Abstract
Objective
Notch signaling is implicated in repressing mesenchymal stem cell (MSC) chondrogenic differentiation. The mechanism of this repression and how this is modulated to permit chondrogenesis has not been elucidated.
Methods
Notch intracellular domain (NICD) protein levels were monitored via western blot throughout chondrogenic differentiation of human in MSC pellet cultures. Overexpression of Notch signaling components and their effect on chondrogenesis was achieved by transfecting plasmids coding for NICD, hairy and enhancer of split 1 (Hes-1) and hairy and enhancer of split related-2 (HERP-2/Hey-1). Col2a1 and aggrecan expression was monitored via quantitative PCR. Chromatin immunoprecipitation (chIP) was utilized to test whether Hes-1 and Hey-1 bind putative N-box domains in intron 1 of Col2a1.
Results
NICD protein levels were reduced during chondrogenesis of hMSC, which was mediated by TGFβ3. Col2a1 gene expression was repressed following overexpression of NICD (2-fold), Hes-1 (3-fold) and markedly by Hey-1 (80-fold). Hey-1 repressed aggrecan expression 10-fold, while NICD and Hes-1 had no effect. chIP studies show that endogenous Hes-1 and Hey-1 bind to two putative N-box domains adjacent to and as part of the Sox9 enhancer binding site located in intron 1 of Col2a1. The Hes-1 co-repressor protein transducin like enhancer (TLE) was displaced during chondrogenic differentiation and following TGFβ3 treatment.
Conclusion
These results reveal novel mechanisms by which Notch signaling represses gene expression and further define the role of TGFβ to promote chondrogenic differentiation.
INTRODUCTION
Notch-1 plays a pivotal role in the differentiation of many tissues; its function is a critical moderator of the fate of all stem cells (1–6). Notch is activated by interaction between the extracellular region of Notch and its ligands, Delta or Jagged/Serrate during direct cell-cell contact (3, 7, 8). Upon ligand binding, the Notch receptor undergoes proteolytic cleavage events in the juxtamembrane domain, leading to the release of the Notch intracellular domain (NICD). Presenilins are important for the proteolytic cleavage of the NICD, and this cleavage is blocked by γ-secretase inhibitors (9, 10). NICD translocates into the nucleus and interacts with the transcriptional regulator CSL (CBF-1, Suppressor of hairless, Lag-1) (11), to convert CSL to a transcriptional activator. Hairy and Enhancer of Split (HES) and HES-related-repressor protein (HERP) genes encoding basic helix-loop-helix (bHLH) transcription regulators are target genes activated by this mechanism (12, 13).
The bHLH family of transcriptional regulators proteins bind DNA as dimers on specific DNA sequences and are classified into several groups. Class A are transcriptional activators and include MyoD and Mash1. These bind class A sites (CANCTG). Myc and Max are class B leucine zipper type proteins and bind class A and B sites. Class B proteins are considered as subtypes of the E-box (CANNTG). HES proteins are class C transcriptional repressors, binding preferentially to class C sites (CACGNG), N-box sequences (CACNAG), but also to class B sites. The HERP family of bHLH proteins bind both class B and C sites (14).
Only six bHLH proteins have been shown to be direct targets of Notch, including Hes-1, Hes-5, Hes-7, HERP-1 (Hey-2), HERP-2 (Hey-1) and HERP-3. Target genes of HES and HERP are not well established, however it has been found that Hes-1 represses itself by binding N-box domains located in the promoter region of Hes-1 (15). Hes-1 over expression leads to N-box dependent repression of CD4 in T-helper cells (16) and Hes-1 binding to E-box domains in the rat lipocalin-type prostaglandin D synthase gene repressed its transcription (17).
Binding of bHLH proteins to DNA involves the recruitment of co-repressor proteins to form functional repressor complexes. HES is known to recruit groucho/transducin like enhancer of split (Gro/TLE) (18), while HERP recruits a co-repressor complex consisting of N-CoR, mSin3A and HDAC (14). Such DNA binding and repressor complex formation represents an active repression mechanism. HES has also been shown to passively repress gene expression by forming non-functional dimers (19).
Studies on over expression or deletion of Notch receptors or ligands suggest a role of this signaling pathway in skeletal development. Misexpression of Delta-1 in chick embryos negatively regulated the transition from prehypertrophic to hypertrophic chondrocytes (20) NICD over expression in ATDC5 cells and murine limb bud micromass cultures strongly suppressed chondrocytic differentiation and cell proliferation. Inhibition of Notch cleavage using a gamma secretase inhibitor led to an increased expression of Sox9, Col2a1 and enhanced proteoglycan deposition. Hes-1 over expression effects were similar but less potent (21, 22).
In contrast to the repressive effects of Notch signaling, the transcription factor Sox9 is central to activate genes that encode cartilage extracellular matrix proteins (23) and binds to high-m group (HMG) DNA domains present in promoter and enhancer regions of these genes (24) has been characterized to activate expression of Col2a1 (25–27), Col9a1 (28), Col11a1 (29), aggrecan (30), cartilage link protein (31) and cartilage-derived retinoic-acid-sensitive protein (CD-RAP) (32).
Stimulation of human mesenchymal chondrogenic differentiation is dependent on a number of factors, principally including dexamethasone and growth factors that are members of the transforming growth factor beta (TGFβ)/ bone morphogenetic protein (BMP) family (33–35).
The opposing roles of Notch and Sox9 signaling systems indicate that mesenchymal stem cell differentiation towards the chondrogenic phenotype requires modification of these signaling events and that pro-chondrogenic factors orchestrate this effect (36). In this study we present evidence that high Notch-1 protein levels are present in MSC, decrease throughout chondrogenic differentiation and that TGFβ3 induces this event. We also show that Hes-1 and Hey-1 bind N-box domains in the enhancer region of Col2a1. Since this site is required for Sox9 mediated transcription of Col2a1, we propose that bHLH proteins negatively compete with Sox9 binding to the enhancer site and prevent Sox9-mediated transcriptional activation of Col2a1 and thus chondrogenic differentiation.
