Abstract
In this report the applicability of an improved method of image segmentation of infrared microspectroscopic data from histological specimens is demonstrated. Fourier transform infrared (FT-IR) microspectroscopy was used to record hyperspectral data sets from human colorectal adenocarcinomas and to build up a database of spatially resolved tissue spectra. This database of colon microspectra comprised 4120 high-quality FT-IR point spectra from 28 patient samples and 12 different histological structures. The spectral information contained in the database was employed to teach and validate multilayer perceptron artificial neural network (MLP-ANN) models. These classification models were then employed for database analysis and utilised to produce false colour images from complete tissue maps of FT-IR microspectra. An important aspect of this study was also to demonstrate how the diagnostic sensitivity and specificity can be specifically optimised. An example is given which shows that changes of the number of teaching patterns per class can be used to modify these two interrelated test parameters. The definition of ANN topology turned out to be crucial to achieve a high degree of correspondence between the gold standard of histopathology and IR spectroscopy. Particularly, a hierarchical scheme of ANN classification proved to be superior for the reliable classification of tissue spectra. It was found that unsupervised methods of clustering, specifically agglomerative hierarchical clustering (AHC), were helpful in the initial phases of model generation. Optimal classification results could be achieved if the class definitions for the ANNs were carried out by considering the classification information provided by cluster analysis.
Keywords: infrared imaging, artificial neural network, hierarchical cluster analysis, image segmentation, FT-IR microspectroscopy, tissue classification
1. INTRODUCTION
Fourier transform infrared (FT-IR) microspectroscopy has been employed since more than a decade to study human tissues and—in particular—of pathological states within tissues [1–5]. The technique may provide spatially resolved structural and compositional information of the histological specimens under investigation and shows in combination with digital imaging techniques a great promise for in-vivo and ex-vivo medical diagnosis. False colour images produced by IR image segmentation methodologies are directly comparable to outcomes of standard histological staining protocols and can be interpreted also by non-spectroscopists. A number of different IR imaging strategies have been proposed in the past. Among them the univariate concept of chemical imaging still enjoys great popularity. Chemical imaging is based on the reduction of an array of infrared spectra to a functional group map [6]. The images are easy to generate and, from a spectroscopic point of view, also easy to interpret. The main drawback of the technique is that only a very small fraction of the available spectral information is used [7,8]. Nevertheless, it is well documented for a number of examples that functional group mapping produces sufficiently high image contrast that permits visualisation the spatial distribution of defined tissue structures [1,2,9].
We found, however, that this technique is insufficient to provide a reasonable and flexible differentiation criterion if chemical mapping is applied to larger data sets that include spectra from many patients [3,5,10]. Several techniques of multivariate imaging attempt to address this shortcoming by analysing large fractions or even the complete spectral information. Among multivariate imaging methods, non-supervised techniques became popular because they allow to produce IR images with the full spectral contrast that often corresponds to the classical histopathological scheme. Specifically, various types of cluster analysis and among them agglomerative hierarchical clustering (AHC) in a combination of a correlation distance measure (D-values) and Ward’s clustering method turned out to be particularly suitable for IR image segmentation. In a comparative study of cluster imaging techniques it was shown, that the highest degree of correspondence between histopathology and IR spectroscopy was achieved when the AHC algorithm was applied [11]. It was found furthermore, that the concepts of k-means and fuzzy-C-means clustering are less effective, but are significantly less CPU intensive image segmentation methods. Drawbacks of the AHC segmentation technique are the very high computing requirements, which become more and more important when large spectral data sets have to be analysed [11].
An important characteristic of non-supervised clustering methods is the tendency to partition the data according to the overall variance. In view of the fact that usually only a small fraction of a samples overall spectral variance is intra-class specific, this strategy is quite efficient (provided spectra from only one tissue sample, i.e. from one patient, are analysed). In these cases the highest spectral variance is found mostly between spectra from different types of tissues (inter-class variance). However, if more than only one sample is examined, the intra-class variance includes now also the much larger variance present between spectra from identical tissue structures but different patients. In these cases, the intra-class variance may be at the order of the inter-class variance. Non-supervised classification methodologies are generally unable to separate between inter- and intra-class variance and consequently, the degree of correspondence with sample histology is decreased. Thus, non-supervised classification is valuable as an explorative technique but may be inappropriate for routine analysis of IR spectral maps of tissues.
It has been noticed that supervised multivariate classification strategies, for example by multilayer perceptron (MLP) artificial neural network (ANN) models with supervised learning are the techniques of choice for the development of effective and robust classifiers for IR-based classification of tissue structures [5]. These supervised techniques can be efficiently optimised by pre-selecting the appropriate spectral features from the spectral data [12,13]. Furthermore, the classification results of the above-mentioned combination of methods will strongly depend on the type of spectral pre-processing. We found that spectral quality criteria, a baseline correction routine (derivatives) and normalisation are essential prerequisites for the development of robust classification models that can be used in practise [3,5].
The outlined classification strategy of a preceding feature selection followed by artificial neural network classification is based on a very high number of free variables, that is over-fitting of the prediction models is a real risk [13,14]. Thus, it is absolutely necessary to evaluate the model by independent sets of patterns not used during the design of the model [13,14]. Consequently, the spectral data should be split in subsets for teaching, a subset for internal validation and a completely independent—ideally blinded—subset for external validation (testing). The process of model development—and this includes also the selection of the spectral features—should therefore be carried out exclusively by using teaching and internal validation data subsets. Therefore, the outlined strategy is the only way to reveal over-fitting of the ANN models and to obtain objective values for statistical test parameters such as accuracy, sensitivity or specificity for the new diagnostic method [12,15].
In the present study we will present new results obtained from the analysis of about 1.5 million FT-IR microspectra from human colorectal adenocarcinoma specimens. We will describe the strategy of extracting single spectra from this huge amount of data and we demonstrate how a database of colorectal reference spectra can be established. The main focus of this report is to present a strategy for the development of classification models for the IR microspectroscopic characterisation of tissues.
2. EXPERIMENTAL
2.1. Sample description and sample preparation
Colorectal adenocarcinoma tissue samples from 28 patients were obtained from the tissue data bank at the Robert-Rössle-Clinic at the Max-Delbrück-Centrum for Molecular Medicine in Berlin. They originated from coecum, colon ascendens, transversum or descendens, sigma and rectum and the histopathological grade of malignancy was established as well differentiated (G1), moderately differentiated (G2) or poorly differentiated (G3). Samples were stored until cryosectioning at a temperature of −80°C. Cryosectioning was performed at temperatures of −18 to −22°C. In order to avoid spectral contaminations associated with the use of embedding medium, the frozen tissue samples were mounted on the cryotome sample holder by means of freezing water. For FT-IR microspectroscopy 8 µm thin tissue slices were thaw-mounted onto CaF2 windows of 1 mm thickness (Korth Kristalle, Germany). The specimens were stained after FT-IR measurements by Haematoxylin/Eosin (H&E) and microphotographs of the imaged areas were obtained to correlate the IR images with histopathology.
