Abstract
Time-of-flight (TOF) PET uses very fast detectors to improve localization of events along coincidence lines-of-response. This information is then utilized to improve the tomographic reconstruction. This work evaluates the effect of TOF upon an observer's performance for detecting and localizing focal warm lesions in noisy PET images.
Methods
An advanced anthropomorphic lesion-detection phantom was scanned 12 times over 3 days on a prototype TOF PET/CT scanner (Siemens Medical Solutions). The phantom was devised to mimic whole-body oncologic 18F-FDG PET imaging, and a number of spheric lesions (diameters 6–16 mm) were distributed throughout the phantom. The data were reconstructed with the baseline line-of-response ordered-subsets expectation-maximization algorithm, with the baseline algorithm plus point spread function model (PSF), baseline plus TOF, and with both PSF+TOF. The lesion-detection performance of each reconstruction was compared and ranked using localization receiver operating characteristics (LROC) analysis with both human and numeric observers. The phantom results were then subjectively compared to 2 illustrative patient scans reconstructed with PSF and with PSF+TOF.
Results
Inclusion of TOF information provides a significant improvement in the area under the LROC curve compared to the baseline algorithm without TOF data (P = 0.002), providing a degree of improvement similar to that obtained with the PSF model. Use of both PSF+TOF together provided a cumulative benefit in lesion-detection performance, significantly outperforming either PSF or TOF alone (P < 0.002). Example patient images reflected the same image characteristics that gave rise to improved performance in the phantom data.
Conclusion
Time-of-flight PET provides a significant improvement in observer performance for detecting focal warm lesions in a noisy background. These improvements in image quality can be expected to improve performance for the clinical tasks of detecting lesions and staging disease. Further study in a large clinical population is warranted to assess the benefit of TOF for various patient sizes and count levels, and to demonstrate effective performance in the clinical environment.
Keywords: positron emission tomography, time-of-flight PET, observer study, LROC
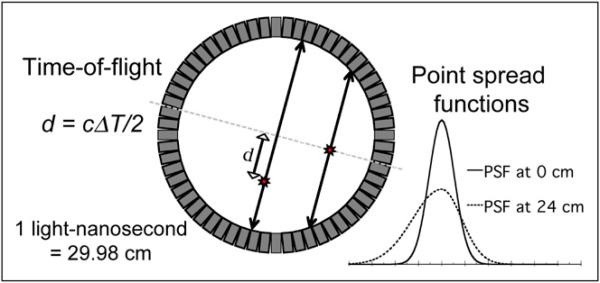
The PET components in the most recent generation of combined PET/CT scanners are equipped with time-of-flight (TOF) capability. The premise for TOF PET is illustrated in Figure 1, which also shows the point spread function (PSF) for 2 source positions. When a PET radioisotope decays, it emits a positron that annihilates with a nearby electron, giving rise to a pair of 511-keV photons emitted simultaneously in (nearly) opposite directions. If both these photons interact with and are detected by the PET tomograph, they give rise to a prompt coincidence event—providing the primary imaging signal measured by the scanner. When the annihilation event occurs at the midpoint of the line-of-response (LOR) between the detector elements, both photons reach the detector at the same instant in time. However, when the annihilation event occurs away from the midpoint of the LOR, one photon travels a shorter distance than the other and reaches the corresponding detector earlier.
FIGURE 1.
Recent advances in PET technology include tomographs with time-of-flight capability and point spread function modeling. TOF PET utilizes very fast detectors and electronics to measure the time difference between detection of each photon of annihilation pair, providing an estimate of depth d along the line-of-response where event originated. When events originate away from central axis of the scanner, the point response function becomes broad and asymmetric due to depth-of-interaction and other effects. Consideration of both of these effects during reconstruction can provide lower noise images with improved spatial resolution.
In TOF PET, the time difference ΔT between detection of each annihilation photon is measured using very fast detectors and electronics. This information is used to estimate the distance along the LOR where the annihilation occurred: d = cΔT/2, where distance d = 0 is the midpoint of the LOR and c is the speed of light (2.998 × 108 m/s). Though TOF PET was first explored in the early 1980s (1–3), the extremely fast timing resolution (sub-nanosecond) required to yield benefits in image quality precluded development of clinical TOF tomographs until many years later. Recent advancements in fast, high-sensitivity scintillators and detector electronics have made clinical TOF PET tomographs a reality (4–7).
At the ultimate limit, TOF PET could potentially localize annihilation events to within a single image voxel—effectively measuring the activity distribution directly and eliminating the need for tomographic reconstruction. However, this would require a timing-resolution of approximately 10 ps to isolate events to within a 3-mm voxel. Current TOF systems have timing resolutions closer to 600 ps, yielding TOF depth resolutions on the order of 9 cm. Time-of-flight PET with these timing resolutions does not directly lead to an improvement in the spatial resolution of the reconstructed image. It does, however, reduce noise propagation by localizing events along segments of each LOR rather than spreading statistical noise across the full length of each LOR. This reduction in noise can be interpreted as an effective gain in signal-to-noise that is dependent on the object size (8–10), and this improvement may support better spatial resolution in the final image.