MATERIALS AND METHODS
Human Tissue Samples and Culture Conditions
Human bone marrow derived mesenchymal stem cells (hMSC) were isolated from the iliac crest bone marrow of normal adult donors (approval by human subjects committee). Bone marrow aspirates were diluted with twice the volume of Hanks Buffered Saline solution (HBSS; Gibco) and filtered through a 70 μm cell strainer (Becton Dickson). The strained samples were gently layered on the top of the Ficoll-Plaque and centrifuged for 30 minutes at 300 × g at room temperature. Cells at the interface were collected, and washed twice with HBSS. After washing, cells were resuspended in Mesenchymal stem cell basal medium (Lonza) and cultured at a density of 2×105 cells/cm2. Culture medium was replaced at 24 h and 72 h and every 3–4 days thereafter. hMSC were subcultured after 10 to 14 days by treatment with Accutase (Innovative Cell Technologies) for 5 min, subsequently washed in medium and seeded into fresh flasks. All samples were screened for multipotential to differentiate into cartilage (see pellet cultures below), bone and fat as previously described (37).
Pellet Cultures
Human MSC pellets (5×105 cells per pellet) were made by centrifuging the cells at 500× g in 15 ml polypropylene conical tubes and cultured in 0.5 ml chondrogenic medium (Lonza), supplemented with BMP6 (500 ng/ml) and TGFβ3 (10 ng/ml). Medium was changed every 2–3 days. To monitor Notch-1 protein levels throughout chondrogenesis, hMSC were processed on day 2, day 4 and day 8. Chondrogenesis was monitored via Col2a1 and aggrecan expression and Safranin O staining.
Gamma Secretase Inhibitor (γSI) Treatments
MSC pellets were made and incubated as described above, but cultured for 14 days. Pellets were exposed to the γSI L685,458 (Calbiochem) at a concentration of 5 μM for either i) the entire 14 days of culture or, ii) only between days 7 and 14. To show specificity of the bTAN20 antibody, we exposed monolayer hMSC to L685,458 for 24 hours and processed for western blot analysis.
Western Blot Analysis
SW1353 human chondrosarcoma cells or hMSC in monolayer culture or in pellet culture were lysed in RIPA buffer (10 mM Tris, pH 7.4; 100 mM NaCl; 1 mM EDTA; 1 mM EGTA; 1 mM NaF; 20 mM Na4P2O7; 0.1% SDS; 0.5% DOC; 1% Triton X-100; 10% glycerol; Fisher Scientific) and protease inhibitor cocktail (Sigma) for 30 min at 4 °C. Insoluble material was removed by centrifugation at 14,000 rpm for 10 min at 4 °C. Protein concentrations were determined by the BCA assay (Pierce) prior to loading either 10 or 20 μg on 6% or 10% SDS-PAGE gels. After protein transfer onto nitrocellulose membranes (Schleicher & Schuell), the membranes were blocked and incubated overnight with an antibody that detects the intracellular domain of human Notch-1 (bTAN 20, Hybridoma Bank, University of Iowa), washed, incubated with anti-rat-HRP conjugated secondary antibody (Santa Cruz) for one hour. The protein was detected using the Supersignal West Pico Chemiluminescent Substrate (Pierce) and exposed to X-Ray film (Pierce).
Over Expression of NICD, Hes-1 and Hey-1
To examine Notch signaling in chondrogenesis, a constitutively active form of human Notch-1 construct lacking the extracellular domain (NICD-pCDNA3.1myc-His, a.a. 1760–2556) (from Dr. Kadesch, University of Pennsylvania, Philadelphia) or control pCDNA3.1 was transfected into hMSC using Human MSC Nucleofector kit (Amaxa) according to manufacturer's instruction. Following transfection, the cells were cultured in 6-well plates for 4 days. The transfected cells were then placed into pellet cultures in chondrogenic medium for 14 days. To directly test the effect of Hes-1 and Hey-1 on chondrogenic differentiation, 5×105 hMSC were transfected with 1μg of pcCMV-FLAG-Hes-1 (from Dr. Stifani, McGill University, Montreal), or pcDNA3-FLAG-Hey-1 (from Dr. Kedes, USC, Los Angeles), or pcDNA3.1 in monolayer culture using FUGENE HD (Roche). After 2 days, the cells were placed into pellet culture with chondrogenic medium. Total RNA was extracted from each pellet after 6 days for detection of Col2a1, aggrecan and GAPDH.
RNA Extraction and Reverse Transcription
Total RNA was isolated from monolayer or pellet cultures using the Qiashredder (Qiagen) and the RNeasy kit (Qiagen) with on column DNA digestion. Complementary DNA (cDNA) was produced using the SuperScript III first strand kit (Invitrogen) with random hexamers.
Semi-quantitative PCR
The Hotstar Taq MasterMix (Qiagen) together 2.5 μl primer mix (end concentration: 1 μM), cDNA and water to a total volume of 25 μl were subjected to PCR amplification for 30 cycles. Primers sets: Col2a1: 5'-ACA CCG TGG CTT CAC TGG TC-3' and 5'-TGG GTT TGC AAC GGA TTG T-3' Notch-1: 5'-CGG ACC CAA CAC TTA CAC CT-3' and 5'-CTC ACA GGC ACA CTC GTA GC-3' GAPDH: 5'-ATCAGCAATGCCTCCTGCAC-3' and 5'-CGTCAAAGGTGGAGGAGTGG-3'.
Quantitative PCR
The level of Col2a1, aggrecan, Sox9 and GAPDH gene expression was assessed using the one-step QuantiTect Probe RT-PCR system (Qiagen) with Taqman primers and probes (Applied Biosystems) at recommended concentrations. All reactions were performed using the iCycler thermocycler (BioRad). Delta Ct values were calculated and all genes were normalized to GAPDH expression levels as previously described (38).
