Abstract
Restriction fragment length polymorphism (RFLP) analysis using a multilocus heat-inducible cytoplasmic heat-shock protein 70 (Hsp70) hybridization probe with EcoRI-digested genomic DNA was used in molecular typing of 129 Trichomonas vaginalis isolates. Results indicate that Trichomonas organisms exhibit considerable polymorphism in their Hsp70 RFLP patterns. Analysis of seven American Type Culture Collection reference strains and 122 clinical isolates, including 84 isolates from Jackson, Mississippi, 18 isolates from Atlanta, Georgia, and 20 isolates from throughout the United States, showed 105 distinct Hsp70 RFLP pattern subtypes for Trichomonas. Phylogenetic analysis of the Hsp70 RFLP data showed that the T. vaginalis isolates were organized into two clonal lineages. These results illustrate the substantial genomic diversity present in T. vaginalis and indicate that a large number of genetically distinct Trichomonas isolates may be responsible for human trichomoniasis in the United States.
Introduction
Trichomonas vaginalis is a flagellated protozoan parasite that infects the urogenital tract of humans, causing the most common non-viral, sexually transmitted disease in the world. The annual global incidence of trichomoniasis is greater than 170 million cases, including 7.4 million in the United States.1,2 The prevalence of trichomoniasis among reproductive-age women in the United States, 3.1% overall, shows a pronounced racial disparity, 1.3% in non-Hispanic white women versus 13.3% in non-Hispanic black women, and can exceed 50% in sexually transmitted disease clinic patients and high-risk adolescents.3,4 Trichomoniasis is associated with vaginitis, cervecitis, dysuria, and urethritis, and is a risk factor for herpes simplex virus type 2 infection, human immunodeficiency virus transmission and acquisition, and possibly pelvic inflammatory disease and cervical neoplasia.5–10 Trichomonas infection in pregnant women increases the incidence of preterm delivery, premature rupture of membranes, low birth weight infants, and can be transmitted to neonates during passage through the birth canal.11–13 In men, trichomoniasis is a cause of chronic prostatitis and non-gonococcal urethritis.14 Despite serious health consequences, efforts to detect and eliminate Trichomonas are confounded by large numbers of asymptomatic infections and the presence of drug-resistant strains.15 In addition, the relationships between Trichomonas strains and the variable clinical manifestations of trichomoniasis are not well understood, in part because of the lack of reliable and widely accepted techniques to type and track individual isolates.
Multiple approaches to typing Trichomonas isolates have been described; antigenic characterization, isozyme analysis, repetitive sequence hybridization, ribosomal gene and intergenic region sequence polymorphisms, pulsed-field gel electrophoresis, random amplified polymorphic DNA analysis, and restriction fragment length polymorphism (RFLP).16–26 These studies produced differing results, even when using similar techniques, in attempting to demonstrate concordance between parasite genotypes and phenotypic expressions during infection, such as virulence and metronidazole resistance. The T. vaginalis genome composition provides a potential explanation for this difficulty in correlating genotype with phenotype.27 The Trichomonas genome is highly repetitive with 65% of the genome consisting of repeated sequences, including 59 repeat families of transposable elements representing 25% of the genome. The movement of these transposable elements through the genome can generate considerable genetic diversity. The population structure of Trichomonas has been proposed to be clonal in nature because there is little evidence of sexual recombination and segregation.28 A consequence of clonality in Trichomonas is that in the presence of active mechanisms to generate diversity, genetic differences can become fixed in the population and lead to a large and diverse population structure.
In this study, we assess the level of genetic diversity in T. vaginalis isolates using restriction fragment length polymorphism of cytoplasmic heat-shock protein 70 (Hsp70) genes.25 The cHsp70s in Trichomonas are a large multilocus gene family present in at least eight nearly identical copies (96–97% sequence identity).27 The use of a multilocus gene family for Trichomonas genome analysis provides a tool with considerable discrimination power to analyze genetic diversity in T. vaginalis isolates.
Materials and Methods
Trichomonas parasites
Trichomonas vaginalis isolates were obtained from the American Type Culture Collection (ATTC, Manassas, VA), the Centers for Disease Control and Prevention (CDC, Atlanta, GA), and urban teaching hospitals in Jackson, Mississippi, and Atlanta, Georgia. Reference strains were obtained from ATCC or provided by the CDC (ATCC and CDC isolates). Twenty isolates were obtained from cultures submitted to the CDC for metronidazole susceptibility testing (MSA isolates) after patients did not respond to a minimum of two courses of metronidazole treatment. Eighteen isolates were cultured from women at a teaching hospital in Atlanta, Georgia (GMH isolates).19 Eighty four isolates came from sexually transmitted disease clinic or emergency department patients at the University of Mississippi Medical Center (UMC), Jackson, Mississippi. This set included isolates from males presenting with urethritis (B isolates, n = 6), women participating in a trial of oral versus intravaginal metronidazole treatment29 (C isolates, n = 9), a study of single versus multiple doses of metronidazole treatment (D isolates, n = 9), women with vaginitis and their sexual partners when available (P, S, and T isolates, n = 8), and random culture of urine specimens, regardless of patient symptoms, submitted to the UMC emergency department (E isolates, n = 52). Informed consent was obtained when required by the UMC Institutional Review Board for Human Investigation. Trichomonas trophozoites were cultured from urethral/penile discharge and posterior vaginal fornix or urine sediment in trypticase-yeast maltose medium and harvested as previously reported.19,25
Nucleic acid isolation
Genomic DNA was isolated from log phase (106 parasites/mL) cultures using a DNA extraction kit (CDC, GMH, and MSA isolates) or a modified DEPC-Triton X-100 method (ATCC, B, C, D, E, P, S, and T isolates).19,25 Purity and concentration of DNA were determined by optical density measurements at 260 nm and 280 nm.