2.2. Data collection
Infrared spectra were collected in transmission mode using a Spectrum Spotlight One FT-IR spectrometer from PerkinElmer coupled to a Spectrum Spotlight 300 infrared microscope. The microscope is equipped with a linear 16 × 1 element (400 × 15 µm2) MCT (HgCdTe) array detector. The microscope optics permits 1:1 or 4:1 imaging, resulting in sample areas of 25 × 25 or 6.25 × 6.25 µm2 projected on each detector element. In this study spectra were recorded in the 4:1 imaging mode. In the 4:1 mode the lateral spatial resolution was found to be approximately 12 µm at 6 µm wavelength (corresponds to 1667 cm−1, amide I region) [16]. A specially designed microscope box was purged by dry air to reduce spectral contributions from atmospheric water vapour and CO2. Nominal spectral resolution was 4 cm−1. Usually, 16 scans were averaged per sample spectrum and apodised applying a Norton-Beer apodisation function for Fourier transformation. Interferograms were zero-filled by a factor of 2. In order to increase the signal-to-noise-ratio background spectra were recorded with 512 scans.
2.3. Data processing
Spectral data were analysed by means of CytoSpec (CytoSpec, Inc. Croton-On-Hudson, NY, USA) and NeuroDeveloper (Synthon GmbH, Heidelberg, Germany). While CytoSpec is a software package specifically designed for the generation of infrared images from large IR mapping data, the NeuroDeveloper software combines modules for spectral feature selection, ANN model development (including modular ANN models) and ANN-based classification. A detailed description of both software packages can be found elsewhere [17,18].
Spectral data were processed using a Fujitsu-Siemens 64 bit Celsius V810 workstation which is equipped with two 2.2 GHz AMD Opteron CPUs and 8 GB of RAM (Fujitsu-Siemens Computers GmbH, Germany). Microsoft Windows XP 64 bit version was chosen as the operating system since it provides significantly enlarged address space for application software.
Pre-processing of the raw spectral data was carried out in CytoSpec’s batch pre-processing mode which permits automation of all steps of data pre-processing. Automated pre-processing included a conversion from transmittance to absorbance spectra, tests for spectral quality and normalisation. The spectra quality test consisted of three separate checks: for water vapour content, for the signal-to-noise ratio (S/N) and for sample thickness. All spectra that have passed these tests were subsequently converted into first derivative spectra (Savitzky-Golay algorithm, seven smoothing points) and vector normalised. Vector normalization was performed in the spectral region of 950–1480 cm−1.
2.4. Extraction of the database spectra from the maps
In order to evaluate the spatial distribution of tissue structures within a given IR data set we have routinely applied the approach of agglomerative hierarchical cluster imaging to all individual spectral maps. Details of this particular image segmentation method can be found in the literature [11,19]. In the present study we used so-called D-values as spectral distance measures (normalised Pearson’s correlation coefficients), and Ward’s algorithm for hierarchical clustering [20,21].
A detailed examination of each of the samples and the assignment of clusters to pre-defined classes of histopathological structures could be subsequently carried out on the basis of photomicrographs of the post-stained tissue specimens and the re-assembled cluster maps. We extracted a representative number of spectra, usually between 10 and 15 per map and cluster. The extracted point spectra were then used to build up a database of IR reference microspectra from all histologically defined structures of the human colon. Since the database contained information from many patients, inter-class as well as the intra-class variance were adequately represented. An overview of the database is given in Table I. This colon database was employed in the following to teach and validate MLP-ANN classifiers.
Table I.
The colon database
| Class name | # of spectra | |
|---|---|---|
| Ia | Crypts | 153 |
| Ib | Mucin | 385 |
| IIa | Necrosis | 169 |
| IIb | Lamina propria mucosae | 128 |
| IIc | Lymphocytes | 41 |
| IIIa | Tunica muscularis | 138 |
| IIIb | Lamina muscularis mucosae | 170 |
| IIIc | Vessel (blood, lymph) | 193 |
| IVa | Submucosa | 34 |
| IVb | Fibrovascular connective tissue | 878 |
| V | Adenocarcinoma | 1805 |
| VI | Fat tissue | 26 |
Overview on sample numbers and sample description.
2.5. ANN analysis
The general strategy of ANN analysis in this study included the procedure of teaching and optimising the MLP network models followed by testing the classifiers with independent (external) validation data sets. Teaching and internal validation were carried out on the basis of IR microspectra with known class assignment, that is with spectra from the colon database. External validation (testing) of the classifier was made be generating ANN images from complete infrared spectral maps. Thus, the classifiers were created with database spectra while model assessment was made by comparing the ANN images and the equivalent photomicrographs of stained tissue specimens.
For network teaching the raw spectral data of the colon database were first pre-processed as already outlined. After this, the effective spectral resolution was reduced by a factor of 6 (averaging) and then 60–85 spectral features were chosen by a covariance analysis procedure implemented in the NeuroDeveloper software package [12]. We used connected three layer feed-forward MLP-ANNs consisting each of a layer of input, hidden and output neurons. Teaching of the ANNs was carried out by utilising the resilient back-propagation (rprop) algorithm [22]. The number of neurons of the input layer corresponds to the length of the input pattern and varied between 60 and 85. Moreover, the number of neurons in the hidden layer was usually set to 4 and the number of output neurons in MLP-ANNs equalled the number of classes.
In the present study a hierarchical system of ANNs was employed. The primary advantage of this method is the capability to separately train and validate small and flexible networks that can be combined afterwards to build up large modular ANN systems [12,14,23]. Furthermore, the use of modular networks permitted to employ specifically optimised combinations of spectral features for each separate ANN module. The ANN model development module of Synthon’s NeuroDeveloper offered the respective software implementation. A description of modular ANN models can be found elsewhere [12,14,23].
ANN imaging on IR mapping data sets was carried out via a software interface between CytoSpec and the NeuroDeveloper software package. Based on NeuroDeveloper-ANN models, this interface can be used to perform image segmentation from the external validation data, that is complete infrared spectral maps. The interface is designed such that spectral pre-processing, feature selection and also ANN classification of the external validation data are automatically performed in the same way as for the teaching data.
3. RESULTS AND DISCUSSION
Infrared microspectroscopic imaging is a data intensive technique which implies storage and processing of enormous amounts of data. While microscopy in the visual range (380–700 nm wavelength) usually provides 24, or 32 bit of spectral information per image pixel, infrared microspectroscopy produces per pixel a complete spectrum of up to several thousands IR intensity values. Therefore, a single point IR spectrum has a size between 5 and 15 kB and an average IR map of 300 × 300 pixel spectra may be as large as 900 MB. This enormous amount of quantitative and qualitative sample information is principally available to define purely local decision criteria for segmentation. Our goal was therefore to develop a computational approach to image segmentation of spatially resolved IR data that is practically useful in histology and histopathology. In order to achieve such a method, it is important that the segmentation method should have the following properties:
IR image segmentation should reflect the classical classification scheme in histology and histopathology. In the final stage of method development precise definitions of parameters reflecting the degree of correspondence between the image segmentation method and histopathology (sensitivity, specificity, etc.) should be available.
