. Since the nuclease domain of RNase R is able to degrade structured RNA, while that from RNase II stalls at a duplex16, we reasoned that it was likely to be one or more of the variant amino acids that was responsible for the ability of RNase R to digest through structured RNA. Thus, we decided to focus on these 24 variant residues. Fig. 1 illustrates the distribution of these residues throughout the nuclease domain channel.
Figure 1. Distribution of Variant Residues in the Nuclease Domain Channel of RNase R.
All panels were generated using PyMOL (DeLano, W. L. The PyMOL Molecular Graphics System (2002). DeLano Scientific, San Carlos, CA. CA.[Online.] http://www.pymol.org) based on the crystal structure of RNase II14 (PDB code: 2IX1). (a) A cartoon representation of RNase R clearly showing the path that the RNA takes between the cold-shock and S1 domains into the nuclease domain. The cold-shock domains are colored in cyan and blue for CSD1 and CSD2, respectively, the nuclease domain is green with the catalytic magnesium ion represented by an orange sphere and the S1 domain is red. The RNA substrate is shown as yellow sticks. (b) An alternative perspective of RNase R obtained by rotation of panel (a). Coloring is the same as in panel (a). (c) The same as panel (b) with residues comprising the nuclease domain channel wall shown in a surface representation. Residues that are conserved between RNase R and RNase II remain in green and residues that differ are in magenta. (d) An enlarged surface representation of the residues which form the nuclease domain channel wall. As in panel (c), residues that are conserved between RNase R and RNase II are shown in green and residues that differ are in magenta. (The black areas are where amino acids have been removed to allow visualization of the channel). The approximate positions of the RNase R mutants constructed in this study are indicated. For orientation purposes, the entrance and exit of the channel are labeled.
Construction of RNase R Nuclease Domain Channel Mutants
To investigate the contribution that the variant residues of the RNase R nuclease domain channel make to substrate binding and exoribonuclease activity, particularly the ability to degrade through double-stranded RNA, a series of mutant proteins was constructed. In each of these mutants, one or more variant residues in the nuclease domain channel of RNase R was replaced by the equivalent amino acid(s) found in RNase II. Using this approach, 23 of the 24 variant residues in the nuclease domain channel of RNase R were mutated. To simplify construction of the mutants, some residues that are not considered to be part of the nuclease domain channel, as defined above, also were mutated in certain cases. A summary of the mutants, detailing which amino acids were changed in each case, is given in Table 1. Although the nuclease domain alone is sufficient for RNase R activity, the level of activity observed with a truncated protein containing this domain alone is extremely low when compared to that of the full-length protein16. Consequently, the channel mutations were made in the context of full-length RNase R.
Table 1. RNase R Nuclease Domain Channel Mutants.
The residue numbers for the mutated residues correspond to the amino acid position in RNase R. Mutations in italics are residues that are not considered part of the nuclease domain channel based on the criteria discussed under “Results”. Nucleotide positions are numbered from the 3′ end of the RNA chain.
| RNase R Mutant | Mutated Residues | Location of the Mutation in the Nuclease Domain Channel |
|---|---|---|
| (273-279) | G273S, E274A, D275S, A276T, R277E, F279M | Base of the nuclease domain channel,near the catalytic center (by the phosphodiester bond between nucleotides 1 and 2) and the exit channel |
| (428-433) | S428F, F429K, E430D, S431R, E432P, E433D | Entrance to the nuclease domain channel, near the base moieties of nucleotides 5 and 6 |
| H456N | H456N | Within the nuclease domain channel, nearest the base moiety of nucleotide 3 |
| (523/545) | L523R, Q536D, L540R, M543Q, K544S, Q545F | Entrance to the nuclease domain channel, near nucleotides 6 and 7 |
| H565T | H565T | Within the nuclease domain channel, near the phosphodiester bond between nucleotides 3 and 4 |
| R572K | R572K | Within the nuclease domain channel, near the phosphodiester bonds between nucleotides 2 and 3 and nucleotides 3 and 4 |
| (620-627) | E620R, R622L, A623N, D624R, E625M, T627E | Entrance to the nuclease domain channel, near nucleotides 5 to 7. E625 also appears to contact the base moiety of nucleotide 1. |
Screening of RNase R Nuclease Domain Channel Mutants
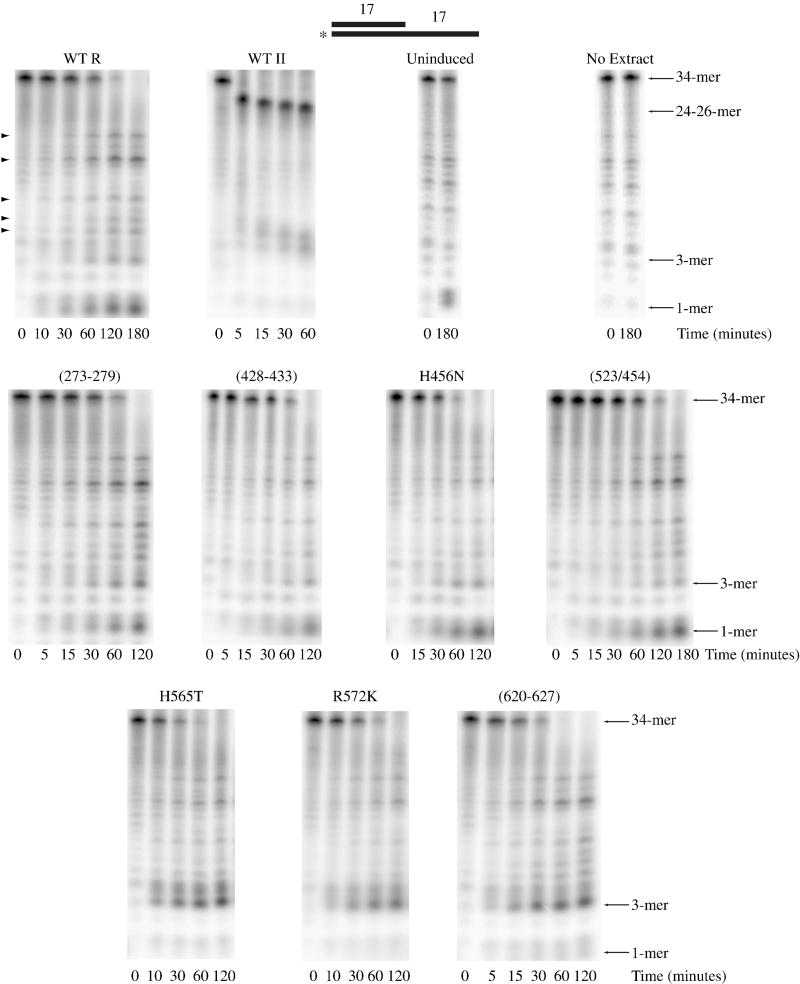
Wild-type RNase R, wild-type RNase II and each of the RNase R nuclease domain channel mutants listed in Table 1 were expressed in an E. coli strain lacking both RNase R and RNase II. Extracts containing the expressed proteins were prepared as described under “Materials and Methods”, and screened for activity using an electrophoretic assay with a substrate consisting of a 5′-[32P]-labeled 34-mer annealed to a complementary 17-mer so as to generate a 17-base-pair duplex with a 17-nucleotide poly(A) 3′ overhang (ds17-A17).