Hsp70 RFLP analysis
Two-microgram aliquots of T. vaginalis genomic DNA were digested with EcoRI, resolved by electrophoresis for at 1,305–1,400 volts/hour in 24 cm × 7 mm, 0.9% agarose gels, transferred to nylon membranes, and hybridized with a radiolabelled 1.8-kb T. vaginalis cytoplasmic Hsp70 cDNA fragment (GenBank U93873, nt 31–1832).30 Blots were washed to a final stringency of 0.2× SSC (30 mM NaCl, 3 mM sodium citrate), 0.5% sodium dodecyl sulfate at 45–55°C and exposed for autoradiography at −70°C.
Phylogenetic analysis
The phylogenetic relatedness of the T. vaginalis isolates was assessed using FreeTree, a software program designed for analysis of binary character data (presence or absence of character state) generated in DNA fingerprinting methods.31 The genetic distances between isolates were calculated using the Nei-Li coefficient of similarity and a phylogenetic tree constructed from the distance matrix using the neighbor-joining method.32,33 Statistical support for topological elements (nodes) within the phylogram was estimated by operational taxonomic unit–based jackknifing with 10,000 re-sampled datasets.31
Results
A total of 129 Trichomonas clinical isolates obtained from 127 individual patients were typed by Hsp70 RFLP of EcoRI-digested genomic DNA. These isolates are listed in Table 1 with the year of initial culture, sex of patient, and geographic origin included when known. Eighty four isolates were obtained in Jackson, Mississippi, 18 in Atlanta, Georgia, and 25 from diverse locations across the United States; the geographic origin of 2 strains was not available. Although most of the Trichomonas isolates were obtained from symptomatic women, the 52 E series isolates (E1–E98, Table 1) were cultured at random and may include asymptomatic patients. The E series study showed an infection rate of 8% (100 of 1,248 samples collected) in UMC emergency room patients, 9.4% in females, and 0.9% in males.
Table 1.
Trichomonas vaginalis isolates analyzed by heat-shock protein 70 restriction fragment length polymorphism*
| Isolate | Year | Sex of patient | Location | Isolate | Year | Sex of patient | Location |
|---|---|---|---|---|---|---|---|
| ATCC30238 | 1963 | F | Baltimore, MD | E45 | 2001 | Jackson, MS | |
| ATCC50140 | 1983 | F | MA | E46 | 2001 | Jackson, MS | |
| ATCC50141 | 1983 | F | CT | E47 | 2001 | Jackson, MS | |
| ATCC50142 | 1983 | F | NY | E49 | 2001 | F | Jackson, MS |
| ATCC50143 | 1980 | F | Columbus, OH | E50 | 2001 | F | Jackson, MS |
| CDC520 | F | E51 | 2001 | F | Jackson, MS | ||
| CDC697 | F | E56 | 2001 | F | Jackson, MS | ||
| B95 | 1993 | M | Jackson, MS | E61 | 2001 | F | Jackson, MS |
| B110 | 1993 | M | Jackson, MS | E62 | 2001 | F | Jackson, MS |
| B114 | 1993 | M | Jackson, MS | E69 | 2001 | F | Jackson, MS |
| B137 | 1993 | M | Jackson, MS | E70 | 2001 | F | Jackson, MS |
| B157 | 1993 | M | Jackson, MS | E77 | 2001 | F | Jackson, MS |
| B161 | 1993 | M | Jackson, MS | E78 | 2001 | M | Jackson, MS |
| C2 | 1993 | F | Jackson, MS | E83 | 2001 | F | Jackson, MS |
| C3 | 1993 | F | Jackson, MS | E86 | 2001 | F | Jackson, MS |
| C4 | 1993 | F | Jackson, MS | E88 | 2001 | F | Jackson, MS |
| C5 | 1993 | F | Jackson, MS | E95 | 2001 | F | Jackson, MS |
| C6 | 1993 | F | Jackson, MS | E98 | 2001 | F | Jackson, MS |
| C7 | 1993 | F | Jackson, MS | P1 | 1995 | F | Jackson, MS |
| C8 | 1993 | F | Jackson, MS | P2 | 1995 | F | Jackson, MS |
| C11 | 1993 | F | Jackson, MS | S1M | 1998 | M | Jackson, MS |
| C14 | 1993 | F | Jackson, MS | S1F | 1998 | F | Jackson, MS |
| D1 | 1994 | F | Jackson, MS | S2 | 1998 | F | Jackson, MS |
| D2 | 1994 | F | Jackson, MS | S3 | 1998 | F | Jackson, MS |
| D4 | 1994 | F | Jackson, MS | T1M | 2000 | M | Jackson, MS |
| D4F | 1994 | F | Jackson, MS | T1F | 2000 | F | Jackson, MS |
| D5 | 1994 | F | Jackson, MS | GMH1 | 1997 | F | Atlanta, GA |
| D6 | 1994 | F | Jackson, MS | GMH2 | 1997 | F | Atlanta, GA |
| D8 | 1994 | F | Jackson, MS | GMH3 | 1997 | F | Atlanta, GA |