Recent work with TOF PET has demonstrated improvement in image characteristics such as spatial resolution, contrast, and voxelwise noise (5–7). Such measures directly relate to quantitative imaging tasks such as computing standardized uptake values (SUVs), but they are less predictive of an observer's ability to detect lesions and distinguish them from a noisy background. Surti et al. assessed a detectability measure in simulations (6), and later in cylindric phantoms (11), demonstrating improvement due to TOF. This work assesses the impact of TOF upon observer performance for detecting focal lesions using localization receiver operating characteristics (LROC) analysis (12,13). An advanced whole-body phantom with distributed lesions was used to mimic whole-body oncologic PET with 18F-FDG, relating to both cancer detection and staging. This custom phantom was previously used to compare the lesion-detection performance of several PET tomographs (14) and reconstruction algorithms (15). The phantom was imaged repeatedly on a prototype TOF PET/CT (Siemens Medical Solutions) scanner with cerium-doped lutetium oxyorthosilicate (LSO) crystals, producing numerous lesion-present and lesion-absent images with known truth. Lesion-detection performance for iterative reconstructions with and without TOF was assessed using both human and model (numeric) observers. The degree of improvement with TOF was compared to that attained by modeling the spatially-variant point spread function (PSF) during reconstruction. The results are discussed, and conclusions drawn regarding the impact of TOF PET for clinical applications. Such applications are illustrated with 2 cancer patient studies acquired on a PET/CT scanner with TOF capability.
MATERIALS AND METHODS
Lesion-Detection Phantom
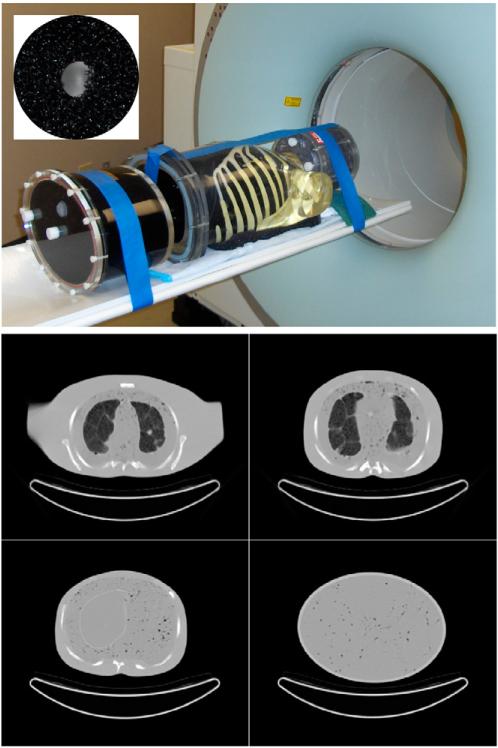
The whole-body phantom, shown in Figure 2, consisted of a 3-dimensional (3D) Hoffman brain phantom (Data Spectrum Corp.); anthropomorphic thorax with lungs, liver, and embedded rib cage and spine attenuating structures (Radiology Support Devices, Inc.); and elliptical cylindric pelvis with bladder (Data Spectrum). The phantom had a number of custom modifications designed to increase realism for modeling whole-body oncologic 18F-FDG PET, including removal of the central cardiac insert and mounting assembly, use of self-filling lungs with Styrofoam beads contained within nylon mesh bags, and use of a low water resistance open cell foam (5 ppi filter foam; New England Foam Products LLC) to introduce tissue heterogeneity. Specific details regarding these modifications can be found in (14,15).
FIGURE 2.
Whole-body phantom (top) consisted of anthropomorphic thorax phantom with lungs and liver, 3D brain phantom, and pelvis compartment with bladder. Inset (top) shows shell-less 68Ge lesion embedded in black open cell foam. Custom modifications to the soft-tissue and lung compartments with open-cell foam and nylon bead bags provided inhomogeneous density and structures as shown by CT (bottom). Such heterogeneities more accurately mimic patient structures, providing a more realistic and challenging lesion-detection task than encountered for phantoms with uniform background compartments.
Twenty-six “shell-less” 68Ge silicone gel lesions (16) were distributed throughout the phantom to model tumors with focal 18F-FDG uptake. These spheric lesions had an activity concentration of 12.8 kBq/cm3 with diameters: 6, 8, 10, 12, and 16 mm. Since the 69Ge radioactivity was infused directly into the body of each lesion, the need for plastic outer walls was eliminated. The lesions were positioned within slits cut into the black foam of the soft tissue compartments (mediastinum, abdomen, pelvis), by packing them into the Styrofoam beads of the lungs, and by mounting them on thin monofilament line in the liver.
The phantom compartments were filled with concentrations of 18F-FDG in water based on 12 oncologic 18F-FDG PET scans performed at our institution. The activity levels were based on 370-MBq administrations with 90-min uptake period, and adjusted downward for the small fillable phantom volume relative to typical patient size. The total activity present at the start of scanning was 180 MBq (Day 1), 222 MBq (Day 2), and 200 MBq (Day 3), with relative activity concentrations in each compartment of: soft tissue 1:1, liver 1.8:1, lungs (average) 0.37:1, brain (average) 6.0:1, and bladder 15:1. Note that 4 scans were acquired each day as the background 18F activity decayed, producing multiple count levels and varying lesion target:background ratios as described in the next section.
PET Scanning
Three days of experiment were performed using a prototype TOF PET/CT scanner. The geometry of this fully-3D scanner matched that of a TruePoint Biograph (Siemens Medical Solutions) with TrueV, with prototype TOF capability and timing resolution of 550 ps. Days 1–2 had 26 lesions distributed throughout the phantom, and Day 3 incorporated no lesions in order to obtain corresponding lesion-absent images. CT scans were performed to obtain attenuation maps for PET attenuation correction, and also to determine the true lesion positions in image space. Note that the CT images were not presented to the observers, and all results are purely from reading the PET images without CT information.
Four whole-body PET scans were acquired on each day, using 4 min per bed over 6 bed positions, at 55-min intervals. Since the 68Ge lesion activity (half-life [T1/2] = 270.8 d) remained essentially constant whereas the 18F background activity (T1/2 = 109.77 min) decayed by a factor of approximately 4 over the scans, the consecutive scans provided 4 different count levels with lesion target:background ratios in the various locations ranging from 1.6:1 up to 37:1. The data were acquired in listmode format (span 11, maximum ring difference 38) with TOF data embedded directly into the listmode records.
