Abstract
The intrinsic resistance of P. aeruginosa PAO1 to the peptide deformylase inhibitor (PDF-I) LBM415 was mediated by the MexAB-OprM and MexXY-OprM efflux pumps, the latter of which was strongly induced by LBM415. Single-step exposure of PAO1 deleted for mexAB-oprM (therefore lacking both MexAB-OprM and MexXY-OprM functions) to PDF-Is selected for nfxB mutants, which express the MexCD-OprJ efflux pump, indicating that these compounds are also substrates for this pump. Selection of resistant mutants by use of levels of LBM415 greater than that accommodated by efflux yielded two additional groups of mutations, in the methionyl-tRNAfmet formyltransferase (fmt) and folD genes. Both mechanisms are known to impose an in vitro growth deficit (also observed here), presumably due to impairment of protein synthesis. We surmised that this inherent impairment of protein synthesis would upregulate expression of mexXY in a fashion similar to upregulation by LBM415 or by ribosome inhibitory compounds. Transcriptional profiling and/or mexX::lux promoter fusion analysis revealed that fmt and folD mutants were strongly upregulated for mexXY and another gene known to be required for upregulation of the pump, PA5471. Complementation of the fmt mutation in trans reversed this constitutive expression. This supports the notion that MexXY has a natural physiological function responding to impairment of ribosome function or protein synthesis and that fmt mutation (Fmt bypass) and folD mutation generate the intracellular mexXY-inducing signal.
Peptide deformylase has been the focus of intense interest as an antibacterial target for nearly 2 decades (1, 2, 8, 9). This has led to the development of numerous potent peptide deformylase inhibitors (PDF-Is), typified by the reverse hydroxamate LBM415 (13), although the spectrum of activity of this compound is limited mainly to gram-positive species and more-intrinsically antibiotic-susceptible gram-negative species, such as Haemophilus influenzae and Moraxella catarrhalis (5, 11, 15, 17, 18, 22-24, 46). Lower levels of activity against other gram-negative species such as Escherichia coli or Pseudomonas aeruginosa might be expected, due to the permeability barrier imposed by the outer membrane combined with active efflux, which affects susceptibility to a very broad range of compounds (25, 36-38). Indeed, even in the case of H. influenzae, where efflux poses less of a problem, the AcrAB-TolC (40, 45) pump is responsible for reduced intrinsic susceptibility to LBM415 (13) compared to gram-positive organisms. Upregulation of AcrAB-TolC due to mutations in the pump repressor AcrR further decreases susceptibility to LBM415 in H. influenzae (13).
The most significant pumps with respect to P. aeruginosa multidrug resistance are the RND family pumps, exemplified by MexAB-OprM, MexXY-OprM, MexCD-OprJ, and MexEF-OprN (39). MexAB-OprM is expressed constitutively, although various mutations can lead to increased expression (e.g., nalB, nalC, and nalD mutants) (7, 42, 44). MexXY is inducible by several structurally unrelated antibiotics that perturb ribosome function/protein synthesis (14, 21, 31, 33). Together, these two pumps cause the majority of intrinsic resistance so far described. Both MexCD-OprJ and MexEF-OprN are not significantly expressed under most laboratory conditions, although mutants expressing these pumps are selected by exposure to antibiotic substrates (25, 39).
For organisms that are highly susceptible to PDF-Is, such as Streptococcus pneumoniae, Chlamydia trachomatis, and Staphylococcus aureus, additional PDF-I resistance mechanisms have been described, including target mutation or upregulation and Fmt bypass (3, 29, 30). The latter is of significant concern since it renders bacteria impervious to all PDF-Is. Mutation in fmt, which encodes methionyl-tRNAfmet formyltransferase, is thought to cause resistance by eliminating the formylation of methionyl-tRNAfmet so that nascent proteins lack the formyl group on their initiating methionine. Therefore, Fmt bypass abrogates entirely the necessity for deformylation (i.e., bypassing the formylation-deformylation cycle). There appears to be different degrees of tolerance for loss of fmt in different bacteria, with Fmt bypass so far unreported for S. pneumoniae, while in S. aureus, it occurs at high frequency but imposes a modest fitness deficit (26, 30). In E. coli and Salmonella enterica, loss of fmt can occur but imposes a significant fitness deficit, and in P. aeruginosa, insertional inactivation of fmt also caused an in vitro fitness deficit (34, 35). Partial suppression of the fitness defect of Fmt bypass in Salmonella enterica can occur through overexpression of initiator tRNA (35). While Fmt bypass-based PDF-I resistance has not been described to occur in P. aeruginosa, the fact that fmt can be insertionally inactivated (34) indicated the likelihood that Fmt bypass could be selected by exposure to PDF-Is. A related but less frequently observed resistance mechanism involves mutation in folD, a component of the folate biosynthesis pathway (35). In this case, interruption of folate synthesis is proposed to reduce or eliminate the synthesis of the formyl group itself, resulting in the same bypassing of the formylation-deformylation cycle resulting from Fmt bypass.
In this study, we showed that intrinsic resistance to LBM415 is mediated by MexAB-OprM and MexXY-OprM and that the latter is inducible by exposure to PDF-Is. Furthermore, Fmt bypass and mutation in folD create an intracellular condition causing constitutive expression of the MexXY-OprM efflux pump in the absence of any compound that inhibits the ribosome. (Portions of this work were presented at the 46th Annual Interscience Conference on Antibacterial Agents and Chemotherapy [ICAAC], 2006.)