| D9 | 1994 | F | Jackson, MS | GMH4 | 1997 | F | Atlanta, GA |
| D10 | 1994 | F | Jackson, MS | GMH5 | 1997 | F | Atlanta, GA |
| E1 | 2000 | F | Jackson, MS | GMH7 | 1997 | F | Atlanta, GA |
| E2 | 2000 | F | Jackson, MS | GMH10 | 1997 | F | Atlanta, GA |
| E3 | 2000 | F | Jackson, MS | GMH11 | 1997 | F | Atlanta, GA |
| E5 | 2000 | Jackson, MS | GMH17 | 1997 | F | Atlanta, GA | |
| E6 | 2000 | Jackson, MS | GMH18 | 1997 | F | Atlanta, GA | |
| E8 | 2000 | Jackson, MS | GMH29 | 1997 | F | Atlanta, GA | |
| E9 | 2001 | F | Jackson, MS | GMH36 | 1997 | F | Atlanta, GA |
| E10 | 2001 | F | Jackson, MS | GMH45 | 1997 | F | Atlanta, GA |
| E12 | 2001 | Jackson, MS | GMH46 | 1997 | F | Atlanta, GA | |
| E16 | 2001 | F | Jackson, MS | GMH47 | 1997 | F | Atlanta, GA |
| E17 | 2001 | F | Jackson, MS | GMH57 | 1997 | F | Atlanta, GA |
| E18 | 2001 | F | Jackson, MS | GMH66 | 1997 | F | Atlanta, GA |
| E19 | 2001 | F | Jackson, MS | GMH67 | 1997 | F | Atlanta, GA |
| E20 | 2001 | Jackson, MS | MSA806 | 1995 | F | Akron, OH | |
| E22 | 2001 | M | Jackson, MS | MSA807 | 1995 | F | Crestview, FL |
| E23 | 2001 | F | Jackson, MS | MSA809B | 1996 | F | Brooklyn, NY |
| E24 | 2001 | M | Jackson, MS | MSA810 | 1996 | F | Gilbert, AZ |
| E25 | 2001 | Jackson, MS | MSA811 | 1996 | F | Benton Harbor, MI | |
| E26 | 2001 | Jackson, MS | MSA812 | 1996 | F | Birmingham, AL | |
| E27 | 2001 | F | Jackson, MS | MSA815 | 1996 | F | Hartford, CT |
| E28 | 2001 | Jackson, MS | MSA816 | 1996 | F | Rochester, NY | |
| E29A | 2001 | Jackson, MS | MSA819 | 1996 | F | Brooklyn, NY | |
| E29B | 2001 | Jackson, MS | MSA822 | 1996 | F | Clearwater, FL | |
| E30 | 2001 | Jackson, MS | MSA823 | 1996 | F | Austin, TX | |
| E34 | 2001 | M | Jackson, MS | MSA824 | 1996 | F | Philadelphia, PA |
| E35 | 2001 | Jackson, MS | MSA826 | 1996 | F | Grover Beach, CA | |
| E36 | 2001 | Jackson, MS | MSA828 | 1997 | F | Minneapolis, MN | |
| E37 | 2001 | M | Jackson, MS | MSA829 | 1997 | F | Columbus, OH |
| E38 | 2001 | F | Jackson, MS | MSA830 | 1997 | F | Great Bend, KS |
| E39 | 2001 | Jackson, MS | MSA831 | 1997 | F | Brooklyn, NY | |
| E40 | 2001 | Jackson, MS | MSA836 | 1997 | F | Greenville, SC | |
| E42 | 2001 | Jackson, MS | MSA844 | 1998 | F | Rochester, NY | |
| E42 | 2001 | Jackson, MS | MSA844 | 1998 | F | Rochester, NY | |
| E43 | 2001 | Jackson, MS | MSA845 | 1998 | F | Nacogloches, TX | |
| E44 | 2001 | Jackson, MS |
ATCC = American Type Culture Collection. A blank space indicates that no information is available.
Figure 1 shows a representative Southern blot with Hsp70 RFLP patterns for 12 Trichomonas isolates. A common set of Trichomonas isolates was included in each analysis to facilitate identification of individual bands and comparison of different Southern blot results. In each isolate tested, 7–11 distinct DNA bands, ranging in size from 1.8 kb to 22.5 kb, were detected. Examples of isolates producing 7 bands (D1, B137, B157, and D10), 8 bands (B114), 9 bands (B110, P1 and T1F), and 10 bands (C3) are shown in Figure 1. More than 50% of T. vaginalis isolates contained 9 bands (n = 68), 35 isolates had 8 hybridizing DNA bands, and isolates with 7 (n = 16), 10 (n = 7), or 11 (n = 3) hybridizing bands were less frequent. Thirty-one DNA fragments were identified; Hsp70 RFLP fragments ranging in size from 1.8 kb to 22.5 kb are shown in Figure 1. The observed frequency for each band in the 129 T. vaginalis isolates is shown in Table 2. Four bands, 2.7 kb, 3.7 kb, 5.6 kb, and 5.8 kb (Figure 1), were present in nearly all isolates tested but many bands were represented in fewer isolates; 10 bands were each present in less than 9 isolates. The Hsp70 RFLP patterns were reproducible because 72 isolates were run multiple times, including reference isolates run 7–15 times each, and in every instance an identical Hsp70 RFLP pattern was produced. The Hsp70 RFLP patterns from individual T. vaginalis isolates were also stable over time, generating identical patterns from DNA samples obtained several years apart.