In order to be of practical use, IR imaging should be a rapid segmentation method that is applicable in routine use also for very large data sets. Ideally, the IR imaging method should run in time nearly linear in the number of image pixels. This is important as the CPU time of unsupervised agglomerative hierarchical cluster imaging which is frequently used in IR imaging scales with the squared number of pixels.
3.1. The strategy
In this study a relatively large number of infrared microspectra has been acquired. In total we recorded more than 1.5 million spectra (35 infrared spectral maps) from 28 patient samples. A complete map usually comprised between 6000 and 100 000 spectra taken from rectangular tissue regions of a size between 500 and 2000 µm edge length. As outlined earlier, cluster images were routinely produced from each of the IR maps. The settings for the number of clusters was chosen such, that a high correlation between histopathology of the tissue section and the IR image was attained. Spectra of the tissue structures of interest were then extracted as outlined above and transferred into our database of IR reference spectra. In general, between 10 and 15 spectra per histological class and patient sample were selected.
An overview of the database composition is given in Table I. This table shows that some of the pre-defined spectral classes such as fat tissue or submucosa are clearly outnumbered by the class of adenocarcinoma. This is not solely due to the availability of the spectra—not all spectral classes can be found in all tissue specimens. One of the main goals for establishing the ‘colon database’ was to systematically investigate the spectral differences between adenocarcinomas at different grading levels. However, the number of patient samples examined so far seems to be still too low to study these grading-related spectral changes. Thus, the high number of spectra of the class ‘adenocarcinoma’ is due to the fact that this class is actually composed of three distinct tumour grading classes G1–G3 each representing lower numbers of spectra. Other spectral classes such ‘fat’, ‘submucosa’ or ‘mucin’ displayed highly distinctive spectral features. Due to their unique mid-infrared spectral patterns, only a few number of spectra were required for an adequate representation of these classes in the database.
3.2. Cluster analysis
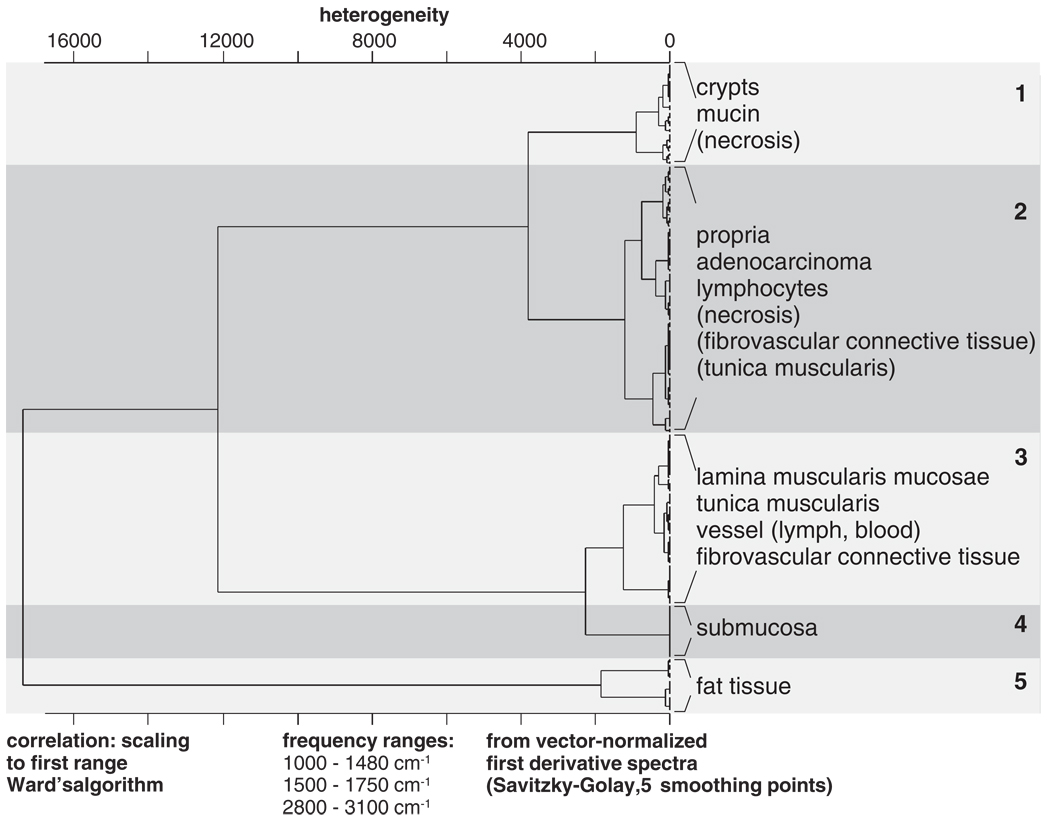
Figure 1 displays the results of AHC of a selection of individual tissue spectra from the database. In this approach at least two spectra per class and patient were used. The dendrogram illustrates that spectra from fat tissue (cluster 5) and also from muscle tissues and the submucosa (clusters 3 and 4) can be easily differentiated from spectra of clusters 1 and 2. Spectra from fat tissues are known to exhibit very intense signals in the CH-stretching region (2800–3050 cm−1). As these compounds show also a prominent carbonyl ester band (νsy > C=OEster, 1737 cm−1), and symmetric deformation bands of methylene (δsy–CH2, ‘scissoring’: 1468cm−1) and methyl groups (δsy–CH3, ‘umbrella’: 1379 cm−1), they can be easily identified already by basic methods of data analysis. The latter statement holds true also for IR spectra of clusters 3 and 4, that is for spectra from distinct smooth muscle structures, fibrovascular connective tissue and the submucosa. These tissue types are known to contain a certain amount of collagen that exhibits a series of highly characteristic IR bands in the spectral range of 1100–1300 cm−1[24]. Obviously, most of the differentiation between clusters 3 + 4 and the remaining spectral classes (1, 2, 5) is due to these collagen bands. On the other hand, spectral signs from contractile elements of smooth muscle fibrils which should be present only in the spectra of cluster 3 are apparently less discriminative.
Figure 1.
Dendrogram produced by agglomerative hierarchical clustering (HCA) of representative FT-IR spectra from the colon database. At least two spectra per spectral class and patient were selected for the analysis. The dendrogram illustrates that spectra from fat tissue and the submucosa can be easily differentiated from the majority of the database spectra. HCA classification of the remaining spectra yielded in a number of cases ambiguous results illustrating that unsupervised cluster analysis alone cannot be used to attain consistent classification results (see text for details).