An extract expressing wild-type RNase R completely degraded the ds17-A17 substrate (Fig. 2). However, in contrast to purified RNase R, which degrades this substrate to limit products of di- and tri-nucleotides3, 12, some exoribonuclease activity in the wild-type extract led to further degradation such that the labeled 5′ residue was released as a mononucleotide. Most likely, RNase R degrades to the usual di- and tri-nucleotides, but oligoribonuclease, present in the extract, extends the degradation to mononucleotides. It should also be noted that bands not typically observed in activity assays carried out with purified RNase R (see arrowheads in Fig. 2) are observed when using extracts, and presumably originate from the activity of enzymes other than RNase R. This could also account for the low level of degradation that occurs with an extract that is not induced with IPTG and therefore does not express either RNase R or RNase II (see Fig. 2).
Figure 2. Activity of Nuclease Domain Channel Mutant Extracts on Structured RNA.
Assays were carried out as described under “Materials and Methods” with 10 μM ds17-A17 substrate and 2.5 μg wild-type RNase R, 0.5 μg wild-type RNase II, 2.5 μg uninduced, 2.5 μg RNase R (273-279), 2.5 μg RNase R (428-433), 5 μg RNase R H456N, 2.5 μg RNase R (523/545), 10 μg RNase R H565T, 2.5 μg RNase R R572K and 2.5 μg RNase R (620-627) cell extract. Aliquots were taken at the indicated times and analyzed by denaturing PAGE. A schematic representation of the substrate is shown at the top of the figure with the position of the 32P label denoted by an asterisk. Non-specific product bands that do not appear to originate from RNase R activity are indicated on the left of the wild-type RNase R panel by arrowheads.
Similar to what is seen with purified RNase II, an extract from cells expressing RNase II is unable to degrade through the double-stranded region of ds17-A17, and stalls 7 to 9 nucleotides before the duplex region of this substrate (Fig. 2). This observation implies that any RNase R mutant which is significantly impaired in its ability to degrade double-stranded RNA, such that it behaves like RNase II, should be detectable within the context of an extract, allowing screening of the mutant proteins without having to purify each one.
However, as shown in Fig. 2, each of the extracts expressing an RNase R mutant protein was still able to degrade through the double-stranded region of ds17-A17 indicating that none of the variant nuclease domain channel residues, by itself, is responsible for the ability of RNase R to degrade structured RNAs. We did note that several mutant extracts (H565T, R572K and (620-627)) generated longer limit products compared to the wild-type extract suggesting that the nuclease domains of these RNase R mutant proteins may bind more weakly to RNA. In fact, RNase II, which also releases longer limit products than RNase R, binds a tetranucleotide RNA molecule less tightly within its nuclease domain as determined by a filter-binding assay16.
In the crystal structure of RNase II, the carboxyl group of E542 appears to be positioned to interact with the 3′ terminal nucleotide where it can disrupt base-stacking, and assist in elimination of the cleaved nucleotide14. Disruption of this interaction would be expected to weaken substrate binding at the base of the nuclease domain channel. The sequence surrounding this amino acid is poorly conserved throughout the RNR family, and we initially identified the corresponding residue in RNase R as T627. However, it is more likely, given its proposed function, that this residue would be conserved between RNase II and RNase R, and that the corresponding residue in RNase R is actually E625. In the RNase R (620-627) mutant, E625 is changed to methionine, and it is possible that it is the mutation of a conserved amino acid that leads to the disruption of a conserved binding interaction and the longer limit products observed with the (620-627) mutant extract. Substitutions of amino acids typically conserved between RNase R and RNase II have been shown to affect the length of the limit products generated by RNase II19. Since E625 does not appear to account for the differences between RNase R and RNase II, the (620-627) mutant was not examined further.
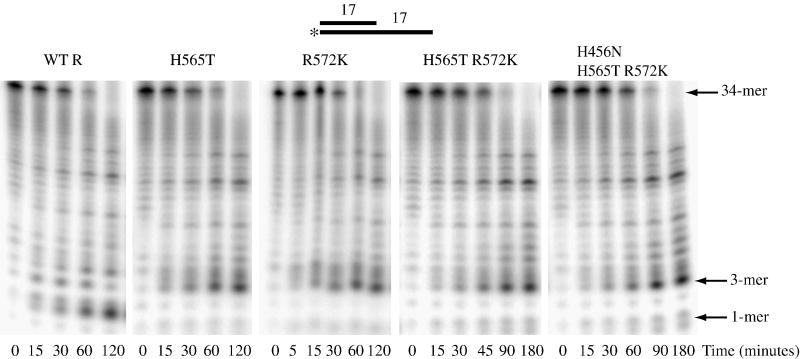
In an attempt to obtain additional information on nuclease domain residues that might be important for RNase R activity on double-stranded RNA, the two other mutations that lead to the release of longer limit products, H565T and R572K, were combined to generate a RNase R H565T R572K double-mutant. A triple-mutant was also constructed that combined the H565T and R572K mutations with H456N. Although an extract with H456N had similar activity to the wild-type extract (Fig. 2), the position of H456 in the nuclease domain channel suggested that further examination was warranted. H456 is near the base moiety of nucleotide 3, whereas H565 and R572 are near the phosphodiester bonds connecting either nucleotides 3 and 4 or connecting both nucleotides 2 and 3 and 3 and 4, respectively.
Cell extracts were prepared from strains expressing either the double- or triple-mutant, and their activity on ds17-A17 was compared to wild-type RNase R and to the single H565T and R572K mutants. As shown in Fig. 3, the double- and triple-mutant extracts also were able to degrade through the double-stranded region of the ds17-A17 substrate. As was found with the single mutants, these extracts also generated longer limit products compared to wild-type RNase R extracts. These data indicate that the effect of the single H565T or R572K mutations is not accentuated by the presence of additional mutations with similar properties. The most likely explanation for this is that mutating either of the neighboring H565 or R572 residues is sufficient to disrupt this region of the channel and eliminate protein-RNA contacts made by one or both of these amino acids.
Figure 3. Activity of RNase R H565T R572K and RNase R H456N H565T R572K on Structured RNA.
Assays were carried out as described under “Materials and Methods” with 10 μM ds17-A17 substrate and 2.5 μg wild-type RNase R, 10 μg H565T, 2.5 μg R572K, 2.5 μg H565T R572K and 5 μg H456N H565T R572K cell extract. Aliquots were taken at the indicated times and analyzed by denaturing PAGE. A schematic representation of the substrate is shown at the top of the figure with the position of the 32P label denoted by an asterisk.