Figure 1.
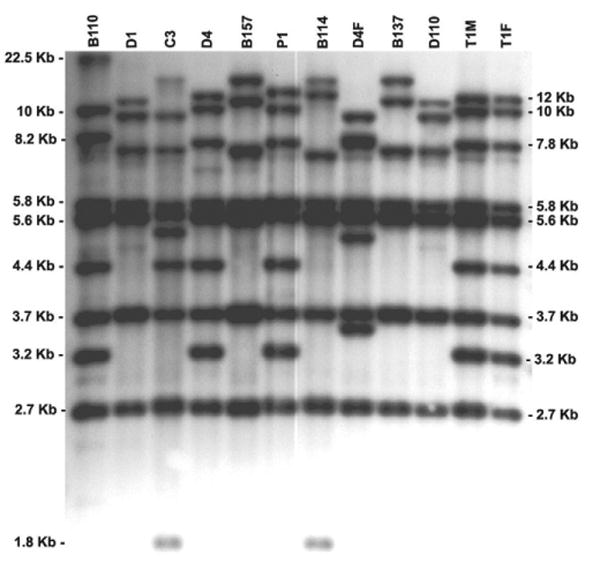
Restriction fragment length polymorphism of Trichomonas vaginalis isolates analyzed on Southern blots containing 1–2 μg EcoRI-digested genomic DNA hybridized with a T. vaginalis cytoplasmic heat-shock protein 70 (HSP70) cDNA probe. Selected Hsp70 RFLP band sizes in kilobases, calculated from the migration of molecular weight markers (not shown), are indicated in the left and right margins. Trichomonas isolates, labeled above each lane are described in Table 1.
Table 2.
Number of Trichomonas isolates possessing individual heat-shock protein 70 hybridizing bands
| Band size (kb) | No. isolates | Band size (kb) | No. isolates | Band size (kb) | No. isolates |
|---|---|---|---|---|---|
| 1.8 | 26 | 5.6 | 6 | 9.6 | 37 |
| 2.0 | 1 | 5.8 | 127 | 10.0 | 24 |
| 2.4 | 3 | 6.5 | 1 | 11.5 | 27 |
| 2.7 | 128 | 7.0 | 34 | 12.0 | 30 |
| 3.2 | 34 | 7.3 | 18 | 12.5 | 16 |
| 3.4 | 2 | 7.5 | 48 | 13.3 | 19 |
| 3.5 | 26 | 7.8 | 36 | 15.0 | 39 |
| 3.7 | 129 | 8.0 | 13 | 16.5 | 5 |
| 4.4 | 103 | 8.2 | 24 | 18.0 | 4 |
| 5.2 | 9 | 9.0 | 8 | 22.5 | 2 |
| 5.5 | 128 |
The Hsp70 RFLP patterns indicate a high degree of diversity among Trichomonas clinical isolates. A total of 105 different Hsp70 RFLP patterns were observed; 85 isolates produced unique patterns and 20 additional Hsp70 RFLP patterns were shared among 44 isolates. Table 3 shows the isolates that comprise these 20 shared patterns; most are paired identical isolates but two groupings contain four isolates each (MSA826, D4, T1F, T1M and D1, D2, D5, D10). In Figure 1, seven Hsp70 RFLP patterns are seen, including matching patterns for D4–T1M–T1F, D1–D10, and B137–B157. Multiple identical Hsp70 RFLP patterns were obtained from isolates widely separated in time of collection and in geographic origin: GMH1 (Atlanta, GA, 1997) and E8 (Jackson, MS, 2000); MSA812 (Birmingham, AL, 1996) and E19 (Jackson, MS, 2001); MSA819 (Brooklyn, NY, 1998) and GMH17 (Atlanta, GA, 1999); MSA826 (Grover Beach, CA, 1996) and three strains from Jackson, MS, D4 (1994) and T1M and T1F (2000); MSA830 (Great Bend, KS, 1997) and E40 (Jackson, MS, 2001); MSA836 (Greenville, SC, 1997) and GMH10 (Atlanta, GA, 1996); and MSA844 (Rochester, NY, 1998), which is identical to E35 (Jackson, MS, 2001). These results illustrate the widespread dispersion of individual isolates as well as their temporal stability over at least six years. One pair of sexual partners in the study (T1F and T1F, Figure 1) had the same Hsp70 RFLP pattern but another partner pair (S1M and S1F) had different patterns. Also of interest, the Trichomonas isolate cultured from patient D4 prior to metronidazole treatment differed from isolate D4F cultured after treatment, indicating that re-infection rather than treatment failure had occurred.
Table 3.
