Abstract
Dysfunction in enzymes involved in one carbon (1-C) metabolism can lead to increased chromosomal strand breaking and abnormal methylation patterns; both are associated with cancer risk. Availability of 1-C units may modify risk. We investigated the association of single nucleotide polymorphisms (SNPs) in 21 genes in the 1-C metabolism pathway among 829 Caucasian cases with primary epithelial ovarian cancer (EOC) and 941 frequency-matched unaffected controls enrolled at Mayo Clinic, Rochester, MN and Duke University, Durham, NC, and examined risk modification by multivitamin supplement use. Multivariable-adjusted SNP-specific logistic regression and haplotype analyses were performed for 180 SNPs and false positive report probabilities (FPRP) were calculated. Each additional copy of the minor allele in the intronic SNP SHMT1 rs9909104 was associated with EOC [odds ratio (OR), 1.2; confidence interval (95% CI), 1.0–1.4; P trend = 0.02; FPRP 0.16] and a 5-SNP SHMT1 haplotype was associated with decreased risk [P = 0.01; FPRP 0.09]. Three SNPs in DNMT3A were associated with risk among multivitamin supplement users: rs13420827 [OR, 0.8; 95% CI, 0.6–1.0; P interaction = 0.006; FPRP 0.54], rs11887120 [OR, 0.8; 95% CI, 0.7–1.0; P interaction = 0.007; FPRP 0.57] and rs11695471 [OR, 1.2; 95% CI, 1.0–1.5; P interaction = 0.01; FPRP 0.66]. These data extend previous findings from other cancers of a role for SHMT1 in ovarian cancer, and provide evidence that SNPs in methylation and DNA synthesis reactions are associated with risk of ovarian cancer. Interventions with modifiable factors such as multivitamin intake may reduce risk.
Keywords: Case-control studies, DNMT3A, MTR, Multivitamins, Ovarian neoplasms, SHMT1
INTRODUCTION
Ovarian cancer is the eighth most common cancer among U.S. women with 22,430 newly diagnosed cases and 15,280 deaths estimated in 2007 (1). The few known risk factors are either reproductive-related (decreased risk from oral contraceptive use (2), parity (3) and long-term breastfeeding (3) or represent inherited mutations in a few high-risk, high-penetrance genes (e.g., BRCA1) (4). It has recently been shown (5) that when predisposing genetic variants are very common in the population (each with prevalence ≥ 25%), a modest number (≤ 20) could explain 50% of the burden of a disease in the population, even if the individual genotype associations are relatively small (e.g., relative risk, 1.2–1.5). These common variants could plausibly interact with common environmental exposures to lower risk among a substantial proportion of individuals.
Perturbation in one-carbon (1-C) metabolism can have pleiotropic consequences that may lead to tumor initiation and progression. One-carbon transfer reactions are important for DNA synthesis and replication, cell division and growth and survival, particularly for rapidly dividing cells (6). One-carbon transfer reactions are also required for the remethylation of homocysteine to methionine, which is important for the biosynthesis of S-adenosyl methionine, an essential supplier of methyl groups for the methylation of many compounds including DNA, RNA, proteins and phospholipids (6). Since folate is a basic component of cell metabolism and is integral to the 1-C transfer pathway, it is not surprising that folate or methyl-donor nutrient deficiency can lead to gene-specific (7) or global DNA (8) hypomethylation, misincorporation of uracil instead of thymine into DNA that predisposes to increased chromosomal strand breaking (9) and alone can act as complete carcinogens or as effective tumor promoters after chemical initiation (10, 11).
Common single nucleotide polymorphisms (SNPs) in genes encoding enzymes that rely on folate or methyl-donor nutrients in 1-C transfer reactions may imitate the outcome of vitamin deficiency by providing insufficient 1-C moieties for methylation or DNA synthesis. SNPs in genes encoding 1-C transfer-associated enzymes have been examined for risk with various tumor types (12–15). Perhaps the most studied is MTHFR: two copies of the rare allele are associated with modest decreased risks of colon cancer, which is most evident among those with higher folate intake (16). Polymorphisms in genes in 1-C metabolism have not been examined extensively with ovarian cancer. Investigation of their association with ovarian cancer can complement and strengthen findings from the dietary-only association studies, which are inconsistent (17, 18), identify novel variants worthy of additional interrogation, locate associated region(s) for future fine-mapping, and lead to functional and interventional studies that examine risk modification within the context of exposure to high or low intakes of folate or methyl-donor nutrients.
Here, we report findings from the association of 180 tag- and putative-functional common SNPs in genes in the 1-C transfer pathway with risk of ovarian cancer using data from two ongoing case-control studies. We also examined effect modification by multivitamin supplement use as an estimate of B-vitamin intake.
MATERIALS AND METHODS
Study Design and Population
Subjects participated in two ongoing case-control studies of epithelial ovarian cancer initiated in January 2000 at Mayo Clinic, Rochester, MN and in May 1999 at Duke University, Durham, NC. Written informed consent was obtained from all participants. For the current analyses, we included participants enrolled during the period June 1999 to March 2006. The Institutional Review Board at both sites approved the study protocols. Details of the study design are described elsewhere (19) and briefly outlined below.
Mayo Clinic Sample
Clinic attendance formed the sampling frame for Mayo cases and controls. Mayo cases were women over age 20 years with histologically-confirmed incident epithelial ovarian cancer (borderline or invasive) and enrolled in the study within one year of date of diagnosis. Cases were identified from the six-state region that defines Mayo Clinic’s primary service population (Minnesota, Iowa, Wisconsin, Illinois, North Dakota, and South Dakota) and comprises approximately 85% of all ovarian cancer cases seen at Mayo Clinic. Controls without ovarian cancer and who had at least one ovary intact were frequency matched on race, age (5-year age categories) and region of residence to cases. Controls were a convenience sample of patients recruited from the outpatient practice of the Division of General Internal Medicine at Mayo Clinic. Women were seen for general medical examinations including common conditions typical of older Americans such as hypertension, diabetes, hyperlipidemia, and coronary artery disease. The response was 83% among cases and 74% among controls.
Duke University Sample
A 48-county area of North Carolina formed the sampling frame for Duke cases and controls. Duke cases were women between the ages 20 and 74 years with histologically-confirmed primary epithelial ovarian cancer (borderline or invasive). Cases were identified using the North Carolina Central Cancer Registry’s rapid case ascertainment system. Controls without ovarian cancer and who had at least one ovary intact were identified from the same 48-county region as the cases using list-assisted random digit dialing. Controls were frequency matched to cases on race and age (5-year age categories). The response was 75% among eligible cases and 64% among the controls.
Risk Factor Questionnaire
Information on demographic data and known and suspected ovarian cancer risk factors were collected through in-person interviews at both sites using similar questionnaires. In January 2003, the Mayo questionnaire was expanded with questions about ‘regular multivitamin’ intake defined as ≥ 4 pills per week during the previous year for controls and one year prior to cancer diagnosis for the cases. The Duke questionnaire elicited this information from study start with three possible responses to any multivitamin use (‘yes, regularly’, ‘yes irregularly’ or ‘no’). We defined users as those who responded ‘yes regularly’ during the past five years for controls and in the five years prior to diagnosis for cases. A common data dictionary was developed for covariates to allow combined analysis of data from both sites.
