Abstract
Very few molecules with biological origins contain the element fluorine. Nature’s inability to incorporate fluorine into biomolecules is related to the low concentration of free fluoride in sea and surface water. However, judicious introduction of fluorine into proteins, nucleic acids, lipids and carbohydrates has allowed mechanistic scrutiny of enzyme catalysis, control of protein oligomerization in membranes, clustered display of ligands on surfaces of living cells, and in increasing the protease stability of protein and peptide therapeutics.
Introduction
For the last forty years, materials that incorporate the element fluorine in their molecular structures have had an enormous impact on society. Indeed, Teflon, Gore-Tex, Nafion and myriad other compounds have become household names. Carbon bound fluorine is key to the material properties of these structures [1,2]. However, only recently have scientists explored the use of fluorine to modify biological molecules for the purposes of perturbing, observing, and controlling biological processes.
The fluorous phase, because of its immiscibility with water and many traditional organic solvents, has gained prominence in catalysis [3,4], reaction acceleration [5], molecular self-assembly [6,7], combinatorial chemistry [8], and organic and biomolecule separation methodology [9–11]. In addition, the supramolecular display of highly fluorinated interfaces has been used in protein design [12–14], in driving selective aggregation in lipid bilayers [15–17] and in modulating phenotypical changes in biological systems [18,19]. We briefly review here work from our own laboratory and related studies in these areas.
Protein Design
Protein folding is predominantly driven by the sequestration of nonpolar side chains into a hydrophobic core. Although many noncovalent forces are responsible for the overall fold, the major driving force is derived from removal of hydrophobic surface area from solvent water. We envisioned that fluorinated side chains would deliver a larger energetic advantage, as trifluoromethyl groups are more hydrophobic than methyl groups [20,21]. Our group at Tufts and that of David Tirrell at Caltech independently designed coiled coils to test this idea [12,22]. Coiled coils consist of two or more intertwined helical strands and have a typical heptad repeat sequence (abcdefg). The a and d residues make up the hydrophobic core (a/d), while the residues at the e and g positions through electrostatic contacts contribute to overall stability and specificity. We replaced three valines at the a position with trifluorovaline, and all four leucines at the d position with trifluoroleucine in a model peptide derived from the coiled coil domain of yeast bZIP transcriptional activator GCN4. The resultant coiled coil ensemble was more stable to heat (ΔTm = 15 °C) and chaotropic denaturation (ΔΔG° ~ 1 kcal/mol). These results were in agreement with those of the Tirrell and co-workers, who replaced all the leucines with trifluoroleucines but left the valines intact and also observed similar increases in stability [22,23]. Marsh and co-workers have further explored the use of hexafluoroleucine in an antiparallel four-helix bundle protein, and determined the contribution of substitution at two, four and six layers in the hydrophobic core [24]. Their results point to a 0.30 kcal/mol stabilization per hexafluoroleucine for the central two layers and 0.12 kcal/mol for the outer layers. The arsenal of proteins and peptides containing fluorinated amino acids has been further expanded and the relative contributions to stability quantified [25].
We extended this paradigm of highly fluorinated interfaces to test whether such supramolecularly displayed fluorous surfaces would self-sort from hydrocarbon surfaces [14a]. Coiled coil peptides were once again used to execute this design paradigm. Peptides adorned with seven leucine or hexafluoroleucine residues at core positions (a and d) were prepared and then linked via a disulfide linkage at the N-terminus. When the heterodimer was allowed to undergo disulfide exchange under redox buffer conditions, it rapidly equilibrated back to the homodimeric disulfide linked constructs. Less than 3% of the heterodimer remained in solution at equilibrium, suggesting that the fluorous and hydrocarbon surfaces did not favor interaction with the unlike strand. Thermodynamic analysis revealed that the disproportionation was driven largely by the hyper-stability of the fluorinated ensemble and the relative instability of the heterodimer [14b].