Trichomonas isolates sharing identical heat-shock protein 70 restriction fragment length polymorphism patterns
| GMH1, E8 | B137, B157 |
| GMH36, GMH47 | D1, D2, D5, D10 |
| MSA809B, MSA831 | E5, E12 |
| MSA812, E19 | E6, E23 |
| MSA819, GMH17 | E9, E36 |
| MSA826, D4, T1F, T1M | E44, E45 |
| MSA830, E40 | E61, E62 |
| MSA836, GMH10 | E70, E78 |
| MSA844, E35 | E83, E95 |
| S2, C8 | E86, E98 |
The phylogenetic relatedness of the 129 Trichomonas isolates was determined by analysis of their Hsp70 RFLP patterns using the program FreeTree.31 A phylogram built from the Nei-Li distance matrix using the neighbor-joining method for tree construction is shown in Figure 2. This phylogenetic analysis shows two major groups of T. vaginalis strains: group I with 78 isolates consisting of three subgroups (A, B, and C) of 37, 33, and 8 T. vaginalis isolates, respectively, and group II with 51 isolates consisting of two subgroups (D and E) containing 28 and 23 isolates, respectively. Each of these groups and subgroups is characterized by deeply diverging branches containing individual T. vaginalis isolates. The operational taxonomic unit–based jackknifing values calculated for each node indicate that there is excellent statistical support for the placement of T. vaginalis isolates within the phylogram, especially at the most distal nodes. The long terminal branches and the high bootstrap support values at the distal nodes are suggestive of a clonal radiation of T. vaginalis with little genetic flow between the different strains. Isolates possessing 7–10 hybridizing bands were distributed throughout the phylogram with representative members present in subgroups A–E, but the three isolates that contained 11 hybridizing bands (E17, E83, E95) were all located in subgroup E. The T. vaginalis isolates are also distributed throughout the figure without apparent regard to their geographical origin or sex of the patient. Although additional phenotypic information such as metronidazole sensitivity and presence of Trichomonas virus was available for some isolates analyzed, these data were not sufficient to permit any determination of concordance between these traits and the phylogenetic relationships shown here.
Figure 2.
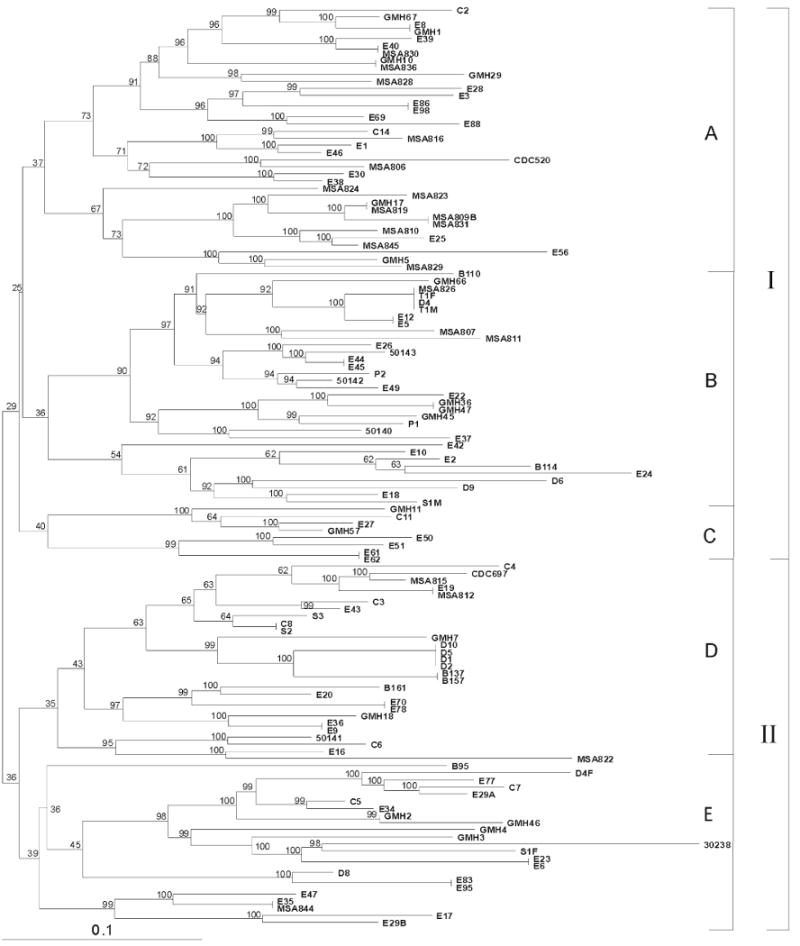
Phylogenetic tree of 129 Trichomonas vaginalis isolates constructed on the basis of the presence or absence of 31 individual heat-shock protein 70 (Hsp70) RFLP bands using the phylogeny inference package FreeTree.33 Numbers at the upper left of each node show the operational taxonomic unit jackknifing percent values based on 10,000 re-sampled datasets, which estimate statistical support for the existence of those branches. The sum of the horizontal branch lengths between individual Trichomonas isolates reflects the relative genetic distance separating the isolates.
Discussion
The genetic diversity among T. vaginalis isolates is considerable with 105 unique Hsp70 RFLP patterns identified in the 129 isolates analyzed. The Hsp70 RFLP patterns are reproducible, temporally stable over six years, and geographically disjunct. The detection of genetically identical isolates from diverse geographic locations is consistent with the theory of clonal propagation proposed for T. vaginalis, suggesting a common clonal descent.28 Significant genetic variability in T. vaginalis can exist even in a localized environment; the 84 isolates from Jackson, Mississippi and 18 isolates from Atlanta, Georgia produced 69 and 17 different Hsp70 RFLP patterns respectively. Much of the observed variability can be attributed to differences in hybridizing bands ≥ 26.5 kb, each present in 37% or less of isolates, and bands present in 80–100% of isolates are ≥ 25.8 kb. The large, diverse set of repeated, mobile genetic elements in the T. vaginalis genome provides a mechanism for generating size diversity.27 Similar repetitive families are present in other human parasites with clonal population structures such as Leishmania, Plasmodium, Giardia, and Trypanosoma.34 The ability of repeat elements to increase in copy number and transpose throughout the genome would naturally have an increased impact on larger DNA fragments. However, the preservation of 2.7 kb, 3.7 kb, 5.5 kb, and 5.8 kb Hsp70 RFLP fragments in nearly all T. vaginalis isolates indicates that transposon-mediated size changes are constrained in the sequences surrounding some T. vaginalis Hsp70 genes. The variable number of Hsp70 hybridizing bands detected in individual isolates, ranging from 7 to 11, predominantly reflects differences in Hsp70 copy number. The Trichomonas genome is highly repetitive with many multi-copy genes, and the existence of gene copy number variation between different T. vaginalis isolates has been demonstrated.20,27 It has also been proposed that Trichomonas has undergone a recent and substantial increase in genome size caused by duplication.27 However, six isolate pairs differ by the conversion of a larger band to two smaller bands of corresponding size, indicating that EcoRI restriction site polymorphism within Hsp70 genes may account for some of this heterogeneity.