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
The bacterial strains and plasmids used in this study are described in Table 1. P. aeruginosa was routinely grown at 37°C in either Mueller-Hinton (Remel) or Luria (Difco) broth or solid medium. Media were supplemented with gentamicin or tetracycline (100 μg/ml for P. aeruginosa and 10 μg/ml of E. coli) or 100 μg/ml ampicillin (E. coli) or carbenicillin (P. aeruginosa) as required. For single-step isolation of mutants with decreased susceptibility to LBM415, P. aeruginosa was grown to mid-log phase (OD600, approximately 0.6) in Mueller-Hinton broth, pelleted by centrifugation, and resuspended in fresh medium. Aliquots were plated on Mueller-Hinton agar containing various levels of LBM415 to select for resistant isolates. Serial dilutions were also plated on Mueller-Hinton agar without compound for enumeration. Resistance frequencies were calculated as the number of CFU on drug-containing plates divided by the number of CFU plated.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| NB52019 | P. aeruginosa PAO1 | K. Poole |
| NB52020 | NB52019 mexAB-oprM; lacks both MexAB-OprM and MexXY-OprM function | K. Poole |
| NB52021 | NB52019 mexB; lacks MexAB-OprM function | K. Poole |
| NB52022 | NB52019 mexX; lacks MexXY-OprM function | K. Poole |
| NB52023 | NB52019 mexB mexX | K. Poole |
| CDS1 | NB52020 derivative with decreased susceptibility to LBM415; nfxB | This study |
| CDS6 | NB52020 derivative with decreased susceptibility to LBM415; nfxB | This study |
| CDR3 | NB52019 mexX::luxCDABE | This study |
| CDS28 | NB52019 derivative with decreased susceptibility to LBM415; fmt | This study |
| CDR5 | CDS28 mexX::luxCDABE | This study |
| CDR1 | NB52019 fmt::aacC1 | This study |
| CDR2 | NB52020 fmt::aacC1 | This study |
| CDR4 | NB52020 mexX::luxCDABE | This study |
| CDS13 | NB52020 derivative with decreased susceptibility to LBM415;fmt | This study |
| CDR6 | CDS13 mexX::luxCDABE | This study |
| CDS30 | NB52019 derivative with decreased susceptibility to LBM415; folD | This study |
| CDS22 | NB52020 derivative with decreased susceptibility to LBM415; folD | This study |
| CDS30R | Fast-growing revertant of CDS30; folD intragenic suppressor | This study |
| E. coli S17-1 | Mobilizer strain | 41b |
| Plasmids | ||
| pAK1900 | E. coli-P. aeruginosa shuttle vector (Apr) | A. Kropinsky |
| pSW1 | pAK1900 containing fmt | This study |
| pRC5 | pAK1900 containing folD | This study |
| Mini CTX luxCDABE | Promoter fusion plasmid, promoterless luxCDABE; Tcr | 4 |
| pRC1 | Mini-CTX mexX::luxCDABE | This study |
| pRC2 | Mini-CTX mexX::luxCDABE | This study |
| pUCGm | Source of aacC1 resistance cartridge; Apr Gmr | 41 |
| pEX18-Tc | Gene replacement vector; Tcr | 20 |
| pFLP2 | Flip recombinase vector; Apr | 20 |
Apr, ampicillin resistance marker; Tcr, tetracycline resistance marker; Gmr, gentamicin resistance marker.
DNA manipulations.
P. aeruginosa genomic DNA was isolated using a Puregene DNA isolation kit (Gentra Systems, Inc., Minneapolis, MN) in accordance with the supplied instructions. The PCR primers used in this study are listed in Table 2. PCRs were carried out using an Accuprime GC-rich DNA polymerase kit (Invitrogen, Carlsbad, CA) in accordance with the supplied instructions. PCR fragments were isolated from agarose gels by using a QIAquick gel extraction kit (Qiagen, Inc., Valencia, CA) in accordance with the supplied instructions. Plasmid pSW1 was constructed as follows. The fmt gene was PCR generated from strain NB52019 by using primers PAfmtF and PAfmtR (Table 2) and cloned into Topo PCR 2.1 (Invitrogen, Carlsbad, CA) in accordance with the protocol provided with the Topo TA cloning kit. The fmt insert was then excised from this construct as a HindIII-XbaI fragment and ligated into HindIII-XbaI-digested pAK1900 to give plasmid pSW1. The folD gene was cloned into pAK1900 as follows. A 961-bp fragment encompassing the P. aeruginosa folD gene was generated from strain NB52019 by using the primers folDfor and folDrev (Table 2), using VENT DNA polymerase, and cloned into SmaI-digested pAK1900 to yield pRC5, which has folD in the same orientation as the lac promoter of pAK1900. Cloned products were sequenced using standard M13 primers. Nucleotide sequencing was done by Agencourt, Inc. (Beverly, MA).
TABLE 2.
Oligonucleotide primers used in this study
| Name | Sequence (5′ to 3′) | Source or reference |
|---|---|---|
| nfxB1 | CGATCCTTCCTATTGCACG | 28 |
| nfxB2 | GCCAAGTGCCAGTATCG | 28 |
| PAfmtF | CATCGACAGCAGGCGTCC | This study |
| PAfmtR | AGCGAGGCGCGTCCGGCGAGCACCG | This study |
| PAfmtF1 | GGCCCGATGCTGCTCAAGGTGAGC | This study |
| PAfmtR1 | CTTCGATCACCGCTTTCGGGCCGA | This study |
| PAfmtF4 | ATCCAGCACGAATGCGACCA | This study |
| PAfmtR4 | TGCAGGCGCGGTTGCCAGCG | This study |
| folDfor | CCTGGCGCCTGGCCGACTGAAC | This study |
| folDrev | CCACCCGATTCGATGGTTAC | This study |
| folDfor2 | GAGTCCGACCGGATTCGCGATCAG | This study |
| folDrev2 | CAGCTATATAAAGGAGTTACGGCAAATC | This study |
| MexXfor1 | TGGGAGAAGTTCACGATCGGA | This study |
| MexZrev1 | TTTCCAGGCCCGGTTCGTAGAT | This study |
| MexZrev2 | AGAGCGATTGCAGATAGA | This study |
| Pser-up | CGAGTGGTTTAAGGCAACGGTCTTGA | 4 |
| Pser-down | AGTTCGGCCTGGTGGAACAACTCG | 4 |
In vitro mutagenesis and gene replacement.