Image Reconstruction
The listmode files from the prototype TOF PET/CT scanner were binned into 2 sets of sinograms with 336 radial bins and 336 azimuthal bins: one set with 15 TOF bins (each 312 ps wide), and the other disregarding the TOF measurements to produce conventional non-TOF sinograms. The CT images were processed using the standard vendor algorithm for conversion to linear attenuation coefficient at 511 keV, from which the PET attenuation correction factors were reprojected. The PET data were then reconstructed by ordered-subsets expectation-maximization (OSEM) in 4 ways using vendor-supplied software: LOR-OSEM (17,18), the “baseline” Ordinary Poisson algorithm; baseline algorithm + PSF model (19), which models the spatially-variant resolution function of the scanner; baseline algorithm + TOF; and baseline algorithm + PSF + TOF (20). We will refer to these algorithms as LOR, PSF, TOF, and PSF+TOF, respectively. In each case, a 168 × 168 image matrix with 4.07 mm pixels and slice thickness of 2.03 mm was used. Each reconstruction used 7 subsets with 24 azimuthal angles per subset; 10 iterations were performed, and the output image from each iteration was stored. Three-dimensional gaussian filters were then applied, with filter kernel widths ranging from 0.0 (no filter) to 1.5 voxels SD in 0.1 voxel increments. The resultant images were stored for subsequent analysis by both model and human observers.
Model Observer Analysis
Objective comparison of performance for the 4 algorithms required that each algorithm be implemented in a near-optimal manner, ensuring that appropriate reconstruction parameters (number of iterations, filter strength) were used for each. In this work, we considered 10 iterations × 16 filters = 160 possible parameter configurations for each algorithm. For each configuration there were 52 lesion-present (26 lesions × 2 days) and 52 corresponding lesion-absent slices, each with 4 background activity levels. This provided a total of 266,240 two-dimensional (2D) test images for LROC analysis. It was not practical for human observers to review all of these images.
Model numeric observers, such as the channelized nonprewhitened (CNPW) observer (21), have been developed to mimic human observers for lesion-detection tasks. Such model observers have certain limitations, but have been found to correlate with human observers under carefully controlled conditions (21,22). We previously used the CNPW observer to guide selection of reconstruction parameters for PET lesion-detection studies (15), and continue that approach here. Our implementation draws heavily upon the work of Gifford et al. (21), and we refer the interested reader to that paper for additional details. Briefly, the CNPW observer computes a perception measurement, zn, at each image voxel n using:
| Eq. 1 |
where is the transpose of the CNPW template image at voxel n, f̂ is the test image, and b̄ is the mean lesion-absent (background) image. The CNPW template wn is the mean 2D lesion profile over a set of training images, mathematically projected onto a set of channel responses (10 difference-of-gaussian channels of (21)), and centered at voxel n. Note that the CNPW observer requires knowledge of the mean lesion profile and background, but it does not require knowledge of higher order statistics. As such, it is well-suited for use with experimental data such as acquired for this study.
Training of the CNPW observer consisted of computing the template images wn and mean lesion-absent images b̄ for each 2D test image. Three approaches to training the CNPW observer for this type of phantom data were previously tested (15) and found to produce identical lesion-detection results. The background images b̄ were computed using the Day 3 scans with no lesions present. Recall that the 4 scans acquired each day provide 4 count levels. Here, the mean background image for a given count level was estimated by averaging the other 3 count levels, then scaled to the target level. In this way, noise structures in the lesion-absent test images were independent from the corresponding mean background images (see (15) for additional details). The template images were computed by averaging the lesion profiles over all lesions (subtracting b̄ from each lesion-present test-case, centering the lesion, then averaging and applying the channels).
The CNPW observer was applied to each voxel of each test image to find the location with the maximum rating. The rating and location data were then analyzed to produce 2 figures-of-merit for lesion-detection performance: the probability of correction localization (PLOC), and the area under the LROC curve (ALROC). The former figure is simply the fraction of lesions found by the observer after applying a localizing-radius tolerance of 2.5 voxels—found to effectively localize lesions while minimizing accidental localization (Supplemental Fig. 1; supplemental materials are available online only at http://jnm.snmjournals.org). Wilcoxon integration of the empiric LROC curve (23) was employed to compute ALROC, using 10,000 bootstrap resampling trials to estimate the uncertainty for each case.
Human Observer Analysis
The CNPW observer results were used to select near-optimal reconstruction parameters (number of iterations, filter strength) for each algorithm, in preparation for the human observer study. The CNPW observer was also used to identify those test images that had lesion sizes, locations, and count levels that resulted in challenging detection tasks across the 4 reconstruction algorithms. Eighty-four of the 208 available lesion-present test cases were selected, along with the corresponding 84 lesion-absent images. These were divided into 2 groups, providing 64 training images and 104 test images for each algorithm. Five experienced Ph.D. medical physics imaging researchers read each set of images (interface shown in Supplemental Fig. 2). Note that these observers were not trained PET clinicians; however, in previous work the performance of such observers was found to correlate with that of trained PET clinicians for the phantom detection task studied herein (14). After initial training to familiarize the reader with the display interface and phantom structure, each observer read 64 training images followed immediately by 104 test images for each algorithm. The observers were unaware of which algorithm they were reading, and both the ordering of the algorithms and images presented for each algorithm were randomized.