The spectral distance levels between the remaining histopathological structures, that is spectra of clusters 1 and 2, permitted further differentiation between histological classes of the colon. As it is illustrated by the dendrogram of Figure 1 spectra from the crypts and mucin-containing structures (such as extracellular ‘mucin lakes’) can be separated from cluster 2. A closer inspection of the spectra of cluster 1 yielded a number of common spectral properties since these spectra exhibited in the carbohydrate region the typical signatures of mucin (data not shown). As it is indicated in Figure 1, a number of spectra from the tunica muscularis and also from fibrovascular connective tissue appear in cluster 2. This finding illustrates that clustering alone, that is without combination of any type of spectral feature selection, cannot be used to attain consistent classification results in IR microspectroscopy of tissues. Thus, HCA is a valuable explorative tool in tissue spectroscopy but certainly not the appropriate technique for routine analysis of IR spectral maps.
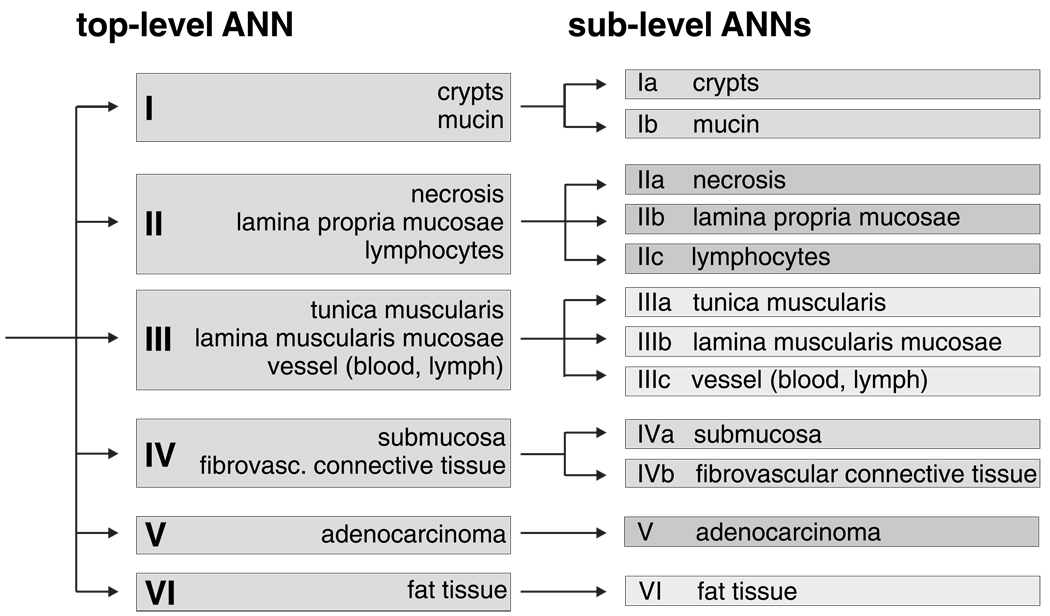
Figure 2 illustrates how the classification results of AHC were utilised to design the system of hierarchically organised (modular) ANNs. A so-called ‘top-level ANN’ which is shown in the left part of Figure 2 assigns FT-IR spectra to one of the main tissue classes I–VI (see inset). Depending upon the activations of the top-level ANN output neurons, more specific ‘sublevel ANNs’ classify then the IR microspectra in a more detailed way. This is demonstrated by the four sublevel ANNs at the right part of Figure 2 (cf. Ia/b–IVa/b). We have used the classification information of AHC particularly for the class definitions of the top-level net. To give an example, cluster 5 (fat tissue) of the dendrogram of Figure 1 corresponds to class VI of the top-level ANN. Also, class I of the top-level ANN was defined on the basis of the class assignment of spectra from crypts and mucin by AHC. On the other hand, practical demands may require the definition of additional top-level classes which usually do not form separate clusters in AHC. In the present work this is the case for spectra of the class ‘adenocarcinoma’. Thus, class definitions for the top-level net were predominantly made in view of spectral similarities of the tissues as revealed by AHC, but considering also practical aspects of the methodology.
Figure 2.
The hierarchical (modular) classification scheme for ANN analysis of IR microspectra from the human colon.
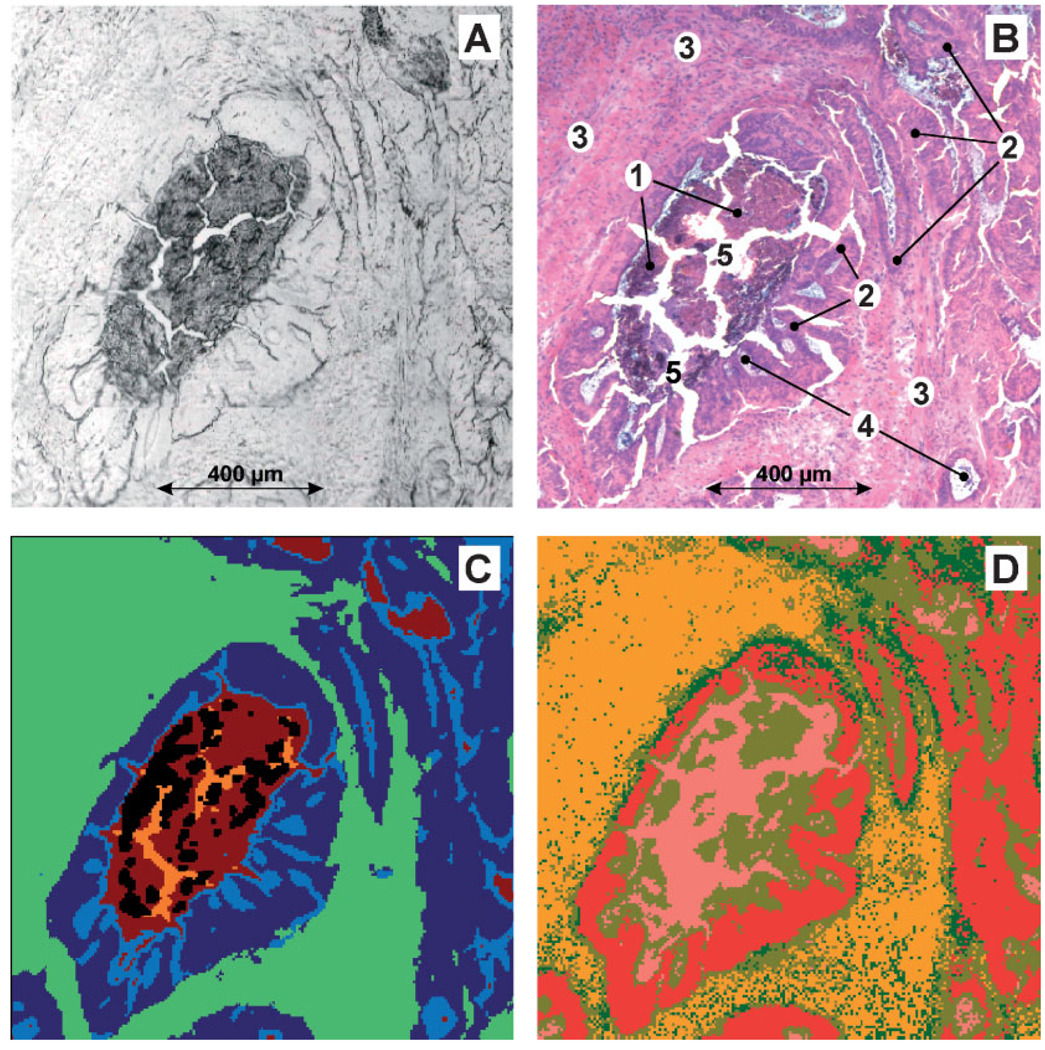
Figure 3 shows the results of image segmentation produced by AHC and the top-level ANN. In this example 194 × 198 IR microspectra from a well differentiated G1 adenocarcinoma from the human rectum were collected and analysed (sample B4205/94). The tissue area was 1206 × 1231 µm2 in size. While panel A of Figure 3 shows the photomicrograph of the unstained cryosection before the IR measurements, panel B displays the same tissue area after staining with H&E (the widening of tissue clefts from A to B is due to water treatment during staining). In panels A–D the central shape is formed by necrotic tumour cells (1). These necrotic cells are surrounded by vital tumour epithelium (2). Neoplastic epithelium can be found also in the right parts of the images. Panels A–D show furthermore non-cancerous fibrovascular connective tissue (3) which is arranged around the central adenocarcinoma structure, extracellular ‘mucin lakes’ (4) and tissue clefts (5).