Biospecimen Collection and Processing
DNA was extracted from blood using the Gentra AutoPure LS Purgene salting out methodology (Gentra, Minneapolis, MN). Due to limited quantity of available DNA from Duke subjects, we performed whole genome amplification (WGA) on all Duke samples (n = 1,282) as a means to enrich DNA quantities. WGA DNA was prepared from 200 ηg genomic DNA using the REPLI-G WGA protocol (Qiagen Inc, Valencia CA). Quantities of 250 ηg genomic and WGA DNA were adjusted to 50 ηg/μl before genotyping and verified using PicoGreen dsDNA quantitation kit (Molecular Probes, Inc., Eugene OR). The samples were bar-coded to ensure accurate and reliable sample processing and storage.
Gene, SNP and tag SNP selection
Genes encoding proteins in the 1-C transfer and metabolism pathway were identified from literature searches and public databases (e.g., Kegg, BioCarta). We focused on genes where there were known or suggestive data of associations with other diseases including cancer (12, 13, 20–23), deficiency syndromes (24, 25), embryonic development (26), neural tube defects (27), cardiovascular disease (28) or functional studies (29), or where the gene-product participated in a rate-limiting step, irreversible direction, affected ligand binding or generated important intermediate substrates to that pathway, with the expectation that polymorphisms in these genes would have the potential to impart the greatest functional impact on outcome according to current knowledge. Twenty-one genes (AHCYL1, ALDH1L1, DHFR, DNMT1, DNMT3A, DNMT3B, DPYD, FOLR1, MAT2B, MBD4, MGMT, MTHFD1, MTHFD1L, MTHFD2, MTHFR, MTHFS, MTR, MTRR, SHMT1, SLC19A1 and TYMS) were selected for their role in 1-C transfer and metabolism, including participation in the folate and methionine cycles, methylation, purine and pyrimidine synthesis and folate transport.
All SNPs within the 21 candidate genes 5 kb of the largest cDNA isoform (genome build 35) were selected from unrelated Caucasian samples within the HapMap Consortium’s release 21 (http://www.hapmap.org) (30), Perlegen Sciences (http://genome.perlegen.com) (31), Seattle SNPs (http://pga.mbt.washington.edu) and Panel 2 of the National Institute for Environmental Health Science SNPs (www.egp.gs.washington.edu). We applied the ld Select program(32) to bin SNPs with minor allele frequency (MAF) ≥ .05 and pair-wise LD (linkage disequilibrium) threshold of r2 ≥ .80. Following binning, we selected tag SNPs for analysis from the source with the greatest number of SNPs with MAF ≥ 0.05 and the greatest number of LD bins that also met criteria for predicted likelihood of successful genotyping using the Illumina Golden Gate Assay™ quality score metrics. We also included all putative functional SNPs (within 1 kb upstream, 5′ UTR, 3′ UTR or non-synonymous) with MAF ≥ .05 identified in Ensembl release 34. Nucleotide positions for SNPs were calculated as the difference between the gene start coordinate and SNP coordinate using Ensembl release 47. The 21 1-C genes contributed 153 tagSNPs (98% from HapMap) representing 2,710 individual SNPs and 35 additional putative functional SNPs for a total of 188 SNPs in the 1-C metabolism pathway. Henceforth, we collectively refer to both tag- and functional SNPs as ‘SNPs’ unless otherwise clarified.
Genotyping
Mayo and Duke samples were plated separately in the Mayo Clinic Cancer Center’s Genotyping Shared Resource with cases and controls randomly mixed within each plate. For the Mayo genomic DNA, each plate contained two subject DNAs in duplicate, a CEPH trio and three known laboratory quality-control samples. For the Duke WGA DNA, 88 samples were duplicated with an aliquot of the same WGA preparation, while 15 were duplicated with a separate WGA preparation. In addition, 124 individuals with WGA samples had sufficient genomic DNA for genotyping in order to understand the performance of WGA compared to genomic DNA (Cunningham JM et al, submitted).
Genotyping of 1,086 genomic and 1,282 WGA DNA samples (total = 2,368 including duplicates and laboratory controls) was performed at Mayo Clinic using the Illumina GoldenGate™ BeadArray assay and BeadStudio software for automated genotype clustering and calling according to a standard protocol (33).
Quality Control and Exclusions
Samples with Illumina GenCall scores (a metric of reliability of called genotypes generated by the BeadStudio software) below 0.25 or call rates below 90%, and SNPs with GenCall scores below 0.4 or call rates below 90%, were failed immediately for both genomic and WGA DNA. Of 2,051 samples genotyped, 10 were found to be ineligible and were excluded and 74 samples failed. These consisted of 72 which clustered especially poorly and were therefore failed for every SNP and 2 confirmed sample errors, resulting in a final sample size of 1,967 subjects. Of 188 SNPs in the 1-C transfer pathway, eight failed leaving 180 SNPs available for analysis listed in Supplementary Table 1.
Among SNPs with an overall call rate ≥ 95%, concordance was 99.99% between duplicates of genomic DNA, 99.97% between duplicates of WGA DNA and 99.16% between genomic and WGA DNA, indicating successful genotyping of WGA DNA for use in this study (Cunningham JM et al, submitted).
Statistical analyses
The present analyses excluded 197 non-Caucasian subjects. Participants’ genotypes were used to estimate allele frequencies. Among control subjects, genotypes were compared with those expected under Hardy-Weinberg equilibrium (HWE).
We compared the distribution of potential risk factors among cases and controls across study sites using ANOVA and χ2 tests. Risk models were adjusted for variables associated with ovarian cancer case-control status (see Table 2 footnotes), but no appreciable differences in risk estimates were observed without their inclusion. Pair-wise LD between SNPs was estimated with r2 values (34) using Haploview (35). Individual SNP associations for ovarian cancer risk were assessed using unconditional logistic regression to estimate odds ratios (ORs) and 95% confidence intervals (CIs). Primary tests for associations assumed an ordinal (log-additive) genotypic relationship with simple tests for trend, as well as separate comparisons of women with one copy and two copies of the minor allele to women with no copies (referent) using a 2 degree-of-freedom test. Haplotype frequencies for each gene were estimated using all SNPs within the gene and a global haplotype score test of no association between haplotypes and ovarian cancer risk was evaluated at the gene level by the method proposed by Schaid et al (36). Individual haplotype associations compared each haplotype to all other haplotypes combined.
Table 2.