Characterization of Purified RNase R R572K and RNase R H456N H565T R572K Mutant Proteins
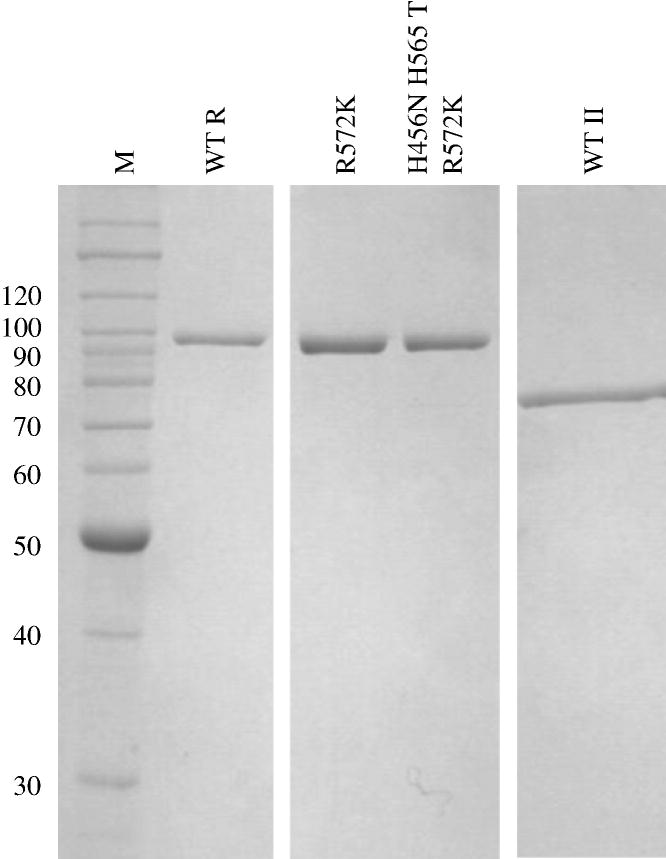
As discussed above, assays carried out with cell extracts generate a number of products that do not appear to originate from RNase R activity. Since these non-specific bands may mask subtle changes in the activity of the RNase R mutants relative to the wild-type enzyme, it seemed worthwhile to purify one single-mutant, R572K, and the triple-mutant, H456N H565T R572K, to more carefully characterize and compare their activities. The purity of these mutant proteins, as well as wild-type RNase R and RNase II, is shown in Fig. 4.
Figure 4. Purity of RNase R Mutants.
One μg of purified wild-type RNase R, RNase R R572K, RNase R H456N H565T R572K and RNase II were resolved by 10% SDS-PAGE and visualized by Coomassie blue staining. The molecular masses, in kDa, of protein standards (M) are indicated on the left. Pure RNase II was obtained from Dr. A. Malhotra (University of Miami, Miami, Florida).
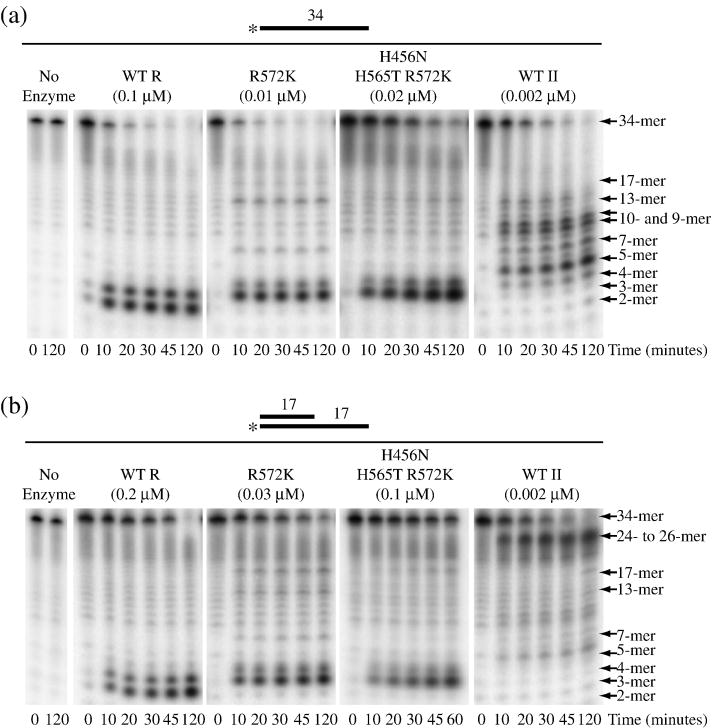
The activities of purified RNase R R572K and H456N H565T R572K were tested with ds17-A17 and the corresponding single-stranded 34-mer, ss17-A17, as substrates. Wild-type RNase R and RNase II were also employed for comparison. As shown in Fig. 5A, all four proteins readily degrade the single-stranded 34-mer. As expected, wild-type RNase R released di- and tri-nucleotides as limit products, whereas RNase II released predominantly penta-nucleotides as the limit of digestion. Confirming the data obtained with cell extracts, purified R572K and H456N H565T R572K released tri- and tetra-nucleotides. This is consistent with the RNase R mutants binding short oligoribonucleotides more weakly than wild-type RNase R, but more tightly than RNase II. We will present evidence below that this is the case.
Figure 5. Activity of Purified RNase R R572K and RNase R H456N H565T R572K.
Activity assays were carried out as described under “Materials and Methods”. Aliquots were taken at the indicated times and analyzed by denaturing PAGE. (a) ss17-A17 was present at 10 μM with 0.1 μM wild-type RNase R, 0.01 μM R572K, 0.02 μM H456N H565T R572K and 0.002 μM RNase II. (b) ds17-A17 was present at 10 μM with 0.2 μM wild-type RNase R, 0.03 μM RNase R R572K, 0.1 μM RNase R H456N H565T R572K and 0.002 μM RNase II. A schematic representation of the substrates is shown above each panel with the position of the 32P label indicated by an asterisk.
Degradation of ss17-A17 by wild-type RNase R is highly processive, since only the original substrate and the di- and tri-nucleotide limit products could be detected (Fig. 5A). This contrasts with RNase II which becomes distributive as the substrate shortens; in addition to the tetra- and penta-nucleotide limit products, bands can also be detected which correspond to 7-, 9-, 10- and 13-mers. Interestingly, RNase R R572K also appears to stall generating 7-, 13- and 17-mers. Similar stalling was observed with the triple-mutant, but the effect appeared to be less pronounced. These data indicate that the R572K mutant behaves more like RNase II and that this amino acid therefore plays an important role in defining the catalytic properties of RNase R and RNase II.