Figure 3.
FT-IR microspectroscopic imaging of a cryostat section from a well differentiated (G1) adenocarcinoma of the rectum. (A) Photomicrograph of the unstained cryostat section. Sample area: 1206 × 1231 µm2. (B) Tissue area shown in A after IR microspectroscopy and staining with H&E. 1, necrotic detritus; 2, vital tumour cells; 3, fibrovascular connective tissue and smooth muscle strands; 4, secretion products (mucin); 5, tissue clefts. (C) IR imaging based on 192 × 194 microspectra of the tissue area shown in panel A and hierarchical cluster analysis (five class classification approach). (D) Imaging based on FT-IR microspectroscopy and ANN analysis (‘top-level net’). See text for details.
These main tissue structures can be successfully differentiated by routine AHC imaging (cf. panel C, ‘routine AHC imaging’ means that only spectra from the analysed map are used). In this five-class-classification approach the formation of clusters for fibrovascular connective tissue and smooth muscle strands (green), for vital tumour epithelium (dark blue) and their secretion products (mostly mucin, light blue) is observed. Clusters of IR microspectra from necrotic tumour cells and tissue clefts are colour-encoded brown or orange, respectively. The results of clustering from this sample correspond quite well with the cluster analysis of the complete database (Figure 1). Again, spectra from smooth muscle structures and fibrovascular connective tissue are closely related, whereas spectra originating from vital or necrotic parts of the adenocarcinoma form separate clusters, respectively. It should be pointed out that this result was obtained by using the complete usable spectral information (no feature selection). Again, the purely data-driven explorative tool of AHC gave a suggestion of how the architecture of an ANN classification system should be designed.
The results of image re-assembling by the top-level net are displayed in Figure 3D. It should be emphasised, that the top-level net was created (taught) without using spectra from sample B4205/94. In image 3D, the class ‘mucin’ (probably secretion products in tissue clefts) is coloured pink, whereas gold and red encode necrotic or vital tumour epithelium (classes II and V). Furthermore, a small number of pixel spectra was identified as smooth muscle structures (class III, dark green). Connective tissue structures from the sub-mucosa or fibrovascular connective tissue (class IV) are coloured orange. Although the class of connective tissue seems to be somewhat over-represented, the example of Figure 3D gives a good impression on the degree of correspondence between IR microspectroscopy and histopathology and demonstrates the potential of ANN imaging for the analysis of tissue sections. The discussion of how this ‘over-representation’ of a defined spectral class can be modified is given later.
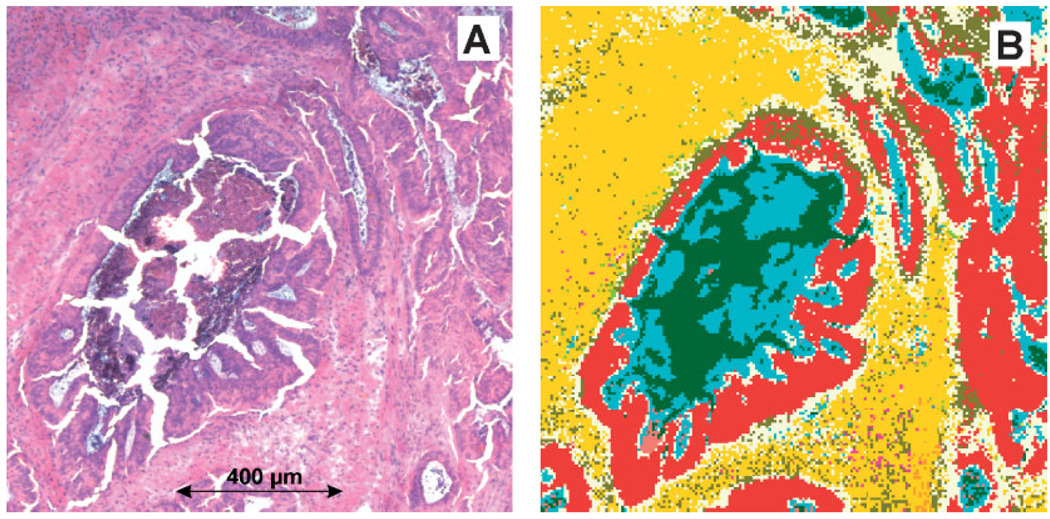
We turn now to the discussion of Figure 4 which shows the classification results obtained by ANNs with a hierarchically organised network architecture. In this example, the network ‘combinet’ consisting of the top-level and four separate sublevel nets (see Figure 2) was used to perform segmentation of the IR imaging data from Figure 3. It is important to note that exactly the same top-level net was used in the examples of Figure 3D and Figure 4B. The colour class assignment as well as the number and percentage of spectra per class can be taken from Table II.
Figure 4.
FT-IR microspectroscopic imaging of a cryostat section from a well differentiated (G1) adenocarcinoma of the rectum. (A) Photomicrograph of the H&E stained cryostat section. Sample area: 1206 × 1231 µm2. (B) Imaging based on FT-IR microspectroscopy and ANN analysis (‘combinet’). See text for details.
Table II.