Multivariable-adjusted* odds ratios (OR) and 95% confidence intervals (CI) between selected polymorphisms in genes in the one-carbon metabolism pathway and ovarian cancer risk among 1,770 Caucasian subjects, Mayo Clinic, MN and Duke University, NC, 1999–2006
| Homozygotes common allele (Referent) | Heterozygotes | Homozygotes rarer allele | Ordinal (Per rare allele) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene/SNP rsID | Cases | Controls | Cases | Controls | OR (95% CI) | Cases | Controls | OR (95% CI) | 2df P | OR (95% CI) | P trend |
| AHCYL1 | |||||||||||
| 17668350 | 684 | 806 | 141 | 127 | 1.4 (1.0–1.8) | 3 | 8 | 0.4 (0.1–1.7) | 0.04 | 1.2 (0.9–1.6) | 0.11 |
| DNMT3A | |||||||||||
| 13420827 | 552 | 602 | 234 | 308 | 0.8 (0.7–1.0) | 38 | 28 | 1.5 (0.9–2.6) | 0.03 | 1.0 (0.8–1.1) | 0.68 |
| DPYD | |||||||||||
| 1801265 | 463 | 585 | 321 | 318 | 1.3 (1.0–1.6) | 44 | 38 | 1.4 (0.9–2.2) | 0.04 | 1.2 (1.0–1.5) | 0.01 |
| MTHFD1 | |||||||||||
| 1950902 | 552 | 586 | 250 | 306 | 0.9 (0.7–1.1) | 17 | 37 | 0.5 (0.3–0.9) | 0.06 | 0.8 (0.7–1.0) | 0.04 |
| 2236225 | 229 | 288 | 421 | 481 | 1.1 (0.9–1.4) | 174 | 166 | 1.3 (1.0–1.8) | 0.15 | 1.1 (1.0–1.3) | 0.05 |
| 11849530 | 508 | 512 | 273 | 375 | 0.7 (0.6–0.9) | 48 | 53 | 0.9 (0.6–1.3) | 0.02 | 0.8 (0.7–1.0) | 0.03 |
| MTHFS | |||||||||||
| 17284990 | 502 | 557 | 262 | 338 | 0.9 (0.7–1.1) | 64 | 45 | 1.6 (1.1–2.5) | 0.01 | 1.1 (0.9–1.2) | 0.49 |
| SHMT1 | |||||||||||
| 9909104 | 437 | 539 | 317 | 340 | 1.2 (0.9–1.4) | 73 | 61 | 1.5 (1.0–2.2) | 0.09 | 1.2 (1.0–1.4) | 0.02 |
| SLC19A1 | |||||||||||
| 3788205 | 422 | 434 | 331 | 409 | 0.8 (0.7–1.0) | 75 | 98 | 0.8 (0.5–1.1) | 0.10 | 0.9 (0.7–1.0) | 0.04 |
| TYMS | |||||||||||
| 495139 | 282 | 352 | 390 | 449 | 1.1 (0.8–1.3) | 154 | 140 | 1.4 (1.0–1.8) | 0.10 | 1.1 (1.0–1.3) | 0.05 |
Adjusted for age (<40, 40–49, 50–59, 60–69, 70+), state (MN, IA, WI, IL, ND/SD, NC), BMI in kg/m2 (<23, 23–25.9, 26–28.9, 29+), postmenopausal hormone use (never, 1–60 months, 60+ months), oral contraceptive use (never, 1–48 months, 48+ months) and parity/age at first birth (nulliparous, 1–2/≤20 y, 1–2/>20 y, 3+/≤20 y, 3+/>20 y).
We also simultaneously modeled the comparison between controls and risk for each of the four main histologic subtypes of epithelial ovarian cancer (serous, endometrioid, clear cell and mucinous) under the ordinal genetic model using polytomous logistic regression and tested for statistical heterogeneity of the SNP-ovarian cancer histology associations (37).
Interactions between multivitamin intake and genotype (and haplotypes for SHMT1 and MTR) were evaluated for all SNPs under an ordinal genotypic relationship, where the association of a “fixed” genotype with ovarian cancer was assumed to depend on the “modifiable” exposure of multivitamin supplement use.
As an adjunct approach to identify genes (and therefore SNPs) that were significantly associated with ovarian cancer, we used principal components analysis to create orthogonal (e.g., uncorrelated) linear combinations of SNP minor allele counts that accounted for at least 90% of the variability in a gene. These were included in multivariable logistic regression models and tested for significance using a likelihood ratio test. By applying this method, we assumed that there would be residual correlation among SNPs (e.g., r2 < 0.8) that, when accounted for, would decrease the dimensionality of the data by reducing the number of independent degrees-of-freedom that comprised the statistical test. Significant associations with ovarian cancer at both the individual SNP level and at the “gene-level” using principal components were interpreted as supportive evidence for the individual SNP-ovarian cancer association.
To account for chance associations from multiple tests of individual SNPs and haplotypes with ovarian cancer risk, we calculated the false positive report probability (FPRP) (38), which depends on the prior probability that the SNP is associated with ovarian cancer, the power of the present study and the observed P value. We set a FPRP threshold of < 0.7 (e.g., 70% or lower probability that the study hypotheses were falsely positive) as ‘noteworthy’ for an initial study of a relatively rare tumor. Assuming a study power of 80%, we assigned a prior probability of 0.01 to detect an odds ratio of 1.5 or 0.67 for an individual SNP or haplotype, and to detect smaller odds ratios of 1.3 or 0.76 for SNP-multivitamin interactions with the expectation that there will be greater power to detect the gene effect among a homogeneous subset of the population exposed to multivitamin use (38). In light of recent reports (39, 40) of altered cancer risk by SHMT1 haplotypes that comprised SNPs similar to or highly correlated with SNPs examined in the present study, we calculated the FPRP for this gene using a higher prior probability of 0.1 for association with ovarian cancer.
Analyses were implemented using Haplo.stats (http://mayoresearch.mayo.edu/mayo/research/biostat/schaid.cfm), SAS (SAS Institute, Cary, NC, Version 8, 1999) and S-Plus (Insightful Corp, Seattle, WA, Version 7.05, 2005) software systems.
RESULTS
Fourteen SNPs showed departures from HWE among control subjects (P < 0.05, Supplementary Table 1); nine would be expected by chance. Although some investigators have discarded SNPs with statistical significance for HWE at P < 0.001 (41), we retained three SNPs in MTR at this level of significance. The MAF among controls ranged from 0.02 to 0.49 and were similar across study site. Cases (n=829) and controls (n=941) at both sites were somewhat different in the distribution of covariates (Table 1). A greater proportion of Mayo cases compared to controls were obese, never-users of oral contraceptives, had not gone beyond high school and fewer were regular multivitamin-users, whereas at Duke a larger proportion of cases compared to controls were post-menopausal, post-menopausal hormone-users and nulliparous. The greater proportion of Mayo compared to Duke subjects with a family history of ovarian cancer might be expected given the older age of the subjects in the Mayo Clinic study, where criteria did not specify an upper age limit. A greater proportion of Mayo compared to Duke controls reported taking multivitamins. Despite these differences, cases were comparable across sites in distribution of tumor histology.
Table 1.