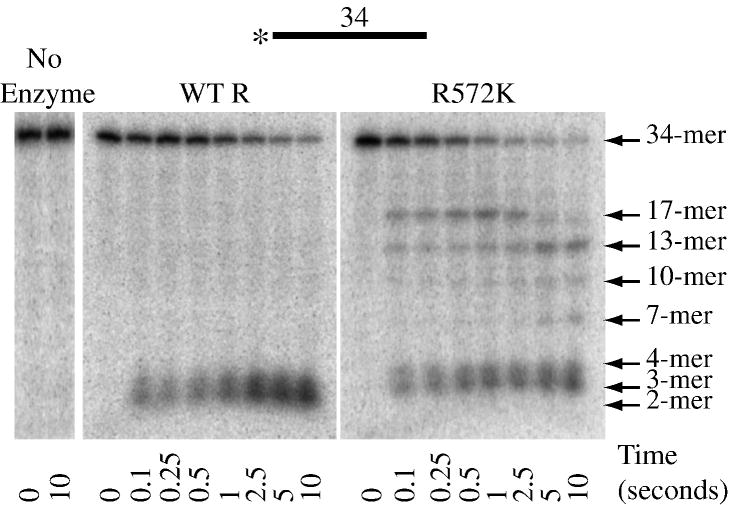
To further investigate the activity of RNase R and RNase R R572K on ss17-A17, the enzymes were compared under single-turnover conditions. This was achieved by carrying out the assays at concentrations of substrate and enzyme above the determined Kd (see Table 3 below) and with the enzyme present at 5-fold excess over substrate20. Reactions were initiated by mixing RNase R with 5′-[32P]-labeled ss17-A17 substrate and quenching the reaction in the millisecond to second timescale using a rapid chemical-quench flow apparatus as described under “Materials and Methods”. Reactions were analyzed by denaturing PAGE as described for the steady-state assays. Such experiments allow clearer visualization of reaction intermediates as degradation of each substrate molecule is initiated at the same time. As shown in Fig. 6, consistent with the steady-state data, for wild-type RNase R, only the initial substrate or the limit products could be detected, even at the shortest time point (0.1 s). This suggests that nucleotide cleavage is fast relative to binding and initiation of degradation. If we define the on-rate as including all possible mechanistic steps up to and including the cleavage of the first phosphodiester bond, this implies that the on-rate is rate-limiting for the reaction (see below). With R572K, again consistent with the steady-state data presented in Fig. 5A, multiple intermediate bands also were detected that corresponded to 7-, 10-, 13- and 17-mers (Fig. 6). However, in the single-turnover experiment the substrate-product relationships are clearer in that the 17-mer intermediate accumulates as the initial substrate disappears and the 13-mer is detected as the 17-mer disappears, and so on. Thus, these bands represent positions at which the enzyme pauses, rather than dissociates from the substrate. Moreover, based on the sequence of the substrate, these pauses occur as the enzyme encounters multiple pyrimidine residues.
Table 3. RNA Binding to RNase R R572K.
The Kd was determined by a filter binding assay as described under “Materials and Methods”. Each value represents the mean of at least two experiments.
| Substrate | Kd (nM) |
|
|---|---|---|
| Wild-Type RNase R | RNase R R572K | |
| A17 | 0.8 ± 0.1 | 0.5 ± 0.1 |
| A4 | 1,200 ± 90 | 5,800 ± 1,500 |
Figure 6. Activity of Purified Wild-Type RNase R and RNase R R572K Under Single-Turnover Conditions.
Single-turnover experiments were carried out as described under “Materials and Methods”. Reactions were allowed to proceed for the indicated times and analyzed by denaturing PAGE. A schematic representation of the substrate is shown at the top of the figure with the position of the 32P label indicated by an asterisk.
On the ds17-A17 substrate, under steady-state conditions, wild-type RNase R, R572K and H456N H565T R572K all were able to degrade to completion, whereas RNase II stalled well before the duplex (Fig. 5B), as was found with cell extracts. Again, the limit products were tri- and tetra-nucleotides for the RNase R mutants compared to di- and tri-nucleotides for wild-type RNase R. The 7-, 13- and 17-mer products detected with the ss17-A17 substrate also were observed with ds17-A17. However, the band at the position of the 17-mer was more pronounced when ds17-A17 was used as the substrate (compare Figs. 5A and 5B). Since this position represents the single-strand/double-strand junction of the ds17-A17 substrate, the data suggest that both R572K and H456N H565T R572K may have more difficulty in initiating degradation of the double-stranded region than does the wild-type protein.
The rates at which ss17-A17 and ds17-A17 were degraded by wild-type RNase R, R572K and H456N H565T R572K are shown in Table 2. The single- and triple-mutant proteins degrade both of these substrates at a considerably faster rate than wild-type RNase R. Although this may at first appear to contradict the data presented in Fig. 5 where the mutants appear to degrade the substrates to the same extent as wild-type, the quantity of enzyme used in the assays shown was actually much lower than for wild-type RNase R. RNase II typically degrades unstructured substrates at a faster rate than RNase R12, suggesting that R572 is, at the least, an important determinant of the catalytic properties specific to RNase R.
Table 2. Comparison of the Activities of Wild-type RNase R, RNase R R572K and RNase R H456N H565T R572K.
Activity assays were performed as described under “Materials and Methods”. Each value represents the mean of at least three experiments.
| Substrate | Activity (nmol min−1 nmol−1 RNase R) |
||
|---|---|---|---|
| Wild-Type | RNase R | RNase R | |
| RNase R | R572K | H456N H565T R572K | |
| ss17-A17 | 5.3 ± 1.6 | 75 ± 24 | 27 ± 12 |
| ds17-A17 | 0.5 ± 0.2 | 5.1 ± 2.1 | 1.5 ± 0.4 |
| A17 | 40 ± 4.8 | 41 ± 7.0 | 33 ± 8.5 |
| C17 | 22 ± 13 | 130 ± 32 | 82 ± 28 |
| U17 | 42 ± 14 | 170 ± 65 | 87 ± 28 |
| ss17 | 29 ± 13 | 700 ± 99 | 200 ± 29 |
ss17-A17 is degraded ~15-fold faster by the single-mutant and ~5-fold faster by the triple-mutant, while ds17-A17 is degraded ~10-fold and ~3-fold more rapidly by the single- and triple-mutant, respectively. Thus, although the mutants are more active than wild-type on both substrates, the relative increase was smaller on the ds17-A17 substrate. This suggests that the mutants have more difficulty degrading double-stranded RNA relative to single-stranded RNA than does the wild-type.
In an attempt to understand the reason for the overall faster rates of degradation by the RNase R mutants, activity also was measured on a variety of other substrates. These included A17, C17, U17 and ss17, which is the 5′ 17 nucleotides of the ss17-A17 and ds17-A17 substrates. As shown in Table 2, wild-type RNase R displayed activities within the same range for all four of these substrates, suggesting that it has little sequence specificity. It was reported previously that RNase R can show some sequence specificity when degrading homopolymers with an order of poly(A) > poly(U) > poly(C)12. However, those assays were carried out at 100 mM KCl rather than the 300 mM KCl used here, which may be responsible for the difference. In fact, RNase II also displays sequence specificity which is lost at elevated salt concentrations13, 21. Both RNase R R572K and RNase R H456N H565T R572K degraded A17 at a rate similar to that of wild-type RNase R (Table 2), whereas C17, U17 and ss17 were degraded at rates between 2- and 25-fold higher by the mutant enzymes (Table 2). Thus, mutation of R572 within the active site channel of RNase R can dramatically increase reaction rates, but this is strongly dependent on the nucleotide composition of the substrate.