The observers performed 2 tasks on each image. First, the location deemed most likely to contain a lesion was selected by mouse. Second, a confidence rating was selected from a 6-point scale ranging from 1 (high confidence lesion absent) to 6 (high confidence lesion present). The observers were informed that approximately half of the test images would contain exactly one lesion, and that approximately half would contain zero lesions. As such, the task amounted to identifying 1 or 0 lesions hidden among multiple noise blobs. For the training images, the observer was immediately provided with the truth regarding lesion presence and location, whereas no such feedback was provided for the test images. The reading sessions were performed in a darkened room with display grayscale calibrated to provide a log-linear relationship between voxel value and displayed luminosity (24), and the observers were permitted to modify the upper and lower grayscale limits on each image as desired. Short breaks were encouraged between reading each algorithm. The location and rating data for each observer were processed in a manner similar to that for the model observer, but used the non-parametric approach of Popescu (25) with Epanechnikov kernel in order to compute the LROC curves.
Clinical Studies
While these phantom studies quantify the impact of TOF PET on lesion detection, the ultimate goal is to assess improvement for clinical tasks. Such an assessment in over 100 patients has been completed and will be the subject of a further publication. For this study, clinical impact is illustrated with 2 typical cancer patient studies comparing PSF reconstructions to PSF+TOF reconstructions for each patient. These clinical examples were acquired on a beta-test clinical TOF PET/CT scanner installed at the University of Tennessee with similar, but not identical, TOF capabilities as the prototype scanner used for the phantom studies.
RESULTS
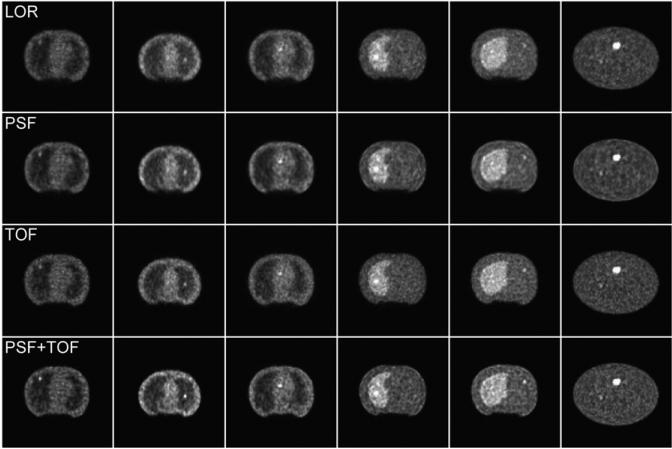
Example images for each of the 4 reconstruction algorithms are shown in Figure 3. Differences in contrast and noise structure can be observed, reflecting the different image characteristics of each algorithm. Previous work by other groups has characterized these differences in terms of spatial resolution, contrast, noise, and signal-to-noise measures (5,6,8–10). The following sections evaluate how these differences affect observer performance for detecting and localizing focal lesions.
FIGURE 3.
Example reconstructed images for (top-to-bottom) LOR, PSF, TOF, and PSF+TOF. Each image slice shown contains exactly 1 lesion, where relatively obvious cases are shown for purposes of this example. Number of iterations and post-reconstruction filter strength used for each algorithm are given in Table 1. Note that while images show broad similarities, differences in lesion depiction and noise texture exist between algorithms. The effect of these differences upon the task of identifying and localizing focal lesions has been evaluated in this study.
CNPW Observer Results
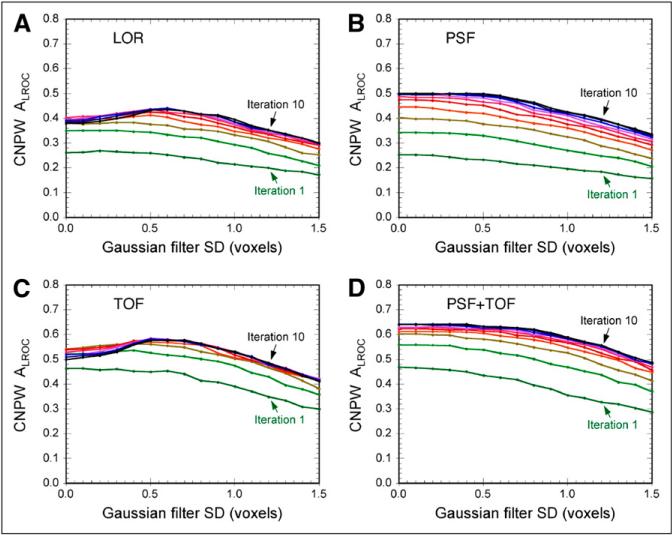
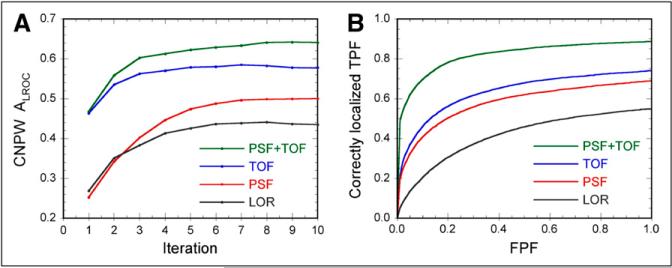
Figure 4 shows the CNPW observer results, plotting ALROC vs. filter strength for each iteration of each algorithm. Similar trends were observed for the PLOC figure-of-merit as well (not shown). The CNPW observer results for the 4 algorithms are directly compared in Figure 5A, which also more clearly shows the effect of increasing the number of iterations for each algorithm.
FIGURE 4.
Results of CNPW model observer analysis, plotting area under the LROC curve versus filter strength for each algorithm. Separate lines are shown on each plot for iterations 1–10. These data were used to select optimal reconstruction parameters for images read by human observers. The algorithms including PSF model (B, D) required little or no filter for maximal performance, whereas other algorithms (A, C) required moderate filtering for peak performance.
FIGURE 5.