The fmt gene was isolated from strain NB52020 by using primers PAfmtF and PAfmtR (Table 2). The product was ligated into pCR2.1-Topo by using a Topo TA cloning kit (Invitrogen, Carlsbad CA) in accordance with the manufacturer's directions. With the pCR2.1-Topo+fmt plasmid used as a template, the fmt gene was amplified using VENT DNA polymerase to create blunt ends, and the product was cloned into SmaI-digested pEX18Tc. The aacC1 gentamicin resistance cartridge was isolated from plasmid pUCGm with SalI and was cloned into the unique XhoI site within fmt in the same orientation as the fmt gene. The in vitro-disrupted fmt gene was introduced into the genome of P. aeruginosa as previously described (12), using the mobilizer strain S17-1. Merodiploid colonies were streaked onto LB plus 5% sucrose plus 256 μg/ml LBM415 to select for loss of the plasmid backbone. Colonies arising on these plates were tested for gentamicin resistance and loss of the plasmid-encoded tetracycline resistance marker. The fmt gene was then amplified with primers PAfmtF1and PAfmtR4 (Table 2) to confirm insertion of aacC1.
Transcriptional profiling.
To test for induction of mexXY by LBM415, duplicate cultures of P. aeruginosa NB52019 were grown in 30 ml Luria broth (1:100 dilution from an overnight culture) in the presence or absence of 12 μg/ml of LBM415 at 37°C with shaking at 180 rpm. Cells were grown to an optical density at 600 nm (OD600) of 0.8 and harvested by centrifugation at 15,000 × g for 10 min. For profiling of fmt or folD mutants, duplicate wild-type and mutant cells were grown in Mueller-Hinton broth to an OD600 of 0.8 and harvested by centrifugation. Total RNA was isolated from the bacteria by using a Gentra RNA isolation kit (Gentra Systems, Minneapolis, MN). Approximately 120 μg of the total RNA isolated from each sample was further purified by use of a Qiagen RNeasy purification kit (Qiagen, Inc., Chatsworth, CA) with an on-column DNase I treatment for 40 min at room temperature. RNA was reverse transcribed, and 1.5 μg of cDNA was fragmented, labeled, and hybridized to Affymetrix P. aeruginosa microarrays in accordance with the manufacturer's protocol (Affymetrix, Santa Clara, CA). Staining and washing were performed with an Affymetrix microfluidic station in accordance with the manufacturer's protocol, and the chips were scanned using an Affymetrix autoloading scanner. Array data were analyzed using Genespring version 6.2 (Invitrogen, Carlsbad, CA).
Construction of mexX::luxCDABE fusions.
Strains CDR3 and CDR5 (Table 1) are wild-type and fmt mutant derivative P. aeruginosa PAO1 (NB52019) strains, respectively, containing a mexX promoter-luxCDABE fusion construct inserted into the attB site on the genome. To construct these reporter strains, primers MexXfor1 and MexZrev2 (Table 2) were used to PCR amplify a 1,316-bp fragment encompassing the mexX promoter region. The resulting product was cloned into pCR2.1-Topo (Invitrogen, Carlsbad, CA) in accordance with the supplied directions and excised by digestion with EcoRI. This fragment was then ligated into EcoRI-cut Mini-CTX-Lux to yield pRC1. Plasmid pRC1 was transformed into E. coli mobilizer strain S17-1, the mexX::luxCDABE fusion was introduced onto the P. aeruginosa chromosomal att site, and the plasmid backbone was excised as previously described (4). Strains CDR4 and CDR6 are efflux pump-deficient (mexAB-oprM) parent and fmt mutant derivative P. aeruginosa strains, respectively, containing a mexX promoter-luxCDABE fusion inserted into the attB site (Table 1). The in vitro construction of this fusion was conducted as described above; however, in this case, the mexX promoter-containing fragment was larger and contained the intact mexZ gene upstream and in the opposite orientation relative to mexX, PCR generated from P. aeruginosa genomic DNA by using primers MexXfor1 and MexZrev1 (Table 2). MexZ is the repressor of mexX expression, and this construct results in two copies of mexZ (the native and one as part of the fusion). The luminescence of fusion strains, as lawns spread on Luria agar plates, was determined qualitatively using a luminescence and fluorescence imaging (IVIS) charge-coupled-device camera system and analyzed with Living Image 2.11 software (Xenogen).
MIC determination.
MIC determinations were carried out using the broth microdilution procedure in accordance with standard CLSI guidelines (10).
RESULTS
Efflux mediated resistance to LBM415 in P. aeruginosa PAO1.
The activity of PDF-Is against gram-negative organisms and the relatively good activity against mutants lacking efflux pumps suggest that broad-spectrum target inhibition is being achieved by certain PDF-Is, including LBM415. In the case of P. aeruginosa, loss of either MexAB-OprM or MexXY-OprM function alone did not substantially increase susceptibility to LBM415 (strains NB52021 and NB52022) (Table 3); however, loss of both simultaneously caused a dramatic increase in susceptibility (NB52020 and NB52023) (Table 3). This indicated that both pumps extrude LBM415 and determine to a large extent the level of intrinsic susceptibility. To examine the role of additional pumps, the PAO1 derivative NB52020, deleted for mexAB-oprM (and therefore lacking both MexAB-OprM and MexXY-OprM functions) was used to select for mutants with reduced susceptibility. Selection experiments were carried out with LBM415 at 32 or 128 μg/ml. Resistant mutants were obtained at a frequency of approximately 10−7. Several of these were tested for reduced sensitivity to trimethoprim and tetracycline, which is indicative of MexCD-OprJ expression (data not shown). To confirm the involvement of MexCD-OprJ, the regulatory gene nfxB, upstream of the mexCD-oprJ operon, was PCR generated from example mutants CDS1 and CDS6 by using primers nfxb1 and nfxb2 (Table 2) and sequenced. Consistent with the resistance profiles, mutations were found in nfxB in both cases (Table 3). This strongly suggests that LBM415 is a substrate for the MexCD-OprJ pump, and the relatively high resistance frequency in the MexAB-OprM-MexXY-OprM-deficient background of strain NB52020 is consistent with loss of function mutations in nfxB.
