Abstract
Mechanisms coupling heart function and cardiac morphogenesis can be accessed in lower vertebrate embryos that can survive to swimming tadpole stages on diffused oxygen. Forward genetic screens in Xenopus tropicalis have identified more than 80 mutations affecting diverse developmental processes, including cardiac morphogenesis and function. In the first positional cloning of a mutation in X. tropicalis, we show that non-contractile hearts in muzak (muz) embryos are caused by a premature stop codon in the cardiac myosin heavy chain gene myh6. The mutation deletes the coiled-coil domain responsible for polymerization into thick filaments, severely disrupting the cardiomyocyte cytoskeleton. Despite the lack of contractile activity and absence of a major structural protein, early stages of cardiac morphogenesis including looping and chamber formation are grossly normal. Muz hearts subsequently develop dilated chambers with compressed endocardium and fail to form identifiable cardiac valves and trabeculae.
Keywords: Xenopus tropicalis, mutation, heart, cardiac, myh6, valve, trabeculation, sarcomere, myosin, genetic mapping
Introduction
Formation of the heart is highly conserved in vertebrate species. Genes relevant to human cardiac development and disease can be studied in lower vertebrate models whose externally-developing embryos are easily accessible during heart forming stages and survive for several days on passively-diffused oxygen if cardiac function is compromised experimentally. Xenopus researchers have combined classical embryological explant and transplant approaches with over- and mis-expression of gene products (Warkman and Krieg, 2007) to examine early steps in heart formation, including specification of the heart field (Sater and Jacobson, 1989), transcriptional regulation of cardiac identity (Evans et al., 1995; Fu et al., 1998; Grow and Krieg, 1998), and signaling pathways underlying cardiac asymmetry (Branford et al., 2000; Hyatt and Yost, 1998; Ramsdell and Yost, 1999). In zebrafish, heart development studies have built on loss-of-function genetic tools, as well as the optical properties of the embryos for microscopy, to analyze cardiac morphogenesis and valve formation (Beis et al., 2005; Sehnert and Stainier, 2002; Stainier, 2001). As teleost fish are the most diverse vertebrates, due in part to the ancestral genome duplication and subsequent shuffling of gene functions (Force et al., 1999; Postlethwait et al., 2000), comparative studies in other models will help identify developmental mechanisms shared broadly among tetrapods. Loss-of-function studies in X. laevis have previously been limited to injection of dominant negative constructs (Grow and Krieg, 1998; Shi et al., 2000) and, more recently, antisense morpholino oligonucleotides (Peterkin et al., 2007; Small et al., 2005). Large-scale genetic approaches are impractical in X. laevis due to its pseudotetraploid genome and long generation time, but are well-suited to its diploid relative Xenopus tropicalis. X. tropicalis reaches maturity in a relatively short 4-6 months, and its small, canonically-organized tetrapod genome (1.5×109 bp in 10 chromosomes) is supported by extensive sequence resources including a high-quality draft genome assembly (http://genome.jgi-psf.org/Xentr4/Xentr4.home.html), over one million ESTs, and a meiotic linkage map of Simple Sequence Length Polymorphisms (SSLPs) (http://tropmap.biology.uh.edu/index.html) (Carruthers and Stemple, 2006; Klein et al., 2006; Klein et al., 2002).
In a pilot screen for chemically-induced mutations in X. tropicalis, we recovered several phenotypes with decreased cardiac function (Goda et al., 2006). Here we show that the lack of cardiac contractility in the muzak mutant is caused by a nonsense mutation truncating the cardiac myosin heavy chain gene myh6. Despite this defect in a major structural component of sarcomeres resulting in absence of myofibrils and contractility, looping and chamber formation appear surprisingly normal. Muz hearts subsequently display dilated ventricles and atria and malformed endocardium, segments of which appear collapsed with little or no lumen. Later steps in cardiac development, such as valve formation and trabeculation, are not detected, but it is beyond the scope of this study to determine whether these are direct or indirect effects of the mutation. This report describes the first positional cloning of a mutation in X. tropicalis.
Experimental Procedures
Frog Strains
The original mutagenesis and fertilization to produce mutant founder F1 animals was performed on the N (Nigerian) strain (kind gift of Enrique Amaya, Manchester University, United Kingdom); polymorphic crosses used for mapping were generated using the IC (Ivory Coast) strain (kind gift of Robert Grainger, University of Virginia, Charlottesville, USA). Mutant and wt embryos used for mapping and phenotyping were generated from a cross of an F2 muz/+ N/IC female and an F3 muz/+ male produced by crossing an F2 N/IC female to an N/PacBio (wild-caught animals of unknown origin obtained from Pacific Biological Supply, Inc.) male carrying the mlc2GFP transgene.
Mapping
Gynogenesis was performed as described previously (Goda et al., 2006). AFLP reactions were performed using the AFLP Analysis System I kit (Invitrogen, 10544-013). PCR products were resolved on 6% denaturing acrylamide gels and visualized by autoradiography. SSLP markers were amplified and resolved as described on the tropmap website (http://tropmap.biology.uh.edu/polyprotocol.html). SSLP markers from the meiotic map 040E09, 018E09, and 026G09 can be found on the tropmap database (http://tropmap.biology.uh.edu/) and have the following sequences:
040E09: F-AAGTTGCCCTAAAGGTAGGC R-GATTATTGCTCCGAATGTGG 018E09: F-CTCAATAATCAGGGCATGTAATC R-GCAGACATAAGCATTGTACCC 026G09: F-TGAAGTGAAGCACAGCACAG R-AGGGACTTTTCCAGATCAAG
Bespoke SSLP markers for scaffold 439 were obtained using Tandem Repeat Finder (http://tandem.bu.edu/trf) and Primer3 (http://primer3.sourceforge.net/). Primers for markers in scaffold 439 were as follows:
439.1: F-TGCCATTTGTATCCCACCTT R-CCAGGGATGACTTTGACACA 439.3: F-TGATCTCAGTGCCAGATGCT R-TGCTCCAGATAGGTGACGTG 439.10: F-TTTCTCCTGTGGGCAACTTT R-GTGCTGGTGGAAGGGAAGTA SSCP439.1 F-GCGCCCTATAGTGAAATCCA R-GCACAAAATTGCAGGAGGTT SSCP439.15 F-CCCTGATCAGTCATGGGTTC R-GTGACATGACAACGCAAACC
Primers to amplify the muz myh6 genomic fragment containing stop mutation:
F-CTCGAGCAACAAGTGGATGA R-GCCCACCATAAAATGACCTG
Whole Mount In Situ Hybridization
Embryos were staged according to Nieuwkoop and Faber. Fixing and WISH were carried out as described previously (Sive et al., 2000).
WISH probes for myh6 and myh6.2 were made by cloning RT-PCR products into the PCRII-TOPO vector using the TOPO TA Cloning Kit (Invitrogen, K4600-40). Probes were prepared by linearizing with XhoI and transcribing with SP6. Primers used were:
myh6 F-GCTAGAGAAGATTCGCAAGCAG R-TCCACAATTGCAGTGTTTTCTT myh6.2 F-TCAGACCTGACAGAGCAACTG R-TCCCCCTCCATCTTCTTTTT
RT-PCR
RNA was prepared using Trizol (Invitrogen). cDNA was prepared and amplified with the Enhanced Avian HS RT-PCR kit (Sigma HSRT-100) using the following primers:
myh6 F-CCAACAAGGGAACTCTGGAA R-CTGCAGTTTCTCGTTGGTGA myh6.2 F-AACCCTGCTGCTATTCCAGA R-TCAAGCTTGGCTTTGGATTT myh7b F-AACTGGACAAGAAGCGGAGA R-GGTCCATTACCCCTGGAGTT myh15 F-ATTCCTCCTCACGGACCTTT R-CGCCCACCTAGAGAGAATGA myh8 F-CCGTCTTGATTACGGGAGAA R-GGGTTTCTTGTTGGTCAGGA odc F-GCCAGTAAGACGGAAATCCA R-CCCATGTCAAAGACACATCG
Immunoblotting
Dissected hearts from st. 40 tadpoles were collected on ice, resuspended in a modified SDS-sample buffer, boiled for 1 minute, resolved by 6% PAGE, transferred to membrane, and immunoblotted as described previously (Ehler et al., 1999)
Silver Staining
Silver staining of proteins on SDS-PAGE gels was performed according to manufacturer’s instructions using the Silver Stain Plus Kit (Bio-Rad, 161-0449)
Morpholino injections
Morpholinos were purchased from GeneTools LLC. A total of 12ng of each morpholino was injected into both cells of a two-cell embryo. Morpholino sequences were as follows:
myh6 translation-blocking morpholino: TCTGCCATCAGGGCATCACCCATTG myh6 morpholino blocking 1st coding exon splice donor: CTTATAAATGTAATACCTTGCCATC Control morpholino: CCTCTTACCTCAGTTACAATTTATA
Immunohistochemistry
Stage 42 tadpoles were fixed in 1% paraformaldehyde for 1 hour, washed in PBS, blocked in PBS+10% sheep serum, 2mg/ml BSA and 0.2% saponin for 1 hour at room temperature (RT), then incubated with primary antibody in block solution at 4°C overnight, washed in PBS containing 0.2% saponin and incubated in block solution containing Alexa Fluor 488-conjugated anti-mouse IgG secondary antibody (Invitrogen, A21202) for 2 hours at RT. After washing in PBS with 0.2% saponin, the tadpoles were incubated with 1:20 dilution of Alexa Fluor 568 phalloidin (Invitrogen, A12380) in block solution, washed again, then hearts were dissected and visualized with a Zeiss LSM5 Pascal confocal microscope.