Growth Factor Treatments
Monolayer cultured hMSC (three donors) were maintained in 6 well plates and subjected to various pro-chondrogenic inducing stimuli for 72 hours including TGFβ3 (10 ng/ml; Lonza), BMP6 (500 ng/ml; R&D Systems), Dexamethasone (10−8M; Sigma), BMP2 (500 ng/ml; R&D Systems) and FGF2 (50 ng/ml; PeproTech Inc.).
Chromatin Immunoprecipitation (chIP) Assays
For each chIP assay 1×106 cells were used. Cells were fixed in 1% formaldehyde, at a density of 5×105 cells per ml, at room temperature for 10 min and glycine was added to terminate cross linking (final concentration 0.125 M). Cells were centrifuged, washed in PBS and re-suspended in 300 μl of low salt lysis buffer (Santa Cruz). The crude nuclear extract was collected via centrifugation, washed in PBS and re-suspended in 100 μl of high salt buffer (Santa Cruz). The lysate was then sonicated (Sonic Dismembrator Model 500; Fisher Scientific) at 4 °C, at a setting of 30% for 15 s on and 30 s off. This cycle was repeated 10 times. Prior to primary antibody incubation, the chromatin solution/cell lysate was pre-cleared by adding 20 μl of Protein A/G PLUS-Agarose (Santa Cruz) for 30 min at 4 °C. The beads were removed by centrifugation and approximately 10% was collected as the input control, while the remaining chromatin solution was transferred into two clean tubes and incubated with either anti-Hes-1 (Santa Cruz), anti-Hey-1 (Avia Systems Biology) or anti-TLE (Santa Cruz) or the species-matched IgG as a negative control. These lysates were incubated at 4 °C overnight on a rotating mixer. Protein A/G PLUS-Agarose beads (20 μl) were added to each tube and incubated for 2 h at 4 °C. Beads were harvested and washed twice in high salt buffer, then four times with the wash buffer (Santa Cruz). The beads were then resuspended in 100 μl of elution buffer (Santa Cruz) and incubated in a 67 °C water bath overnight to reverse crosslinks. The beads were removed by centrifugation and DNA was isolated from the supernatant using the Qiaquick gel extraction kit (Qiagen). PCR was then performed using specific primer sets deigned to amplify putative N-box regions located in intron 1 of the Col2a1 enhancer sequence (5'-ACACCCCTCCTCTCCATCTT-3' and 5'TCATGAATGGGGCTTTTCTC-3'). The amplified PCR products (30–36 cycles) were visualized via agarose electrophoresis in the presence of ethidium bromide. Input control lysates were included as well as genomic DNA as positive controls. PCR products were initially extracted from the agarose and sequenced to confirm identity.
Statistical Analysis
Values are reported as the mean ± SD. Only means are reported for quantitative PCR (triplicate readings).
RESULTS
NICD Levels Decrease in hMSC During Chondrogenesis and this is Mediated by TGFβ3
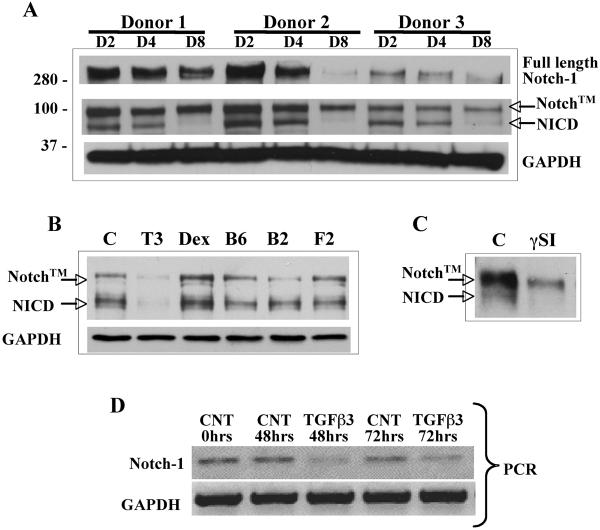
Notch-1 protein levels were monitored by western blot for the first 8 days of chondrogenic differentiation of hMSC in a pellet culture system. Active Notch signaling as indicated by the presence of NICD was detectable in monolayer hMSC cultures (not shown) and for the first 4 days of pellet culture, but reduced to low or almost undetectable levels after this time (Fig. 1A). This observation is consistent with Notch's repressive role in differentiation of many tissues (5, 20, 39, 40) and the requirement for Notch signaling cessation to allow chondrogenic differentiation (21). While various factors are essential to permit chondrogenesis, we hypothesized that factors added to chondrogenic medium regulate Notch protein expression levels and remove Notch signaling mediated suppression. hMSC in monolayer culture, were treated with TGFβ3 (10 ng/ml), dexamethasone (10−8 M), BMP6 (500 ng/ml), BMP2 (500 ng/ml) and FGF2 (50 ng/ml) for 72 h. Western blot analyses showed that only TGFβ3 treatment reduced NICD protein levels (Fig. 1B), which was reproduced in two other hMSC donors treated with TGFβ3 (data not shown). The specificity of the bTAN20 antibody to detect NICD was tested in monolayer cultured hMSC in the presence of L685,458 (a y secretase inhibitor). Botch Notch transmembrane (Notch™) and NICD protein bands were detected in control cells, but the NICD band was absent following L685,458 exposure (Fig. 1C). TGFβ3 treatment (10 ng/ml) reduced the levels of Notch-1 mRNA in monolayer cultured hMSC after 48 hr and 72 hrs (Fig. 1D) indicating the mechanism by which TGFβ3 alters Notch protein levels during chondrogenesis. These results demonstrate that TGFβ3 is an important inhibitor of Notch signaling to permit chondrogenesis.
Figure 1. Reduced Notch-1 activity during chondrogenic differentiation is mediated by TGFβ exposure.