Final results of model observer study (A) and human observer study (B), providing comparison and ranking of 4 algorithms studied. Model observer results plot area under LROC curve versus number of iterations, where optimal filter for each iteration and algorithm was applied (as determined from results shown in Fig. 4). Note that reconstructions with TOF provided markedly higher performance at earliest iterations, showing that the most important image features for lesion detection are recovered more quickly when using TOF. Human observer LROC curves demonstrate distinctions in performance for 4 algorithms, and confirm model observer results. Quantitative results for these curves are provided in Table 2.
Several interesting trends can be observed in these results. The best lesion-detection performance for the LOR and TOF algorithms was obtained when applying a post-reconstruction gaussian filter with an SD of 0.6 voxels, whereas maximal performance for the 2 algorithms with PSF model was achieved with little or no filtering. It is well known that modeling the PSF during iterative reconstruction introduces correlations between neighboring voxels in a manner similar to smoothing filters, and our result suggests that little or no additional smoothing was required to maximize lesion-detection performance when modeling the PSF. Also note that performance increased out to 10 iterations (and potentially beyond) for the 2 algorithms with PSF model, whereas performance for the other algorithms peaked by approximately 6 iterations. This is suggestive of two phenomena: the rate of iterative convergence is slowed when using the broader reconstruction kernel that arises through PSF modeling; and the use of resolution modeling permits additional information about the object to be recovered, which requires additional iterations. The result that higher performance was obtained with PSF model than without further supports the latter observation.
The inclusion of TOF produced higher lesion-detection performance than the reconstructions without TOF measurements, suggesting that the improved signal-to-noise ratio (SNR) provided by TOF PET translates into improved CNPW observer performance. Also note from Figure 5A that the TOF reconstructions provided markedly higher performance at early iterations, suggesting that TOF also benefits the rate of iterative recovery of important image features. This may be a significant consideration in the clinic, where extended fully-3D iterative reconstruction times may be of concern.
These CNPW observer results provide our first-pass evaluation of the impact of TOF upon lesion-detection performance, and they will be validated against human observer results from reading a subset of the images. Table 1 shows the reconstruction parameters for each algorithm that nearly-maximized performance for the CNPW observer without using excessive iterations that could result in unacceptably long reconstruction times. These parameters were used for the images read by the human observers. The model observer results were also used to select 84 test cases (lesion size, location, and count levels) that were near the verge of detectability to be read by the human observers, thereby maximizing statistical power in the human observer study for ranking the 4 algorithms.
TABLE 1.
CNPW Observer Results*
| Optimal parameters |
CNPW observer results |
|||
|---|---|---|---|---|
| Algorithm | No. of iterations† | Filter SD (voxels) | PLOC ± SD | ALROC ± SD |
| LOR | 6 | 0.5 | 0.588 ± 0.068 | 0.418 ± 0.051 |
| PSF | 8 | 0.1 | 0.706 ± 0.063 | 0.516 ± 0.052 |
| TOF | 6 | 0.5 | 0.804 ± 0.056 | 0.673 ± 0.054 |
| PSF+TOF | 8 | 0.1 | 0.882 ± 0.045 | 0.813 ± 0.046 |
Results are for subset of images read by human observers.
Ordinary Poisson LOR-OSEM with 7 subsets.
Human Observer Results
Each of the 5 human observers successfully completed their reading sessions within 2–4 h. Table 2 presents the LROC results for the human observers, with corresponding LROC curves shown in Figure 5B. A 2-way ANOVA with Tukey all-pairs comparison was performed to test for statistically-significant differences between observers and between algorithms. While differences in the magnitudes of PLOC and ALROC were identified between observers, each observer obtained identical rankings among algorithms. This reflects differences in inter-observer performance, but confirms consistent results as to the relative performance of each algorithm.
TABLE 2.
Human Observer Results
| Human observer results |
Tukey all-pairs comparison (P)* |
|||||
|---|---|---|---|---|---|---|
| Algorithm | PLOC ± SD | ALROC ± SD | LOR | PSF | TOF | PSF+TOF |
| LOR | 0.549 ± 0.039 | 0.486 ± 0.045 | N/A | 0.002 | 0.001 | <0.001 |
| PSF | 0.690 ± 0.064 | 0.662 ± 0.087 | 0.002 | N/A | 0.882 | 0.001 |
| TOF | 0.741 ± 0.043 | 0.691 ± 0.046 | 0.001 | 0.882 | N/A | 0.002 |
| PSF+TOF | 0.886 ± 0.056 | 0.873 ± 0.062 | <0.001 | 0.001 | 0.002 | N/A |
Tukey all-pairs comparison performed on ALROC figure-of-merit.
N/A = not applicable.
The fully-3D LOR-OSEM reconstructions without- and with-PSF model provide a context for assessing the degree of improvement obtained through inclusion of TOF information. Note that LOR-OSEM without PSF model produced the lowest lesion-detection performance of the 4 algorithms studied, and that a statistically significant improvement was obtained when modeling the spatially-variant PSF (P = 0.002). The inclusion of TOF data improved lesion-detection performance somewhat more than that obtained through modeling the PSF (ALROC = 0.512 and 0.673 for PSF and TOF, respectively). In both cases the addition of the PSF model or TOF information alone provided a statistically-significant improvement in performance (P = 0.002 and 0.001, respectively), but the difference between PSF and TOF was not statistically significant (P = 0.882). The use of both the PSF model and TOF together provided markedly improved performance (P < 0.001), outperforming either algorithmic advance alone. This suggests that the fundamental processes providing improved lesion-detection performance (e.g., improved imaging signal recovery, or reduced noise propagation) are different for PSF modeling and TOF; and the use of both approaches together provides the greatest advantage for the lesion-detection task.