TABLE 3.
Characteristics of P. aeruginosa LBM415 resistant mutants
| Strain | Description | LBM415 MIC (μg/ml) | Fold increase relative to parent |
|||
|---|---|---|---|---|---|---|
| mexX | mexY | mexZ | PA5471 | |||
| NB52019 | Wild type | 128 | ||||
| NB52021 | mexB | 64 | ||||
| NB52022 | mexX | 128 | ||||
| NB52023 | mexB mexX | 8 | ||||
| NB52020 | mexAB-oprM | 4 | ||||
| CDS1 | nfxB | >128 | ||||
| 14-bp deletion, base 92 | ||||||
| CDS6 | nfxB(A115C, T39P)b | >128 | ||||
| CDS28 | fmt; 16-bp deletion, position 833 | >1,024 | 13.5 | 18 | 8.4 | 13.9 |
| CDS28(pSW1)a | 128 | |||||
| CDS13 | mexAB-oprM fmt(C127T, R43C)b | >128 | 11 | 11.2 | 5.6 | 10.7 |
| CDS13(pSW1)a | 4 | |||||
| CDR1 | fmt::aacC1 | >1,024 | 8.3 | 7.2 | 4.9 | 6.5 |
| CDR2 | mexAB-oprM | >1,024 | 6.2 | 5.6 | 4.6 | 6.7 |
| fmt::aacC1 | ||||||
| CDS30 | folD(G493T, V165R)b | >1,024 | 5.6 | 8.9 | 5.4 | 4.4 |
| CDS30(pRC5)a | 128 | |||||
| CDS22 | mexAB-oprM folD(A424C, T142P)b | >128 | 3.5 | 4.8 | 3.3 | 4.7 |
| CDS22(pRC5)a | 4 | |||||
Plasmid pAK1900 alone did not alter susceptibility.
Nucleotide and amino acid changes are shown in parentheses.
LBM415 induces expression of MexXY, MexZ, and PA5471.
MexXY-OprM is involved in the extrusion of several antibiotics and, notably, is involved in aminoglycoside resistance (43). Unlike for most other RND family pumps, MexXY expression is highly inducible by antibiotics such as tetracycline and tigecycline, which target the ribosome (14, 21, 31, 33). As described above, both MexAB-OprM and MexXY-OprM mediate intrinsic resistance to LBM415. Since LBM415 presumably disturbs protein synthesis, and resistance to LBM415 is partly mediated by MexXY-OprM, we used transcriptional profiling to test whether mexXY expression was induced by LBM415. Total RNA isolated from P. aeruginosa NB52019 grown in the presence of LBM415 had an approximately 11.3-fold increase in mexXY transcript titer (Table 4) in comparison to the level for untreated bacteria, confirming that PDF-Is are strong inducers of this pump, on par with tetracycline and tigecycline (14). Consistent with previous experiments with tetracycline and tigecycline (14), expression of the gene encoding the putative negative repressor of mexXY expression, mexZ (PA2020), was also upregulated (Table 4), further indicating that it is autoregulated, and expression of oprM, carrying the outer membrane channel component as part of the mexAB-oprM operon, was not responsive to the conditions inducing mexXY expression. LBM415 also strongly induced expression of another gene, PA5471, previously shown to be required for upregulation of mexXY during exposure to ribosome inhibitors (33).
TABLE 4.
Induction of mexXY expression in P. aeruginosa NB52019 by LBM415
| GeneID | Gene | Flag (−/+ LBM415)a | Fold change |
|---|---|---|---|
| PA2018 | mexY | A/P | +11.7 |
| PA2019 | mexX | P/P | +11.3 |
| PA2020 | mexZ | P/P | +4.8 |
| PA5471 | P/P | +3.1 | |
| PA0427 | oprM | P/P | 0.78 |
Transcript absence (A) and presence (P), as determined by Affymetrix microarray suite software, are indicated.
Fmt bypass causes resistance to PDF-Is in P. aeruginosa PAO1.
As described in the introduction, the fmt gene in P. aeruginosa can be insertionally inactivated (34). Because of this, we surmised that, primarily, Fmt bypass would emerge in P. aeruginosa under selection by PDF-Is at levels predicted to exceed those accommodated by efflux pumps. To examine this, LBM415 was included at a high 1,024 μg/ml into solid medium to select resistant isolates from PAO1 strain NB52019. At these levels, resistant mutants were obtained again at a frequency of approximately 10−7; however, these grew slowly, even in the absence of selection, requiring extended incubation for at least 48 h to obtain reasonably sized colonies. This slow growth was consistent with the growth defect described for fmt knockouts constructed in P. aeruginosa (34). The fmt genes from several representative isolates were PCR generated and sequenced using the primer pairs PAfmtR/PAfmtF1 and PAfmtR1/PAfmtF4 (Table 2), revealing a preponderance of fmt mutations. One of these mutants, designated CDS28, was selected for further study. Strain CDS28 showed high resistance to LBM415 (MICs of ≥1,024 μg/ml versus 128 μg/ml for wild-type PAO1) (Table 3), as would be expected for Fmt bypass mutants, and resistance extended to several additional related PDF-Is (data not shown). Transforming CDS28 with plasmid pSW1 (harboring fmt) complemented the growth defect (data not shown) and restored PDF-I sensitivity (Table 3), whereas the vector alone did not. The fmt gene, PCR generated in two overlapping fragments from the genome of CDS28(pSW1) by using primers PAfmtR4/PafmtF1 and PafmtR1/PAfmtF4 (Table 2), was sequenced to ensure that fmt had not reverted to the wild type and that no intragenic suppressor mutations which could account for the restoration of growth/sensitivity were occurring. Similar selection experiments with LBM415 and related PDF-Is, using the mexAB-oprM-deficient strain NB52020 (Table 1), yielded additional mutants with very high resistance to LBM415, and one of these, CDS13, was confirmed as an fmt mutant by sequencing of fmt and complementation with pSW1 (Table 3). Again, sequencing of the native copy of fmt was done for CDS13(pSW1) to support complementation rather than selection of an fmt revertant or intragenic suppressor.