Plastic Sections and 3-D modeling
Embryos were fixed o/n in Bouin’s fixative (BDH Laboratory Supplies,28087 4V), dehydrated in ethanol, embedded in JB-4 resin (Polysciences Inc.), 3μm sections cut with a Leica RM 2165 microtome, and stained with Hematoxylin and Eosin (both Sigma). Sections were visualized on a Zeiss axiocam microscope, serial images were converted into 8bit greyscale stacks and loaded in Amira 3D Visualisation software Mercury Computer Systems, Germany) and heart structures were manually outlined and annotated. 3D models were generated using the surface rendition tool in Amira.
Results
The muzak mutation affects heart function
Homozygous muz embryos were identified by complete lack of cardiac contractility at heart looping stages (Movie S1). Embryonic blood fails to circulate in muz tadpoles, and erythrocytes pool in the ventral blood islands where they form. The tadpoles swim normally, indicating that the mutation does not affect skeletal muscle, and other tissues are not visibly affected. By stage 43 (3 days post fertilization), muz embryos develop cardiac edema, and absence of heart function persists until at least feeding tadpole stage (5 days post fertilization). No phenotype was observed in heterozygotes, suggesting that the muz allele behaves in a simple recessive fashion.
Muz maps to an interval containing cardiac myosin heavy chain gene
When we began linkage studies to identify the gene underlying the muz phenotype, no meiotic map was available. In a map-independent initial strategy, bulk segregant pools of DNA from gynogenetic muz and wild type siblings were used to obtain a set of Amplified Fragment Length Polymorphism (AFLP (Vos et al., 1995) markers linked to the mutant locus. 5 bands which amplified from wild type but not muz DNA (Figure S1A) were extracted, reamplified, sequenced, and placed on the X. tropicalis genome assembly in Version 4 scaffolds 554, 91, 567, 289, 158 (http://genome.jgi-psf.org/Xentr4/Xentr4.home.html). The subsequent release of an X. tropicalis meiotic map of SSLP markers (http://tropmap.biology.uh.edu) located these scaffolds in a ~12 cM interval on Linkage Group 1 (LG1). Linkage of the mutation to SSLP markers in these scaffolds was confirmed by bulk segregant analysis of pools of mutant and wild type embryos from a conventional cross of heterozygous carrier siblings (see Figure S1B for an example).
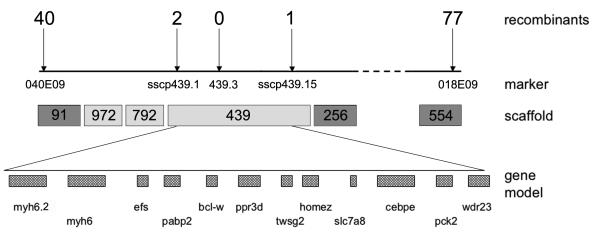
To define the genetic interval containing the muz locus, individual muz embryos from a conventional sibling cross were genotyped with SSLP markers from LG1 of the meiotic map. Analysis of 3200 meioses placed muz between two flanking markers, 040E09 in scaffold 91 (40 recombination events, Figure 1A) and 018E09 in scaffold 554 (77 recombination events). We tested the set of recombinant embryo DNAs further with a marker between the flanking markers, 026G09 (scaffold 256), and found a subset of the recombinants with 018E09 were still recombinant with this polymorphism, whereas all the recombinants with 040E09 were homozygous for the wild type 026G09 allele, suggesting that muz was located between the latter two markers. As the X. tropicalis genomic sequence assembly was fragmented in this region, and many scaffolds are not represented on the meiotic linkage map, we compared syntenic regions in well-characterized mammalian genomes to generate an in silico hypothetical local scaffold assembly. By examining syntenic human and mouse genomic regions that overlapped the termini of scaffolds 256 and 91, we identified candidate intervening scaffolds 439, 792 and 972 in the muz interval. Analysis of SSLP markers 1.439.1 (two recombination events), 1.439.3 (no recombination events) and 1.439.10 (1 recombination event) confirmed this local assembly and placed the mutation in scaffold 439. Further analysis refined the muz interval to a 370kb region between Single Strand Conformation Polymorphism (SSCP) markers SSCP439.1 (two recombination events) and SSCP439.15 (one recombination event) on scaffold 439 containing 12 gene models on the JGI assembly (Figure 1A and Table S1). The sequence interval containing muz was then inspected for candidate genes.
Figure 1. Muz maps to an interval containing cardiac myosin heavy chain gene.
A. Individual muz embryos were genotyped with SSLP markers from scaffold 91 and scaffold 554. Mapping was refined with SSCP markers (sscp439.1 and sscp439.15) and an SSLP marker (1.439.3) from scaffold 439; number of recombination events detected in 3200 meioses shown above each marker. Dark grey scaffolds are present on the tropicalis meiotic map; intervening light grey scaffolds were obtained by analysis of synteny to reference genomes and confirmed by linkage. Muz maps to a 370 kb genomic interval between sscp439.1 and sscp439.15 containing 12 gene models in the JGI assembly, including myh6 and myh6.2.
Compellingly, two gene models in this interval, myh6 and myh6.2, were annotated as myosin heavy chain (MHC), with >88% identity to the human cardiac MYH6 and MYH7 proteins, the major MHC genes expressed in mammalian hearts. These genes are known to be required for normal heart function in humans, with mutations in MYH6 and MYH7 implicated in atrial-septal defects and familial hypertrophic cardiomyopathies respectively (Ching et al., 2005; Geisterfer-Lowrance et al., 1990). In human, mouse, and rat these gene pairs are chromosomally adjacent, and are thought to have arisen by tandem duplication before these species diverged, some 70 million years ago (Mahdavi et al., 1984; Mahdavi et al., 1982). Of the two X. tropicalis MHC genes on scaffold 439, the centromere-proximal is orthologous to MYH6 based on mutual best BLAST as well as its strong expression in wild type hearts (Figure 2A, black arrow); weaker expression is also seen in jaw muscles (Figure 2A, white arrow). The distal gene, annotated myh6.2, is expressed in developing jaw muscle but not heart (Figure 2C), and hence is unlikely to be responsible for the muz phenotype.
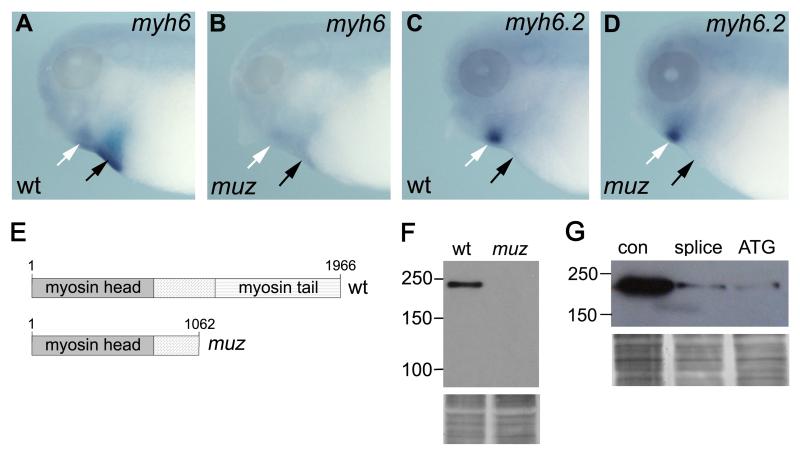
Figure 2. muzak is encoded by myh6.