Cell lysates were prepared from pellet cultured human MSC at days 2, 4 or 8 from three donors (panel A). Western blots showed a reduction in NICD and full length Notch-1 protein levels were consistently seen by 8 days of pellet culture. To test whether the prochondrogenic factors added during chondrogenesis induced reduction in NICD levels, hMSC were exposed to TGFβ3 (T3; 10 ng/ml), Dexamethasone (Dex; 10−8 M), BMP6 (B6; 500 ng/ml), BMP2 (B2; 500 ng/ml) and FGF2 (F2; 50 ng/ml) in monolayer culture for 72 hours. Western blot of NICD indicated that TGFβ3 was responsible for the reduction in Notch-1 protein levels (panel B). Notch™ = Transmembrane portion of Notch-1 plus the uncleaved NICD fragment. In monolayer cultured hMSC addition the gamma secreatase inhibitor (γSI) L685,458 at a concentration of 5 μM for 24 hours prevented cleavage of NICD, indicating that the bTAN20 antibody detects both Notch™ and cleaved Notch-1 intracellular domain (NICD) proteins (panel C). RT-PCR (panel D) showing expression levels of Notch-1 mRNA in monolayer cultured hMSC incubated with TGFβ3 (10 ng/ml) for 48 and 72 hrs. Reduction in Notch-1 protein levels is regulated by TGFβ3 at the transcription level.
NICD, Hes-1 and Hey-1 Over Expression and Timing of γ-SI Inhibitor Treatment Repress Chondrogenic Differentiation
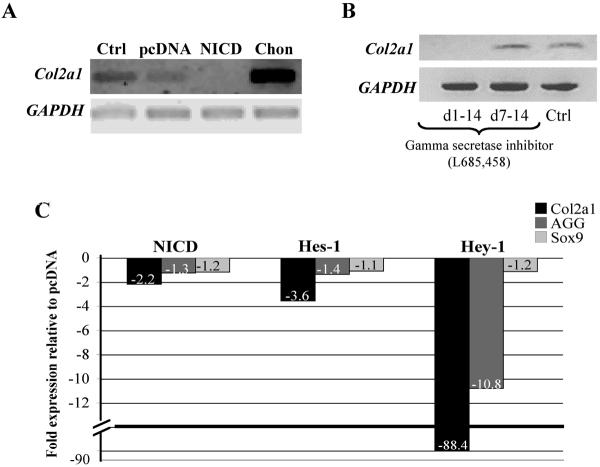
To test whether continued activation of Notch signaling may alter chondrogenic differentiation, we transfected NICD into hMSC and placed them into pellet culture for 14 days. NICD over expression prevented chondrogenesis as measured by a reduction in Col2a1 mRNA levels (Fig. 2A). To determine whether blocking Notch signaling at different stages of chondrogenic differentiation would effect Col2a1 expression, we incubated pellets with the γSI, L685–458 (5 μM), for the entire 14 days or between day 7 to day 14. Col2a1 expression was abolished in pellets treated for the entire culture period, but not for the second week. This indicates that Notch signaling is essential in early chondrogenic differentiation (Fig. 2B) and is consistent with recent observations by Vujovic et al. (41).
Figure 2. NICD, Hes-1 and Hey-1 over expression and timing of γ-SI inhibitor treatment prevents chondrogenic differentiation.
A constitutively active form of human Notch-1 construct or control pcDNA3.1 was transfected into monolayer cultured hMSC and then placed into pellet culture for 14 days. NICD over expression reduced Col2a1 expression levels (panel A). The gamma secretase inhibitor (γSI; L685–458) was either added to hMSC pellet cultures from days 1 to 14 or added on day 7 to day 14. Col2a1 expression was abolished in pellets treated for the entire culture period, but not for the second week, indicating that Notch signaling is essential in early chondrogenic differentiation (panel B). NICD, Hes-1 and Hey-1 plasmids were transfected into human MSC and maintained in pellet culture for 6 days. In comparison to pcDNA transfected pellets, NICD, Hes-1 and especially Hey-1 overexpression led to a repression of Col2a1 gene expression levels (panel B). Hey-1 over expression reduced aggrecan (AGG) expression while NICD and Hes-1 did not. Sox9 expression was not altered by these treatments (panel C). Ctrl: Mock nucleofection without plasmid. Chon: Chondrocyte positive control.
To examine the effect of Notch signaling further, we over expressed Hes-1 and Hey-1 in hMSC since these are known effectors of Notch's repressive action (12, 14, 18, 19). Gene expression levels of Col2a1, aggrecan and Sox9 were measured via quantitative PCR following 6 days in pellet culture and normalized to GAPDH. NICD and Hes-1 overexpression led to a 2- to 3-fold repression in Col2a1 transcription relative to the pcDNA controls. A surprisingly strong (over 80-fold) repression was found in Hey-1 transfected pellet cultures (Fig. 2C). Interestingly, NICD and Hes-1 did not alter aggrecan transcription, yet Hey-1 showed a 10-fold repression (Fig. 2C). Sox9 expression was not altered following any of these treatments (Fig. 2C). These results show that Notch signaling plays an essential role in chondrogenic differentiation, which is mediated by Hes-1 and Hey-1.
Hes-1 and Hey-1 Bind to N-box Domains Located in Intron I ofCol2a1
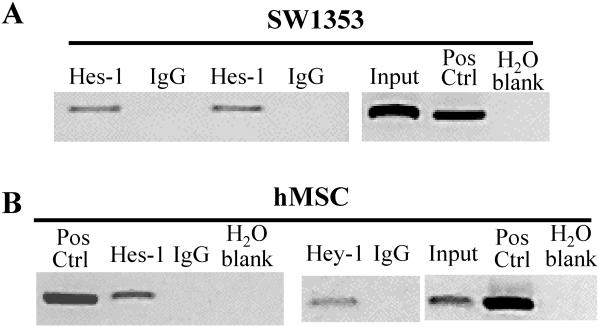
For transcriptional activation of Col2a1 binding of Sox9 to the enhancer sequence, located in intron 1, is essential (26). Since Notch signaling appears to be involved in suppressing Col2a1 expression, we hypothesized that bHLH consensus sites may be present in close proximity to the Sox9 enhancer binding domains. We located two putative binding sites, one −190bp (CACGAG) from the Sox9 HMG domains and the second included the first known Sox9 binding site (CTCGCG). To establish whether Hes-1 and Hey-1 bind to these sites, we performed chIP analysis. Using primers that specifically amplified the region spanning both N-box sites, we co-immunoprecipitated DNA bound Hes-1 to demonstrate that endogenous Hes-1 in SW1353 cells binds to the predicted sites (Fig. 3A). Both Hes-1 and Hey-1 show binding in hMSC (Fig. 3B).