Mutations in folD cause resistance to PDF-Is in P. aeruginosa PAO1.
During our screening for the fmt mutants described above, we encountered mutants CDS30 (from NB52019) and CDS22 (from NB52020), which were resistant to PDF-Is and had growth defects but lacked mutations in the fmt structural gene (Table 3). Recently, mutations in folD were reported to confer resistance to PDF-Is in Salmonella enterica (35), and further examination of CDS30 and CDS22 revealed mutations in the folD gene (Table 3). The growth defect of these isolates and sensitivity to PDF-Is were also complemented by plasmid pRC5, containing wild-type folD (Table 3). Similar to previous reports with folD mutants of S. enterica (35), fast-growing revertants appeared readily in the absence of PDF-I selection, and sequencing of the folD gene from an example revertant (CDS30R derived from CDS30) (Table 3) revealed an additional mutation in folD (C to T at position 566), suggesting that intragenic suppression was occurring. As above, retention of the mutations in folD on the chromosome were confirmed for isolates complemented with pRC5 by PCR and sequencing using primers folDfor2 and folDrev2 (Table 2), which lie outside the region cloned into pRC5.
Fmt bypass and folD mutation induce expression of the MexXY efflux pump.
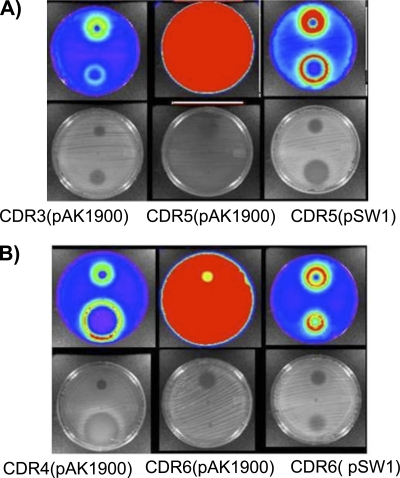
It has been clearly shown that mexXY is induced by traditional ribosome inhibitors, such as tetracycline and aminoglycosides (21, 33). Since the PDF-I LBM415 also induces expression of this pump, we surmised that the growth defect imposed by Fmt bypass, which presumably impacts the ribosome/protein synthesis, would cause constitutive expression of mexXY. Transcriptional profiling revealed that mexXY transcript titer was significantly higher in the fmt mutants CDS28 and CDS13 than in their corresponding parent strains (Table 3), and this level of increase was similar to that observed for drug-treated cells (Table 4). Similar results were obtained using mutants with fmt insertionally inactivated (CDR1 and CDR2) (Table 3), although the increase in mexXY expression was somewhat lower than that observed for the drug-selected mutants. We do not currently understand the reason for the difference in expression levels between selected and engineered fmt mutants. The induction of mexXY by LBM415 in wild-type strain NB52019 and constitutive expression in the Fmt bypass mutants CDS28 and CDS13 were further confirmed using a mexX promoter-driven luxCDABE fusion placed on the genome. By use of a xenogen charge-coupled-device camera, induction of luminescence from cells treated with tetracycline or LBM415 can be clearly seen in the cells surrounding the tetracycline or LBM415 zones of clearance on solid agar plates (Fig. 1, left panels). The fmt mutant strains CDR5 and CDR6, which also contain the mexX::luxCDABE fusion, are constitutively luminescent, confirming constitutive expression from the mexX promoter (Fig. 1, middle panels [entire plate is red]). Constitutive expression in these mutants is reversed, and drug inducibility is restored, upon transformation with plasmid pSW1 containing the fmt gene (Fig. 1, right panels). This confirms the contribution of Fmt bypass, and presumably the resulting growth defect, in creating the intracellular condition leading to mexXY expression. Transcriptional profiling of the folD mutants CDS30 and CDS22 also revealed increased expression of mexXY, mexZ, and PA5471 relative to the levels for their parent strains, confirming that the growth defect arising from this separate but related resistance mechanism also creates an intracellular condition leading to pump expression. The induction was somewhat less for folD mutants and varied noticeably between experiments, suggesting that fast-growing intragenic suppressors might have been emerging during growth of cells for profiling experiments. Alternatively, the folD mutation may reduce formylation rather than eliminating it, as would be predicted for fmt mutants, leading to a less severe defect and consequently less pump induction; however, additional experiments would be required to clearly define this.
FIG. 1.
Impact of LBM415 exposure or Fmt bypass on expression of a mexX::luxCDABE fusion in strain NB52019 (A) and the mexAB-oprM pump-deficient mutant NB52020 (B). Left panels, inducibility of mexX::luxCDABE fusion by tetracycline (top spot) and LBM415 (bottom spot). Center panels, corresponding fmt mutants showing constitutive expression. Right panels, fmt mutants complemented in trans with wild-type fmt, showing reversal of constitutive mexX expression and restoration of drug inducibility. Both luminescent and visible pictures are shown. Note zones of clearance for LBM415 in right panels indicating restoration of sensitivity upon complementation of fmt mutation.