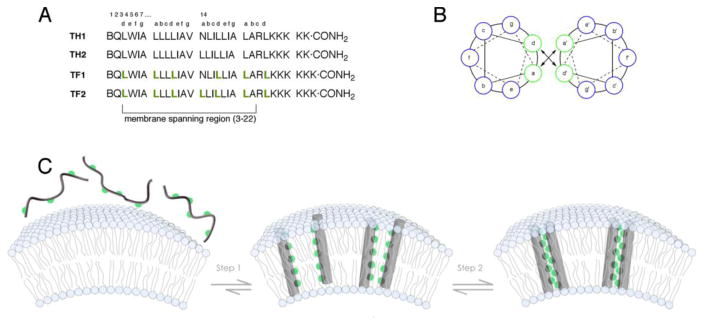
These experiments suggested a solution to a hitherto unsolved problem – specific control of oligomeric structures in membrane environments that the simultaneously hydrophobic and lipophobic character of fluorocarbons could be used to drive higher order assemblies. We envisioned a two-step orchestrated self-assembly process (Figure 1). First, the designed transmembrane peptides would partition into micelles or vesicles and adopt α-helical structures by main chain hydrogen bonding. These helical structures would have a fluorous stripe running down one face. Second, solvophobic sequestration of this interface away from the lipid would result in helical bundles. Fluorinated peptides were designed both with (TF1) and without (TF2) a central asparagine, that has hydrogen bonding capability in the side chain primary amide [13,26,27]. The membrane embedded ensembles were characterized by a battery of biophysical techniques and compared to control peptides (TH1 and TH2) containing residues with natural hydrocarbon side chains. Circular dichroism in micellar solutions showed that all peptides adopted -helical conformations. Equilibrium analytical ultracentrifugation of peptides in solutions of micelles revealed that TF1 and TF2 are both capable of forming higher order assemblies, forming a tetramer and dimer respectively. In contrast, among the hydrocarbon peptides, only the one capable of hydrogen bonding using the side chain of asparagine (TH1) formed a dimer, while the other (TH2) failed to oligomerize.
Figure 1.
Schematic illustration depicting the two-step self-assembly process in vesicles of membrane soluble peptides. (A) Sequences of the peptides TH1, TH2, TF1 and TF2. (B) Helical wheel diagram indicating the position of residues at the a and d positions. (C) The hydrophobic peptides partition into vesicles (or micelles) and form α-helices to avoid exposure of the backbone amides. This step exposes a helical face adorned with fluorinated groups in the case of TF1 and TF2 (fluorinated residues are depicted as green spheres) that further segregates away from the lipid yielding higher order assemblies.
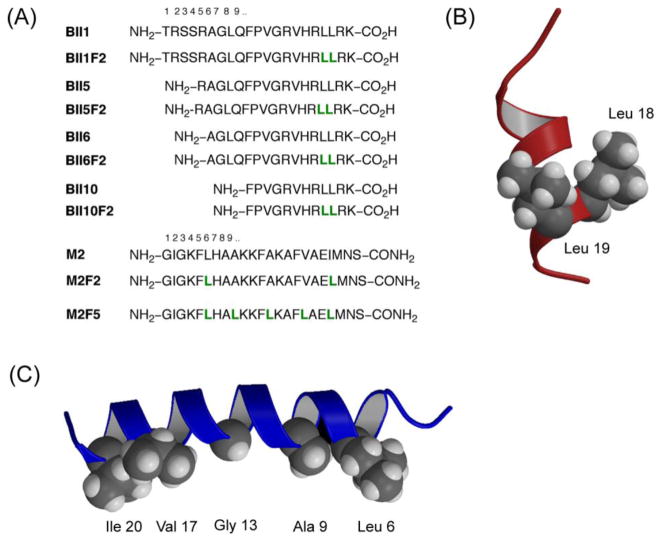
This control of structure and higher order assembly within membrane environments bodes well for applications possible with fluorinated peptides. Over half the drug targets are thought to be membrane proteins [28] and many antimicrobial peptides function by forming oligomeric structures [29] compromising the integrity of target cell membranes. Peptides however, suffer from limited metabolic stability [30], and are usually degraded rapidly by proteases. Because of the higher structural stabilities and greater membrane affinities of fluorinated peptides [31], we envisioned that they could function as more potent antimicrobials with longer half-lives. We chose to make variants of two known antimicrobial peptides: magainin 2 amide (M2) that lyses bacterial cells by forming toroidal pores in the membrane and buforin II (BII) which exerts its action by binding intracellular nucleic acid components thus disrupting crucial cellular function. Hexafluoroleucine residues were introduced on the nonpolar face of each peptide, resulting in four analogues of buforin, and two in the magainin series (Figure 2). Most variants (five of six, with the exception of M2F5) had significantly enhanced (upto 25-fold) or similar antimicrobial activity compared to the parent peptides [32]. It is crucial that antimicrobials discriminate between bacterial and mammalian cells. To this end, they must display low hemolytic activity in order to be useful. Fluorinated buforin analogues showed essentially zero lytic activity when challenged with erythrocytes, while the magainin analogues showed an increase in hemolysis compared to the parent peptide M2. These constructs were further characterized in membrane-mimetic environments (addition of trifluoroethanol). All fluorinated peptides showed greater secondary structure content and were more hydrophobic than the parent peptides. Likely due to these factors, the fluorinated antimicrobials were also more resistant to hydrolytic cleavage catalyzed by the protease trypsin than their hydrocarbon counterparts. These results point to the value of fluorination as a strategy to increase potency of known antimicrobials and also in modulating proteolytic stability.