Characteristics of 1,770 Caucasian subjects, Mayo Clinic, MN and Duke University, NC, 1999–2006
| Mayo | Duke | ||||||
|---|---|---|---|---|---|---|---|
| Characteristic* | Level | Cases | Controls | P† | Cases | Controls | P† |
| N | 385 | 462 | 444 | 479 | |||
| Age, y | Mean (S.D.) | 59.9 (13.4) | 60.0 (13.0) | 0.91 | 54.6 (11.3) | 54.8 (12.0) | 0.82 |
| Body mass index‡, kg/m2 | < 23 | 76 (20.5) | 107 (24.8) | 0.03 | 120 (27.8) | 129 (27.6) | 0.54 |
| 23–26 | 88 (23.2) | 121 (28.0) | 106 (24.5) | 117 (25.1) | |||
| 26–29 | 96 (25.9) | 110 (25.5) | 88 (20.4) | 110 (23.6) | |||
| ≥ 29 | 112 (30.3) | 94 (21.8) | 118 (27.3) | 111 (23.8) | |||
| Age at menarche, y | < 12 | 52 (18.2) | 67 (15.8) | 0.47 | 110 (24.8) | 85 (17.7) | 0.05 |
| 12 | 75 (26.3) | 97 (22.9) | 129 (29.1) | 143 (29.9) | |||
| 13 | 78 (27.4) | 124 (29.2) | 106 (23.9) | 139 (29.0) | |||
| ≥ 14 | 80 (28.1) | 136 (32.1) | 98 (22.1) | 112 (23.4) | |||
| Oral contraceptive use, months | Never | 171 (47.6) | 164 (38.6) | <0.001 | 137 (31.4) | 147 (30.9) | 0.65 |
| 1–48 | 94 (26.2) | 90 (21.2) | 133 (30.5) | 134 (28.2) | |||
| ≥ 48 | 94 (26.2) | 171 (40.2) | 166 (38.1) | 194 (40.8) | |||
| Postmenopausal status | Yes | 262 (71.2) | 327 (75.2) | 0.20 | 302 (73.5) | 316 (67.2) | 0.04 |
| Postmenopausal hormone use, months | Never | 231 (63.1) | 243 (58.4) | 0.40 | 151 (35.0) | 278 (59.5) | <0.001 |
| 1–60 | 63 (17.2) | 79 (19.0) | 168 (38.9) | 99 (21.2) | |||
| ≥ 60 | 72 (19.7) | 94 (22.6) | 113 (26.2) | 90 (19.3) | |||
| Parity, n/Age at first birth, y | Nulliparous | 68 (18.2) | 64 (14.8) | 0.07 | 95 (21.4) | 62 (12.9) | 0.01 |
| 1–2/≤ 20 y | 28 (7.5) | 25 (5.8) | 56 (12.6) | 56 (11.7) | |||
| 1–2/>20 y | 102 (27.3) | 128 (29.6) | 171 (38.5) | 212 (44.3) | |||
| ≥ 3/≤ 20 y | 71 (19.0) | 63 (14.5) | 59 (13.3) | 62 (12.9) | |||
| ≥ 3/>20 y | 104 (27.9) | 153 (35.3) | 63 (14.2) | 87 (18.2) | |||
| Family history of ovarian cancer§ | Yes | 50 (13.4) | 32 (7.3) | 0.004 | 33 (7.4) | 20 (4.2) | 0.03 |
| Smoking, pack y | None | 227 (65.2) | 279 (68.0) | 0.42 | 245 (57.4) | 248 (54.0) | 0.60 |
| ≤ 20 | 69 (19.8) | 83 (20.2) | 102 (23.9) | 120 (26.1) | |||
| > 20 | 52 (14.9) | 48 (11.7) | 80 (18.7) | 91 (19.8) | |||
| Education | No diploma | 23 (6.5) | 19 (4.4) | <0.001 | 35 (7.9) | 43 (9.0) | 0.43 |
| High school diploma | 133 (37.7) | 114 (26.1) | 139 (31.3) | 132 (27.6) | |||
| Post high school education | 197 (55.8) | 303 (69.5) | 270 (60.8) | 304 (63.5) | |||
| Multivitamin use|| | Yes | 105 (51.7) | 271 (64.7) | 231 (52.3) | 238 (49.8) | ||
| Tumor histology, cases | Serous | 230 (59.7) | 270 (60.8) | ||||
| Mucinous | 28 (7.3) | 52 (11.7) | |||||
| Endometrioid | 64 (16.6) | 56 (12.6) | |||||
| Clear cell | 22 (5.7) | 29 (6.5) | |||||
| Other | 40 (10.4) | 36 (8.1) | |||||
Data are counts (percentage) unless otherwise indicated. Counts do not total to 1,967 subjects due to missing data for some variables.
Continuous variables (t-test) and categorical variables (Chi square test).
BMI, body mass index.
In first or second degree relative.
≥ 4 pills per week during the previous year (Mayo subjects) or ‘regular multivitamin’ use (Duke subjects).
Selected multivariable-adjusted SNP associations are shown in Table 2; 10 SNPs in eight genes showed significance at P ≤ 0.05 (ordinal or general model). Of these, only SNPs in DPYD (P = 0.05) and SHMT1 (P = 0.03) were significant at the gene-centric level using principal components analysis (data not shown). Two copies of the minor allele in both DPYD Arg29Cys (rs1801265) and SHMT1 Intron5 A>G (rs9909104) were associated with increased risk in a dose-response manner. Results from the SNPs in the genes AHCYL1, DNMT3A, MTHFS, MTHFD1, SLC19A1 and TYMS also showed associations with ovarian cancer risk, but in the absence of a significant gene-level test.
Only the SHMT1 and MTR genes were significant using global haplotype score tests for association with ovarian cancer risk (Table 3). Of five individual haplotypes estimated in SHMT1, the 5-SNP haplotype #1 accounting for 33% of all estimated haplotypes was associated with decreased risk (P = 0.01), while the 5-SNP haplotype #5 with 25% frequency was associated with increased risk (P = 0.03). The difference in risks associated with the two SHMT1 haplotypes was seemingly attributable to the single locus Intron5 A>G (rs9909104) for which we observed a significant individual effect (Table 2). Of 11 individual haplotypes estimated in MTR, the 8-SNP haplotype #1 with 12% frequency was associated with decreased risk (P = 0.02), while the 8-SNP haplotype #11 with 2% frequency was associated with increased risk (P = 0.01). The difference between the two MTR haplotypes seemed to be attributable to two loci (3′ UTR C>A [rs2853523] and 3′ UTR C>T [rs1050993]). Both MTR loci, in addition to three other MTR loci (Intron4 A>G [rs12759827], Intron5 C>T [rs4659724] and 3′ UTR G>T [rs6676866]) that comprised the 8-SNP haplotypes, had genotypic distributions among control subjects that were significantly different than expected under HWE (P < 0.002) (Supplementary Table 1). When Mayo and Duke samples were examined separately, all but one of the eight SNPs (Intron5 C>T [rs4659724], P = 0.01) was in HWE among Duke subjects; however, all SNPs remained out of HWE (P < 0.001) among Mayo subjects. This was apparent despite adequate clustering of genotypes for these SNPs (data not shown) and despite no heterogeneity in genotype distributions between Mayo and Duke samples for all MTR loci when samples were examined separately. Thus, we cannot be certain of a spurious haplotype association.