Figure 3. Hes-1 and Hey-1 bind to putative N-box domains located within intron 1 of Col2a1.
Chromatin Immuno-Precipitation (chIP) assay was performed using SW1353 and hMSC. Immunoprecipitated DNA was amplified via PCR with primers specific for intron 1 of Col2a1. These agarose gel images depict a single PCR product in chromatin lysates incubated with the Hes-1 and Hey-1 antibodies corresponding to the targeted DNA sequence. This product was not amplified in the non-specific isotype controls (goat and rabbit IgG). Input control corresponds to lysate collected before antibody incubation. Human genomic DNA was used as a positive control for PCR amplification (Pos Ctrl). chIP assays with SW1353 cells (panel A) and hMSC (panel B).
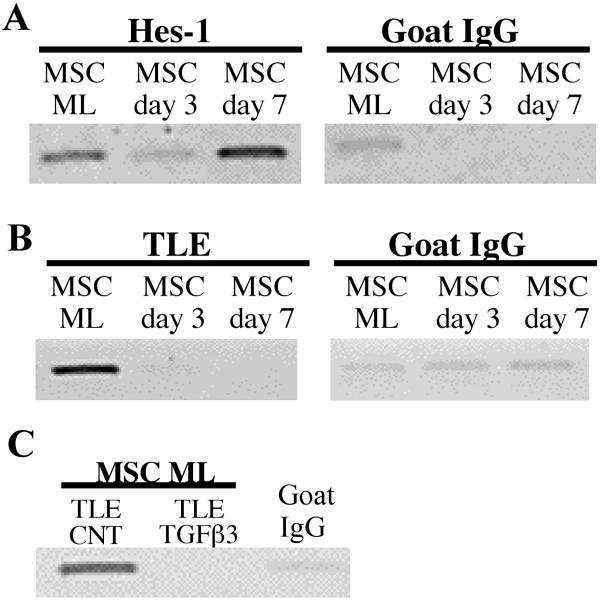
Hes-1 and TLE are Displaced in the Presence of TGFβ3 and During Chondrogenic Differentiation
Further chIP assays on hMSC undergoing chondrogenic differentiation in pellet cultures indicate that Hes-1 binding to DNA was inhibited by day 3 of pellet culture (Fig. 4A), yet this was restored by day 7. To explore co-repressor proteins that are recruited by Hes-1, we performed chIP analysis using antibodies specific for the co-repressor protein transducin like enhancer (TLE) and demonstrated that TLE was no longer present after 3 or 7 days of pellet culture (Fig. 4B). To establish whether TLE dismissal is mediated by TGFβ3 exposure, monolayer cultured hMSC were subjected to 72 h of TGFβ3. In control conditions TLE was present, indicating that it was associated with Hes-1 still bound to DNA (Fig. 4C). However, TLE was not present following TGFβ3 exposure, indicating that this growth factor is involved with the dismissal of TLE from the repressor complex.
Figure 4. TLE displacement from DNA during chondrogenesis and following TGFβ3 exposure.
Hes-1 and TLE are bound to monolayer cultured hMSC (ML), however during chondrogenic differentiation in pellet cultures both Hes-1 and TLE binding is reduced or inhibited by day 3, but Hes-1 binding is restored by day 7 (panel A and panel B). TLE does not co-immunoprecipitate following exposure of monolayer cultured hMSC to TGFβ3 for 72 hours (panel C).
DISCUSSION
The current study provides new insights into the mechanisms of gene repression by Notch and how this is abrogated in the presence of TGFβ3 in the model of chondrogenic differentiation of hMSC. Notch signaling was known to suppress chondrocytic differentiation (20–22), but the corresponding mechanisms remained to be determined.
Notch signaling is active in undifferentiated MSC and during the early phase of chondrogenesis. When chondrocyte differentiation ensues in pellet cultures, Notch signaling is down regulated as indicated by a loss of the Notch signaling component NICD. Correspondingly, over expression of NICD and of the Notch target genes Hes-1 and HERP-2/Hey-1 prevented chondrogenesis and Col2a1 gene expression. The present results suggest that Notch signaling and Hes-1 and HERP-2/Hey-1 may mediate active Col2a1 transcriptional repression.
In active repression or DNA-binding-dependent-transcriptional repression, HES binds to class C or N-box consensus DNA sites. Our results show that N-box domains are present in intron 1 of Col2a1 and that Hes-1 and Hey-1 bind to these sites. To effect transcriptional repression, Hes-1 recruits the co-repressor protein TLE (18) and Hey-1 or N-CoR mSin3A and HDAC (14) and bind as a complex to DNA to effect transcriptional repression. In this study, we co-immunoprecipitated TLE indirectly bound to DNA via chIP analysis. In accordance to Grbavec and Stifani (18), we propose that a Hes-1/TLE complex is present since TLE does not directly bind to DNA. The Hes-1/TLE complex bound to the Sox9 enhancer site may then block Sox9 binding to this site and thus active Col2a1 transcription. Hes-1 and TLE interaction are also important in regulating preadipocyte differentiation (40). In other cells systems, bHLH proteins are also known to repress transcription passively (19). For instance, competition between Sox10 and Hes5 has been demonstrated to control myelin gene transcription (42). Thus, it is plausible that Hes-1 and Hey-1 may physically interact and form non-viable bHLH/Sox9 complexes to passively control Sox9 mediated Col2a1 transcription.