DISCUSSION
The PDF-Is described to date have not shown promising activity against more-recalcitrant gram negative species such as P. aeruginosa. Nonetheless, they do show some activity indicating that broad-spectrum target inhibition is being achieved. We show here that in P. aeruginosa PAO1, intrinsic resistance to the PDF-I LBM415 is largely due to the overlapping effect of MexAB-OprM- and MexXY-OprM-mediated efflux. Additionally, the MexCD-OprJ efflux pump, if expressed due to mutation in nfxB, is likely to also extrude this inhibitor. The contribution of MexXY-OprM to intrinsic resistance suggested that this pump was likely induced by inhibition of peptide deformylase, and this was confirmed for the deformylase inhibitor LBM415 by using transcriptional profiling and mexX::luxCDABE fusion experiments. LBM415 treatment also upregulated expression of mexZ, encoding the repressor of mexXY, and PA5471, now known to be involved in the control of mexXY expression (33). Upregulation of mexXY by LBM415 is intriguing in that PDF-Is are thought to inhibit the removal of formyl groups from nascent proteins emerging from the ribosome, as opposed to more-commonly examined protein synthesis inhibitors, such as tetracycline, tigecycline, and aminoglycosides, which bind ribosomes tightly, specifically interfere with protein synthesis, and/or generate aberrant proteins. However, although PDF-Is have been studied for years, surprisingly little is known specifically about the mechanism of growth inhibition (27), and the interaction of the deformylase protein itself with ribosomes has only recently been investigated in detail (6). Nonetheless, the induction of MexXY via this apparently novel mechanism interfering with protein synthesis supports the notion that MexXY pump levels are controlled in response to intracellular conditions generated during interference with the ribosome (21) or protein synthesis generally.
Beyond efflux, resistance to peptide deformylase can arise from mutations in fmt (Fmt bypass) (27) and mutations in folD (35). These mechanisms serve to eliminate (Fmt bypass) or reduce (folD) the formylation of initiating methionines, abrogating the need for deformylation entirely in those organisms that can still initiate protein synthesis in the absence of formylation. Despite the ability to grow without the formylation-deformylation cycle, these mutants exhibit a growth defect, presumably reflecting impaired protein synthesis. Again, however, it is not clear whether Fmt bypass results in a simple slowing of protein synthesis due to less-efficient initiation or whether there are other effects on the ribosome or protein synthesis per se, leading to, for example, the accumulation of aberrant proteins. Further supporting a physiological role for MexXY, possibly in enhancing survival of cells in the face of protein synthesis inhibition or its downstream effects, these mutants exhibited constitutive expression of mexXY. Moreover, they had increased expression of mexZ, encoding the negatively autoregulated repressor of mexXY expression, and another gene, encoding a protein of unknown function critical to the regulatory circuit controlling mexXY expression, PA5741 (33). This shows that the intracellular condition imposed by mutations in fmt and folD (or treatment with LBM415) recruits the overall regulatory circuitry mediating MexXY expression, presumably including the recently described translational control of PA5471 expression (32). Under these conditions, MexXY-OprM may be continuously extruding at least one of its physiological substrates, and overall, this further supports the idea that MexXY may have a specific physiological function and substrate(s) other than direct antibiotic extrusion. Intriguingly, Fmt bypass itself differs from treatment of cells with LBM415 in that it would be expected to slow initiation by virtue of lacking the formyl group on initiating methionyl-tRNAfmet whereas the compound treatment presumably interferes with synthesis at a different stage (i.e., the removal of the formyl group from nascent proteins emerging from the ribosome). Therefore, these two mechanisms of interference differ from each other as well as from those of tetracyclines and aminoglycosides, again suggesting that mexXY expression is responsive to general interference with ribosomes/protein synthesis. This may also be consistent with a recent report that MeXY-mediated efflux was increased by a transposon insertion in ribosomal protein gene rplY (16). Whether MexXY induction is specific to protein synthesis disruption per se or to another underlying downstream scenario resulting from interference with protein synthesis remains to be determined. As expected, upregulation of mexXY transcription was also observed in fmt or folD mutants generated in a mexAB-oprM-deficient background. The lack of OprM in this case may eliminate MexXY-OprM function, possibly offsetting any physiological benefit of increased MexXY expression in these mutants. However, MexXY may be interacting with a different outer membrane channel in this genetic background and therefore still providing some benefit, or alternatively, MexXY-OprM might have only a marginal impact on survival in the face of general protein synthesis inhibition or may function to extrude natural protein synthesis inhibitors (e.g., aminoglycosides and natural PDF inhibitors, such as actinonin), and the signal to turn on the pump might simply be the inhibition of protein synthesis itself. Suggesting that the MexXY pump itself is not essential for the survival of fmt mutants, we were able to readily select mutants by using plates containing 500 μg/ml LBM415, having the typical slow-growth phenotype observed for Fmt bypass mutants, from a mexXY deletion strain (strain K1525) (19) and another P. aeruginosa derivative with unmarked deletions of mexAB, mexXY, mexCD, and mexEF (strain K2424; K. Poole, unpublished). PCR and sequencing of fmt from two example mutants derived from each pump-deficient strain revealed nucleotide deletions in fmt (data not shown). Therefore, although MexXY is responding to Fmt bypass, it does not appear to specifically provide for the survival of these mutants. More work will hopefully shed light on the natural physiological relationship between MexXY and protein synthesis. Finally, the constitutive expression of MexXY in fmt and folD mutants is reminiscent of clinical isolates that have been described and that express MexXY at high levels but lack mutations in the repressor gene mexZ (43). Whether any of these or other clinical MexXY overexpressors are fmt or folD mutants remains to be seen; however, this provides additional support for the emerging idea that multiple mutations, perhaps affecting protein synthesis, conferring resistance to antibiotics, or resulting from adaptations to the lung environment during chronic infection, may account for the MexXY overexpression phenotype in clinical isolates. Furthermore, at least one clinical isolate of S. aureus with mutations in fmt and with high-level resistance to LBM415 has been identified, indicating that preexisting Fmt bypass mutants may exist in the clinic (46).
Acknowledgments
Plasmid pAK1900 was kindly provided by A. Kropinski (Queen's University), Kingston, ON, Canada. We thank K. Poole (Queen's University) and H. Schweizer (University of Colorado) for bacterial strains and helpful discussion.
D.M.D. is a Novartis presidential fellow.
Footnotes
Published ahead of print on 28 September 2009.