WISH shows myh6 expression in wild type heart (A, black arrow) and jaw muscle (white arrow) is diminished in muz (B). (C, D) myh6.2 is expressed in jaw muscle (white arrow) but not heart (black arrow), and is unaffected by the mutation. (E) Schematic showing domain structure of wild type X. tropicalis myh6 and the truncated protein lacking the myosin coiled-coil tail encoded by the muz allele. (F) Western blot analysis does not detect sarcomeric MHC protein in extracts of muz heart; silver stained loading control below. (Movie S2 and G) myh6 morphant hearts do not beat and show strong depletion of sarcomeric MHC protein relative to control morpholino-injected tadpoles; silver stained loading control below.
To assess whether a defect in myh6 might underlie the muz phenotype, we sequenced cDNA from mutant and unrelated wild type embryos, and found a C to T transition creating a premature stop codon at position 3187 of the coding sequence. Genomic DNA from adult muz carrier animals was also found to be heterozygous for this lesion. The resulting truncated protein (1062 aa vs 1996 aa wild type, Figure 2E) is likely to be nonfunctional as it deletes the coiled-coil tail required for dimerization and aggregation into functional thick filaments.
Myh6 expression is strongly reduced in muzak hearts
We then evaluated how the mutation affected expression of the two MHC genes in the interval. Whole Mount In Situ Hybridization (WISH) showed a significant decrease in myh6 expression in muz embryos compared to wild type (Figure 2A, B), possibly due to nonsense-mediated decay (Peltz et al., 1993; Whitfield et al., 1994). Expression of the neighboring paralog myh6.2 in jaw muscle was unaffected by the mutation (Figure 2C, D, black arrow).
Levels of cardiac MHC protein were assayed by immunoblotting with the A4.1025 antibody, which recognizes an epitope shared by sarcomeric myosin heavy chain head domains (Dan-Goor et al., 1990) retained in the muz allele. A band of ~220kDa is observed in extracts of dissected wild type but not muz hearts (Figure 2F). The mutant protein of predicted size ~120kDa is not detected, possibly due to depletion of the mRNA by nonsense-mediated decay, as suggested by WISH. Given the deletion of the tail domain required for thick filament formation and the severe reduction in expression levels, muz is likely to be a strong hypomorph or null allele of myh6.
Myh6 antisense morpholinos phenocopy the muz mutation
To confirm that a defect in myh6 could produce the muz cardiac phenotype, we designed morpholino antisense oligonucleotides to deplete the endogenous protein. Both translation-blocking and splice-blocking morpholinos, when injected into both blastomeres of a two-cell embryo, affected cardiac contractility with high penetrance (76/79 and 94/100 injected embryos respectively). In contrast, heart looping and chamber formation were unaffected. Approximately 50% of myh6-depleted embryos had no detectable heartbeat, mimicking the muzak phenotype, while the remainder exhibited faint twitching insufficient for blood circulation (Movie S2). Injected embryos were otherwise morphologically normal, with tadpole motility unaffected, indicating that the morpholinos did not interfere with off-target skeletal MHCs. Control morpholino injections had no effect on cardiac function (85/85 wild type). Knockdown efficacy was assayed by immunoblotting protein extracts from dissected morphant hearts with the A4.1025 antibody. Both myh6 morpholinos strongly depleted cardiac MHC compared to control morpholino (Figure 2G). These gene knockdown data confirm a requirement for myh6 in cardiac function, strongly supporting the conclusion that a defect in this gene underlies the muz phenotype.
Myh6 is the major cardiac sarcomeric MHC at swimming tadpole stages and is necessary for myofibril formation
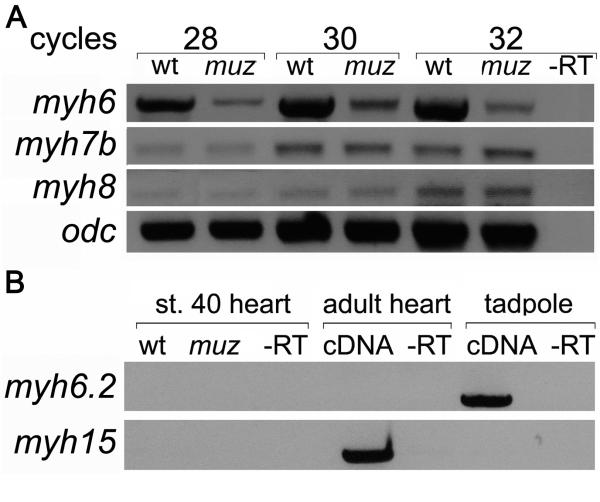
Myh6 is likely to be the principal functional sarcomeric MHC in tadpole hearts, based both on the failure of the A4.1025 antibody to detect any immunoreactive species in muz heart extracts and the penetrance of the morphant phenotype. However, since the antibody may not recognize all Xenopus MHC proteins, and some morpholino-injected embryos retained faint twitching, we asked whether other sarcomeric MHC mRNAs were expressed in stage 40 hearts or upregulated in muz. RT-PCR of dissected stage 40 hearts confirms that myh6 is expressed strongly in wild type and at much reduced levels in muz hearts (Figure 3A). Myh6.2 was amplified from stage 40 whole embryo mRNA, consistent with its expression in jaw muscle, but not from wild type or muz embryonic hearts, nor from adult heart (Figure 3B). The Xenopus genome is not thought to contain an ortholog of mammalian MYH7 (Garriock et al., 2005), and it is likely that myh6.2 derives from a separate tandem duplication from the one which gave rise to mammalian MYH6 and MYH7. A third cardiac MHC, myh15/vMHC (an inactive pseudogene in human), has been found in chicken (Oana et al., 1998), as well as X. laevis (Garriock et al., 2005) where it is not expressed until after chamber formation. We found no myh15/vMHC expression in hearts of either wild type or muz stage 40 embryos by RT-PCR, although it is detected in adult heart (Figure 3B), consistent with previously described onset of expression in X. laevis at stage 43. Similarly, no expression in embryonic heart was observed for the skeletal MHCs myh1,2,3 or 4 (data not shown). However, two MHCs present in mammalian heart EST collections, the slow-tonic myh7B and myh8, were detected at comparable levels in both wild type and muz dissected hearts (Figure 3A). Absence of myh6 protein in muz does not appear to induce expression of non-cardiac MHCs or up-regulate myh7B and myh8 mRNAs which, although present in muz hearts, are not sufficient to rescue the phenotype.
Figure 3. MHC genes expressed in stage 40 wild type and muz hearts.
RT-PCR from isolated stage 40 hearts shows lower levels of myh6 in muz; myh7B and myh8 are unaffected. (A) myh6.2 mRNA is not detected in wild type or mutant tadpole hearts or wild type adult heart, although it is amplified from whole-embryo mRNA; myh15 is expressed in adult but not stage 40 tadpole heart(B).
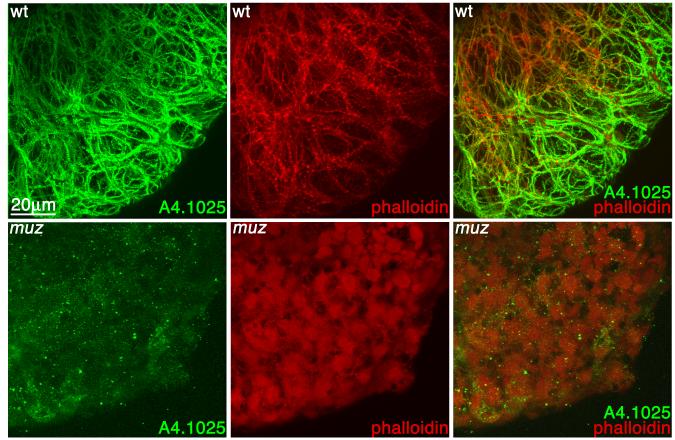
We then examined sarcomere formation in muz to see whether the remaining myh7B and myh8 could organize myofibrillar structures. Stage 42 wild type and muz embryos were stained with the A4.1025 antibody, counterstained with phalloidin, and their hearts dissected and visualized by confocal microscopy. Consistent with the depletion of myh6 mRNA and protein levels, anti-MHC immunostaining is greatly diminished in muz and is not organized in striated myofibrils (Figure 4). Significantly, phalloidin staining shows that actin does not form myofibrils in the mutant heart confirming the lack of any cardiac MHC proteins capable of assembling into sarcomeres in muz embryos. Myofibrils were absent at stage 35 (data not shown) when contractions begin as well as stage 40, making it unlikely that muz hearts have sarcomeres at any stage of development.