It is important to note that these results are for a single phantom size, and that this study has not investigated the effect of changing patient sizes or count levels. Time-offlight PET provides greater benefit for large patients (8,9), where the improved depth-localization of coincident events provides more gain than for smaller patients. As such, TOF may prove more valuable than PSF modeling for larger patients. On the other hand, PSF modeling is capable of improving spatial resolution—provided that sufficient imaging statistics are present to support such resolution. Thus, the relative benefit of TOF and PSF modeling is dependent on both the noise level and the patient size; however, since each approach exploits different tomographic processes, the combined improvement upon image quality is cumulative.
Clinical Studies
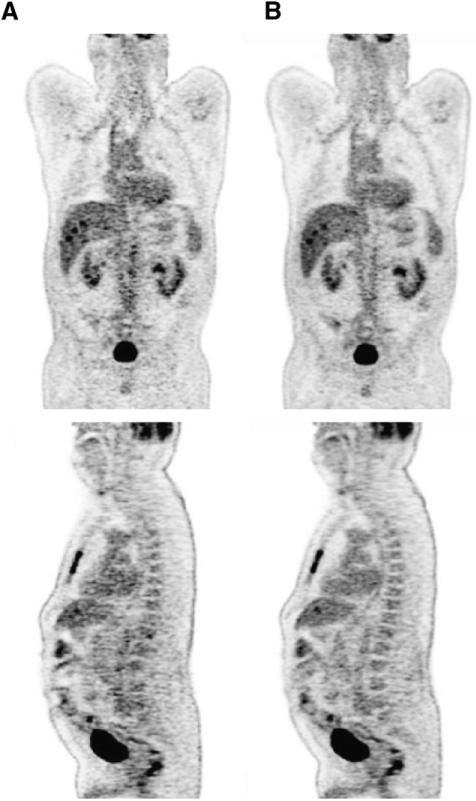
While it is beyond the scope of this article to include a full assessment of the impact of TOF on patient studies, it is instructive to include 2 illustrative examples. Figure 6 shows coronal and sagittal sections through a patient with a history of colon cancer. The 71-y-old male patient had a body mass index (BMI) of 33.5 and was injected with 396 MBq of 18F-FDG. After a 90-min uptake period the patient underwent a CT scan (130 kV; 180 mAs) with both intravenous and oral contrast, followed by a PET scan acquired at 6 bed positions for 3 min per bed. The scan was acquired on a prototype Biograph 6 with TOF capability (Siemens Molecular Imaging). The data were reconstructed onto a 168 × 168 matrix using 3D LOR-OSEM with 14 subsets and incorporating PSF (Fig. 6A) and PSF+TOF (Fig. 6B). The PSF image was reconstructed with 4 iterations, whereas the PSF+TOF image used 2 iterations based on the noted faster iterative convergence when TOF is incorporated. Neither image was smoothed after reconstruction. Comparing Figures 6A and 6B, it is evident that the TOF reconstruction demonstrates visually lower noise and improved SNR; the uptake in the mediastinum and liver is more uniform with TOF, and the spinal column is more clearly depicted. The scan highlights lesions in the sternum, liver, and pelvic region metastatic to the patient's primary colon cancer. The liver lesions in particular are more clearly evident in the PSF+TOF images (Fig. 6B).
FIGURE 6.
Coronal (top) and sagittal (bottom) sections through 18F-FDG PET/CT scan of a patient with history of colon cancer. The 71-y-old male patient had BMI of 33.5 and was injected with 396 MBq (10.7 mCi) of 18F-FDG. After 90-min uptake period, patient underwent CT scan (130 kV; 180 mAs) with both intravenous and oral contrast, followed by PET scan acquired at 6 bed positions for 3 min per bed. PET data were reconstructed into 168 × 168 matrix with (A) baseline algorithm and PSF (4 iterations, 14 subsets, no smoothing), and (B) baseline algorithm with PSF and TOF (2 iterations, 14 subsets, no smoothing) and time resolution kernel of 590 ps.
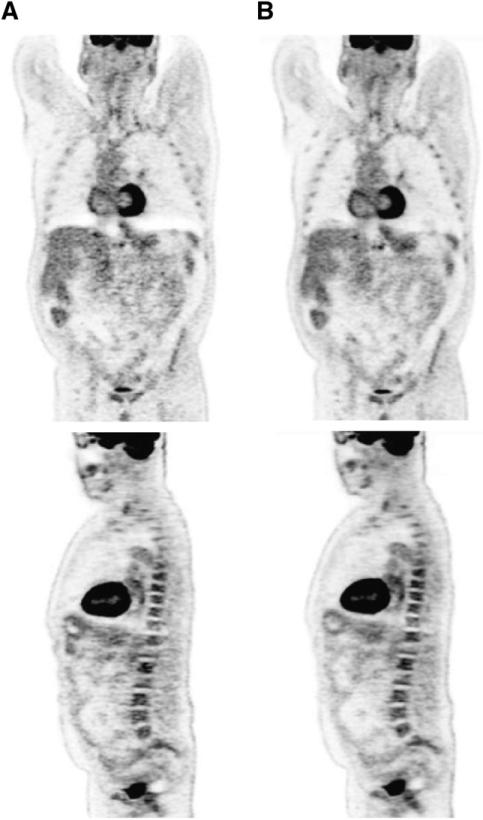
A second example is illustrated in Figure 7, where coronal and sagittal sections through a patient with esophageal cancer are presented for PSF (Fig. 7A) and for PSF+TOF (Fig. 7B) reconstructions. The 61-y-old patient with a BMI of 27 was injected with 366 MBq of 18F-FDG. After a 90-min uptake period, the patient underwent CT, followed by PET for = bed positions at 3 min per bed. The same reconstruction approach was followed as for Figure 6. Again, the PSF+TOF images show improved uniformity in the liver and mediastinal region and better delineation of the bone marrow in the ribs and spine as compared to PSF alone. An additional benefit of TOF appears to be a reduction in the photopenic lesions above the liver and spleen (Fig. 7A) caused by a spatial mismatch between the CT and PET images due to respiratory motion. As a result of TOF information, noise propagation was limited to a restricted region, thereby reducing the effect of the incorrect attenuation correction factors originating from this mismatch.