Classification results of the hierarchical network (‘combinet’) on FT-IR microspectroscopic imaging data obtained from a cryosection through a human colorectal adenocarcinoma (B4205/94)
| Class name | Colour code | # of spectra | Percentage [%] | Mean activation | |
|---|---|---|---|---|---|
| Ia | Crypts | Salmon | 116 | 0.3 | 0.97 |
| Ib | Mucin | Dark green | 3313 | 8.62 | 1.00 |
| IIa | Necrosis | Aqua | 4567 | 11.89 | 0.98 |
| IIb | Lamina propria mucosae | Light yellow | 6189 | 16.11 | 0.98 |
| IIc | Lymphocytes | Green yellow | 201 | 0.52 | 0.90 |
| IIIa | Tunica muscularis | Orange | 73 | 0.19 | 0.91 |
| IIIb | Lamina muscularis mucosae | Olive | 3012 | 7.84 | 0.99 |
| IIIc | Vessel (blood, lymph) | Deep pink | 132 | 0.34 | 0.82 |
| IVa | Submucosa | Blue | 1 | 0 | 0.01 |
| IVb | Fibrovascular connective tissue | Gold | 11 323 | 29.48 | 1.00 |
| V | Adenocarcinoma | Red | 9485 | 24.69 | 0.73 |
| VI | Fat tissue | Beige | 0 | 0 | — |
| Unclassified spectra | Black | 0 | 0 | — |
The corresponding IR image is given in Figure 4B. Colour encoding in this figure was made according to the table content of columns 2 and 3.
Panel A of Figure 4 shows for comparative purposes again the histoarchitecture of the post-stained tissue of specimen B4205/94. A comparative examination of Figure 3D and Figure 4B reveals that the classification results for the classes ‘adenocarcinoma’ (red) and ‘fat tissue’ (beige) remain unchanged. This is not surprising as no sublevel nets were defined to further evaluate the output activation values of these particular classes (cf. Figure 2). As it is shown in Figure 3D and Figure 4B most of the predictions by the sublevel nets correlate quite well with histopathology. To give an example, spectra from the tissue clefts in the central shape of Figure 4B were classified by sublevel net #I as ‘mucin’ (dark green). This is correct, we have stated earlier that these clefts contain mostly secretion products such as mucin, and cell debris. Furthermore, areas of necrotic tumour cells have been identified by sublevel net #II in accordance with histopathology as belonging to the class ‘necrosis’ (aqua). On the other hand, sublevel net #II classified a large fraction of the spectra into the class ‘lamina propria mucosae’ (light yellow). The classification of this particular class is probably incorrect. As it can be taken from Figure 4B, the class ‘lamina propria mucosae’ is mostly found as a ‘transitional state’ between the classes ‘adenocarcinoma’ (red) and fibrovascular connective tissue, a class that is once again highly correlated with histology (gold). Thus, the results in Figure 4B demonstrate in an exemplary manner not only the potentials, but also the problems of the ANN imaging methodology. It turned out that an adequate representation of all pre-defined spectral classes in the network’s teaching phase is crucial for model optimisation. With reference to the example of Figure 4B, only sublevel net #II (and not the complete combinet-classifier) should be re-trained with additional data from the classes ‘necrosis’, ‘lamina propria mucosae’ and ‘lymphocytes’.
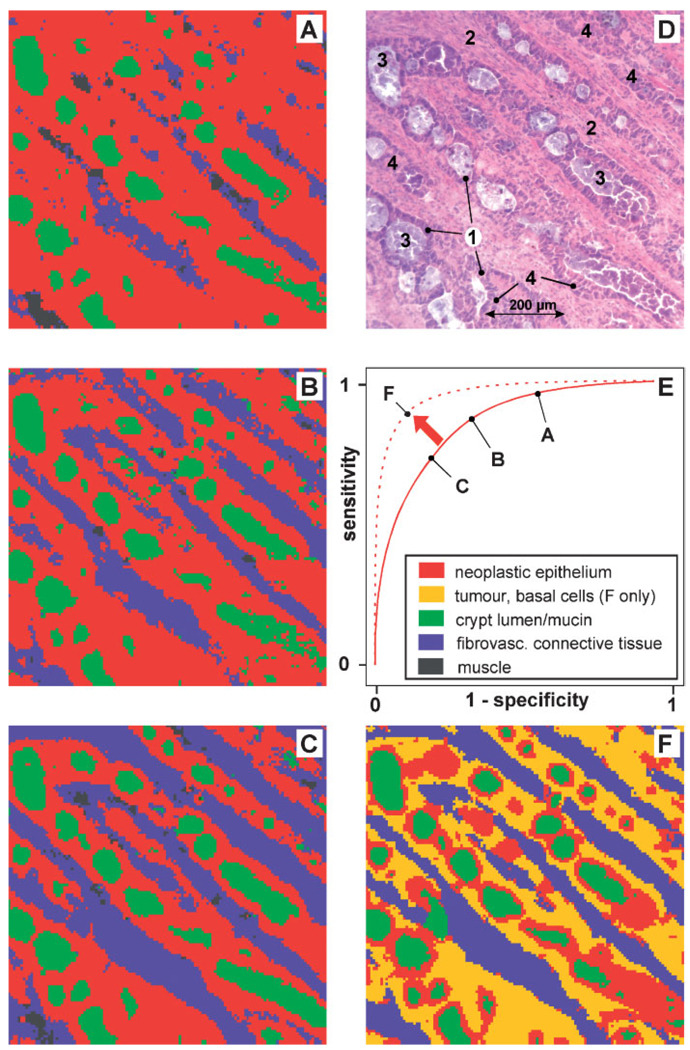
In the following we will give now a number of examples that illustrate how MLP-ANN models can be optimised in terms of diagnostic sensitivity or specificity. Figure 5 shows IR maps re-assembled on the basis of IR data of a well differentiated (G1) rectal adenocarcinoma (B279/01) and optimised ANN classification models. For the example of Figure 5, 128 × 123 IR microspectra from a tissue section area of a size of 794 × 762 µm2 were obtained. For a comparison with histology the tissue specimen was routinely stained by H&E after the IR measurements. The corresponding microphotograph (panel D) displays neoplastic crypts composed of absorptive epithelia (1) and basal cells (4). The neoplastic crypts are separated by fibrovascular connective tissue (2) and filled with detritus (loose material after cell death) and products of secretion (3). Secretion product could be spectroscopically identified as mucin (not shown).
Figure 5.
Optimisation of ANNs: illustration of the dependency of sensitivity/specificity on the number of spectra from mucosa structures used for network teaching. (A) ANN image reassembled from FT-IR microspectra of the colon database (four-class classification trial). Very high sensitivity, but low specificity for spectra from the class ‘adenocarcinoma’. (B) Same as A. Moderately improved specificity and high sensitivity for the class ‘adenocarcinoma’ (C) same as A and B. Relatively high specificity and sensitivity for the class ‘adenocarcinoma’ (D) photomicrograph of the adenocarcinoma cryosection after post-staining by H&E. Neoplastic crypts are composed of absorptive epithelia (1) and basal cells (4). The crypts are separated by proliferated fibrovascular connective tissue (2) and filled with detritus and products of secretion (3). (E) Theoretical interrelationship of sensitivity and specificity (receiver operating characteristics, ROC) (F) five-class-classification approach. Sensitivity and specificity can be increased by introducing new spectral classes (see inset).