Figure 2.
Fluorinated antimicrobial peptides – analogues of buforin II (BII) and magainin 2 amide (M2) show increased proteolytic stability, and in six out of seven cases retention or enhanced activity. The highly fluorinated peptide M2F5 forms a helical bundle in water and is inactive against bacterial strains. (A) Peptide sequences in one letter code. Underlined residues were chosen for replacement (L = hexafluoroleucine). (b) Model structure of buforin indicating the location of residues that were replaced by hexafluoroleucine. (c) NMR structure of magainin 2 amide indicating the hydrophobic face where substitutions with fluorinated residues were made (PDB code: 2mag).
The quintessential case for a peptide therapeutic with enormous medical potential, but one that is thwarted by its low metabolic stability is the gut hormone, glucagon-like peptide-1 (GLP-1) [33]. It modulates glucose dependent insulin release, enhances β-cell mass and activity, curbs appetite, and lowers glucagon secretion. These properties make GLP-1 an attractive lead compound for the treatment of type 2 diabetes. However, the use of native GLP-1 in clinical settings is limited because it is rapidly (~2 mins) degraded by the serine protease DPP IV [34]. The greater membrane affinity and structural stability of fluorinated peptides prompted us to design analogues of GLP-1 containing hexafluoroleucine [35]. Sites were chosen such that they would have the greatest effect on binding to its cognate receptor (GLP-1R) or in protection from protease catalyzed cleavage (Figure 3). Seven fluorinated analogues were prepared and tested for their ability to: (a) bind the receptor using a radioligand exchange assay; (b) activate GLP-1R by following the production of cAMP and (c) to resist proteolytic cleavage by DPP IV. Every fluorinated peptide was more stable to cleavage by the protease, and while the peptides showed a moderate decrease in the in vitro binding affinity and signal transduction ability, the efficacy of cAMP production was retained in 6 out of 7 fluorinated analogues. These results suggest that fluorination may be a useful method to improve stability of bioactive peptides where low metabolic stability limits therapeutic value.
Figure 3.
Fluorinated analogues of GLP-1. (A) Sequences of the peptides. All fluorinated analogues were more stable to proteolysis by DPP IV. L denotes hexafluoroleucine. (B) Efficacy of cAMP production shown as percentage of the level produced by GLP-1 (95% confidence intervals). All peptides with the exception of F89 showed ≥ 88% stimulatory action.
Affinity Purification and Enrichment
Fluorous affinity purification burst onto the scene as a method for the removal of catalysts from complex mixtures [3]. A perfluoroalkyl tail, typically no shorter than — C6F13 is attached to the compound of interest. Once the reaction is complete, liquid-liquid or liquid-solid phase extractions can then rapidly separate tagged products. This strategy has now been applied to microarray preparation [36] and in biomolecule purification and enrichment [37,38].