FIGURE 7.
Coronal (top) and sagittal (bottom) sections through 18F-FDG PET/CT scan of a patient with esophageal cancer. The 61-y-old male patient had a BMI of 27 and was injected with 366.3 MBq (9.9 mCi) of 18F-FDG. After 90-min uptake period, patient underwent CT scan (130 kV; 180 mAs), followed by PET scan that was acquired at = bed positions for 3 min per bed. PET data were reconstructed into 168 × 168 matrix by (A) baseline algorithm with PSF (4 iterations, 14 subsets, no smoothing), and (B) baseline algorithm with PSF and TOF (2 iterations, 14 subsets, no smoothing) and time resolution kernel of 590 ps.
These 2 patient studies, even though the assessment is subjective, provide further evidence for improvement in image clarity and reduction in noise offered by TOF and validated quantitatively by the phantom studies. As noted, these improvements are more evident in larger patients with BMI greater than 30.
DISCUSSION
One of the most significant challenges in performing lesion-detection studies is the need to obtain sufficient statistical power to differentiate between the algorithms with good confidence. The phantom experiments presented here were specifically designed to provide a number of images with lesion contrast and count levels near the verge of detectability, thereby maximizing statistical power for differentiating the algorithms. The results are thus valuable for ranking the relative performance of the test algorithms. Note, however, that one consequence of this approach is that the lesion sizes/contrasts were not representative of the full range and distribution of lesions encountered in typical clinical practice. As such, the quantitative results of this study should not be interpreted as quantifying the sensitivity and specificity that would be encountered in clinical practice. However, the improvements observed in this study will translate to improvements in observer performance for reading images with challenging lesion presentations, and thus it is reasonable to conclude that improved overall clinical performance would also result.
Another limitation is that the task of identifying and localizing a single lesion in a single 2D slice differs from the broader free-response clinical task for 3D images. Moreover, the clinical task involves significant effort in differentiating focal 18F-FDG uptake in tumors from the normal (complex and variable) 18F-FDG distribution, as well as distinguishing tumors from inflammatory processes. Performance for these tasks is largely dependent on the training and experience of the reading physician, and was not studied in this work. However, the differences in image characteristics that provided improvements in lesion-detection performance in this study can also be expected to improve visualization of other normal and inflammatory structures. In addition, both the mathematical and human observers provided identical rankings among algorithms as to detection performance, although each observer type gave different statistical-power. This study, coupled with previous work showing improved resolution, contrast, and noise measures, together provide strong evidence that the reconstruction advances of PSF modeling and TOF PET provide significant improvements in clinical performance. These results are consistent with and build upon those obtained by Surti et al. in simulations (6) and simpler phantoms (11g).
CONCLUSION
Time-of-flight PET provides improved identification of focal lesions resulting from reduced background noise, and also affects the rate of recovery of image features allowing for a reduction in the number of iterations required for iterative reconstruction algorithms. Within the limitations of this study, inclusion of TOF information resulted in improved lesion-detection performance with high statistical power for both human and model observers. Cumulative improvement was observed when including TOF in conjunction with PSF modeling, suggesting that combined use of these newest techniques may provide the best image quality. The same image characteristics that led to improved observer performance for the phantom data are also observed in actual clinical images of patients, as illustrated here in Figures 6 and 7. Further study is needed to conclusively demonstrate that these improved images translate into better diagnosis and staging for the general clinical population.
Supplementary Material
SUPPLEMENTAL FIGURE 1
Relationship between the probability of correct localization and radius threshold used to determine a “hit”. A non-zero radius is required because the lesions have finite size and are subject to the spatial resolution limitations of the image. A radius threshold of 2.5 voxels was found to fall on the plateau between correct localizations and chance hits, and was used for this study.
SUPPLEMENTAL FIGURE 2
The display interface used for the human observer study included a single slice display with user-adjustable grayscale limits. The reader was asked to identify the location deemed most likely to contain a lesion with a mouse click (red crosshairs), and then asked to make a confidence determination based on a six-point rating scale. For training images, feedback as to the truth regarding lesion presence and location was provided immediately after clicking “next” to accept the reader's input.
ACKNOWLEDGMENTS
We thank Noel Black, Frederic Noo, John Roberts, and Craig Goodrich for their diligent efforts as observers, and James Hamill for supplying the shell-less 68Ge-spheres. We also thank Misty Long for acquisition of the clinical images and Karl Hubner for his interpretation and helpful discussions. This project was supported by award R01 CA107353 from the National Cancer Institute (NCI), and the clinical studies by NCI grant R33 CA94317. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCI or NIH.