REFERENCES
- 1.Apfel, C., D. W. Banner, D. Bur, M. Dietz, T. Hirata, C. Hubschwerlen, H. Locher, M. G. Page, W. Pirson, G. Rosse, and J. L. Specklin. 2000. Hydroxamic acid derivatives as potent peptide deformylase inhibitors and antibacterial agents. J. Med. Chem. 43:2324-2331. [DOI] [PubMed] [Google Scholar]
- 2.Apfel, C. M., H. Locher, S. Evers, B. Takacs, C. Hubschwerlen, W. Pirson, M. G. Page, and W. Keck. 2001. Peptide deformylase as an antibacterial drug target: target validation and resistance development. Antimicrob. Agents Chemother. 45:1058-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balakrishnan, A., B. Patel, S. A. Sieber, D. Chen, N. Pachikara, G. Zhong, B. F. Cravatt, and H. Fan. 2006. Metalloprotease inhibitors GM6001 and TAPI-0 inhibit the obligate intracellular human pathogen Chlamydia trachomatis by targeting peptide deformylase of the bacterium. J. Biol. Chem. 281:16691-16699. [DOI] [PubMed] [Google Scholar]
- 4.Becher, A., and H. P. Schweizer. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. BioTechniques 29:948-950, 952. [DOI] [PubMed] [Google Scholar]
- 5.Bell, J. M., J. D. Turnidge, M. Inoue, S. Kohno, Y. Hirakata, Y. Ono, and R. N. Jones. 2005. Activity of a peptide deformylase inhibitor LBM415 (NVP PDF-713) tested against recent clinical isolates from Japan. J. Antimicrob. Chemother. 55:276-278. [DOI] [PubMed] [Google Scholar]
- 6.Bingel-Erlenmeyer, R., R. Kohler, G. Kramer, A. Sandikci, S. Antolic, T. Maier, C. Schaffitzel, B. Wiedmann, B. Bukau, and N. Ban. 2008. A peptide deformylase-ribosome complex reveals mechanism of nascent chain processing. Nature 452:108-111. [DOI] [PubMed] [Google Scholar]
- 7.Cao, L., R. Srikumar, and K. Poole. 2004. MexAB-OprM hyperexpression in NalC-type multidrug-resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol. Microbiol. 53:1423-1436. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D., C. Hackbarth, Z. J. Ni, C. Wu, W. Wang, R. Jain, Y. He, K. Bracken, B. Weidmann, D. V. Patel, J. Trias, R. J. White, and Z. Yuan. 2004. Peptide deformylase inhibitors as antibacterial agents: identification of VRC3375, a proline-3-alkylsuccinyl hydroxamate derivative, by using an integrated combinatorial and medicinal chemistry approach. Antimicrob. Agents Chemother. 48:250-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, D. Z., D. V. Patel, C. J. Hackbarth, W. Wang, G. Dreyer, D. C. Young, P. S. Margolis, C. Wu, Z. J. Ni, J. Trias, R. J. White, and Z. Yuan. 2000. Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry 39:1256-1262. [DOI] [PubMed] [Google Scholar]
- 10.CLSI. 2006. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically (M7-A7). CLSI, Wayne, PA.
- 11.Credito, K., G. Lin, L. M. Ednie, and P. C. Appelbaum. 2004. Antistaphylococcal activity of LBM415, a new peptide deformylase inhibitor, compared with those of other agents. Antimicrob. Agents Chemother. 48:4033-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean, C. R., C. V. Franklund, J. D. Retief, M. J. Coyne, Jr., K. Hatano, D. J. Evans, G. B. Pier, and J. B. Goldberg. 1999. Characterization of the serogroup O11 O-antigen locus of Pseudomonas aeruginosa PA103. J. Bacteriol. 181:4275-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean, C. R., S. Narayan, D. M. Daigle, J. L. Dzink-Fox, X. Puyang, K. R. Bracken, K. E. Dean, B. Weidmann, Z. Yuan, R. Jain, and N. S. Ryder. 2005. Role of the AcrAB-TolC efflux pump in determining susceptibility of Haemophilus influenzae to the novel peptide deformylase inhibitor LBM415. Antimicrob. Agents Chemother. 49:3129-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean, C. R., M. A. Visalli, S. J. Projan, P. E. Sum, and P. A. Bradford. 2003. Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 47:972-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ednie, L. M., G. Pankuch, and P. C. Appelbaum. 2004. Antipneumococcal activity of LBM415, a new peptide deformylase inhibitor, compared with those of other agents. Antimicrob. Agents Chemother. 48:4027-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El'Garch, F., K. Jeannot, D. Hocquet, C. Llanes-Barakat, and P. Plesiat. 2007. Cumulative effects of several nonenzymatic mechanisms on the resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 51:1016-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fritsche, T. R., H. S. Sader, R. Cleeland, and R. N. Jones. 2005. Comparative antimicrobial characterization of LBM415 (NVP PDF-713), a new peptide deformylase inhibitor of clinical importance. Antimicrob. Agents Chemother. 49:1468-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hackbarth, C. J., D. Z. Chen, J. G. Lewis, K. Clark, J. B. Mangold, J. A. Cramer, P. S. Margolis, W. Wang, J. Koehn, C. Wu, S. Lopez, G. Withers III, H. Gu, E. Dunn, R. Kulathila, S. H. Pan, W. L. Porter, J. Jacobs, J. Trias, D. V. Patel, B. Weidmann, R. J. White, and Z. Yuan. 2002. N-Alkyl urea hydroxamic acids as a new class of peptide deformylase inhibitors with antibacterial activity. Antimicrob. Agents Chemother. 46:2752-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirakata, Y., R. Srikumar, K. Poole, N. Gotoh, T. Suematsu, S. Kohno, S. Kamihira, R. E. Hancock, and D. P. Speert. 2002. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 196:109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 21.Jeannot, K., M. L. Sobel, F. El Garch, K. Poole, and P. Plesiat. 2005. Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J. Bacteriol. 187:5341-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, R. N., T. R. Fritsche, and H. S. Sader. 2004. Antimicrobial spectrum and activity of NVP PDF-713, a novel peptide deformylase inhibitor, tested against 1,837 recent Gram-positive clinical isolates. Diagn. Microbiol. Infect. Dis. 49:63-65. [DOI] [PubMed] [Google Scholar]