Moreover, the experiments carried out under single-turnover conditions indicated that there is a significant increase in the on-rate (defined as the rate including all mechanistic steps up to and including the cleavage of the first phosphodiester bond) for RNA-binding by RNase R R572K compared to the wild-type enzyme. By measuring the rate of disappearance of the substrate, the on-rates were found to be 0.38 ± 0.1 s−1 for wild-type RNase R and 4.6 ± 0.3 s−1 for RNase R R572K with the ss17-A17 substrate at 15°C. This large difference in on-rates may explain the faster degradation rates observed for the R572K mutant compared to the wild-type enzyme under steady-state conditions which involve multiple binding/initiation events over the course of an assay.
The finding that the R572K and H456N H565T R572K mutants stall more frequently and produce longer limit products than the wild-type protein suggests that they may bind RNA more weakly. To test this directly, the Kd for binding of A17 and A4 was measured for wild-type RNase R and R572K. As shown in Table 3, both proteins bound A17 with a similar Kd. However, the Kd value for A4, which should be bound entirely within the nuclease domain channel, was ~5-fold higher for the R572K mutant. This result indicates that R572 does contribute to substrate binding within the channel. Therefore, the mutation of R572 to lysine brings the catalytic properties of RNase R closer to those of RNase II which binds to A17 with a Kd ~7-fold greater than RNase R and is unable to bind to A4 in a filter-binding assay16.
Effect of Temperature on the Degradation of Structured RNA
One possibility for how RNase R degrades through structured RNA is that it exploits the transient opening of the RNA duplex that occurs during thermal breathing. For many enzyme-catalyzed reactions the temperature coefficient, or Q10, (the factor by which the rate of a reaction increases for every 10 degree rise in temperature) is approximately two. However, for biological processes requiring large conformational changes, such as thermal breathing of the RNA substrate, Q10 is often greater than two. Consequently, it is possible to determine whether other factors may contribute to the mechanism of a reaction by investigating the change in reaction rate with increasing temperature.
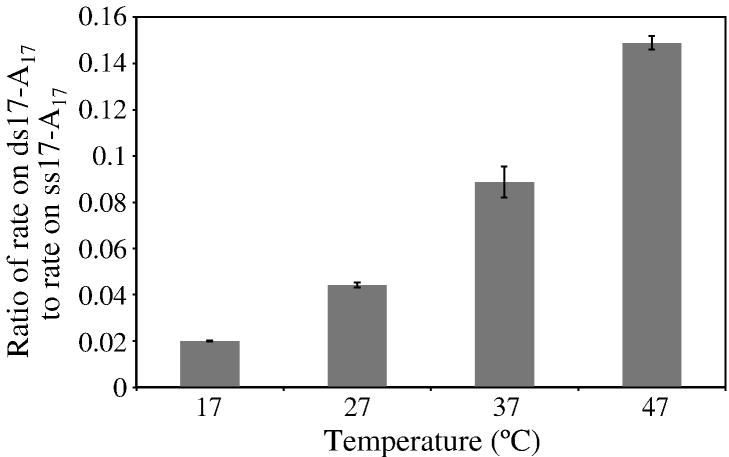
To determine whether thermal breathing might contribute to the ability of RNase R to degrade structured RNA, rates of degradation of ss17-A17 and ds17-A17 were measured at 17°, 27°, 37° and 47°C. If thermal breathing were involved, it would be expected that the rate on a structured substrate would be affected more than the rate on a single-stranded substrate by increasing the temperature of the reaction. This was found to be the case. For the single-stranded ss17-A17 substrate the Q10 was determined to be 1.8, consistent with the increase in temperature leading to an increase in the rate of the reaction chemistry. In contrast, the Q10 for the structured ds17-A17 substrate was found to be 3.6. The importance of this step to the mechanism is even more apparent when we compare the ratio of the rate on the structured substrate to that on the single-stranded substrate, thereby eliminating Q10 effects on reaction chemistry. As shown in Fig. 7, this ratio dramatically increases with increasing temperature from 0.02 at 17°C to 0.15 at 47°C, indicating that there is an additional step, specific to the structured RNA, that contributes to the overall reaction. This step alone also has a Q10 of ~2.
Figure 7. Effect of Temperature on the Ability of RNase R to Degrade Structured RNA.
Assays were carried out as described under “Materials and Methods” except they were performed at the indicated temperatures. ss17-A17 was present at 10 μM with 0.07 μM wild-type RNase R or ds17-A17 was present at 10 μM with 0.2 μM RNase R. Aliquots were taken during an appropriate time-course to measure linear degradation of the substrate and analyzed by denaturing PAGE. Results are plotted as a ratio of the rate on double-stranded RNA to the rate on single-stranded RNA. The values represent the mean of two experiments and the standard deviation is indicated.
Overall, these data are most consistent with a temperature-dependent conformational change, most likely thermal breathing of the RNA duplex, playing an important role in the degradation of double-stranded RNA by RNase R.
Discussion
Although both RNase R and RNase II belong to the RNR family of exoribonucleases6, 7, there are significant differences in their catalytic properties. RNase II is limited to single-stranded regions3, 8–10 while RNase R readily degrades through double-stranded RNA when provided with a single-stranded overhang to bind and initiate degradation3, 11. We have recently shown that the nuclease domain alone of RNase R is sufficient to perform this function16. Furthermore, RNase R binds RNA more tightly within its nuclease domain than RNase II16. This could explain the slower rates of degradation of single-stranded RNA relative to RNase II and the maintenance of processivity even on very short oligoribonucleotides resulting in the release of shorter limit products than RNase II.
The available crystal structures for RNase II provide an explanation for the single-strand specificity of this enzyme. In these structures, the path an RNA substrate must take between the cold-shock domains (CSDs) and S1 domain and into the nuclease domain is only wide enough to accommodate single-stranded RNA14, 15. Thus, although the RNA-binding domains constitute the initial barrier to double-stranded RNA8, 11, 16, the nuclease domain itself also is unable to degrade duplex RNA8, 16.
RNase R requires a 3′ single-stranded overhang at least 5 nucleotides long3, 11, 12 (H. A. Vincent and M. P. Deutscher, unpublished observation) in order to degrade through a double-strand. Likewise, the yeast homologue of RNase R, Rrp44, also requires a 3′ overhang to degrade structured RNA17, 22. Based upon the crystal structure of this latter enzyme, its nuclease domain channel also is too narrow to allow entry of an RNA duplex17. This explains the requirement for a 3′ single-stranded overhang on RNase R/Rrp44 substrates. Recent studies with Rrp44 have revealed that in addition to acting as an exoribonuclease it also contains an endonuclease activity within its N-terminal PIN domain23–25. This activity may allow the generation of RNA fragments with the required single-stranded overhang for exoribonucleolytic degradation23–25. Furthermore, in contrast to RNase II, the nuclease domain alone of RNase R is able to degrade through double-stranded RNA16, suggesting that the nuclease domain of RNase R must unwind the duplex prior to its entrance into the channel in order to degrade the RNA.