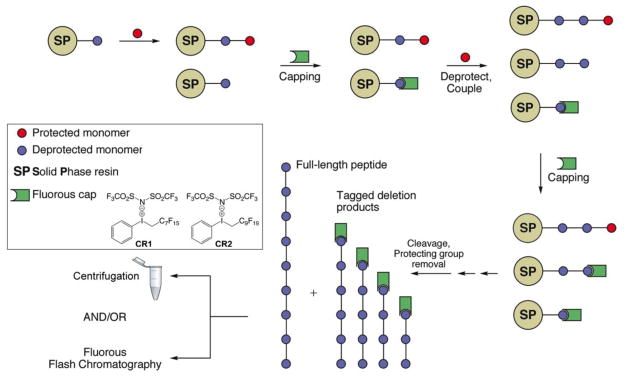
Automated solid-phase synthesis has greatly facilitated the facile and reliable construction of oligonucleotides and peptides. The tedium remains in the purification of the final product away from the other components that are structurally similar, resulting from incomplete couplings during monomer addition. Furthermore, purification is expensive and relies on solvents that are detrimental to the environment. Given the unique miscibility of perfluoroalkyl groups, we have developed a capping reagent that can substitute for acetic anhydride in tagging residual amines that have failed to react during a coupling step. Two variants of the trivalent iodonium salt, with —C7F15 (CR1) and —C9F19 (CR2) groups have been employed [39–41]. The reaction with amines results in the formation of an alkylated amine that is unreactive in all subsequent couplings, deprotections and cleavage conditions usually employed during SPPS (Figure 4). Both t-Boc and Fmoc chemistries are compatible and the reagents can be used as in automated synthesizers. The capped deletion products are easily removed by simple centrifugation or by fluorous solid phase extraction. Other strategies have also been devised for Fmoc synthesis of peptides, and separately for oligonucleotides [42–45].
Figure 4.
General strategy of fluorous capping by reagents CR1 and CR2. SPPS requires the removal of side products resulting from incomplete couplings. Fluorous trivalent iodonium salts efficiently tag all deletion products permitting facile separation from the full-length peptide simply by centrifugation, or fluorous flash filtration.
An analogous strategy has been applied in enriching peptides of interest from a complex cellular mixture. Enzymatic digestion of the crude extracts followed by selective labeling of peptides by reaction of cysteines with fluorous tags or by β-elimination/Michael addition to phosphorylated peptides. Upon extraction using fluorous solid phase, the tagged peptides were significantly enriched and were amenable to mass spectral analysis directly [46,47].
The phase properties of fluorocarbons and the prudent placement of fluorine in biological molecules offer avenues to modulate function and structure in unprecedented ways. The paucity of fluorine in naturally occurring molecules often means that background free non-invasive imaging (19F MRI) [48] and spectroscopy are possible with excellent signal to noise ratios. Fluorination has allowed design of hyper-stable structural folds in proteins, probing of ligand-receptor interactions, and improving the protease stability of peptide therapeutics. The potential applications of fluorine containing biomolecules continue to surprise and grow in number.
Acknowledgments
This work was funded in part by the NIH (GM65500 and CA125033). We are thankful to present and past co-workers in the Kumar laboratory for stimulating discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dunitz JD. ChemBioChem. 2004;5:614. doi: 10.1002/cbic.200300801. [DOI] [PubMed] [Google Scholar]
- 2.Lemal DM. J Org Chem. 2004;69:1. doi: 10.1021/jo0302556. [DOI] [PubMed] [Google Scholar]
- 3.Horváth IT, Rabai J. Science. 1994;266:72. doi: 10.1126/science.266.5182.72. [DOI] [PubMed] [Google Scholar]