Figure 4. Muz hearts lack myofibrils.
3D confocal projections of wild type (A) and muz (B) hearts immunostained with the pan-sarcomeric MHC A4.1025 antibody (green) and counterstained with phalloidin (red). In wild type hearts, MHC and actin colocalize to myofibrils, while muz hearts show very little A4.1025 immunostaining and no fibrillar structures.
Cardiac chamber morphology and valve development in muzak
In addition to depletion of the myh6 protein, a major structural component of myocardial cells, the muz mutation results in abrogation of contractile activity (thought to be required for various steps in cardiac morphogenesis, as well as loss of sarcomeres (known to play signaling as well as mechanical roles in cardiac function (Nicol et al., 2000)). We wished to describe how these deficits affect the major morphogenetic steps in heart development.
As in other vertebrates, the Xenopus heart initially forms as a linear cardiac tube comprising a muscular myocardial layer surrounding an inner endocardial channel. After undergoing rightward looping, this tube balloons out into chambers separated by cardiac valves. The final stages of heart development in Xenopus include trabeculation of the ventricular myocardium and septation of the atrium into two chambers (Kolker et al., 2000; Mohun et al., 2000). To characterize how these processes are affected by absence of myh6 and the resulting lack of sarcomeres and contractility, muz hearts were subjected to histological analysis. Plastic sections of the cardiac region of wild type and mutant embryos were obtained at stages relevant to specific tissue formation processes: stage 35 (heart looping), 40 (onset of chamber formation), and 42 (valve formation).
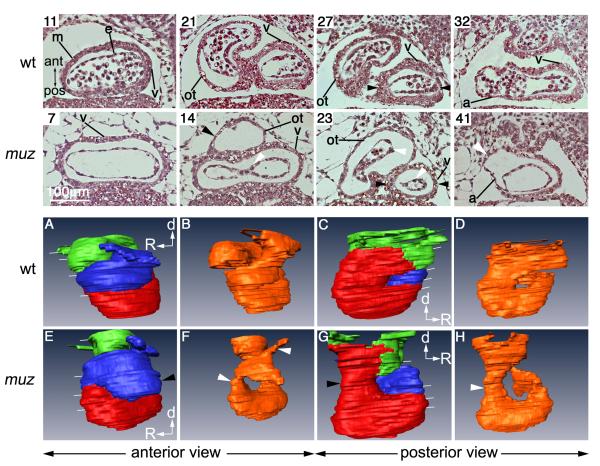
Figure 5 shows stage 40 wild type and muz hearts; sections are numbered to indicate their position in the stack beginning at the ventral side of the cardiac cavity. Outlines of myocardial and endocardial layers in the image stacks were then used to generate 3D projections (Figure 5A-H, see also movies S3, S4 for a rotating view). Regions of the heart are indicated by colour: the myocardium of the outflow tract (blue) was defined by morphological position; thinner myocardium in dorsal sections (green) forms the atrium; thicker myocardium (red) in ventral sections is clearly ventricular (e.g. Figure 5 section 32 ‘v’); however, in malformed mutant hearts, where atrial and ventricular chambers showed little difference in wall thickness, the precise border was assigned arbitrarily.
Figure 5. Altered chamber morphology in muz hearts.
Coronal plastic sections of stage 40 wild type and muz hearts (top rows) numbered from ventral side of cardiac cavity, and indicated by white lines in 3D models (bottom rows). m= myocardium, e= inner endocardial tube, v= ventricle, ot= outflow tract , a= atrium. No blood cells are seen in the muz sections due to lack of circulation, and myocardial layer appears thinner throughout the muz heart compared to wild type. The muz ventricle is wider than in wild type (sections 7 and 11), while outflow tract and atrium are dilated (sections 14, 23 and 41). Abnormal muz chamber morphology is highlighted in 3D projections of outlines of myocardium (A, C, E, G, red=ventricle, blue=outflow tract, green=atrium) and endocardium (B, D, F, H, orange), including elongated ventricle, dilated outflow tract (black arrowhead in E) and narrow cardiac tube at AVC level (black arrow in G). muz endocardium is very compressed with drastically reduced lumen (white arrows in 23, F and H)
Cardiac chambers in muz are dilated, and at stage 40 the myocardial wall appears thinner than wild type throughout. Segments of the endocardial tube, notably in outflow tract and atrioventricular canal (AVC), appear constricted with little lumen (white arrowheads, section 23 and F, H). The expanded ‘peri-endocardial’ region between the distended myocardium and the constricted endocardium distorts their alignment (white arrowhead, section 14). The cardiac tube at AVC level, spanned by black arrowheads in sections 27 and 23, is narrower in the mutant (black arrowhead in C and G). Dorsally the muz atrium is usually distended (white arrowhead, section 41). No blood cells are seen in muz hearts at this stage due to lack of circulation. Many of these abnormalities are already present prior to chamber differentiation in earlier looped cardiac tube (stage 35) muz tadpoles, including the dilated outflow tract, collapsed endocardial tube, and the narrow cardiac tube at the level of the AVC (Figure S2).
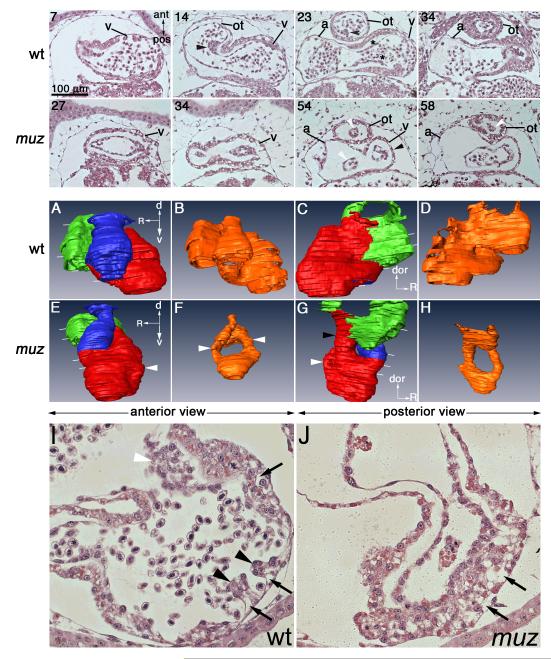
At slightly later stages, valve formation begins in Xenopus; this process is not thought to occur in the absence of contraction in zebrafish (Bartman et al., 2004). We therefore examined plastic sections of stage 42 wild type and muz tadpoles (Figure 6). A spiral valve can be distinguished in the outflow tract of wild type embryos (black arrowhead, sections 14 and 23), and the ‘endocardial cushion’ valve precursors are forming in the AVC (black asterisks, section 23). In stage 42 muz hearts, as at earlier stages, the endocardial tube is often narrower (white arrowheads, sections 54, 58 and F, H) and no valve formation can be discerned. Transverse sections more clearly show endocardial cushions forming in the AVC region of wild type (Figure 6I, white arrowhead) but not muz hearts (Figure 6J). Since it is difficult to unambiguously identify valve-forming AVC and outflow tract positions in the morphologically-distorted mutant hearts, we have also examined complete stacks of cardiac-level sections from 10 muz embryos without detecting identifiable cushions at any position (data not shown). Endocardial cushions were clearly visible in 10/10 sibling wild type embryos.
Figure 6. Muz hearts become dilated and lack valves and trabeculae.
Coronal plastic sections of stage 42 wt and muz hearts (top rows) numbered from ventral side of cardiac cavity, and indicated by white lines in 3D models (middle rows). v= ventricle, ot= outflow tract , a= atrium. Wild type hearts show a spiral valve in the outflow tract (sections 14, 23, black arrows), and thickening of endocardium preceding atrioventricular valve formation (section 23, black asterisk). Valve formation is not detected in muz hearts, and endocardial lumen is drastically reduced in outflow tract and AVC regions (white arrowheads sections 54, 58, also compare models B and F). Endocardial cushion formation in AVC can also be seen in transverse sections of stage 42 wild type (I, white arrowhead) hearts but not in muz (J). Trabeculation has initiated in the wild type ventricle (I, black arrowheads) but is absent in muz (J). At this stage the ventricular myocardium has a vacuolated appearance in both wt and mutant embryos (I, J black arrows). Middle two rows: 3D projections of outlines of myocardium (A, C, E, G) and endocardium (B, D, F, H) highlight abnormal muz chamber morphology; red = ventricle, green = atrium, blue = outflow tract, orange = endocardium. Muz ventricles are elongated relative to wild type (E, G white arrows). A narrow tube connects muz ventricle and atrium (section 54 and G, black arrowheads; compare to 23, C).