In this study, we identified 24 residues within the nuclease domain channel of RNase R and RNase II that vary between the two enzymes, and are therefore, likely candidates for defining the catalytic properties of these enzymes and contributing to the ability of RNase R to degrade through double-stranded RNA. The majority of these variant residues were found around the entrance to the nuclease domain channel where they could assist in duplex unwinding by intercalating between the bases of a double-strand, a strategy employed by cold-shock proteins26–28, or form a structural wedge which separates the strands as the RNA is pulled across it during translocation, analogous to the “pin” region of RecBCD29. However, none of the proteins that were mutated in this region impaired the ability of RNase R to degrade through double-stranded RNA. Nevertheless, we cannot rule out the possibility that residues conserved between RNase R and RNase II adopt alternative conformations in the two proteins. In fact, three arginine residues which are conserved in RNase R, RNase II and Rrpp44, are in a different position in the Rrp44 crystal structure compared with RNase II such that they bind to the RNA substrate in Rrp44, but do not appear to do so in RNase II17.
We did identify at least one residue within the nuclease domain channel of RNase R that affected RNA binding and the ability of RNase R to initiate the degradation of structured substrates. Mutation of R572 or the neighboring H565 resulted in proteins which produced longer limit products, consistent with the fact that they bind to short oligoribonucleotides more weakly. Direct measurement of binding of A4 to purified RNase R R572K confirmed that binding was weaker than to the wild-type protein and, therefore, that the R572K mutation impairs substrate binding within the nuclease domain channel. The purified R572K mutant also stalled more frequently than wild-type RNase R, including when approaching an RNA duplex. These data suggest that the ability to degrade through double-stranded RNA is related to the tight binding of the substrate in the nuclease domain channel of RNase R.
A mutational analysis of residues within the nuclease domain of RNase II found that a number of residues at the catalytic center of this enzyme affect both the length of the limit products produced and the affinity of the enzyme for RNA19. In this study the amino acids that were mutated were those that are conserved between RNase R and RNase II19. Likewise, we found that mutation of the conserved E625 residue in RNase R led to the production of longer limit products.
The nuclease domain of RNase R binds considerably more tightly to RNA than the nuclease domain of RNase II16, and we believe that this property is important for RNase R to degrade structured RNA. As RNase R digests and moves into a double-stranded region, the very tight binding of single-stranded RNA within the channel may favor strand separation and translocation forward rather than dissociation of the substrate and reannealing of the opened base-pairs. This would be analogous to the role of single-stranded binding proteins in DNA recombination and repair30. Such a mechanism would allow RNase R to capitalize on the natural thermal breathing of the RNA duplex to move through the double-stranded region. The dramatic increase in activity on a structured substrate relative to single-stranded RNA as temperature is increased suggests that thermal breathing does play a role in the ability of RNase R to degrade structured RNA.
It is also important to consider what happens to the displaced strand as RNase R degrades through a duplex. Binding and sequestration of this strand could assist in the unwinding process by preventing reannealing of the unwound bases. A region on the exterior of the nuclease domain may be able to perform this function. For example, it is possible that the alternate path between the nuclease domain and the CSDs that was proposed for RNase II15, and was observed in the Rrp44 crystal structure17, actually serves as an exit channel for the displaced strand of a duplex.
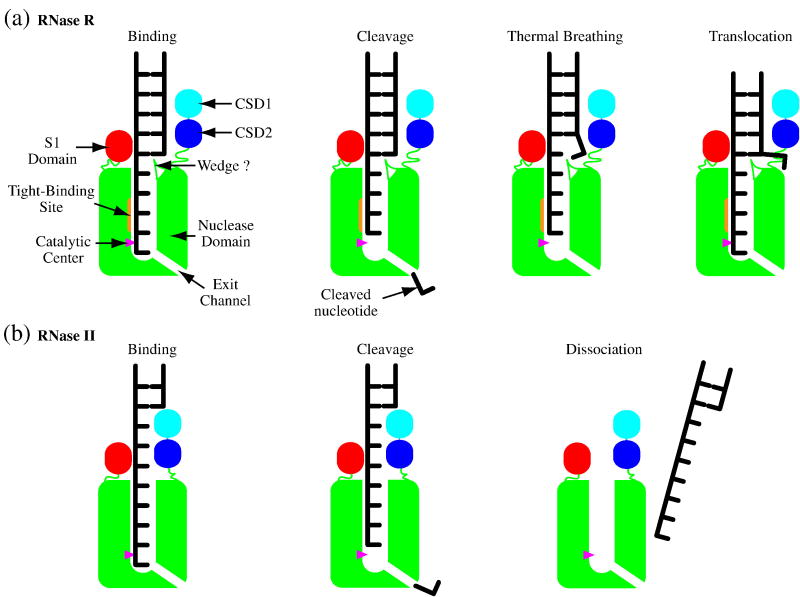
Based on the data currently available, we propose the following model to explain the ability of RNase R to degrade structured RNAs (Fig. 8A). RNase R initiates degradation of a structured substrate with a minimal 3′ overhang of 5 nucleotides by binding the single-stranded region within its nuclease domain channel. Following cleavage and release of the 3′ terminal nucleotide, translocation along the RNA substrate would be blocked by the RNA duplex. However, tight binding within the channel allows RNase R to remain bound at the single-strand/double-strand junction long enough to take advantage of thermal breathing events and thereby advance into the duplex. This process may be enhanced by residues at the entrance to the nuclease domain channel that maintain the separation of the opened base-pairs. Translocation of RNase R along the RNA during this transient opening of the terminal base-pair of the duplex would be favored because it restores the binding interactions between the 3′ terminal nucleotide and the catalytic center that were lost by the previous cleavage event. In contrast, RNase II (Fig. 8B), which binds RNA more weakly within its channel, would dissociate from the RNA too rapidly to allow such a mechanism to operate. In addition, the closer proximity of the CSDs and the S1 domain in RNase II prevents the double-stranded region from even approaching the channel.
Figure 8. A Model for the Mechanism of Degradation of Double-Stranded RNA by RNase R.
(a) RNase R. (b) RNase II. The RNA-binding domains are shown in cyan, blue and red for the CSD1s, CSD2s and S1 domains, respectively. The nuclease domains are in green with the catalytic centers shown as magenta triangles. In the binding step, RNase R or RNase II bind to the 3′ single-stranded overhang of a structured RNA molecule. The 3′ terminal nucleotide is then cleaved and released. Translocation of both enzymes is blocked by the double-stranded region of the RNA. RNase R remains bound to the RNA substrate as the end of the duplex is opened by thermal breathing and/or a wedge at the entrance to the nuclease domain channel. RNase R translocates forward to position the RNA for the cleavage of the next nucleotide. In contrast, following nucleotide cleavage, the RNA dissociates from RNase II preventing further degradation of the substrate.
This model, together with the supporting data presented here, suggest a mechanism for the degradation of structured RNAs by RNase R. Moreover, they suggest an explanation for RNase R having the ability to degrade through double-stranded RNA while the closely related protein, RNase II, stalls as it approaches a duplex. We anticipate that future studies will further clarify the differences between these two related enzymes, and lead to an even more detailed description of RNase R catalysis.