Diversity among Trichomonas isolates has been observed in data from prior studies characterizing antigenic, enzymatic, or genetic markers.16–26 However, those analyses either used a limited number of strains and were not informative of the larger T. vaginalis population structure, or were based on random amplified polymorphic DNA analysis, which can be compromised by problems with reproducibility caused by sensitivity of the PCR and contaminating DNA from mycoplasma infection.35 The Hsp70 RFLP technique used here, coupled with the large number of isolates examined, offers a reliable and reproducible approach for demonstrating genetic diversity in Trichomonas. Although there are potential biases inherent in sample selection for this study, E series isolates cultured from randomly selected urine samples could represent T. vaginalis strains with increased survivability in urine and random selection could also produce multiple isolates from the same patient, it is difficult to envision how these biases could significantly impact our findings. In the United States, T. vaginalis isolates are diverse and are comprised of two major clonal lineages. Whether this population structure proves to be representative of T. vaginalis isolates across the globe is an interesting objective for future studies.
Trichomoniasis is characterized by significant heterogeneity in its clinical manifestations such as pathogenicity, metronidazole sensitivity, sequelae to infection, and susceptibility to acquisition of other infectious agents. Whether this clinical variability is caused by host factors or to differences in the phenotypic expression of individual T. vaginalis isolates is not clear. However, the genetic diversity among Trichomonas isolates demonstrated here suggests that an association between Trichomonas genotypes and highly variable clinical pathology may exist. Understanding this relationship will be a critical factor in devising strategies to control this disease and its clinical sequelae. Similar phenotypic variation in the course of human infection has been noted for other pathogens, which exhibit substantial genomic variation, including Leishmania species, Trypanosoma cruzi, Toxoplasma gondii, and Plasmodium falciparum.36–39
Genetic diversity in Trichomonas is an attribute that will need to be carefully considered in future investigations of trichomoniasis. The recent advent of proteomic and genomic techniques based on the T. vaginalis sequence presents an opportunity to elucidate the genetic factors controlling clinical manifestations, association with other disease entities, and drug resistance. The selection of Trichomonas isolates used to characterize the basis for its biologic variability will need to focus on those that are genetically similar to maximize the discovery of relevant genetic elements and avoid differences rooted in genetic diversity. In addition, efforts to design a vaccine for trichomoniasis will benefit from an understanding of the genetic diversity present in Trichomonas by focusing efforts on antigenic determinants which are broadly representative of the T. vaginalis population.
Contributor Information
John C. Meade, Department of Microbiology, University of Mississippi Medical Center, 2500 North State Street, Jackson, MS 39216-4505.
Jacqueline de Mestral, Department of Biochemistry and Molecular Biology, Dalhousie University, 5850 College Street, Halifax, Nova Scotia, Canada, B3H 1X5.
Jonathan K. Stiles, Department of Microbiology and Immunology, Morehouse School of Medicine, 720 Westview Drive, Atlanta, GA 30310-1495
W. Evan Secor, Division of Parasitic Diseases, Centers for Disease Control and Prevention, 4770 Buford Highway NE, Mailstop F-13, Atlanta, GA 30341.