ANN classification models were taught and validated with database microspectra. Again, teaching and internal validation were carried out by the exclusive use of spectra from other patients. For the examples of IR imaging of the left column of Figure 5 monolithic four-class-classification ANN models were generated (see inset of panel E for colour class assignment). A closer examination of panels A–C reveals decreasing probabilities for classification of a given pixel spectrum as ‘adenocarcinoma’ and vice versa increasing probabilities to fall into another class such as ‘fibrovascular connective tissue’. While in example panel 5A about 76% of all spectra were classified as ‘adenocarcinoma’ this number drops down in panel 5B to 60% and reaches 45% in panel 5C. On the other hand, the fraction of spectra classified as fibrovascular connective tissue is increasing at the same time from 10% (panel 5A) to 25% (panel 5B) and finally 36% (panel 5C). In terms of statistical classification criteria, the sensitivity of the diagnosis ‘adenocarcinoma’ made in panel 5A is fairly high, because almost all of the adenocarcinoma spectra were classified as adenocarcinoma and the number of false negatives is close to zero. On the other hand, we observe a high number of false positives since many spectra from other spectral classes were classified as adenocarcinoma. Thus, the image of panel 5A illustrates an example with high sensitivity but poor diagnostic specificity. The situation depicted in panel 5B differs from panel 5A in a reduced number of false positive spectral diagnoses. Interestingly, the number of false negatives still seems to be low. Obviously, Figure 5B illustrates an example of increased specificity at a still high sensitivity. The tendency of increasing specificity is continued in panel 5C. Now a relatively high correlation between the gold standard of histopathology and the spectral diagnoses is observed. Further increase of the specificity of the MLP-ANN models result in an increase of false negative rates, that is in a significantly decreased sensitivity (not shown). The observed interdependence of sensitivity and specificity is known in medicine and psychology as the receiver operating characteristic (ROC) and is often used for the comparison of diagnostic tests. A schematic ROC curve is shown in panel 5E. Please note that the examples of panels 5A–C are indicated as points along the solid line.
What are the variables that allow to specifically design and optimise ANN models? It turned out that false negative/positive rates, that is the sensitivity or specificity of ANN classification can be modified by varying only one single parameter: the number of teaching pattern per class. For example, the ANN classifier which was used to produce image panel 5A was taught with 1658 spectra of the class ‘adenocarcinoma’, 812 spectra for the class ‘fibrovascular connective tissue’, 335 spectra of the class ‘mucin’ and 286 spectra for the class of ‘smooth muscles structures’. In the example of Figure 5B,C the number of spectra of classes 2–4 was kept constant and only the number of spectra of class 1 (adenocarcinoma) was varied (panel 5B: 826 and panel 5C: 389 spectra). As described above, and shown in panels 5A–C this change directly resulted in the very different classification results of the test data.
In order to understand this phenomenon it is important to realise that the criterion that is usually minimised in the teaching phase of a backpropagation network is a sum-squared error (SSE). This SSE is obtained as the squared difference between the desired (pdes,i) and the actual output patterns (pcalc,i) added up over all teaching patterns:
| (1) |
Alternatively, the global SSE can be described as the sum of SSE’s obtained for each individual (pre-defined) class:
| (2) |
Obviously, the global SSE depends mainly not only on the error rates (pdes,i–pcalc,i)2, but also on the number of samples present in the individual classes. If there is no easy-to-find global error minimum available—which is the case in most of the more complex classification tasks—then the global SSE can be principally minimised in two ways: firstly, in the desired way by minimising the error rates for all samples from all classes. A second way is the more efficient the more the pre-defined classes differ in the number of samples. In these cases discriminant functions are computed which produce reduced error rates for classes with high numerical representation and in turn, increased error rates for classes with poor representation. These findings can be synonymously described as increased numbers of false classifications for over-represented classes (high sensitivity), but at the expenses of high rates of misclassifications for the other classes (low specificity). As a result, the SSEx of classes with low sample numbers will be responsible for the most part of the global SSE. We wish to emphasise that an increase of the overall prediction accuracy, that is of sensitivity and specificity can be achieved by several measures. First, we believe it is important to perform class definitions such that the internal data structure is represented. Thus, in order to achieve classification models which show at the same time decreased false negative and false positive rates one have to create models that ideally account for all relevant spectral classes. We have illustrated this situation in the example of Figure 5F where the heterogeneous class ‘adenocarcinoma’ was subdivided into a class of basal parts of the cancerous epithelia (dark yellow) and the diagnostically more important class of apical sides of the adenocarcinoma structures (red). As it can be taken from Figure 5F the modified ANN classification approach now provides a higher degree of correspondence between histopathology and the segmentation technique (see also the dotted line in Figure 5E).
Another important aspect with significant impact on the accuracy of prediction of the ANN models is the representation of the intra- and inter-class variance in the teaching data. An ideal teaching data set should contain a sufficient amount of patterns assuring that both types of variances are represented. Aside from the fact that we often do not know how many patterns per class would be required, an increase of sample numbers is for practical reasons sometimes not possible. As outlined earlier, also in this study the number of samples is still too low to be comprehensive. We are planning therefore to extend the study by adding more samples and to increase the number of database spectra.
Aside from class definitions and sample numbers also the type of spectral pre-processing interferes with the accuracy of classification. It has been shown several times that standardised tests for spectral quality, the calculation of derivative spectra and a normalisation procedure (vector) improve the accuracy of classification.
Finally, it should be pointed out that also the type and the design of the classification model strongly influences the accuracy of prediction. In the present study a combination of small modular artificial neural networks is suggested to give best performance. These classifiers are fast and the time which is required for classification scales linearly with the number of input pattern. To give an example, the segmentation of a complete IR map containing 100 000 spectra takes acceptable 20 s. Furthermore, modular ANNs can be taught and validated independently and are extendable at a later stage for additional or more specialised classification tasks. Another advantage of modular ANN models is the fact, that individual ANNs (i.e. the sublevel nets) can be specifically optimised to identify only a few (down to two) classes. This is particularly important since our model development strategy usually includes a procedure of detecting sets of discriminative spectral features. Thus, each of the individual ANNs can be optimised on the basis of pre-selected specific combinations of spectral features which will further increase the overall classification accuracy. Finally, each of the individual ANNs is principally adaptable in terms of sensitivity and specificity, that is the classifier can be specifically optimised to meet the particular needs of the application. This high level of flexibility and the above-mentioned advantages make modular ANNs to ideal tools for routine use in IR microspectroscopic imaging of tissues.
4. CONCLUSIONS
The combination of infrared microspectroscopy and artificial neural network analysis has great potentials for rapid and reliable identification of tissue structures not only for scientific research purposes but also in a real clinical setup. In this paper we exemplarily showed how optimised network models can be utilised to re-assemble false colour images from infrared spectral maps of tissue sections and to visualise the spatial distribution of tissue structures of interest. Pre-conditions for a successful application of the IR-based methodology are adequate data pre-treatment strategies (i), feature selection (ii) and the use of dedicated classification models (iii). Particularly, the concept of hierarchical (modular) network classification in combination with feature selection methods dramatically enhances the capabilities of the method. Compared to ‘monolithic’ networks modular ANNs provide enhanced flexibility and permit the design of classifiers with improved accuracy of prediction also for high numbers of classes. It was shown furthermore, that unsupervised methods of cluster analysis may be helpful for class definitions in the design phase of the models. We believe that the technique outlined here may be successfully applied to a great variety of applications in biomedical spectroscopy, specifically in histopathology.