Materials and Methods
Materials
Mutagenic primers were synthesized and purified by Sigma Genosys. KOD Hot Start DNA Polymerase was from Novagen. DpnI and T4 polynucleotide kinase were purchased from New England Biolabs, Inc. pETRNB15 and purified RNase II15 were obtained from Dr. A. Malhotra (University of Miami, Miami, Florida). Protein assay dye reagent concentrate for Bradford assays was obtained from Bio-Rad Laboratories. Bovine serum albumin was purchased from EM Science. RNA oligoribonucleotide substrates were synthesized and purified by Dharmacon Inc. [γ-32P]ATP was from PerkinElmer Life and Analytical Sciences. SequaGel solutions for preparation of denaturing urea-polyacrylamide gels were purchased from National Diagnostics. Nitrocellulose and Biodyne Plus nylon membranes were from Pall Corp. All other chemicals were reagent grade.
Cloning of RNase R Mutant Constructs
pET44R(273-279), pET44R(428-433), pET44R(H456N), pET44R(H565T), and pET44R(R572K) were constructed by standard site-directed mutagenesis of pET44R11 using the corresponding primer pairs listed in Table 4. The primers required to create pET44R(523/545), pET44R(H565T R572K) and pET44R(620-627) were generated by annealing and extending the respective oligodeoxyribonucleotide pairs listed in Table 4. The extended primers were then used to mutate pET44R11 using standard site-directed mutagenesis protocols. pET44R(H456N H565T R572K) was constructed through site-directed mutagenesis of pET44R(H565T R572K) using the H456N primers (Table 4).
Table 4. Site-Directed Mutagenesis Primers.
Nucleotides corresponding to the mutations are in bold. Complementary nucleotides between the forward and reverse primers are in italics. F = forward primer and R = reverse primer.
| Mutant Primer | Primer Sequence |
|---|---|
| (273-279) | F: 5′ cgc gat tta ccg ctg gtc acc att gat agt gcc agc aca gaa gat atg gac gat gca gtt tac tgc gag aaa aaa cgc 3′ |
| R: 5′ gcg ttt ttt ctc gca gta aac tgc atc gtc cat atc ttc tgt gct ggc act atc aat ggt gac cag cgg taa atc gcg 3′ | |
| (428-433) | F: 5′ c cgt gaa gaa cgc ggt ggg atc ttt aaa gat cgc ccg gat gcg aag ttc att ttc aac gct gaa cgc cg 3′ |
| R: 5′ cg gcg ttc agc gtt gaa aat gaa ctt cgc atc cgg gcg atc ttt aaa gat ccc acc gcg ttc ttc acg g 3′ | |
| H456N | F: 5′ gaa cag acc cag cgt aac gac gcg aac aaa tta att gaa gag tgc atg att ctg gcg aat atc tcg gc 3′ |
| R: 5′ gc cga gat att cgc cag aat cat gca ctc ttc aat taa ttt gtt cgc gtc gtt acg ctg ggt ctg ttc 3′ | |
| (523/545) | F: 5′ g cgt gac tac gcg gag ctg cgc gag tcg gtt gcc gat cgt cct gat gca gaa atg 3′ |
| R: 5′ cc acg gtt ttc ttg atc gta aat cgc aaa gct ctg cga gcg gcg cag cat ggt atc cag cat ttc tgc atc agg acg atc g 3′ | |
| H565T | F: 5′ gc ctg gca ttg cag tcc tat gcg acc ttt act tcg ccg att cgt cgt tat cca gac3′ |
| R: 5′ gtc tgg ata acg acg aat cgg cga agt aaa ggt cgc ata gga ctg caa tgc cag gc 3′ | |
| R572K | F: 5′ gcg cac ttt act tcg ccg att cgt aaa tat cca gac ctg acg ctg cac cgc 3′ |
| R: 5′ gcg gtg cag cgt cag gtc tgg ata ttt acg aat cgg cga agt aaa gtg cgc 3′ | |
| H565T R572K | F: 5′ gc ctg gca ttg cag tcc tat gcg acc ttt act tcg ccg att cgt 3′ |
| R: 3′ cg gtg cag cgt cag gtc tgg ata ttt acg aat cgg cga agt aaa ggt cg 3′ | |
| (620-627) | F: 5′ ggt cag cac tgt tcg atg gcg cgc cgt ctc aac cgc atg gca g 3′ |
| R: 3′ ctt cag cca gtc agc cac atc gcg ttc tgc cat gcg gtt gag acg g 3′ |
Overexpression of RNase R Mutants for Preparation of Cell Extracts
BL21II−R−(DE3)pLysS harboring pET44R11, pETRNB15, pET44R(273-279), pET44R(428-433), pET44R(H456N), pET44R(523/545), pET44R(H565T), pET44R(R572K), pET44R(620-627), pET44R(H565T R572K) or pET44R(H456N H565T R572K), were grown at 37°C with shaking to an A600 ≈ 0.6 in 50 ml yeast-Tryptone medium supplemented with 100 μg/ml ampicillin, 34 μg/ml chloramphenicol, 25 μg/ml kanamycin and 10 μg/ml tetracycline. Expression was induced by the addition of IPTG to a final concentration of 1 mM. Cells were grown for a further two hours at 37°C. After induction, cells from 5 ml were harvested by centrifugation at 5,000 × g for 10 minutes at 4°C. The resulting cell pellet was stored frozen at −20°C. Control uninduced extracts were prepared by growing BL21II−R− (DE3)pLysS harboring pET44R11 as described above except that no IPTG was added.
Overexpression of RNase R Mutants for Purification
Wild-type RNase R, RNase R R572K and RNase R H456N H565T R572K were overexpressed from BL21II−R−(DE3)pLysS harboring pET44R11, pET44R(R572K) or pET44R(H456N H565T R572K) were grown as described above for the preparation of cell extracts except that they were grown in 500 ml yeast-Tryptone medium supplemented with 100 μg/ml ampicillin, 34 μg/ml chloramphenicol, 25 μg/ml kanamycin and 10 μg/ml tetracycline and the resulting cell pellet was stored frozen at −80°C.
Preparation of Cell Extracts
Cell pellets prepared as described above were thawed on ice and resuspended in 1.5 ml 50 mM Tris-HCl (pH 8.0), 300 mM KCl, 0.5 mM EDTA and 5 mM DTT. Cells were lysed by sonication using two 20-second pulses. The lysate was centrifuged at 16,000 × g for 10 minutes at 4°C and the resulting supernatant was retained as the cell extract. The protein concentration of the cell extracts were determined by Bradford assay 31 using bovine serum albumin as the standard.
Purification of RNase R Mutants
Wild-type RNase R, RNase R R572K and RNase R H456N H565T R572K were purified as previously described for wild-type RNase R11.
Substrate Preparation
The oligoribonucleotide substrates, supplied with the 2′ hydroxyl protected by an acid-labile O-orthoester group to prevent degradation, were deprotected according to the manufacturer’s instructions. The single-stranded oligoribonucleotide substrates used were A17, C17, U17, ss17 (5′ CCCCACCACCAUCACUU 3′) and ss17-A17. These were 5′-labeled with 32P using T4 polynucleotide kinase and [γ-32P]ATP. The ds17-A17 substrate was prepared by heating a mixture containing 5′-labeled ss17-A17 and the non-radioactive complementary oligoribonucleotide (5′ AAGUGAUGGUGGUGGGG 3′) in a 1:1.2 molar ratio, 10 mM Tris-HCl (pH 8.0) and 20 mM KCl in a boiling water bath and then allowing it to cool slowly to room temperature.