Table 3.
Multivariable adjusted* haplotype analysis of genes in the one-carbon metabolism pathway and ovarian cancer risk among 1,770 Caucasian subjects, Mayo Clinic, MN and Duke University, NC, 1999–2006
| Haplotype | Global P† | Estimated haplotype frequency | Score test ‡ | Haplotype P§ | |
|---|---|---|---|---|---|
| No. | Allele combinations | ||||
| MTR|| | 0.04 | ||||
| 1 | AGCTAATT | 0.12 | −2.29 | 0.02 | |
| 2 | AACCGCCG | 0.14 | −1.45 | 0.14 | |
| 3 | AGTCACCG | 0.002 | −0.69 | 0.49 | |
| 4 | AGCTACCG | 0.004 | −0.07 | 0.94 | |
| 5 | GGCTAATT | 0.26 | 0.26 | 0.79 | |
| 6 | AGCCGCCG | 0.04 | 0.40 | 0.69 | |
| 7 | AGTTACCG | 0.38 | 0.79 | 0.43 | |
| 8 | AGTTAATT | 0.01 | 1.03 | 0.30 | |
| 9 | AGTTACCT | 0.002 | 1.07 | 0.28 | |
| 10 | GGCTACCG | 0.01 | 1.17 | 0.24 | |
| 11 | AGCTACCT | 0.02 | 2.47 | 0.01 | |
| SHMT1** | 0.05 | ||||
| 1 | GTCAG | 0.33 | −2.56 | 0.01 | |
| 2 | ATCGG | 0.01 | −0.44 | 0.66 | |
| 3 | ACTAG | 0.31 | −0.01 | 0.99 | |
| 4 | GTCAA | 0.10 | 1.21 | 0.23 | |
| 5 | GTCGG | 0.25 | 2.14 | 0.03 | |
Adjusted for age (<40, 40–49, 50–59, 60–69, 70+), state (MN, IA, WI, IL, ND/SD, NC), BMI in kg/m2 (<23, 23–25.9, 26–28.9, 29+), postmenopausal hormone use (never, 1–60 months, 60+ months), oral contraceptive use (never, 1–48 months, 48+ months) and parity/age at first birth (nulliparous, 1–2/≤20 yrs, 1–2/>20 yrs, 3+/≤20 yrs, 3+/>20 yrs)
P value from the global score test of Schaid et al (36) across haplotypes.
Score statistics comparing haplotype of interest with all other haplotypes combined. Negative values imply decreased risk of ovarian cancer, whereas positive values imply increased risk.
P value comparing haplotype of interest with all other haplotypes combined.
Haplotype-forming SNPs in MTR are rs12759827 (A>G), rs4659723 (G>A), rs4659724 (C>T), rs10925250 (T>C), rs1805087 (A>G), rs2853523 (C>A), rs1050993 (C>T), rs6676866 (G>T). Different alleles between significant haplotypes are underlined.
Haplotype-forming SNPs in SHMT1 are rs921986 (G>A), rs12952556 (T>C), rs1979277 (C>T), rs9909104 (A>G), rs2273026 (G>A). Different alleles between significant haplotypes are underlined.
Analyses by histologic subtype revealed statistical heterogeneity (P = 0.01) for the association of DNMT3B Intron1 G>A [rs6119954] with ovarian cancer. Compared to controls, the ordinal genetic model estimated increased risk for endometrioid (OR, 1.6; 95% CI, 1.1–2.2; 120 cases) and clear cell (OR, 1.6; 95% CI, 1.0–2.8; 51 cases) tumors, but not for serous (OR, 0.9; 95% CI, 0.7–1.2; 500 cases) or mucinous (OR, 0.9; 95% CI, 0.5–1.4; 80 cases) tumors, although the findings may be from chance due to small numbers of cases.
SNP-specific associations with ovarian cancer and modified by multivitamin use under the ordinal genetic model are shown in Table 4. Among women who took multivitamins regularly, the per-minor allele risk was decreased for SNPs in DNMT3A (3′ UTR C>G [rs13420827] and Intron6 G>A [rs11887120]), DNMT1 Intron23 C>T (rs9305012) and MTHFR 3′ UTR A>G (rs2184226), but risk was increased for SNPs in DNMT3A Intron22 A>T (rs11695471) and MTHFD1 Intron17 C>T (rs17101854). Only the DNMT3A 3′ UTR C>G [rs13420827] was significantly associated with risk in main effects models (Table 2). In subsequent analyses of SHMT1 and MTR haplotypes, an interaction with multivitamin use was not significant at the global haplotype score test for SHMT1 (P = 0.11), although the 5-SNP SHMT1 haplotype #1 (GTCAG) was associated with decreased risk among users (P = 0.03). Interactions were significant at the global test for MTR (P = 0.03), with decreased risk (P = 0.004) associated with the 8-SNP MTR haplotype #1 (AGCTAATT) among supplement users (data not shown).
Table 4.
Age and residence-adjusted odds ratios (OR) and 95% confidence intervals (CI) for the joint effect of selected polymorphisms in genes in the one-carbon metabolism pathway and multivitamin supplement use using an ordinal model for ovarian cancer risk among 1,770 Caucasian subjects, Mayo Clinic, MN and Duke University, NC, 1999–2006
| Multivitamin Non Users | Multivitamin Users | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Homozygous common allele (Referent) | Heterozygous | Homozygous rare allele | Ordinal (Per rare allele) | Homozygous common allele (Referent) | Heterozygous | Homozygous rare allele | Ordinal (Per rare allele) | ||
| Gene/SNP rsID | Cases/Controls | Cases/Controls | Cases/Controls | OR (95% CI) | Cases/Controls | Cases/Controls | Cases/Controls | OR (95% CI) | P inter-action |
| DNMT3A | |||||||||
| 11887120 | 93/149 | 142/175 | 70/64 | 1.3 (1.0–1.6) | 124/157 | 151/264 | 58/88 | 0.8 (0.7–1.0) | 0.007 |
| 11695471 | 154/165 | 123/163 | 30/58 | 0.8 (0.6–1.0) | 126/224 | 170/234 | 39/51 | 1.2 (1.0–1.5) | 0.01 |
| 13420827 | 192/252 | 95/129 | 19/6 | 1.3 (1.0–1.7) | 237/321 | 84/166 | 13/20 | 0.8 (0.6–1.0) | 0.006 |
| DNMT1 | |||||||||
| 9305012 | 268/345 | 36/42 | 4/1 | 1.3 (0.9–2.1) | 306/442 | 30/65 | 0/2 | 0.6 (0.4–1.0) | 0.03 |
| MTHFR | |||||||||
| 2184226 | 252/337 | 53/48 | 2/3 | 1.3 (0.9–1.9) | 418/288 | 44/80 | 3/9 | 0.7 (0.5–1.1) | 0.05 |
| MTHFD1 | |||||||||
| 17101854 | 296/367 | 13/21 | 0 | 0.6 (0.3–1.3) | 311/488 | 25/21 | 0 | 2.0 (1.0–3.7) | 0.02 |
The calculated FPRPs were well below our preset value of 0.7 for the main effect of SHMT1 Intron5 A>G (rs9909104; FPRP = 0.16) and for the 5-SNP SHMT1 haplotype #1 (0.09 for GTCAG). FPRPs were also lower for the 8-SNP MTR haplotypes #1 and #11 (0.52 for AGCTAATT and 0.67 for AGCTACCT, respectfully) and for three SNPs in DNMT3A when examined within the context of multivitamin use (0.54 for 3′ UTR C>G [rs13420827], 0.57 for Intron6 G>A [rs11887120] and 0.66 for Intron22 A>T [rs11695471]). These calculations suggest that the probability of our findings being falsely positive is 9–16 % for the SHMT1 SNPs and higher for the other SNPs.