- 23.Jones, R. N., G. J. Moet, H. S. Sader, and T. R. Fritsche. 2004. Potential utility of a peptide deformylase inhibitor (NVP PDF-713) against oxazolidinone-resistant or streptogramin-resistant Gram-positive organism isolates. J. Antimicrob. Chemother. 53:804-807. [DOI] [PubMed] [Google Scholar]
- 24.Jones, R. N., H. S. Sader, and T. R. Fritsche. 2005. Antimicrobial activity of LBM415 (NVP PDF-713) tested against pathogenic Neisseria spp. (Neisseria gonorrhoeae and Neisseria meningitidis). Diagn. Microbiol. Infect. Dis. 51:139-141. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, A., and H. P. Schweizer. 2005. Bacterial resistance to antibiotics: active efflux and reduced uptake. Adv. Drug Deliv. Rev. 57:1486-1513. [DOI] [PubMed] [Google Scholar]
- 26.Leeds, J., C. Dean, B. Favre, J. Dzink-Fox, M. Sachdeva, S. Narayan, J. Medeiros, and N. Ryder. 2004. In vitro selection of decreased susceptibility to the novel peptide deformylase inhibitor LBM415 in three pathogens, abstr. C1-1880, p. 88. Abstr. 44th Annu. Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 27.Leeds, J. A., and C. R. Dean. 2006. Peptide deformylase as an antibacterial target: a critical assessment. Curr. Opin. Pharmacol. 6:445-452. [DOI] [PubMed] [Google Scholar]
- 28.Linares, J. F., J. A. Lopez, E. Camafeita, J. P. Albar, F. Rojo, and J. L. Martinez. 2005. Overexpression of the multidrug efflux pumps MexCD-OprJ and MexEF-OprN is associated with a reduction of type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187:1384-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margolis, P., C. Hackbarth, S. Lopez, M. Maniar, W. Wang, Z. Yuan, R. White, and J. Trias. 2001. Resistance of Streptococcus pneumoniae to deformylase inhibitors is due to mutations in defB. Antimicrob. Agents Chemother. 45:2432-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margolis, P. S., C. J. Hackbarth, D. C. Young, W. Wang, D. Chen, Z. Yuan, R. White, and J. Trias. 2000. Peptide deformylase in Staphylococcus aureus: resistance to inhibition is mediated by mutations in the formyltransferase gene. Antimicrob. Agents Chemother. 44:1825-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-oprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morita, Y., C. Gilmour, D. Metcalf, and K. Poole. 2009. Translational control of the antibiotic inducibility of the PA5471 gene required for mexXY multidrug efflux gene expression in Pseudomonas aeruginosa. J. Bacteriol. 191:4966-4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morita, Y., M. L. Sobel, and K. Poole. 2006. Antibiotic inducibility of the MexXY multidrug efflux system of Pseudomonas aeruginosa: involvement of the antibiotic-inducible PA5471 gene product. J. Bacteriol. 188:1847-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newton, D. T., C. Creuzenet, and D. Mangroo. 1999. Formylation is not essential for initiation of protein synthesis in all eubacteria. J. Biol. Chem. 274:22143-22146. [DOI] [PubMed] [Google Scholar]
- 35.Nilsson, A. I., A. Zorzet, A. Kanth, S. Dahlstrom, O. G. Berg, and D. I. Andersson. 2006. Reducing the fitness cost of antibiotic resistance by amplification of initiator tRNA genes. Proc. Natl. Acad. Sci. USA 103:6976-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poole, K. 2004. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 10:12-26. [DOI] [PubMed] [Google Scholar]
- 37.Poole, K. 2001. Multidrug resistance in Gram-negative bacteria. Curr. Opin. Microbiol. 4:500-508. [DOI] [PubMed] [Google Scholar]
- 38.Poole, K. 2002. Outer membranes and efflux: the path to multidrug resistance in Gram-negative bacteria. Curr. Pharm. Biotechnol. 3:77-98. [DOI] [PubMed] [Google Scholar]
- 39.Poole, K., and R. Srikumar. 2001. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top. Med. Chem. 1:59-71. [DOI] [PubMed] [Google Scholar]
- 40.Sánchez, L., W. Pan, M. Viñas, and H. Nikaido. 1997. The acrAB homolog of Haemophilus influenzae codes for a functional multidrug efflux pump. J. Bacteriol. 179:6855-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweizer, H. D. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-834. [PubMed] [Google Scholar]
- 41b.Simon, R., M. O'Connel, M. Iabes, and A. Puhler. 1986. Plasmid vectors for the genetic analysis and manipulation of Rhizobia and other gram negative bacteria. Methods. Enzymol. 111:640-649. [DOI] [PubMed] [Google Scholar]
- 42.Sobel, M. L., D. Hocquet, L. Cao, P. Plesiat, and K. Poole. 2005. Mutations in PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:1782-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sobel, M. L., G. A. McKay, and K. Poole. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 47:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trepod, C. M., and J. E. Mott. 2004. Identification of the Haemophilus influenzae tolC gene by susceptibility profiles of insertionally inactivated efflux pump mutants. Antimicrob. Agents Chemother. 48:1416-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watters, A. A., R. N. Jones, J. A. Leeds, G. Denys, H. S. Sader, and T. R. Fritsche. 2006. Antimicrobial activity of a novel peptide deformylase inhibitor, LBM415, tested against respiratory tract and cutaneous infection pathogens: a global surveillance report (2003-2004). J. Antimicrob. Chemother. 57:914-923. [DOI] [PubMed] [Google Scholar]