Following TGFβ3 exposure or during chondrogenesis, both Hes-1 and TLE are dismissed and are no longer bound to the enhancer site to permit Col2a1 transcription and chondrogenesis. This scenario is consistent the removal of Hes-1/TLE following Nerve Growth Factor treatment to permit neurite outgrowth (43). Since Hey-1 was more potent in its repression of Col2a1 gene expression, future studies should focus on this bHLH and its co-repressor proteins. We also observed Hes-1 binding after 7 days in pellet culture and the absence of TLE, which may be compatible with Hes-1's dual role of repressor and activator. Ju et al (44) showed that TLE was dismissed from Hes-1 as a consequence of PDGF treatment to permit neuronal stem cell differentiation, however Hes-1 remained bound to the DNA and was then part of an activation complex. It is possible that this also occurs during chondrogenesis.
We have located putative N-box and/or E-box domains in several chondrocyte marker genes including the proximal promoter region of Col9a1, Col11a2, CD-RAP, cartilage link protein and several domains in intron 1 of aggrecan in close proximity to an Sry/Sox consensus sequence (AACAAAG) (data not shown). Interestingly, one putative N-box domain in Col11a2 is identical to the Sox9 binding site that is important for Sox9 transcriptional activation (29). These observations suggest that the Notch and Sox9 signaling pathways are coordinated in an exquisite manner to repress or permit chondrogenesis.
Some cell types only express HES while other express HERP (14, 45). In these instances, homodimers are formed. However, cells expressing both bHLH proteins form heterodimers that show higher binding affinities compared to their homodimer counterparts (14). In this current study, Col2a1 is most likely repressed by a Hes-1/Hey-1 heterodimer complex, yet the presence of a Hey-1 homodimer may be more potent. Since aggrecan expression was only repressed following Hey-1 over expression, this indicates that both homo- and heterodimers may be present in hMSC and the ratio of these dimer combinations could modulate the extent by which genes are repressed.
In the chondrogenesis model, TGFβ induces removal of Notch signaling by two distinct mechanisms: by reducing Notch protein levels and by mediating removal of DNA bound Hes-1/TLE repressor complexes. In addition, TGFβ signaling promotes translocation of Smad2/3 from the cytosol into the nucleus, which effectively increases the concentration of functional Sox9, since this transcription factor is recruited as part of co-activator complex, that binds to the enhancer site of Col2a1 to initiate transcription (46). The formation of such active transcription complexes in concert with lower Notch signaling shifts the balance from repression to activation of Col2a1 transcription.
While understanding mechanisms that induce chondrogenic differentiation is essential, expanding knowledge related to systems that repress this process are required. The data presented in this current work may be useful for elucidating the role of these pathways in normal development and for application in regenerative medicine. Controlled modulation of cellular response will aid in regenerating tissue in vitro and may also be applied in modulating cellular responses in pathological conditions in vivo, such as osteoarthritis (OA). Notch signaling is important for many cellular processes such as cell proliferation, apoptosis and differentiation (47). Furthermore, Notch-1 is up regulated in OA affected tissues (48) and OA is characterized by inappropriate differentiation states (49). Notch signaling is already a candidate target for cancer therapy (50), and therefore could be modulated to initiate or repress differentiation state and disease progression.
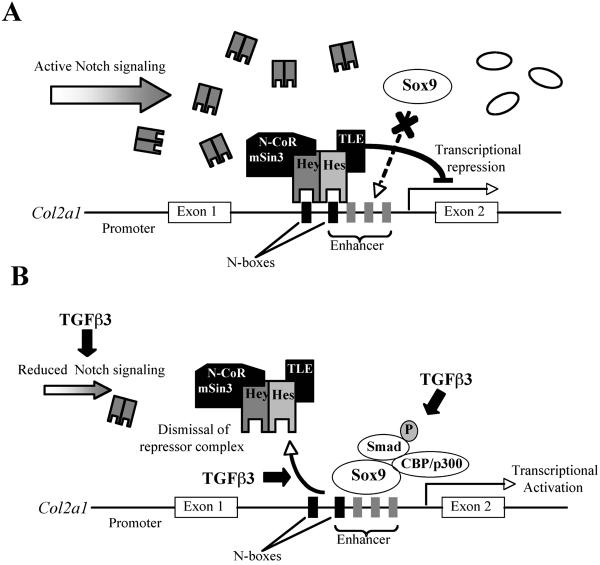
A summary of the proposed interactions between Notch signaling, Sox9 and the effect of TGFβ3 treatment on Col2a1 transcription and Hes-1/TLE complex dismissal is presented in Figure 5. The present study demonstrates that TGFβ-induced chondrogenesis is in part mediated by removal of the Notch transcriptional repression. TGFβ abrogates Notch suppression by reducing Notch-1 gene expression levels and by inhibiting the formation of active NICD and NICD-dependent events. The suppressive effects of Notch signaling are mediated by binding of the Notch targets Hes-1 and HERP-2/Hey-1. Hes-1 and Hey-1 actively suppress Col2a1 expression by binding to the Sox9 enhancer site located in intron 1 of Col2a1 and recruiting co-repressor proteins.
Figure 5. Proposed model of Notch signaling repression of Col2a1 gene transcription and the effect of TGFβ3 to permit Col2a1 expression.
In undifferentiated Mesenchymal stem cells Col2a1 is repressed by Notch signaling effectors such as Hes-1 and Hey-1, which bind to the enhancer site of Col2a1 and recruits co-repressor proteins such as TLE (Hes-1) and N-CoR (Hey-1) (panel A). To remove Notch's repressive influence, TGFβ3 stimulation (10 ng/ml) leads to a reduction in Notch gene expression and protein levels, initiates dismissal of the Hes-1/TLE repressor complex from DNA and leads to the formation of a Sox9 activation complex (46) that binds to the enhancer site to activate Col2a1 transcription (panel B).