DISCUSSION
To our knowledge, we are the first to examine a large number of SNPs (n=180) in genes (n=21) in the 1-C metabolism pathway for ovarian cancer risk among Caucasians in the U.S., and the findings provide an initial report of potential causal variants, most of which are novel for their previously-unexamined association with ovarian cancer. We extend findings of the recently-reported association of SHMT1 SNPs in other cancers and confirm their relevance to ovarian cancer. Interactions with multivitamin intake are suggestive as are haplotypes in MTR, but the absence of HWE could have resulted in spurious associations. No definitive differences were observed across histologic subtypes.
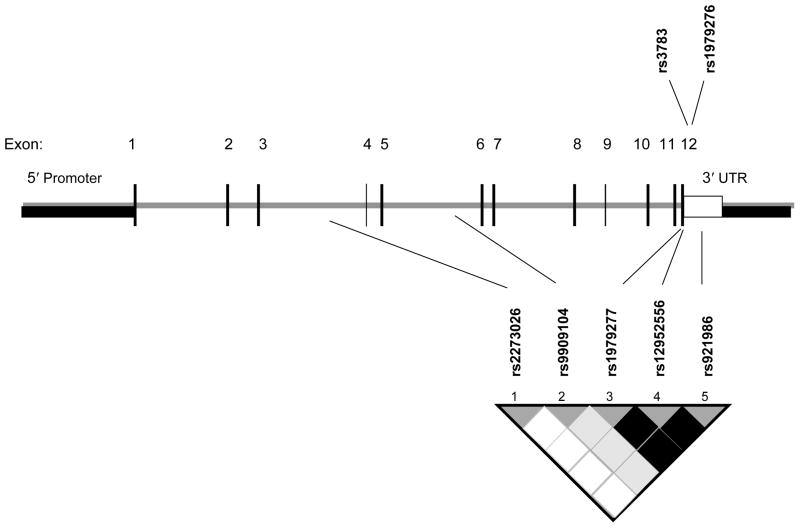
The vitamin B6-dependent SHMT1 enzyme catalyzes the reversible conversion of serine and tetrahydrofolate to glycine and 5,10 methylenetetrahydrofolate in the cytoplasm for the synthesis of methionine, thymidylate and purines (42). Incorporation of the β-carbon of serine into DNA and SHMT1 activity are increased when cells are stimulated to proliferate (43). SHMT1 activity is also elevated in tumor tissues (44). In our study, the two significant haplotypes in SHMT1 differed only at a single locus, Intron5 A>G (rs9909104), which was also independently associated with ovarian cancer. Intron5 A>G (rs9909104) was in low LD (r2 = 0.21) with SHMT1 Leu435Phe (rs1979277), also in the haplotype, and not significantly associated with ovarian cancer in our study. Others also observed generally null associations with SHMT1 Leu435Phe (20, 22, 23, 28, 40, 45, 46), SHMT1 Exon12 C>T [rs1979276] (13, 40, 46) and 3′ UTR C>G [rs3783] (40). The two latter SNPs were not genotyped in our study but were tagged at pair-wise r2 ≥ 0.8 by the assayed SNPs that comprise the 5-SNP SHMT1 haplotype (Leu435Phe, 3′ UTR C>T [rs12952556] and 3′ UTR [rs921986]) (Figure 1). Although different yet correlated loci were examined, our findings are supported by those from Zhang et al (40), who reported significantly altered risk of squamous cell carcinoma of the head and neck from haplotypes comprising three SHMT1 SNPs (Leu435Phe, Exon12 C>T [rs1979276] and 3′ UTR C>G [rs3783]), and by the same group (39) of significantly altered risk of lung cancer associated with carrying an increasing number of variant genotypes in five SHMT1 SNPs (Leu435Phe, Exon12 C>T [rs1979276], 3′ UTR C>G [rs3783], promoter SNP C>A [rs643333] and promoter SNP G>C [rs638416]). The observed associations from the same SNPs or SNPs that are highly correlated in each of our studies strongly suggests that these SHMT1 SNPs may themselves be or are in strong LD with putative causal alleles. Mutations in the C-terminal region of SHMT1 leads to incorrect protein folding (47) and the location of the SNPs near or in the 3′ UTR region of the gene suggests they may impact enzyme conformation and activity. Fine-mapping of this chromosomal region with further association testing is therefore a recommended priority for future studies.
Figure 1. Gene structure and location of SNPs in the SHMT1 gene.
tagSNPs that comprise the haplotype assayed among 1,770 Caucasian subjects (Mayo Clinic, MN and Duke University, NC, 1999–2006) are shown below the gene. SNPs previously reported to be associated with cancer (39, 40) in the same region are shown above the gene. Adapted from (39).
Shaded regions in the linkage disequilibrium (LD) plot indicate the strength of LD between pair-wise combinations of SNPs (white: r2 = low LD, black: r2 = high LD).
Suggestive findings were observed with SNPs in DNMT3A, particularly when stratified by multivitamin use. In eukaryote cells, the addition of methyl groups to the carbon-5 position of cytosines within CpG-rich DNA sequences (‘CpG islands’) in gene promoters is facilitated by the DNA methyltransferase (DNMT) enzymes. Genes with methylated CpG islands are incapable of transcription initiation unless the methylation signal can be overridden (48). DNMT1 is thought to function primarily to maintain the inherited DNA methylation pattern during DNA synthesis, while DNMT3 alpha and DNMT3 beta are believed to function principally in de novo methylation (49). In one of the few studies to examine DNMT3A SNPs for cancer risk, Cebrian et al (14) did not find significant associations of 13 tagSNPs with breast cancer risk. Eight tagSNPs in that study were also genotyped in the present analysis or were represented in our data by tagged SNPs. None of these eight were the three SNPs in DNMT3A for which we observed significant associations by multivitamin supplement use. Our findings could be due to chance, or there may have been greater power to detect the gene effect among the homogeneous subset of the population defined by multivitamin intake. As these analyses were secondary and comprised small numbers of subjects, confirmation of our findings is necessary.
We observed fourteen SNPs with genotypic distributions significantly different from that expected under HWE; nine would be expected by chance. Following additional review of cluster plots and genotype calls for these SNPs, we could not find obvious deviations to explain disequilibrium statistics. Nor was there evidence for statistical heterogeneity in genotypic distributions between sites for these SNPs. Further, we might have expected the genotypes among Mayo controls to be in HWE because settlers of the upper mid-West tend to represent a fairly homogeneous sample of Scandinavian and northern European ancestries whose genotypes may be less likely to be biased by population stratification. We conclude that the findings may be due to chance or random error, which would attenuate observed associations.