Another important process, trabeculation, in which the ventricular myocardium takes on a spongiform appearance, is also occurring at this stage. In wild type hearts, myocardial cells can be seen proliferating and protruding into the lumen (Figure 6I, black arrowheads); interestingly, these cells also take on a vacuolated appearance that may be integral to the mechanism of trabeculation (black arrows). No trabeculae are seen in muz ventricular myocardium, which is very thick but retains abundant vacuole-like structures similar to wild type (Figure 6J, black arrows).
3-D modeling at stage 42 reveals that mutant cardiac morphology is becoming progressively more distorted (Figure 6, see also movies S5, S6 for a rotating view). Whereas in wild type the outflow tract rises sharply out of the ventricle towards the dorsal side of the embryo (Figure 6 section 14 and A), in muz hearts it gently loops out of the end of the elongated ventricle (Figure 6, 34 and E). The narrow cardiac tube at AVC level seen at stages 35 and 40 becomes more pronounced at stage 42 (Black arrowhead, G). Again, blood cells are absent in the muz heart except for a few in the atrium and inflow tract (Figure 6 section 58). The qualitative morphological abnormalities described here are consistently present in muz embryos at stages 40-42 (>10 mutant and wild type hearts examined in plastic sections, and 6 mutant and wild type examined by High Resolution Episcopic Microscopy (HREM)(data not shown)).
Analysis of histological sections of muz hearts demonstrates that later steps in heart development such as valve formation and trabeculation do not occur in the absence of myh6/contractility and sarcomeres. The morphology of heart chambers is altered; dilated ventricles and atria are observed as early as stage 35, and become progressively more pronounced. The endocardium is likewise severely malformed, with segments of lumen highly constricted. It is beyond the scope of this analysis to conclude that these late effects are direct consequences of the mutation in myh6. However, early steps in cardiogenesis, such as looping and chamber formation, are relatively unaffected by absence of contractility and blood flow.
Discussion
The mapping of muzak marks the first identification of a sequence lesion underlying an induced mutation in X. tropicalis, an important step in establishing this species as a genetic model organism. The non-contractile heart phenotype is tightly linked to a nonsense mutation in the myh6 gene deleting the coiled-coil tail domain required for aggregation into functional thick filaments. This nonfunctional peptide, associated with severe reduction of mRNA and absence of detectable MHC protein and myofibrils, suggests that the muz allele is a strong hypomorph or null of myh6. Loss-of-function studies in Xenopus have previously been limited to morpholino knockdown and dominant negative strategies, where it can be difficult to obtain reproducible and complete deletions of specific activities. Precision loss-of-function tools are available in genetic systems such as mice and zebrafish. However, mutational analysis of cardiac development can be challenging in mammals, where heart function is required early in gestation; indeed, the null phenotype of mouse Myh6 has not been characterized due to early lethality (Jones et al., 1996). Genetic screens in fish have uncovered a large number of cardiac gene functions, but the basic structure of the two-chambered fish heart differs significantly from the four-chambered mammalian heart. The ancestral teleost genome duplication has also led to wholesale reassignment and shuffling of gene functions (Force et al., 1999; Postlethwait et al., 2000), complicating orthology assignment and contributing to the diversity of developmental mechanisms. For example, zebrafish cardiac valves are thought to form by an atypical direct invagination of endocardial epithelia into leaflet structures (Scherz et al., 2008) rather than via a mesenchymal ‘endocardial cushion’ intermediate as has been described in other vertebrates (Armstrong and Bischoff, 2004; Eisenberg and Markwald, 1995) and indeed other fish (Gallego et al., 1997; Icardo et al., 2004). Genetic analysis of X. tropicalis, with its more conventionally-organized tetrapod genome and array of functional assays, will help bridge studies of cardiac development from teleost models to amniotes.
In muzak embryos, the early processes of heart looping and chamber formation are remarkably successful despite the lack of myh6 protein and consequent absence of myofilaments, sarcomeres, heartbeat and blood flow. We have not ascertained which of these deficits is responsible for the later defects observed in chamber morphology, valve formation, and trabeculation, or whether these are direct or indirect consequences of the mutation. However, it is worth noting that mutant hearts never initiate detectable contraction and beating, and hence develop in the complete absence of blood flow-mediated pressure load and shear stress. The role of mechanical forces in cardiac morphogenesis has been studied extensively, with conflicting results (Taber, 2006). In diverse vertebrates, beating begins substantially prior to requirements for transport of blood-borne oxygen and nutrients, consistent with a role as a physical influence on early steps such as looping and chamber formation (Burggren et al., 2000; Mellish, 1994; Pelster and Burggren, 1996; Territo and Burggren, 1998); indeed, heart looping begins when the first myofibrils appear (Manasek et al., 1978). Mechanical or genetic perturbation of contraction and blood flow have supported a role in these early steps in some cases (Hove et al., 2003; Huang et al., 2003; Nishii et al., 2008), but not in others (Sehnert et al., 2002). Our histological analysis and 3-D modelling of muz hearts demonstrates that contractility and blood flow are not required for the key early steps of looping and chamber formation in this tetrapod.
Slightly later in heart development, chamber outgrowth or ‘ballooning’ is thought to be shaped by mechanical forces. Analysis of the chamber-specific MHC mutations weak atrium (atrial MHC, myh6) and half hearted (ventricular MHC, vmhc) show that blood flow promotes cardiomyocyte elongation in specific regions of the linear heart tube in the zebrafish embryo, while contractility restricts cell size and elongation (Auman et al., 2007). The muzak cardiac tube still undergoes ballooning into ventricle and atrium, suggesting that factors other than fluid shear forces can initiate chamber outgrowth. Another striking feature of muz hearts is the constriction of the lumen seen in the atrioventricular canal and outflow tract segments of the endocardial tube. The developing heart has been compared to a specialized blood vessel; arteries are thought to remodel their lumen diameters to maintain shear stress near an optimal set point, decreasing diameter in response to decreased shear (Taber et al., 1995). It is possible that morphogenesis and inflation of these heart regions are particularly shear-dependent.
Another key step in cardiac development, remodeling of the ventricular myocardium to form trabeculae, is critical for increasing the surface area through which the muscle mass of the ventricle can diffuse oxygen prior to the development of coronary circulation (Sedmera, 2005). Trabeculation does not occur in muz; instead the non-trabeculating regions of the ventricular myocardial wall become very thick. Wild type myocardium undergoing trabeculation displays a vacuolated appearance that we also observe in muz. Failure to form trabeculae could be simply due to lower oxygen requirements of the inactive mutant heart; trabeculation could also depend structurally on sarcomere integrity, or require signals from the overlying endocardium (Gassmann et al., 1995; Grego-Bessa et al., 2007; Meyer and Birchmeier, 1995), some of which regulate myocyte proliferation. Interestingly, the non-trabeculating muz myocardial wall appears as thick as its wild type counterpart, suggesting that proliferation may still occur. Although endocardium does not express myh6, it is known to alter its gene expression in response to haemodynamic changes (Groenendijk et al., 2005); it remains to be seen whether specific trabeculation signals are affected in the mutant.
As the embryonic heart matures, efficient function depends on the formation of endocardial valves to prevent retrograde blood flow between chambers. Studies in Danio suggest that when contraction and/or blood flow is disrupted mechanically (Hove et al., 2003) or genetically (Bartman et al., 2004), valve formation is impaired, but this process is now thought to occur by an atypical mechanism of direct leaflet invagination in zebrafish (Scherz et al., 2008). We have seen no evidence of precursors or differentiated valves in muz embryos, consistent with a requirement for blood flow in valve formation mediated by more conventional endocardial cushion intermediates. However, in the absence of cushion-specific markers, which have not been described in Xenopus, morphological distortion of the muz endocardium makes it difficult for us to conclusively rule out the presence of ectopic cushion precursors.