- 4.Studer A, Hadida S, Ferritto R, Kim SY, Jeger P, Wipf P, Curran DP. Science. 1997;275:823. doi: 10.1126/science.275.5301.823. [DOI] [PubMed] [Google Scholar]
- 5.Myers KE, Kumar K. J Am Chem Soc. 2000;122:12025. [Google Scholar]
- 6.Ishikawa Y, Kuwahara H, Kunitake T. J Am Chem Soc. 1994;116:5579. [Google Scholar]
- 7.Percec V, Johansson G, Ungar G, Zhou J. J Am Chem Soc. 1996;118:9855. [Google Scholar]
- 8.Horvath IT. Acc Chem Res. 1998;31:641. [Google Scholar]
- 9.Curran DP, Luo Z. J Am Chem Soc. 1999;121:9069. [Google Scholar]
- 10.Fish RH. Chem--Eur J. 1999;5:1677. [Google Scholar]
- 11.Gladysz JA. Science. 1994;266:55. doi: 10.1126/science.266.5182.55. [DOI] [PubMed] [Google Scholar]
- 12.(a) Bilgiçer B, Fichera A, Kumar K. J Am Chem Soc. 2001;123:4393. doi: 10.1021/ja002961j. [DOI] [PubMed] [Google Scholar]; (b) Montclare JK, Son S, Clark GA, Kumar K, Tirrell DA. ChemBioChem. 2009;10:84. doi: 10.1002/cbic.200800164. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Jackel C, Salwiczek M, Koksch B. Angew Chem, Int Ed. 2006;45:4198. doi: 10.1002/anie.200504387. [DOI] [PubMed] [Google Scholar]
- 13.Bilgiçer B, Kumar K. Proc Natl Acad Sci U S A. 2004;101:15324. doi: 10.1073/pnas.0403314101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Bilgiçer B, Xing X, Kumar K. J Am Chem Soc. 2001;123:11815. doi: 10.1021/ja016767o. [DOI] [PubMed] [Google Scholar]; (b) Bilgiçer B, Kumar K. Tetrahedron. 2002;58:4105. [Google Scholar]
- 15.Dafik L, Kalsani V, Leung AKL, Kumar K. J Am Chem Soc. 2009;131 doi: 10.1021/ja902777d. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Yoder NC, Kalsani V, Schuy S, Vogel R, Janshoff A, Kumar K. J Am Chem Soc. 2007;129:9037. doi: 10.1021/ja070950l. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Schuy S, Faiss S, Yoder NC, Kalsani V, Kumar K, Janshoff A, Vogel R. J Phys Chem B. 2008;112:8250. doi: 10.1021/jp800711j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoder NC, Kumar K. Chem Soc Rev. 2002;31:335. doi: 10.1039/b201097f. [DOI] [PubMed] [Google Scholar]
- 18.Dafik L, d’Alarcao M, Kumar K. Bioorganic and Medicinal Chemistry Letters. 2008;18:5945. doi: 10.1016/j.bmcl.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dafik L, Kalsani V, Leung AKL, Kumar K. J Am Chem Soc. 2009;131 doi: 10.1021/ja902777d. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Resnati G. Tetrahedron. 1993;49:9385. [Google Scholar]
- 21.Seebach D. Angew Chem, Int Ed Engl. 1990;29:1320. [Google Scholar]
- 22.Tang Y, Ghirlanda G, Vaidehi N, Kua J, Mainz DT, Goddard WA, III, DeGrado WF, Tirrell DA. Biochemistry. 2001;40:2790. doi: 10.1021/bi0022588. [DOI] [PubMed] [Google Scholar]
- 23.Son S, Tanrikulu IC, Tirrell DA. ChemBioChem. 2006;7:1251. doi: 10.1002/cbic.200500420. [DOI] [PubMed] [Google Scholar]
- 24.Lee KH, Lee HY, Slutsky MM, Anderson JT, Marsh ENG. Biochemistry. 2004;43:16277. doi: 10.1021/bi049086p. [DOI] [PubMed] [Google Scholar]