The strengths of our study include the large sample size and coordinated data collection across study sites, the investigation of a large number of SNPs across 21 genes with well-defined roles in 1-C transfer, the observance of noteworthy associations with SNPs not previously examined with ovarian cancer and the use of the FPRP to account for falsely positive findings. Although preliminary, the genes that interacted with multivitamin use in our study potentially supports a role of 1-C donor units in pathways that influence both genetic expression via methylation (e.g., DNMTs) and enzyme function via disruption of transfer of 1-C units (e.g, MTR). The comparable associations of the SHMT1 haplotype in this study and those by the Spitz investigative team for risk of other cancers (39, 40) are an important finding given the rather null associations observed with the candidate SNP Leu435Phe (rs1979277) in this gene in earlier studies (12, 20, 23, 28). This strengthens the utility of tagSNPs, bioinformatics tools and haplotype analyses for identifying common genetic variants in disease risk.
Some potential limitations of our study warrant discussion. First, this was not a comprehensive examination of 1-C metabolism genes. Second, different reference periods defined regular multivitamin use between sites, complicating our measure of exposure time. We did not have information on dietary intake and could not verify which nutrient(s) was related to the modifying effects of multivitamins. Also, a greater proportion of both Mayo and Duke controls reported taking multivitamins compared to 38–40% of women in the US (50), and the greater prevalence among Mayo compared to Duke controls could be attributable to fewer smokers, higher level of education and the older age of Mayo controls, which are factors associated with multivitamin use (50). Third, in our analyses of effect modification by multivitamin use, the sample was too small to examine genetic models other than log-additive where the relationship with ovarian cancer of the main effect of genotype appeared to deviate from an ordinal relationship.
In conclusion, our data provide evidence for genetic variation in SHMT1 with ovarian cancer risk, and suggestive associations of DNMT3A, MTR and possibly the modifying effects of multivitamins as suppliers of 1-C units. Replication of these findings should be pursued by other investigators in other populations, including those with detailed information on diet.
Supplementary Material
Acknowledgments
Financial Support: National Institutes of Health grants R01 CA88868 (TAS) and 2-R01-CA76016 (JMS); Department of Defense DAMD17-02-1-0666 (JMS); Fraternal Order of Eagles (ELG) and the Minnesota Ovarian Cancer Alliance (ELG). LEK was supported by National Institutes of Health grant R25 CA92049-03 (TAS).
References
- 1.American Cancer Society. Cancer facts and figures 2007. Estimated new cancer cases and deaths by sex for all sites, US, 2007. 2005 [cited April 13, 2007.] Available from: www.cancer.org/docroot/med/content.
- 2.Hankinson SE, Colditz GA, Hunter DJ, Spencer TL, Rosner B, Stampfer MJ. A quantitative assessment of oral contraceptive use and risk of ovarian cancer. Obstetrics and gynecology. 1992;80(4):708–14. [PubMed] [Google Scholar]
- 3.Whittemore AS, Harris R, Itnyre J. Characteristics relating to ovarian cancer risk: collaborative analysis of 12 US case-control studies. II. Invasive epithelial ovarian cancers in white women. Collaborative Ovarian Cancer Group. Am J Epidemiol. 1992;136(10):1184–203. doi: 10.1093/oxfordjournals.aje.a116427. [DOI] [PubMed] [Google Scholar]
- 4.Antoniou A, Pharoah PD, Narod S, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–30. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Q, Khoury MJ, Friedman J, Little J, Flanders WD. How many genes underlie the occurrence of common complex diseases in the population? International journal of epidemiology. 2005;34(5):1129–37. doi: 10.1093/ije/dyi130. [DOI] [PubMed] [Google Scholar]
- 6.Choi SW, Mason JB. Folate and carcinogenesis: an integrated scheme. J Nutr. 2000;130:129–32. doi: 10.1093/jn/130.2.129. [DOI] [PubMed] [Google Scholar]
- 7.Kim YI, Pogribny IP, Basnakian AG, et al. Folate deficiency in rats induces DNA strand breaks and hypomethylation within the p53 tumor suppressor gene. Am J Clin Nutr. 1997;65(1):46–52. doi: 10.1093/ajcn/65.1.46. [DOI] [PubMed] [Google Scholar]
- 8.Jacob RA, Gretz DM, Taylor PC, et al. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J Nutr. 1998;128(7):1204–12. doi: 10.1093/jn/128.7.1204. [DOI] [PubMed] [Google Scholar]
- 9.Blount BC, Mack MM, Wehr CM, et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci USA. 1997;94:3290–5. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mikol YB, Hoover KL, Creasia D, Poirier LA. Hepatocarcinogenesis in rats fed methyl-deficient, amino acid-defined diets. Carcinogenesis. 1983;4(12):1619–29. doi: 10.1093/carcin/4.12.1619. [DOI] [PubMed] [Google Scholar]
- 11.Ghoshal AK, Farber E. The induction of liver cancer by dietary deficiency of choline and methionine without added carcinogens. Carcinogenesis. 1984;5(10):1367–70. doi: 10.1093/carcin/5.10.1367. [DOI] [PubMed] [Google Scholar]
- 12.Lim U, Wang SS, Hartge P, et al. Gene-nutrient interactions among determinants of folate and one-carbon metabolism on the risk of non-Hodgkin lymphoma: NCI-SEER case-control study. Blood. 2007;109(7):3050–9. doi: 10.1182/blood-2006-07-034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Shi Q, Liu Z, Sturgis EM, Spitz MR, Wei Q. Polymorphisms of methionine synthase and methionine synthase reductase and risk of squamous cell carcinoma of the head and neck: a case-control analysis. Cancer Epidemiol Biomarkers Prev. 2005;14(5):1188–93. doi: 10.1158/1055-9965.EPI-04-0501. [DOI] [PubMed] [Google Scholar]
- 14.Cebrian A, Pharoah PD, Ahmed S, et al. Genetic variants in epigenetic genes and breast cancer risk. Carcinogenesis. 2006;27(8):1661–9. doi: 10.1093/carcin/bgi375. [DOI] [PubMed] [Google Scholar]
- 15.Shen H, Wang L, Spitz MR, Hong WK, Mao L, Wei Q. A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer research. 2002;62(17):4992–5. [PubMed] [Google Scholar]
- 16.Giovannucci E. Epidemiologic studies of folate and colorectal neoplasia: a review. Journal of Nutrition. 2002;132(8 Suppl):2350S–5S. doi: 10.1093/jn/132.8.2350S. [DOI] [PubMed] [Google Scholar]
- 17.Larsson SC, Giovannucci E, Wolk A. Dietary folate intake and incidence of ovarian cancer: the Swedish Mammography Cohort. Journal of the National Cancer Institute. 2004;96(5):396–402. doi: 10.1093/jnci/djh061. [DOI] [PubMed] [Google Scholar]