ACKNOWLEDGEMENTS
We appreciate the support by Drs. Hiroshi Asahara and Masano Tsuda and Noboru Taniguchi. We are grateful to Ms Jean Valbracht and Ms Jianfen Chen for their technical support. The NICD antibody (bTAN 20) developed by Dr. S. Artavanis-Tsakonas, was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242.
Grant support: This work was supported by NIH Grant AG07996.
REFERENCES
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284(5415):770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Baron M. An overview of the Notch signalling pathway. Semin Cell Dev Biol. 2003;14(2):113–9. doi: 10.1016/s1084-9521(02)00179-9. [DOI] [PubMed] [Google Scholar]
- 3.Chiba S. Notch signaling in stem cell systems. Stem Cells. 2006;24(11):2437–47. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- 4.Frisen J, Lendahl U. Oh no, Notch again! Bioessays. 2001;23(1):3–7. doi: 10.1002/1521-1878(200101)23:1<3::AID-BIES1001>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 5.Kuroda K, Tani S, Tamura K, Minoguchi S, Kurooka H, Honjo T. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J Biol Chem. 1999;274(11):7238–44. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 6.Mizutani T, Taniguchi Y, Aoki T, Hashimoto N, Honjo T. Conservation of the biochemical mechanisms of signal transduction among mammalian Notch family members. Proc Natl Acad Sci U S A. 2001;98(16):9026–31. doi: 10.1073/pnas.161269998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egan SE, St-Pierre B, Leow CC. Notch receptors, partners and regulators: from conserved domains to powerful functions. Curr Top Microbiol Immunol. 1998;228:273–324. doi: 10.1007/978-3-642-80481-6_11. [DOI] [PubMed] [Google Scholar]
- 8.Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7(2):93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 9.Berezovska O, Jack C, McLean P, Aster JC, Hicks C, Xia W, et al. Aspartate mutations in presenilin and gamma-secretase inhibitors both impair notch1 proteolysis and nuclear translocation with relative preservation of notch1 signaling. J Neurochem. 2000;75(2):583–93. doi: 10.1046/j.1471-4159.2000.0750583.x. [DOI] [PubMed] [Google Scholar]
- 10.Karlstrom H, Bergman A, Lendahl U, Naslund J, Lundkvist J. A sensitive and quantitative assay for measuring cleavage of presenilin substrates. J Biol Chem. 2002;277(9):6763–6. doi: 10.1074/jbc.C100649200. [DOI] [PubMed] [Google Scholar]
- 11.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377(6547):355–8. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 12.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194(3):237–55. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 13.Maier MM, Gessler M. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem Biophys Res Commun. 2000;275(2):652–60. doi: 10.1006/bbrc.2000.3354. [DOI] [PubMed] [Google Scholar]
- 14.Iso T, Sartorelli V, Poizat C, Iezzi S, Wu HY, Chung G, et al. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol Cell Biol. 2001;21(17):6080–9. doi: 10.1128/MCB.21.17.6080-6089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J Biol Chem. 1994;269(7):5150–6. [PubMed] [Google Scholar]
- 16.Kim HK, Siu G. The notch pathway intermediate HES-1 silences CD4 gene expression. Mol Cell Biol. 1998;18(12):7166–75. doi: 10.1128/mcb.18.12.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujimori K, Fujitani Y, Kadoyama K, Kumanogoh H, Ishikawa K, Urade Y. Regulation of lipocalin-type prostaglandin D synthase gene expression by Hes-1 through E-box and interleukin-1 beta via two NF-kappa B elements in rat leptomeningeal cells. J Biol Chem. 2003;278(8):6018–26. doi: 10.1074/jbc.M208288200. [DOI] [PubMed] [Google Scholar]
- 18.Grbavec D, Stifani S. Molecular interaction between TLE1 and the carboxyl-terminal domain of HES-1 containing the WRPW motif. Biochem Biophys Res Commun. 1996;223(3):701–5. doi: 10.1006/bbrc.1996.0959. [DOI] [PubMed] [Google Scholar]
- 19.Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6(12B):2620–34. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 20.Crowe R, Zikherman J, Niswander L. Delta-1 negatively regulates the transition from prehypertrophic to hypertrophic chondrocytes during cartilage formation. Development. 1999;126(5):987–98. doi: 10.1242/dev.126.5.987. [DOI] [PubMed] [Google Scholar]
- 21.Fujimaki R, Toyama Y, Hozumi N, Tezuka K. Involvement of Notch signaling in initiation of prechondrogenic condensation and nodule formation in limb bud micromass cultures. J Bone Miner Metab. 2006;24(3):191–8. doi: 10.1007/s00774-005-0671-y. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe N, Tezuka Y, Matsuno K, Miyatani S, Morimura N, Yasuda M, et al. Suppression of differentiation and proliferation of early chondrogenic cells by Notch. J Bone Miner Metab. 2003;21(6):344–52. doi: 10.1007/s00774-003-0428-4. [DOI] [PubMed] [Google Scholar]
- 23.Goldring MB, Tsuchimochi K, Ijiri K. The control of chondrogenesis. J Cell Biochem. 2006;97(1):33–44. doi: 10.1002/jcb.20652. [DOI] [PubMed] [Google Scholar]
- 24.Harley VR, Clarkson MJ, Argentaro A. The molecular action and regulation of the testis-determining factors, SRY (sex-determining region on the Y chromosome) and SOX9 [SRY-related high-mobility group (HMG) box 9] Endocr Rev. 2003;24(4):466–87. doi: 10.1210/er.2002-0025. [DOI] [PubMed] [Google Scholar]