Several other mutations affecting heart function have been identified in pilot genetic screens in X. tropicalis (Goda et al., 2006; Grammer et al., 2005; Noramly et al., 2005), rapid mapping strategies have been established ((Khokha et al., 2009); see also Supplemental Figure S3 for an X. tropicalis genetic mapping strategy flowchart), and reverse genetic resources are being developed ((Goda et al., 2006), http://www.sanger.ac.uk/Teams/Team31/xtmr.shtml) from which mutants in known genes can be obtained. Heart development in X. tropicalis genetic models can be analyzed with a broad array of molecular, genomic, and embryological tools, including gain-of-function mRNA expression screens (Smith and Harland, 1992) to identify interacting suppressor or enhancer functions and sophisticated explant assays modeling differentiation to diverse tissue fates including beating cardiac muscle (Latinkic et al., 2003). Reinforced by these robust functional assays, genetic approaches in amphibians complement rapidly-advancing genomics technologies for dissecting tetrapod developmental processes. The work presented here demonstrates the feasibility of positionally cloning mutations in X. tropicalis, greatly increasing the range of genetic studies.
Supplementary Material
Figure S1. AFLP and SSLP markers define the muz-containing interval on LG1. (A) AFLP reactions on bulk segregant wild type and muz genomic DNA produced five polymorphic markers linked to the mutation (white boxes). (B, left panel) Linked AFLP markers were placed on genomic sequence scaffolds, from which SSLP markers were tested on bulk segregant (BS) wt and muz DNA, confirming linkage to these scaffolds. (B, right panel) Genotyping of individual muz embryos with SSLP markers 040E09 and 018E09 defined the muz-containing interval. Recombinant embryos are indicated by asterisks.
Movie S6. 3-D model of a stage 42 muz heart rotating about it’s dorsal-ventral axis. Ventral is at bottom. Red=ventricle, blue=outflow tract, green=atrium.
Table S1. Gene models in the muz-containing genetic interval and their official full names as provided by the HUGO gene nomenclature committee.
Figure S2. Abnormal morphology of muz hearts is evident as early as the looping cardiac tube stage.
Coronal plastic sections of stage 35 wt and muz hearts (top rows), numbered from ventral side of cardiac cavity, and indicated by white lines in 3D models (bottom rows). V=ventricle, ot=outflow tract, a=atrium. Bottom two rows: 3D projections of outlines of myocardium (A, C, E, G, red=ventricle, blue=outflow tract, green=atrium) and endocardium (B, D, F, H, orange). Abnormal cardiac morphology is already evident in muz hearts at the looped cardiac tube stage. The muz ventricle is enlarged (E and G), except at the level of the AVC where a narrow cardiac tube connects the ventricular and atrial chambers (24 and G, black arrowhead). The outflow tract is dilated (25 and E, black arrowhead). The myocardial layer is thinner throughout the mutant heart and the endocardial tubes appear much narrower, with little lumen (24, 39 and F, white arrowheads).
Figure S3. Flowchart for Genetic Mapping in X. tropicalis
A recessive mutation (asterisk) is induced on one strain (for our screens this is an outbred N (Nigerian) stock) represented by red chromosomes, top left. Polymorphisms for mapping are introduced by crossing to strain(s) which differ from N at many sequence loci (blue chromosomes, top right (for muz, depending on availability of appropriate genders for crosses, these included both IC and PacBio)) to obtain a hybrid map cross generation. Meiotic recombination generates crossovers between red and blue strain DNA. In phenotypically mutant embryos, regions close to the homozygous mutant locus are likely to be homozygous ‘red’; with increasing distance, intervening crossovers produce heterozygous red:blue. Rapid assignment of mutations to chromosome/linkage group can often be accomplished by analysis of gynogenetic embryos with polymorphic markers from each of the 10 tropicalis centromeres (see Khokha et al. 2009); the ratio of mutant to wild type in gynogenetic embryos also provides an estimate of the mutation’s distance from the centromere. Representative chromosomes from two mutant (left) and wild type (right) gynogenetic embryos are shown; linkage is detected to red strain centromere (red circles) of the large chromosome; wild type or unlinked chromosomes show both blue and red centromere alleles. In cases where mutant loci are far from centromeres, mutations can be placed on a linkage group by assaying more distal polymorphisms from the meiotic map (whole genome marker scanning), or using the more cumbersome AFLP (used for initial steps of muz mapping predating the meiotic map (Vos et al., 1995)) to obtain linked sequences in map regions where markers are at low density. For further intermediate- and high-resolution mapping, embryos from natural matings are preferable (right column). To define the interval containing the mutation, polymorphisms derived from the meiotic map ~3-10cM apart flanking the locus are identified in mutant embryos: if markers are on opposite sides of the mutation, mutant embryos with crossovers between one marker and the mutant locus will not be recombinant for the marker on the other side and vice versa. High-resolution mapping involves typing a large number (>500) of mutant embryos with the flanking markers to identify the mutant set containing proximal crossovers. The small set can then be analyzed with subdividing polymorphisms to refine the interval, ideally placing the mutation on a single sequence scaffold that can be inspected for candidate genes. Candidate genes can be evaluated by expression in affected tissue, sequence lesions in the mutant, and the ability to phenocopy or rescue the mutation.
Movie S1. Muz embryos have no heartbeat.
Stage 37 muz embryo has no cardiac contractility (top), a wt sibling shows a strong regular heartbeat (bottom).
Movie S2. Myh6 morpholino phenocopies the muz mutation.
An myh6 morpholino injected embryo is morphologically normal but has almost no visible heartbeat, although at high magnification very weak twitching can be seen (bottom). In contrast the control morpholino has no effect on cardiac function (top).
Movie S3. 3-D model of a stage 40 wild type heart rotating about it’s dorsal-ventral axis. Ventral is at bottom. Red=ventricle, blue=outflow tract, green=atrium.
Movie S4. 3-D model of a stage 40 muz heart rotating about it’s dorsal-ventral axis. Ventral is at bottom. Red=ventricle, blue=outflow tract, green=atrium.
Movie S5. 3-D model of a stage 42 wild type heart rotating about it’s dorsal-ventral axis. Ventral is at bottom. Red=ventricle, blue=outflow tract, green=atrium.
Acknowledgements
We are very grateful to Elisabeth Ehler for the A4.1025 antibody, Elke Ober for comments on the manuscript, and members of the Zimmerman lab and Division of Developmental Biology for many helpful discussions. This work was supported by the Medical Research Council and NIH grant 1 RO1 HD4 2276-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong EJ, Bischoff J. Heart valve development: endothelial cell signaling and differentiation. Circ Res. 2004;95:459–70. doi: 10.1161/01.RES.0000141146.95728.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auman HJ, Coleman H, Riley HE, Olale F, Tsai HJ, Yelon D. Functional modulation of cardiac form through regionally confined cell shape changes. PLoS Biol. 2007;5:e53. doi: 10.1371/journal.pbio.0050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartman T, Walsh EC, Wen KK, McKane M, Ren J, Alexander J, Rubenstein PA, Stainier DY. Early myocardial function affects endocardial cushion development in zebrafish. PLoS Biol. 2004;2:E129. doi: 10.1371/journal.pbio.0020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beis D, et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132:4193–204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- Branford WW, Essner JJ, Yost HJ. Regulation of gut and heart left-right asymmetry by context-dependent interactions between xenopus lefty and BMP4 signaling. Dev Biol. 2000;223:291–306. doi: 10.1006/dbio.2000.9739. [DOI] [PubMed] [Google Scholar]
- Burggren WW, Warburton SJ, Slivkoff MD. Interruption of cardiac output does not affect short-term growth and metabolic rate in day 3 and 4 chick embryos. J Exp Biol. 2000;203:3831–8. doi: 10.1242/jeb.203.24.3831. [DOI] [PubMed] [Google Scholar]