Activity Assays
RNase R assays were carried out as described previously16. Briefly, 30 μl reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 300 mM KCl, 0.25 mM MgCl2, 5 mM DTT and the amount of substrate and cell extract or purified enzyme as indicated in the Figure legends, or for calculation of the rates, 10 μM substrate and an amount of enzyme to ensure that less than 25% of the substrate was degraded. Reactions were incubated at 37°C and samples were taken at the indicated times, or at regular intervals for determination of the rates, and analyzed by denaturing 7.5 M urea-20% polyacrylamide gel electrophoresis.
Binding Assays
The binding assays were carried out based on the double-filter nucleic acid-binding assay developed by Wong and Lohman32 as described previously16. Briefly, 20 μl reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 100 mM KCl, 5 mM DTT, 10 mM EDTA, 10% glycerol, 200 pM 32P-labeled substrate and varying amounts of purified enzyme were incubated on ice for 30 minutes. The reaction was applied to a nitrocellulose membrane placed above a nylon membrane in a 96-well dot-blot apparatus (Bio-Rad Laboratories). The fraction of RNA bound at each protein concentration was determined from signalnitrocellulose/(signalnitrocellulose + signalnylon). The Kd was determined by non-linear regression analysis using a one-site binding hyperbola in Prism (GraphPad Software, Inc).
Single-turnover Experiments
Reactions were carried out essentially as described by Johnson20 at 15°C. They were initiated by mixing 15 μl of 200 nM RNase R in 50 mM Tris-HCl, 300 mM KCl, 0.25 mM MgCl2 and 5 mM DTT with 15 μl of 40 nM 5′-[32P]-labeled ss17-A17 substrate, also in 50 mM Tris-HCl, 300 mM KCl, 0.25 mM MgCl2 and 5 mM DTT, using an RQF-3 chemical-quench flow apparatus (KinTek Corporation). The reaction was quenched by the addition of 0.5 M EDTA after a time ranging from 2 ms to 20 s. Reaction products were analyzed by denaturing PAGE.
Acknowledgments
This work was supported by Grant GM16317 from the National Institutes of Health.
We thank Dr. A. Malhotra, Dr. Y. Zuo and Y. Wang for purified RNase II. We thank Dr. T. K. Harris for the use of his FPLC and chemical-quench flow apparatus. We thank Dr. A. Malhotra, Dr. Y. Zuo and Dr. T. K. Harris for helpful discussions and comments.
Abbreviations
- RNase
ribonuclease
- PNPase
polynucleotide phosphorylase
- CSD
cold-shock domain
- Kd
dissociation constant
- EDTA
ethlylenediaminetetraacetic acid
- DTT
dithiothreitol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Donovan WP, Kushner SR. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci USA. 1986;83:120–4. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khemici V, Carpousis AJ. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol Microbiol. 2004;51:777–90. doi: 10.1046/j.1365-2958.2003.03862.x. [DOI] [PubMed] [Google Scholar]
- 3.Cheng ZF, Deutscher MP. An important role for RNase R in mRNA decay. Mol Cell. 2005;17:313–8. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 4.Cheng ZF, Deutscher MP. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc Natl Acad Sci USA. 2003;100:6388–93. doi: 10.1073/pnas.1231041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, Reimers S, Pandit S, Deutscher MP. RNA quality control: degradation of defective transfer RNA. EMBO J. 2002;21:1132–8. doi: 10.1093/emboj/21.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mian IS. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res. 1997;25:3187–95. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–26. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amblar M, Barbas A, Fialho AM, Arraiano CM. Characterization of the functional domains of Escherichia coli RNase II. J Mol Biol. 2006;360:921–33. doi: 10.1016/j.jmb.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 9.Coburn GA, Mackie GA. Overexpression, purification, and properties of Escherichia coli ribonuclease II. J Biol Chem. 1996;271:1048–53. doi: 10.1074/jbc.271.2.1048. [DOI] [PubMed] [Google Scholar]
- 10.Spickler C, Mackie GA. Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J Bacteriol. 2000;182:2422–7. doi: 10.1128/jb.182.9.2422-2427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281:29769–75. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 12.Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. 2002;277:21624–9. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 13.Cannistraro VJ, Kennell D. The processive reaction mechanism of ribonuclease II. J Mol Biol. 1994;243:930–43. doi: 10.1006/jmbi.1994.1693. [DOI] [PubMed] [Google Scholar]
- 14.Frazão C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–4. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- 15.Zuo Y, Vincent HA, Zhang J, Wang Y, Deutscher MP, Malhotra A. Structural basis for processivity and single-strand specificity of RNase II. Mol Cell. 2006;24:149–56. doi: 10.1016/j.molcel.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Vincent HA, Deutscher MP. The roles of individual domains of RNase R in substrate binding and exoribonuclease activity: The nuclease domain is sufficient for digestion of structured RNA. J Biol Chem. 2008;284:486–94. doi: 10.1074/jbc.M806468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell. 2008;29:717–28. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 18.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 19.Barbas A, Matos RG, Amblar M, López-Viñas E, Gomez-Puertas P, Arraiano CM. New insights into the mechanism of RNA degradation by ribonuclease II: identification of the residue responsible for setting the RNase II end product. J Biol Chem. 2008;283:13070–6. doi: 10.1074/jbc.M709989200. [DOI] [PubMed] [Google Scholar]
- 20.Johnson KA. Rapid quench kinetic analysis of polymerases, adenosinetriphosphatases, and enzyme intermediates. Methods Enzymol. 1995;249:38–61. doi: 10.1016/0076-6879(95)49030-2. [DOI] [PubMed] [Google Scholar]
- 21.Singer MF, Tolbert G. Purification and properties of a potassium-activated phosphodiesterase (RNAase II) from Escherichia coli. Biochemistry. 1965;4:1319–30. doi: 10.1021/bi00883a016. [DOI] [PubMed] [Google Scholar]
- 22.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–37. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 23.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–6. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 24.Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkn1020. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phadtare S, Inouye M, Severinov K. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J Biol Chem. 2002;277:7239–45. doi: 10.1074/jbc.M111496200. [DOI] [PubMed] [Google Scholar]
- 27.Phadtare S, Inouye M, Severinov K. The mechanism of nucleic acid melting by a CspA family protein. J Mol Biol. 2004;337:147–55. doi: 10.1016/j.jmb.2004.01.023. [DOI] [PubMed] [Google Scholar]
- 28.Phadtare S, Tyagi S, Inouye M, Severinov K. Three amino acids in Escherichia coli CspE surface-exposed aromatic patch are critical for nucleic acid melting activity leading to transcription antitermination and cold acclimation of cells. J Biol Chem. 2002;277:46706–11. doi: 10.1074/jbc.M208118200. [DOI] [PubMed] [Google Scholar]
- 29.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–93. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 30.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu Rev Biochem. 1994;63:527–70. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 31.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 32.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc Natl Acad Sci USA. 1993;90:5428–32. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]