Acknowledgements
We are grateful to Miloš Miljkowić, Melissa Romeo (Hunter College, New York, USA) and Heinz Fabian (Robert-Koch-Institut, Berlin, Germany) for fruitful discussions and support. Furthermore, we would like to thank Jürgen Schmitt, Thomas Udelhoven and particularly, Mark S. Novozhilov from Synthon GmbH (Heidelberg, Germany) for the excellent collaboration.
Abbreviations
- ANN
artificial neural network
- FT-IR
Fourier transform infrared
- IR
infrared
- AHC
agglomerative hierarchical clustering
- H&E
Haematoxylin-Eosin
- MCT
mercury cadmium telluride
- MLP
multilayer perceptron
- ROC
receiver operating characteristic
- S/N
signal-to-noise ratio
REFERENCES
- 1.Choo LP, Wetzel DL, Halliday WC, Jackson M, LeVine SM, Mantsch HH. In situ characterization of β-amyloid in Alzheimer’s diseased tissue by synchrotron FTIR microspectroscopy. Biophys. J. 1996;71:1672–1679. doi: 10.1016/S0006-3495(96)79411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kidder LH, Kalasinsky VF, Luke JL, Levin IW, Lewis EN. Visualization of Silicone Gel in Human Breast Tissue using new Infrared Imaging Spectroscopy. Nat. Med. 1997;3:235–237. doi: 10.1038/nm0297-235. [DOI] [PubMed] [Google Scholar]
- 3.Lasch P, Naumann D. FT-IR microspectroscopic imaging of human carcinoma thin sections based on pattern recognition techniques. Cell. Mol. Biol. 1998;44(1):189–202. [PubMed] [Google Scholar]
- 4.Chiriboga L, Xie P, Yee H, Zarou D, Zakim W, Diem M. Cell. Mol. Biol. 1998;44(1):219. [PubMed] [Google Scholar]
- 5.Lasch P, Haensch W, Kidder L, Lewis EN, Naumann D. Colorectal adenocarcinoma characterization by spatially resolved FT-IR microspectroscopy. Appl. Spectrosc. 2002;56(1):1–9. [Google Scholar]
- 6.Harthcock MA, Atkin SC. Imaging with functional group maps using infrared microspectroscopy. Appl. Spectrosc. 1988;42(3):449–455. [Google Scholar]
- 7.Guilment J, Markel S, Windig W. Infrared chemical micro-imaging assisted by interactive self-modeling multivariate analysis. Appl. Spectrosc. 1994;48:320–326. [Google Scholar]
- 8.Duponchel L, Elmi-Rayaleh W, Ruckebusch C, Huvenne JP. Multivariate curve resolution methods in imaging spectroscopy: influence of extraction methods and instrumental perturbations. J. Chem. Inf. Comput. Sci. 2003;43:2057–2067. doi: 10.1021/ci034097v. [DOI] [PubMed] [Google Scholar]
- 9.Kneipp J, Beekes M, Lasch P, Naumann D. Molecular changes of preclinical scrapie can be detected by infrared spectroscopy. J. Neurosci. 2002;22(8):2989–2997. doi: 10.1523/JNEUROSCI.22-08-02989.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lasch P, Wäsche W, Müller G, Naumann D. In: de Haseth JA, editor. FT-IR microspectroscopic imaging of human carcinoma thin sections; "Fourier Transform Spectroscopy: 11th International Conference" AIP Conference Proceedings; Woodbury: New York. 1998. pp. 308–311. [Google Scholar]
- 11.Lasch P, Haensch W, Naumann D, Diem M. Cluster Analysis of Colorectal Adenocarcinoma Imaging Data: A FT-IR Microspectroscopic Study. Biochim. Biophys. Acta. (BBA)—Mol. Basis Dis. 2004;1688(2):176–186. doi: 10.1016/j.bbadis.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Udelhoven T, Naumann D, Schmitt J. Development of a hierarchical classification system with artificial neural networks and FT-IR spectra for the identification of bacteria. Appl. Spectrosc. 2000;54(10):1471–1479. [Google Scholar]
- 13.Zell A. Simulation Neuronaler Netze. Bonn, Paris: Addison-Welsey; 1994. Reading. [Google Scholar]
- 14.Marques de Sa JP. Pattern Recognition: Concepts, Methods and Applications. Berlin, Heidelberg, New York: Springer-Verlag; 2001. [Google Scholar]
- 15.Lasch P, Schmitt J, Beekes M, Udelhoven T, Eiden M, Fabian H, Petrich H, Naumann D. Ante mortem identification of BSE from serum using infrared spectroscopy. Anal. Chem. 2003;75(23):6673–6678. doi: 10.1021/ac030259a. [DOI] [PubMed] [Google Scholar]
- 16.Lasch P, Naumann D. Spatial resolution in infrared microspectroscopic imaging of tissues. Biochim. Biophys. Acta. 2006;1758(7):814–829. doi: 10.1016/j.bbamem.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 17.NeuroDeveloper Website. http://www.synthon-analytics. de [01. October 2005]
- 18.CytoSpec Website. http://www.cytospec.com [01. October 2005]
- 19.Wood BR, Chiriboga L, Yee H, Quinn MA, McNaughton D, Diem M. Fourier transform infrared (FTIR) spectral mapping of the cervical transformation zone, and dysplastic squamous epithelium. Gynecol. Oncol. 2004;93(1):59–68. doi: 10.1016/j.ygyno.2003.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward JH. Hierarchical grouping to optimise an objective function. J. Americ. Stat Assoc. 1963;58:236–244. [Google Scholar]
- 21.Helm D, Labischinski H, Schallehn G, Naumann D. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. J. Gen. Microbiol. 1991;137:69–79. doi: 10.1099/00221287-137-1-69. [DOI] [PubMed] [Google Scholar]
- 22.Riedmiller M, Braun H. A direct adaptive method for faster backpropagation learning: the RPROP algorithm; ICNN-93, IEEE International Conference on Neural Networks; San Francisco, CA. 1993. pp. 586–591. [Google Scholar]
- 23.Gasser A, Kamel M. Modular network classifiers: a comparative study. J. Intell. Robotics Syst. 1998;21:117–129. [Google Scholar]
- 24.Jackson M, Choo L-P, Watson PH, Halliday WC, Mantsch HH. Beware of connective tissue proteins: assignments and implications of collagen absorptions in infrared spectra of human tissues. Biochim. Biophys. Acta. 1995;1270:1–5. doi: 10.1016/0925-4439(94)00056-v. [DOI] [PubMed] [Google Scholar]