- Carruthers S, Stemple DL. Genetic and genomic prospects for Xenopus tropicalis research. Semin Cell Dev Biol. 2006;17:146–53. doi: 10.1016/j.semcdb.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Ching YH, et al. Mutation in myosin heavy chain 6 causes atrial septal defect. Nat Genet. 2005;37:423–8. doi: 10.1038/ng1526. [DOI] [PubMed] [Google Scholar]
- Dan-Goor M, Silberstein L, Kessel M, Muhlrad A. Localization of epitopes and functional effects of two novel monoclonal antibodies against skeletal muscle myosin. J Muscle Res Cell Motil. 1990;11:216–26. doi: 10.1007/BF01843575. [DOI] [PubMed] [Google Scholar]
- Ehler E, Rothen BM, Hammerle SP, Komiyama M, Perriard JC. Myofibrillogenesis in the developing chicken heart: assembly of Z-disk, M-line and the thick filaments. J Cell Sci. 1999;112(Pt 10):1529–39. doi: 10.1242/jcs.112.10.1529. [DOI] [PubMed] [Google Scholar]
- Eisenberg LM, Markwald RR. Molecular regulation of atrioventricular valvuloseptal morphogenesis. Circ Res. 1995;77:1–6. doi: 10.1161/01.res.77.1.1. [DOI] [PubMed] [Google Scholar]
- Evans SM, Yan W, Murillo MP, Ponce J, Papalopulu N. tinman, a Drosophila homeobox gene required for heart and visceral mesoderm specification, may be represented by a family of genes in vertebrates: XNkx-2.3, a second vertebrate homologue of tinman. Development. 1995;121:3889–99. doi: 10.1242/dev.121.11.3889. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–45. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Yan W, Mohun TJ, Evans SM. Vertebrate tinman homologues XNkx2-3 and XNkx2-5 are required for heart formation in a functionally redundant manner. Development. 1998;125:4439–49. doi: 10.1242/dev.125.22.4439. [DOI] [PubMed] [Google Scholar]
- Gallego A, Duran AC, De Andres AV, Navarro P, Munoz-Chapuli R. Anatomy and development of the sinoatrial valves in the dogfish (Scyliorhinus canicula) Anat Rec. 1997;248:224–32. doi: 10.1002/(SICI)1097-0185(199706)248:2<224::AID-AR9>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Garriock RJ, Meadows SM, Krieg PA. Developmental expression and comparative genomic analysis of Xenopus cardiac myosin heavy chain genes. Dev Dyn. 2005;233:1287–93. doi: 10.1002/dvdy.20460. [DOI] [PubMed] [Google Scholar]
- Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klein R, Lemke G. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature. 1995;378:390–4. doi: 10.1038/378390a0. [DOI] [PubMed] [Google Scholar]
- Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, Seidman JG. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- Goda T, Abu-Daya A, Carruthers S, Clark MD, Stemple DL, Zimmerman LB. Genetic screens for mutations affecting development of Xenopus tropicalis. PLoS Genet. 2006;2:e91. doi: 10.1371/journal.pgen.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammer TC, Khokha MK, Lane MA, Lam K, Harland RM. Identification of mutants in inbred Xenopus tropicalis. Mech Dev. 2005;122:263–72. doi: 10.1016/j.mod.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Grego-Bessa J, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12:415–29. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenendijk BC, Hierck BP, Vrolijk J, Baiker M, Pourquie MJ, Gittenberger-de Groot AC, Poelmann RE. Changes in shear stress-related gene expression after experimentally altered venous return in the chicken embryo. Circ Res. 2005;96:1291–8. doi: 10.1161/01.RES.0000171901.40952.0d. [DOI] [PubMed] [Google Scholar]
- Grow MW, Krieg PA. Tinman function is essential for vertebrate heart development: elimination of cardiac differentiation by dominant inhibitory mutants of the tinman-related genes, XNkx2-3 and XNkx2-5. Dev Biol. 1998;204:187–96. doi: 10.1006/dbio.1998.9080. [DOI] [PubMed] [Google Scholar]
- Hove JR, Koster RW, Forouhar AS, Acevedo-Bolton G, Fraser SE, Gharib M. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature. 2003;421:172–7. doi: 10.1038/nature01282. [DOI] [PubMed] [Google Scholar]
- Huang C, Sheikh F, Hollander M, Cai C, Becker D, Chu PH, Evans S, Chen J. Embryonic atrial function is essential for mouse embryogenesis, cardiac morphogenesis and angiogenesis. Development. 2003;130:6111–9. doi: 10.1242/dev.00831. [DOI] [PubMed] [Google Scholar]
- Hyatt BA, Yost HJ. The left-right coordinator: the role of Vg1 in organizing left-right axis formation. Cell. 1998;93:37–46. doi: 10.1016/s0092-8674(00)81144-7. [DOI] [PubMed] [Google Scholar]
- Icardo JM, Guerrero A, Duran AC, Domezain A, Colvee E, Sans-Coma V. The development of the sturgeon heart. Anat Embryol (Berl) 2004;208:439–49. doi: 10.1007/s00429-004-0418-x. [DOI] [PubMed] [Google Scholar]
- Jones WK, Grupp IL, Doetschman T, Grupp G, Osinska H, Hewett TE, Boivin G, Gulick J, Ng WA, Robbins J. Ablation of the murine alpha myosin heavy chain gene leads to dosage effects and functional deficits in the heart. J Clin Invest. 1996;98:1906–17. doi: 10.1172/JCI118992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha MK, et al. Rapid gynogenetic mapping of Xenopus tropicalis mutations to chromosomes. Dev Dyn. 2009;238:1398–46. doi: 10.1002/dvdy.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein SL, Gerhard DS, Wagner L, Richardson P, Schriml LM, Sater AK, Warren WC, McPherson JD. Resources for genetic and genomic studies of Xenopus. Methods Mol Biol. 2006;322:1–16. doi: 10.1007/978-1-59745-000-3_1. [DOI] [PubMed] [Google Scholar]
- Klein SL, Strausberg RL, Wagner L, Pontius J, Clifton SW, Richardson P. Genetic and genomic tools for Xenopus research: The NIH Xenopus initiative. Dev Dyn. 2002;225:384–91. doi: 10.1002/dvdy.10174. [DOI] [PubMed] [Google Scholar]
- Kolker SJ, Tajchman U, Weeks DL. Confocal imaging of early heart development in Xenopus laevis. Dev Biol. 2000;218:64–73. doi: 10.1006/dbio.1999.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latinkic BV, Kotecha S, Mohun TJ. Induction of cardiomyocytes by GATA4 in Xenopus ectodermal explants. Development. 2003;130:3865–76. doi: 10.1242/dev.00599. [DOI] [PubMed] [Google Scholar]
- Mahdavi V, Chambers AP, Nadal-Ginard B. Cardiac alpha- and beta-myosin heavy chain genes are organized in tandem. Proc Natl Acad Sci U S A. 1984;81:2626–30. doi: 10.1073/pnas.81.9.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdavi V, Periasamy M, Nadal-Ginard B. Molecular characterization of two myosin heavy chain genes expressed in the adult heart. Nature. 1982;297:659–64. doi: 10.1038/297659a0. [DOI] [PubMed] [Google Scholar]
- Manasek FJ, Kulikowski RR, Fitzpatrick L. Cytodifferentiation: a causal antecedent of looping? Birth Defects Orig Artic Ser. 1978;14:161–78. [PubMed] [Google Scholar]
- Mellish J-AE, Pinder AW, Smith SC. You’ve got to have heart… or do you? Axolotl Newsletter. 1994;23:34–38. [Google Scholar]
- Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378:386–90. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- Mohun TJ, Leong LM, Weninger WJ, Sparrow DB. The morphology of heart development in Xenopus laevis. Dev Biol. 2000;218:74–88. doi: 10.1006/dbio.1999.9559. [DOI] [PubMed] [Google Scholar]
- Nicol RL, Frey N, Olson EN. From the sarcomere to the nucleus: role of genetics and signaling in structural heart disease. Annu Rev Genomics Hum Genet. 2000;1:179–223. doi: 10.1146/annurev.genom.1.1.179. [DOI] [PubMed] [Google Scholar]
- Nishii K, Morimoto S, Minakami R, Miyano Y, Hashizume K, Ohta M, Zhan DY, Lu QW, Shibata Y. Targeted disruption of the cardiac troponin T gene causes sarcomere disassembly and defects in heartbeat within the early mouse embryo. Dev Biol. 2008;322:65–73. doi: 10.1016/j.ydbio.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Noramly S, Zimmerman L, Cox A, Aloise R, Fisher M, Grainger RM. A gynogenetic screen to isolate naturally occurring recessive mutations in Xenopus tropicalis. Mech Dev. 2005;122:273–87. doi: 10.1016/j.mod.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Oana S, Machida S, Hiratsuka E, Furutani Y, Momma K, Takao A, Matsuoka R. The complete sequence and expression patterns of the atrial myosin heavy chain in the developing chick. Biol Cell. 1998;90:605–13. [PubMed] [Google Scholar]
- Pelster B, Burggren WW. Disruption of hemoglobin oxygen transport does not impact oxygen-dependent physiological processes in developing embryos of zebra fish (Danio rerio) Circ Res. 1996;79:358–62. doi: 10.1161/01.res.79.2.358. [DOI] [PubMed] [Google Scholar]
- Peltz SW, Brown AH, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993;7:1737–54. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- Peterkin T, Gibson A, Patient R. Redundancy and evolution of GATA factor requirements in development of the myocardium. Dev Biol. 2007;311:623–35. doi: 10.1016/j.ydbio.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan YL, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- Ramsdell AF, Yost HJ. Cardiac looping and the vertebrate left-right axis: antagonism of left-sided Vg1 activity by a right-sided ALK2-dependent BMP pathway. Development. 1999;126:5195–205. doi: 10.1242/dev.126.23.5195. [DOI] [PubMed] [Google Scholar]