- 18.Kelemen LE, Sellers TA, Vierkant RA, Harnack L, Cerhan JR. Association of folate and alcohol with risk of ovarian cancer in a prospective study of postmenopausal women. Cancer Causes & Control. 2004;15(10):1085–93. doi: 10.1007/s10552-004-1546-6. [DOI] [PubMed] [Google Scholar]
- 19.Sellers TA, Schildkraut JM, Pankratz VS, et al. Estrogen bioactivation, genetic polymorphisms, and ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(11 Pt 1):2536–43. doi: 10.1158/1055-9965.EPI-05-0142. [DOI] [PubMed] [Google Scholar]
- 20.Lissowska J, Gaudet MM, Brinton LA, et al. Genetic polymorphisms in the one-carbon metabolism pathway and breast cancer risk: a population-based case-control study and meta-analyses. Int J Cancer. 2007;120(12):2696–703. doi: 10.1002/ijc.22604. [DOI] [PubMed] [Google Scholar]
- 21.Kelemen LE. The role of folate receptor alpha in cancer development, progression and treatment: Cause, consequence or innocent bystander? Int J Cancer. 2006;119(2):243–50. doi: 10.1002/ijc.21712. [DOI] [PubMed] [Google Scholar]
- 22.Skibola CF, Forrest MS, Coppede F, et al. Polymorphisms and haplotypes in folate-metabolizing genes and risk of non-Hodgkin lymphoma. Blood. 2004;104(7):2155–62. doi: 10.1182/blood-2004-02-0557. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Kyte C, Valcin M, et al. Polymorphisms in the one-carbon metabolic pathway, plasma folate levels and colorectal cancer in a prospective study. Int J Cancer. 2004;110(4):617–20. doi: 10.1002/ijc.20148. [DOI] [PubMed] [Google Scholar]
- 24.Van Kuilenburg AB, Vreken P, Abeling NG, et al. Genotype and phenotype in patients with dihydropyrimidine dehydrogenase deficiency. Human genetics. 1999;104(1):1–9. doi: 10.1007/pl00008711. [DOI] [PubMed] [Google Scholar]
- 25.Prasannan P, Pike S, Peng K, Shane B, Appling DR. Human mitochondrial C1-tetrahydrofolate synthase: gene structure, tissue distribution of the mRNA, and immunolocalization in Chinese hamster ovary calls. The Journal of biological chemistry. 2003;278(44):43178–87. doi: 10.1074/jbc.M304319200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Pietro E, Sirois J, Tremblay ML, MacKenzie RE. Mitochondrial NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase is essential for embryonic development. Molecular and cellular biology. 2002;22(12):4158–66. doi: 10.1128/MCB.22.12.4158-4166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaw GM, Lammer EJ, Zhu H, Baker MW, Neri E, Finnell RH. Maternal periconceptional vitamin use, genetic variation of infant reduced folate carrier (A80G), and risk of spina bifida. Am J Med Genet. 2002;108:1–6. doi: 10.1002/ajmg.10195. [DOI] [PubMed] [Google Scholar]
- 28.Lim U, Peng K, Shane B, et al. Polymorphisms in cytoplasmic serine hydroxymethyltransferase and methylenetetrahydrofolate reductase affect the risk of cardiovascular disease in men. J Nutr. 2005;135(8):1989–94. doi: 10.1093/jn/135.8.1989. [DOI] [PubMed] [Google Scholar]
- 29.Ulrich CM, Bigler J, Velicer CM, Greene EA, Farin FM, Potter JD. Searching expressed sequence tag databases: discovery and confirmation of a common polymorphism in the thymidylate synthase gene. Cancer Epidemiol Biomarkers Prev. 2000;9(12):1381–5. [PubMed] [Google Scholar]
- 30.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426(6968):789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 31.Hinds DA, Stuve LL, Nilsen GB, et al. Whole-genome patterns of common DNA variation in three human populations. Science (New York, NY. 2005;307(5712):1072–9. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 32.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74(1):106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques. 2002;(Suppl):56–8. 60–1. [PubMed] [Google Scholar]
- 34.Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 1995;29(2):311–22. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- 35.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (Oxford, England) 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 36.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosmer DW, Lameshow SL, editors. Applied Logistic Regression. New York, NY: John Wiley and Sons, Inc; 1989. [Google Scholar]
- 38.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96(6):434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Lu J, An J, Shi Q, Spitz MR, Wei Q. Polymorphisms of cytosolic serine hydroxymethyltransferase and risk of lung cancer: A case-control analysis. Lung cancer (Amsterdam, Netherlands) 2007;57(2):143–51. doi: 10.1016/j.lungcan.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z, Shi Q, Sturgis EM, Spitz MR, Wei Q. Polymorphisms and haplotypes of serine hydroxymethyltransferase and risk of squamous cell carcinoma of the head and neck: a case-control analysis. Pharmacogenetics and genomics. 2005;15(8):557–64. doi: 10.1097/01.fpc.0000170915.19522.b2. [DOI] [PubMed] [Google Scholar]
- 41.Balding DJ. A tutorial on statistical methods for population association studies. Nature reviews. 2006;7(10):781–91. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 42.Schirch L. Serine hydroxymethyltransferase. Advances in enzymology and related areas of molecular biology. 1982;53:83–112. doi: 10.1002/9780470122983.ch3. [DOI] [PubMed] [Google Scholar]
- 43.Snell K, Natsumeda Y, Weber G. The modulation of serine metabolism in hepatoma 3924A during different phases of cellular proliferation in culture. The Biochemical journal. 1987;245(2):609–12. doi: 10.1042/bj2450609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snell K, Natsumeda Y, Eble JN, Glover JL, Weber G. Enzymic imbalance in serine metabolism in human colon carcinoma and rat sarcoma. Br J Cancer. 1988;57(1):87–90. doi: 10.1038/bjc.1988.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Guo W, He Y, et al. Association of MTHFR C677T and SHMT(1) C1420T with susceptibility to ESCC and GCA in a high incident region of Northern China. Cancer Causes Control. 2007;18(2):143–52. doi: 10.1007/s10552-006-0097-4. [DOI] [PubMed] [Google Scholar]
- 46.Moore LE, Malats N, Rothman N, et al. Polymorphisms in one-carbon metabolism and trans-sulfuration pathway genes and susceptibility to bladder cancer. Int J Cancer. 2007;120(11):2452–8. doi: 10.1002/ijc.22565. [DOI] [PubMed] [Google Scholar]
- 47.Snell K, Baumann U, Byrne PC, et al. The genetic organization and protein crystallographic structure of human serine hydroxymethyltransferase. Advances in enzyme regulation. 2000;40:353–403. doi: 10.1016/s0065-2571(99)00035-7. [DOI] [PubMed] [Google Scholar]
- 48.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 50.Radimer K, Bindewald B, Hughes J, Ervin B, Swanson C, Picciano MF. Dietary supplement use by US adults: data from the National Health and Nutrition Examination Survey, 1999–2000. Am J Epidemiol. 2004;160(4):339–49. doi: 10.1093/aje/kwh207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.