Richard W. Finley, Department of Medicine, Division of Infectious Diseases, University of Mississippi Medical Center, 2500 North State Street, Jackson, MS 39216-4505
John D. Cleary, Department of Medicine, Division of Infectious Diseases, University of Mississippi Medical Center, 2500 North State Street, Jackson, MS 39216-4505
William B. Lushbaugh, Department of Microbiology, University of Mississippi Medical Center, 2500 North State Street, Jackson, MS 39216-4505
References
- 1.World Health Organization. Global Prevalence and Incidence of Selected Curable Sexually Transmitted Infections: Overview and Estimates. Geneva: World Health Organization; 2001. [Google Scholar]
- 2.Weinstock H, Berman S, Cates W., Jr Sexually transmitted diseases among American youth: incidence and prevalence estimates, 2000. Perspect Sex Reprod Health. 2004;36:6–10. doi: 10.1363/psrh.36.6.04. [DOI] [PubMed] [Google Scholar]
- 3.Soper D. Trichomoniasis: under control or undercontrolled? Am J Obstet Gynecol. 2004;190:281–290. doi: 10.1016/j.ajog.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 4.Sutton M, Sternberg M, Koumans EH, McQuillan G, Berman S, Markowitz L. The prevalence of Trichomonas vaginalis infection among reproductive-age women in the United States, 2001–2004. Clin Infect Dis. 2007;45:1319–1326. doi: 10.1086/522532. [DOI] [PubMed] [Google Scholar]
- 5.Swygard H, Sena AC, Hobbs MM, Cohen MS. Trichomoniasis: clinical manifestations, diagnosis and management. Sex Transm Infect. 2003;80:91–95. doi: 10.1136/sti.2003.005124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobel JD. Vaginitis. N Engl J Med. 1997;337:1896–1903. doi: 10.1056/NEJM199712253372607. [DOI] [PubMed] [Google Scholar]
- 7.Viikki M, Pukkala E, Nieminen P, Hakama M. Gynaecological infections as risk determinants of subsequent cervical neoplasia. Acta Oncol. 2000;39:71–75. doi: 10.1080/028418600431003. [DOI] [PubMed] [Google Scholar]
- 8.Cherpes TL, Wiesenfeld HC, Melan MA, Kant JA, Cosentino LA, Meyn LA, Hillier SL. The associations between pelvic inflammatory disease, Trichomonas vaginalis infection, and positive herpes simplex virus type 2 serology. Sex Transm Dis. 2006;33:747–752. doi: 10.1097/01.olq.0000218869.52753.c7. [DOI] [PubMed] [Google Scholar]
- 9.Sorvillo F, Smith L, Kerndt P, Ash L. Trichomonas vaginalis, HIV, and African-Americans. Emerg Infect Dis. 2001;7:927–932. doi: 10.3201/eid0706.010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McClelland RS, Sangare L, Hassan WM, Lavreys L, Mandaliya K, Kiarie J, Ndinya-Achola J, Jaoko W, Baeten JM. Infection with Trichomonas vaginalis increases the risk of HIV-1 acquisition. J Infect Dis. 2007;195:698–702. doi: 10.1086/511278. [DOI] [PubMed] [Google Scholar]
- 11.Cotch MF, Pastorek JG, Nugent RP, Hillier SL, Gibbs RS, Martin DH, Eschenbach DA, Edelman R, Carey JC, Regan JA, Krohn MA, Klebanoff MA, Rao AV, Rhoads GG. Trichomonas vaginalis associated with low birth weight and preterm delivery. The Vaginal Infections and Prematurity Study Group. Sex Transm Dis. 1997;24:353–360. doi: 10.1097/00007435-199707000-00008. [DOI] [PubMed] [Google Scholar]
- 12.Morton K, Regan L, Spring J, Houang F. A further look at infection at the time of therapeutic abortion. Eur J Obstet Gynecol Reprod Biol. 1990;37:231–236. doi: 10.1016/0028-2243(90)90029-z. [DOI] [PubMed] [Google Scholar]
- 13.Smith LM, Wang M, Zangwill K, Yeh S. Trichomonas vaginalis infection in a premature newborn. J Perinatol. 2002;22:502–503. doi: 10.1038/sj.jp.7210714. [DOI] [PubMed] [Google Scholar]
- 14.Krieger JN. Consider diagnosis and treatment of trichomoniasis in men. Sex Transm Dis. 1999;27:241–242. doi: 10.1097/00007435-200004000-00011. [DOI] [PubMed] [Google Scholar]
- 15.Sobel JD, Nagappan V, Nyirjesy P. Metronidazole-resistant vaginal trichomoniasis—An emerging problem. N Engl J Med. 1999;341:292–293. doi: 10.1056/NEJM199907223410417. [DOI] [PubMed] [Google Scholar]
- 16.Kreiger JN, Holmes KK, Spence MR, Rein MF, McCormack WM, Tam MR. Geographic variation among isolates of Trichomonas vaginalis: demonstration of antigenic heterogeneity by using monoclonal antibodies and the indirect immunofluorescence technique. J Infect Dis. 1985;152:979–984. doi: 10.1093/infdis/152.5.979. [DOI] [PubMed] [Google Scholar]
- 17.Proctor EM, Naaykens W, Wong Q, Bowie WR. Isoenzyme patterns of isolates of Trichomonas vaginalis from Vancouver. Sex Transm Dis. 1988;15:181–185. doi: 10.1097/00007435-198810000-00001. [DOI] [PubMed] [Google Scholar]
- 18.Ryu JS, Min DY, Shin MH, Cho YH. Genetic variance of Trichomonas vaginalis isolates by Southern hybridization. Korean J Parasitol. 1998;36:207–211. doi: 10.3347/kjp.1998.36.3.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snipes LJ, Gamard PM, Narcisi EM, Beard CB, Lehman T, Secor WE. Molecular epidemiology of metronidazole resistance in a population of Trichomonas vaginalis clinical isolates. J Clin Microbiol. 2000;38:3004–3009. doi: 10.1128/jcm.38.8.3004-3009.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Upcroft JA, Delgadillo-Correa MG, Dunne RL, Sturm AW, Johnson PJ, Upcroft P. Genotyping Trichomonas vaginalis. Int J Parasitol. 2006;36:821–828. doi: 10.1016/j.ijpara.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 21.Vaňá čová S, Tachezy J, Kulda J, Flegr J. Characterization of trichomonad species and strains by PCR fingerprinting. J Eukaryot Microbiol. 1997;44:545–552. doi: 10.1111/j.1550-7408.1997.tb05960.x. [DOI] [PubMed] [Google Scholar]