REFERENCES
- 1.Allemand R, Gresset C, Vacher J. Potential advantages of a cesium fluoride scintillator for a time-of-flight positron camera. J Nucl Med. 1980;21:153–155. [PubMed] [Google Scholar]
- 2.Mullani NA, Markham J, Ter-Pogossian MM. Feasibility of time-of-flight reconstruction in positron emission tomography. J Nucl Med. 1980;21:1095–1097. [PubMed] [Google Scholar]
- 3.Ter-Pogossian MM, Mullani NA, Ficke DC, Markham J, Snyder DL. Photon time-of-flight-assisted positron emission tomography. J Comput Assist Tomogr. 1981;5:227–239. doi: 10.1097/00004728-198104000-00014. [DOI] [PubMed] [Google Scholar]
- 4.Conti M, Bendriem B, Casey M, et al. First experimental results of time-of-flight reconstruction on an LSO PET scanner. Phys Med Biol. 2005;50:4507–4526. doi: 10.1088/0031-9155/50/19/006. [DOI] [PubMed] [Google Scholar]
- 5.Karp JS, Surti S, Daube-Witherspoon ME, Muehllehner G. Benefit of time-of-flight in PET: experimental and clinical results. J Nucl Med. 2008;49:462–470. doi: 10.2967/jnumed.107.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Surti S, Karp JS, Popescu LM, Daube-Witherspoon ME, Werner M. Investigation of time-of-flight benefit for fully 3-D PET. IEEE Trans Med Imaging. 2006;25:529–538. doi: 10.1109/TMI.2006.871419. [DOI] [PubMed] [Google Scholar]
- 7.Surti S, Kuhn A, Werner ME, Perkins AE, Kolthammer J, Karp JS. Performance of Philips Gemini TF PET/CT scanner with special consideration for its time-of-flight imaging capabilities. J Nucl Med. 2007;48:471–480. [PubMed] [Google Scholar]
- 8.Budinger TF. Time-of-flight positron emission tomography: status relative to conventional PET. J Nucl Med. 1983;24:73–78. [PubMed] [Google Scholar]
- 9.Tomitani T. Image-reconstruction and noise evaluation in photon time-of-flight assisted positron emission tomography. IEEE Trans Nucl Sci. 1981;28:4581–4589. [Google Scholar]
- 10.Watson CC. An evaluation of image noise variance for time-of-flight PET. IEEE Trans Nucl Sci. 2007;54:1639–1647. [Google Scholar]
- 11.Surti S, Karp JS. Experimental evaluation of a simple lesion detection task with time-of-flight PET. Phys Med Biol. 2009;54:373–384. doi: 10.1088/0031-9155/54/2/013. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farquhar TH, Llacer J, Sayre J, Tai YC, Hoffman EJ. ROC and LROC analyses of the effects of lesion contrast, size, and signal-to-noise ratio on detectability in PET images. J Nucl Med. 2000;41:745–754. [PubMed] [Google Scholar]
- 13.Swensson RG. Unified measurement of observer performance in detecting and localizing target objects on images. Med Phys. 1996;23:1709–1725. doi: 10.1118/1.597758. [DOI] [PubMed] [Google Scholar]
- 14.Kadrmas DJ, Christian PE. Comparative evaluation of lesion detectability for 6 PET imaging platforms using a highly reproducible whole-body phantom with 22Na lesions and localization ROC analysis. J Nucl Med. 2002;43:1545–1554. [PubMed] [Google Scholar]
- 15.Kadrmas DJ, Casey ME, Black NF, Hamill JJ, Panin VY, Conti M. Experimental comparison of lesion detectability for four fully-3D PET reconstruction schemes. IEEE Trans Med Imaging. 2009;28:523–534. doi: 10.1109/TMI.2008.2006520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamill JJ, Arnsdorff E, Casey ME, Liu X, Raulstron WJA. Ge-68 PET hot sphere phantom with no cold shells. Nucl Sci Symp Conf Rec. 2005;3:1609–1613. [Google Scholar]
- 17.Shepp LA, Vardi Y. Maximum likelihood estimation for emission tomography. IEEE Trans Med Imaging. 1982;1:113–121. doi: 10.1109/TMI.1982.4307558. [DOI] [PubMed] [Google Scholar]
- 18.Kadrmas DJ. LOR-OSEM: statistical PET reconstruction from raw line-of-response histograms. Phys Med Biol. 2004;49:4731–4744. doi: 10.1088/0031-9155/49/20/005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panin VY, Kehren F, Michel C, Casey M. Fully 3-D PET reconstruction with system matrix derived from point source measurements. IEEE Trans Med Imaging. 2006;25:907–921. doi: 10.1109/tmi.2006.876171. [DOI] [PubMed] [Google Scholar]
- 20.Casey ME. Improving PET with HD-PET 1 Time of Flight. Siemens Molecular Imaging; Knoxville, TN: 2008. [Google Scholar]
- 21.Gifford HC, Kinahan PE, Lartizien C, King MA. Evaluation of multiclass model observers in PET LROC studies. IEEE Trans Nucl Sci. 2007;54:116–123. doi: 10.1109/TNS.2006.889163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gifford HC, King MA, Pretorius PH, Wells RG. A comparison of human and model observers in multislice LROC studies. IEEE Trans Med Imaging. 2005;24:160–169. doi: 10.1109/tmi.2004.839362. [DOI] [PubMed] [Google Scholar]
- 23.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 24.Nawfel RD, Chan KH, Wagenaar DJ, Judy PF. Evaluation of video gray-scale display. Med Phys. 1992;19:561–567. doi: 10.1118/1.596846. [DOI] [PubMed] [Google Scholar]
- 25.Popescu LM. Nonparametric ROC and LROC analysis. Med Phys. 2007;34:1556–1564. doi: 10.1118/1.2717407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTAL FIGURE 1
Relationship between the probability of correct localization and radius threshold used to determine a “hit”. A non-zero radius is required because the lesions have finite size and are subject to the spatial resolution limitations of the image. A radius threshold of 2.5 voxels was found to fall on the plateau between correct localizations and chance hits, and was used for this study.
SUPPLEMENTAL FIGURE 2
The display interface used for the human observer study included a single slice display with user-adjustable grayscale limits. The reader was asked to identify the location deemed most likely to contain a lesion with a mouse click (red crosshairs), and then asked to make a confidence determination based on a six-point rating scale. For training images, feedback as to the truth regarding lesion presence and location was provided immediately after clicking “next” to accept the reader's input.