- 25.Bell DM, Leung KK, Wheatley SC, Ng LJ, Zhou S, Ling KW, et al. SOX9 directly regulates the type-II collagen gene. Nat Genet. 1997;16(2):174–8. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- 26.Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 1997;17(4):2336–46. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng LJ, Wheatley S, Muscat GE, Conway-Campbell J, Bowles J, Wright E, et al. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol. 1997;183(1):108–21. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- 28.Zhang P, Jimenez SA, Stokes DG. Regulation of human COL9A1 gene expression. Activation of the proximal promoter region by SOX9. J Biol Chem. 2003;278(1):117–23. doi: 10.1074/jbc.M208049200. [DOI] [PubMed] [Google Scholar]
- 29.Bridgewater LC, Walker MD, Miller GC, Ellison TA, Holsinger LD, Potter JL, et al. Adjacent DNA sequences modulate Sox9 transcriptional activation at paired Sox sites in three chondrocyte-specific enhancer elements. Nucleic Acids Res. 2003;31(5):1541–53. doi: 10.1093/nar/gkg230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekiya I, Tsuji K, Koopman P, Watanabe H, Yamada Y, Shinomiya K, et al. SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J Biol Chem. 2000;275(15):10738–44. doi: 10.1074/jbc.275.15.10738. [DOI] [PubMed] [Google Scholar]
- 31.Kou I, Ikegawa S. SOX9-dependent and -independent transcriptional regulation of human cartilage link protein. J Biol Chem. 2004;279(49):50942–8. doi: 10.1074/jbc.M406786200. [DOI] [PubMed] [Google Scholar]
- 32.Xie WF, Zhang X, Sakano S, Lefebvre V, Sandell LJ. Trans-activation of the mouse cartilage-derived retinoic acid-sensitive protein gene by Sox9. J Bone Miner Res. 1999;14(5):757–63. doi: 10.1359/jbmr.1999.14.5.757. [DOI] [PubMed] [Google Scholar]
- 33.Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res. 1998;238(1):265–72. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 34.Lee JW, Kim YH, Kim SH, Han SH, Hahn SB. Chondrogenic differentiation of mesenchymal stem cells and its clinical applications. Yonsei Med J. 2004;45(Suppl):41–7. doi: 10.3349/ymj.2004.45.Suppl.41. [DOI] [PubMed] [Google Scholar]
- 35.Sekiya I, Vuoristo JT, Larson BL, Prockop DJ. In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. Proc Natl Acad Sci U S A. 2002;99(7):4397–402. doi: 10.1073/pnas.052716199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardingham TE, Oldershaw RA, Tew SR. Cartilage, SOX9 and Notch signals in chondrogenesis. J Anat. 2006;209(4):469–480. doi: 10.1111/j.1469-7580.2006.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbero A, Ploegert S, Heberer M, Martin I. Plasticity of clonal populations of dedifferentiated adult human articular chondrocytes. Arthritis Rheum. 2003;48(5):1315–25. doi: 10.1002/art.10950. [DOI] [PubMed] [Google Scholar]
- 38.Martin I, Jakob M, Schafer D, Dick W, Spagnoli G, Heberer M. Quantitative analysis of gene expression in human articular cartilage from normal and osteoarthritic joints. Osteoarthritis Cartilage. 2001;9(2):112–8. doi: 10.1053/joca.2000.0366. [DOI] [PubMed] [Google Scholar]
- 39.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134(7):1243–51. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 40.Ross DA, Hannenhalli S, Tobias JW, Cooch N, Shiekhattar R, Kadesch T. Functional analysis of Hes-1 in preadipocytes. Mol Endocrinol. 2006;20(3):698–705. doi: 10.1210/me.2005-0325. [DOI] [PubMed] [Google Scholar]
- 41.Vujovic S, Henderson SR, Flanagan AM, Clements MO. Inhibition of gamma-secretases alters both proliferation and differentiation of mesenchymal stem cells. Cell Prolif. 2007;40(2):185–95. doi: 10.1111/j.1365-2184.2007.00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, et al. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. Embo J. 2006;25(20):4833–42. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strom A, Castella P, Rockwood J, Wagner J, Caudy M. Mediation of NGF signaling by post-translational inhibition of HES-1, a basic helix-loop-helix repressor of neuronal differentiation. Genes Dev. 1997;11(23):3168–81. doi: 10.1101/gad.11.23.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ju BG, Solum D, Song EJ, Lee KJ, Rose DW, Glass CK, et al. Activating the PARP-1 sensor component of the groucho/ TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell. 2004;119(6):815–29. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 45.Chin MT, Maemura K, Fukumoto S, Jain MK, Layne MD, Watanabe M, et al. Cardiovascular basic helix loop helix factor 1, a novel transcriptional repressor expressed preferentially in the developing and adult cardiovascular system. J Biol Chem. 2000;275(9):6381–7. doi: 10.1074/jbc.275.9.6381. [DOI] [PubMed] [Google Scholar]
- 46.Furumatsu T, Tsuda M, Yoshida K, Taniguchi N, Ito T, Hashimoto M, et al. Sox9 and p300 cooperatively regulate chromatin-mediated transcription. J Biol Chem. 2005;280(42):35203–8. doi: 10.1074/jbc.M502409200. [DOI] [PubMed] [Google Scholar]
- 47.Zlobin A, Jang M, Miele L. Toward the rational design of cell fate modifiers: notch signaling as a target for novel biopharmaceuticals. Curr Pharm Biotechnol. 2000;1(1):83–106. doi: 10.2174/1389201003379013. [DOI] [PubMed] [Google Scholar]
- 48.Hiraoka K, Grogan S, Olee T, Lotz M. Mesenchymal progenitor cells in adult human articular cartilage. Biorheology. 2006;43(3–4):447–54. [PubMed] [Google Scholar]
- 49.Tchetina EV, Squires G, Poole AR. Increased type II collagen degradation and very early focal cartilage degeneration is associated with upregulation of chondrocyte differentiation related genes in early human articular cartilage lesions. J Rheumatol. 2005;32(5):876–86. [PubMed] [Google Scholar]
- 50.Shih Ie M, Wang TL. Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res. 2007;67(5):1879–82. doi: 10.1158/0008-5472.CAN-06-3958. [DOI] [PubMed] [Google Scholar]