- Sater AK, Jacobson AG. The specification of heart mesoderm occurs during gastrulation in Xenopus laevis. Development. 1989;105:821–30. doi: 10.1242/dev.105.4.821. [DOI] [PubMed] [Google Scholar]
- Scherz PJ, Huisken J, Sahai-Hernandez P, Stainier DY. High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development. 2008;135:1179–87. doi: 10.1242/dev.010694. [DOI] [PubMed] [Google Scholar]
- Sedmera D. Form follows function: developmental and physiological view on ventricular myocardial architecture. Eur J Cardiothorac Surg. 2005;28:526–8. doi: 10.1016/j.ejcts.2005.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnert AJ, Huq A, Weinstein BM, Walker C, Fishman M, Stainier DY. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31:106–10. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- Sehnert AJ, Stainier DY. A window to the heart: can zebrafish mutants help us understand heart disease in humans? Trends Genet. 2002;18:491–4. doi: 10.1016/s0168-9525(02)02766-x. [DOI] [PubMed] [Google Scholar]
- Shi Y, Katsev S, Cai C, Evans S. BMP signaling is required for heart formation in vertebrates. Dev Biol. 2000;224:226–37. doi: 10.1006/dbio.2000.9802. [DOI] [PubMed] [Google Scholar]
- Sive H, Grainger RM, Harland RM. Early development of Xenopus laevis: A laboratory manual. Cold Spring Harbor Laboratory Press; Woodbury (NY): 2000. p. 338. [Google Scholar]
- Small EM, Warkman AS, Wang DZ, Sutherland LB, Olson EN, Krieg PA. Myocardin is sufficient and necessary for cardiac gene expression in Xenopus. Development. 2005;132:987–97. doi: 10.1242/dev.01684. [DOI] [PubMed] [Google Scholar]
- Smith WC, Harland RM. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell. 1992;70:829–40. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- Stainier DY. Zebrafish genetics and vertebrate heart formation. Nat Rev Genet. 2001;2:39–48. doi: 10.1038/35047564. [DOI] [PubMed] [Google Scholar]
- Taber LA. Biophysical mechanisms of cardiac looping. Int J Dev Biol. 2006;50:323–32. doi: 10.1387/ijdb.052045lt. [DOI] [PubMed] [Google Scholar]
- Taber LA, Lin IE, Clark EB. Mechanics of cardiac looping. Dev Dyn. 1995;203:42–50. doi: 10.1002/aja.1002030105. [DOI] [PubMed] [Google Scholar]
- Territo PR, Burggren WW. Cardio-respiratory ontogeny during chronic carbon monoxide exposure in the clawed frog Xenopus laevis. J Exp Biol. 1998;201:1461–72. doi: 10.1242/jeb.201.9.1461. [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–14. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkman AS, Krieg PA. Xenopus as a model system for vertebrate heart development. Semin Cell Dev Biol. 2007;18:46–53. doi: 10.1016/j.semcdb.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield TT, Sharpe CR, Wylie CC. Nonsense-mediated mRNA decay in Xenopus oocytes and embryos. Dev Biol. 1994;165:731–4. doi: 10.1006/dbio.1994.1289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. AFLP and SSLP markers define the muz-containing interval on LG1. (A) AFLP reactions on bulk segregant wild type and muz genomic DNA produced five polymorphic markers linked to the mutation (white boxes). (B, left panel) Linked AFLP markers were placed on genomic sequence scaffolds, from which SSLP markers were tested on bulk segregant (BS) wt and muz DNA, confirming linkage to these scaffolds. (B, right panel) Genotyping of individual muz embryos with SSLP markers 040E09 and 018E09 defined the muz-containing interval. Recombinant embryos are indicated by asterisks.
Movie S6. 3-D model of a stage 42 muz heart rotating about it’s dorsal-ventral axis. Ventral is at bottom. Red=ventricle, blue=outflow tract, green=atrium.
Table S1. Gene models in the muz-containing genetic interval and their official full names as provided by the HUGO gene nomenclature committee.
Figure S2. Abnormal morphology of muz hearts is evident as early as the looping cardiac tube stage.
Coronal plastic sections of stage 35 wt and muz hearts (top rows), numbered from ventral side of cardiac cavity, and indicated by white lines in 3D models (bottom rows). V=ventricle, ot=outflow tract, a=atrium. Bottom two rows: 3D projections of outlines of myocardium (A, C, E, G, red=ventricle, blue=outflow tract, green=atrium) and endocardium (B, D, F, H, orange). Abnormal cardiac morphology is already evident in muz hearts at the looped cardiac tube stage. The muz ventricle is enlarged (E and G), except at the level of the AVC where a narrow cardiac tube connects the ventricular and atrial chambers (24 and G, black arrowhead). The outflow tract is dilated (25 and E, black arrowhead). The myocardial layer is thinner throughout the mutant heart and the endocardial tubes appear much narrower, with little lumen (24, 39 and F, white arrowheads).
Figure S3. Flowchart for Genetic Mapping in X. tropicalis
A recessive mutation (asterisk) is induced on one strain (for our screens this is an outbred N (Nigerian) stock) represented by red chromosomes, top left. Polymorphisms for mapping are introduced by crossing to strain(s) which differ from N at many sequence loci (blue chromosomes, top right (for muz, depending on availability of appropriate genders for crosses, these included both IC and PacBio)) to obtain a hybrid map cross generation. Meiotic recombination generates crossovers between red and blue strain DNA. In phenotypically mutant embryos, regions close to the homozygous mutant locus are likely to be homozygous ‘red’; with increasing distance, intervening crossovers produce heterozygous red:blue. Rapid assignment of mutations to chromosome/linkage group can often be accomplished by analysis of gynogenetic embryos with polymorphic markers from each of the 10 tropicalis centromeres (see Khokha et al. 2009); the ratio of mutant to wild type in gynogenetic embryos also provides an estimate of the mutation’s distance from the centromere. Representative chromosomes from two mutant (left) and wild type (right) gynogenetic embryos are shown; linkage is detected to red strain centromere (red circles) of the large chromosome; wild type or unlinked chromosomes show both blue and red centromere alleles. In cases where mutant loci are far from centromeres, mutations can be placed on a linkage group by assaying more distal polymorphisms from the meiotic map (whole genome marker scanning), or using the more cumbersome AFLP (used for initial steps of muz mapping predating the meiotic map (Vos et al., 1995)) to obtain linked sequences in map regions where markers are at low density. For further intermediate- and high-resolution mapping, embryos from natural matings are preferable (right column). To define the interval containing the mutation, polymorphisms derived from the meiotic map ~3-10cM apart flanking the locus are identified in mutant embryos: if markers are on opposite sides of the mutation, mutant embryos with crossovers between one marker and the mutant locus will not be recombinant for the marker on the other side and vice versa. High-resolution mapping involves typing a large number (>500) of mutant embryos with the flanking markers to identify the mutant set containing proximal crossovers. The small set can then be analyzed with subdividing polymorphisms to refine the interval, ideally placing the mutation on a single sequence scaffold that can be inspected for candidate genes. Candidate genes can be evaluated by expression in affected tissue, sequence lesions in the mutant, and the ability to phenocopy or rescue the mutation.
Movie S1. Muz embryos have no heartbeat.
Stage 37 muz embryo has no cardiac contractility (top), a wt sibling shows a strong regular heartbeat (bottom).
Movie S2. Myh6 morpholino phenocopies the muz mutation.
An myh6 morpholino injected embryo is morphologically normal but has almost no visible heartbeat, although at high magnification very weak twitching can be seen (bottom). In contrast the control morpholino has no effect on cardiac function (top).
Movie S3. 3-D model of a stage 40 wild type heart rotating about it’s dorsal-ventral axis. Ventral is at bottom. Red=ventricle, blue=outflow tract, green=atrium.
Movie S4. 3-D model of a stage 40 muz heart rotating about it’s dorsal-ventral axis. Ventral is at bottom. Red=ventricle, blue=outflow tract, green=atrium.
Movie S5. 3-D model of a stage 42 wild type heart rotating about it’s dorsal-ventral axis. Ventral is at bottom. Red=ventricle, blue=outflow tract, green=atrium.