- 22.Hampl V, Vaňá čová S, Kulda J, Flegr J. Concordance between genetic relatedness and phenotypic similarities of Trichomonas vaginalis strains. BMC Evol Biol. 2001;1:11. doi: 10.1186/1471-2148-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rojas L, Fraga J, Sariego I. Genetic variability between Trichomonas vaginalis isolates and correlation with clinical presentation. Infect Genet Evol. 2004;4:53–58. doi: 10.1016/j.meegid.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Kaul P, Gupta I, Sehgal R, Malla N. Trichomonas vaginalis: random amplified polymorphic DNA analysis of isolates from symptomatic and asymptomatic women in India. Parasitol Int. 2004;53:255–262. doi: 10.1016/j.parint.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Stiles JK, Shah PH, Xue L, Meade JC, Lushbaugh WB, Cleary JD, Finley RW. Molecular typing of Trichomonas vaginalis isolates by Hsp70 restriction fragment length polymorphism. Am J Trop Med Hyg. 2000;62:441–445. doi: 10.4269/ajtmh.2000.62.441. [DOI] [PubMed] [Google Scholar]
- 26.Simões-Barbosa A, Lobo TT, Xavier J, Carvalho SE, Leornadecz E. Trichomonas vaginalis: intrastrain polymorphisms within the ribosomal intergenic spacer do not correlate with clinical presentation. Exp Parasitol. 2005;110:108–113. doi: 10.1016/j.exppara.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, Sicheritz-Ponten T, Noel CJ, Dacks JB, Foster PG, Simillion C, Van de Peer Y, Miranda-Saavedra D, Barton GJ, Westrop GD, Muller S, Dessi D, Fiori PL, Ren Q, Paulsen I, Zhang H, Bastida-Corcuera FD, Simoes-Barbosa A, Brown MT, Hayes RD, Mukherjee M, Okumura CY, Schneider R, Smith AJ, Vanacova S, Villalvazo M, Haas BJ, Pertea M, Feldblyum TV, Utterback TR, Shu CL, Osoegawa K, de Jong PJ, Hrdy I, Horvathova L, Zubacova Z, Dolezal P, Malik SB, Logsdon JM, Jr, Henze K, Gupta A, Wang CC, Dunne RL, Upcroft JA, Upcroft P, White O, Salzberg SL, Tang P, Chiu CH, Lee YS, Embley TM, Coombs GH, Mottram JC, Tachezy J, Fraser-Liggett CM, Johnson PJ. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tibayrenc M, Kjellberg F, Ayala FJ. A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas, and Trypanosoma and their medical and taxonomical consequences. Proc Natl Acad Sci USA. 1990;87:2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tidwell BH, Lushbaugh WB, Laughlin MD, Cleary JD, Finley RW. A double blind placebo-controlled trial of single-dose intravaginal versus single dose oral metronidazole in the treatment of trichomonal vaginitis. J Infect Dis. 1994;170:242–246. doi: 10.1093/infdis/170.1.242. [DOI] [PubMed] [Google Scholar]
- 30.Davis-Hayman SR, Shah PH, Finley RW, Lushbaugh WB, Meade JC. Trichomonas vaginalis: analysis of a heat inducible member of the cytosolic HSP70 multigene family. Parasitol Res. 2000;86:608–612. doi: 10.1007/pl00008538. [DOI] [PubMed] [Google Scholar]
- 31.Hampl V, Pavlíček A, Flegr J. Construction and bootstrap analysis of DNA fingerprinting-based phylogenetic trees with the freeware program FreeTree: application to trichomonad parasites. Int J Syst Evol Microbiol. 2001;51:731–735. doi: 10.1099/00207713-51-3-731. [DOI] [PubMed] [Google Scholar]
- 32.Nei M, Li W-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitou N, Nei M. The neighbor-joining method: a new method for reconstruction of phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 34.Wickstead B, Ersfeld K, Gull K. Repetitive elements in genomes of parasitic protozoa. Microbiol Mol Biol Rev. 2003;67:360–375. doi: 10.1128/MMBR.67.3.360-375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao JC, Xie LF, Zhao L, Fang SL, Lun ZR. The presence of Mycoplasma hominis in isolates of Trichomonas vaginalis impacts significantly on DNA fingerprinting results. Parasitol Res. 2008;102:613–619. doi: 10.1007/s00436-007-0796-0. [DOI] [PubMed] [Google Scholar]
- 36.Bañuls AL, Hide M, Prugnolle F. Leishmania and the leishmaniases: a parasite genetic update and advances in taxonomy, epidemiology and pathogenicity in humans. Adv Parasitol. 2007;64:1–109. doi: 10.1016/S0065-308X(06)64001-3. [DOI] [PubMed] [Google Scholar]
- 37.Meyer CG, May J, Arez AP, Gil JP, do Rosario V. Genetic diversity of Plasmodium falciparum: asexual stages. Trop Med Int Health. 2002;7:395–408. doi: 10.1046/j.1365-3156.2002.00875.x. [DOI] [PubMed] [Google Scholar]
- 38.Buscaglia CA, Di Noia JM. Trypanosoma cruzi clonal diversity and the epidemiology of Chagas' disease. Microbes Infect. 2003;5:419–427. doi: 10.1016/s1286-4579(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 39.Ajzenberg D, Bañuls AL, Tibayrenc M, Dardé ML. Microsatellite analysis of Toxoplasma gondii shows considerable polymorphism structured into two main clonal groups. Int J Parasitol. 2002;32:27–38. doi: 10.1016/s0020-7519(01)00301-0. [DOI] [PubMed] [Google Scholar]