- 25.(a) Montclare JK, Son S, Clark GA, Kumar K, Tirrell DA. ChemBioChem. 2009;10:84. doi: 10.1002/cbic.200800164. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Woll MG, Hadley EB, Mecozzi S, Gellman SH. J Am Chem Soc. 2006;128:15932. doi: 10.1021/ja0634573. [DOI] [PubMed] [Google Scholar]; (c) Lee HY, Lee KH, Al-Hashimi HM, Marsh ENG. J Am Chem Soc. 2006;128:337. doi: 10.1021/ja0563410. [DOI] [PubMed] [Google Scholar]; (d) Chiu HP, Suzuki Y, Gullickson D, Ahmad R, Kokona B, Fairman R, Cheng RP. J Am Chem Soc. 2006;128:15556. doi: 10.1021/ja0640445. [DOI] [PubMed] [Google Scholar]; (e) Zheng H, Comeforo K, Gao JM. J Am Chem Soc. 2009;131:18. doi: 10.1021/ja8062309. [DOI] [PubMed] [Google Scholar]
- 26.Naarmann N, Bilgiçer B, Kumar K, Steinem C. Biochemistry. 2005;44:5188. doi: 10.1021/bi050096f. [DOI] [PubMed] [Google Scholar]
- 27.Naarmann N, Bilgiçer B, Meng H, Kumar K, Steinem C. Angew Chem, Int Ed. 2006;45:2588. doi: 10.1002/anie.200503567. [DOI] [PubMed] [Google Scholar]
- 28.Bakheet TM, Doig AJ. Bioinformatics. 2009;25:451. doi: 10.1093/bioinformatics/btp002. [DOI] [PubMed] [Google Scholar]
- 29.Yeaman MR, Yount NY. Pharmacological Reviews. 2003;55:27. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 30.Adessi C, Soto C. Curr Med Chem. 2002;9:963. doi: 10.2174/0929867024606731. [DOI] [PubMed] [Google Scholar]
- 31.Niemz A, Tirrell DA. J Am Chem Soc. 2001;123:7407. doi: 10.1021/ja004351p. [DOI] [PubMed] [Google Scholar]
- 32.Meng H, Kumar K. J Am Chem Soc. 2007;129:15615. doi: 10.1021/ja075373f. [DOI] [PubMed] [Google Scholar]
- 33.Meier JJ, Nauck MA. Diabetes-Metabolism Research and Reviews. 2005;21:91. doi: 10.1002/dmrr.538. [DOI] [PubMed] [Google Scholar]
- 34.Drucker DJ. Curr Pharm Des. 2001;7:1399. doi: 10.2174/1381612013397401. [DOI] [PubMed] [Google Scholar]
- 35.Meng H, Krishnaji ST, Beinborn M, Kumar K. J Med Chem. 2008;51:7303. doi: 10.1021/jm8008579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ko KS, Jaipuri FA, Pohl NL. J Am Chem Soc. 2005;127:13162. doi: 10.1021/ja054811k. [DOI] [PubMed] [Google Scholar]
- 37.Matsugi M, Curran DP. Org Lett. 2004;6:2717. doi: 10.1021/ol049040o. [DOI] [PubMed] [Google Scholar]
- 38.Zhang W, Curran DP. Tetrahedron. 2006;62:11837. doi: 10.1016/j.tet.2006.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montanari V, Kumar K. J Am Chem Soc. 2004;126:9528. doi: 10.1021/ja0479033. [DOI] [PubMed] [Google Scholar]
- 40.Montanari V, Kumar K. J Fluorine Chem. 2006;127:565. [Google Scholar]
- 41.Montanari V, Kumar K. Eur J Org Chem. 2006:874. [Google Scholar]
- 42.De Visser PC, Van Helden M, Filippov DV, Van Der Marel GA, Drijfhout JW, Van Boom JH, Noort D, Overkleeft HS. Tetrahedron Lett. 2003;44:9013. [Google Scholar]
- 43.Del Pozo C, Keller AI, Nagashima T, Curran DP. Org Lett. 2007;9:4167. doi: 10.1021/ol701631m. [DOI] [PubMed] [Google Scholar]
- 44.Filippov DV, Van Zoelen DJ, Oldfield SP, Van Der Marel GA, Overkleeft HS, Drijfhout JW, Van Boom JH. Tetrahedron Lett. 2002;43:7809. [Google Scholar]
- 45.Pearson WH, Berry DA, Stoy P, Jung KY, Sercel AD. J Org Chem. 2005;70:7114. doi: 10.1021/jo050795y. [DOI] [PubMed] [Google Scholar]
- 46.Brittain SM, Ficarro SB, Brock A, Peters EC. Nat Biotechnol. 2005;23:463. doi: 10.1038/nbt1076. [DOI] [PubMed] [Google Scholar]
- 47.Evanko D. Nat Methods. 2005;2:406. doi: 10.1038/nmeth0905-650. [DOI] [PubMed] [Google Scholar]
- 48.Yu JX, Kodibagkar VD, Cui WN, Mason RP. Curr Med Chem. 2005;12:819. doi: 10.2174/0929867053507342. [DOI] [PubMed] [Google Scholar]