Abstract
Hygromycin A (HA) is an aminocyclitol antibiotic produced and excreted by Streptomyces hygroscopicus. Deletion of hyg26 from the hygromycin A biosynthetic gene cluster has previously been shown to result in a mutant that produces 5″-dihydrohygromycin A (DHHA). We report herein on the purification and characterization of Hyg26 expressed in Escherichia coli. The enzyme catalyzes an NAD(H)-dependent reversible interconversion of HA and DHHA, supporting the role of the reduced HA as the penultimate biosynthetic pathway intermediate and not a shunt product. The equilibrium for the Hyg26-catalyzed reaction heavily favors the DHHA intermediate. The high-titer production of the HA product by S. hygroscopicus must be dependent upon a subsequent energetically favorable enzyme-catalyzed process, such as the selective and efficient export of HA. hyg19 encodes a putative proton gradient-dependent transporter, and a mutant lacking this gene was observed to produce less HA and to produce the DHHA intermediate. The DHHA produced by either the Δhyg19 or the Δhyg26 mutant had slightly reduced activity against E. coli and reduced protein synthesis-inhibitory activity in vitro. The data indicate that Hyg26 and Hyg19 have evolved to produce and export the final potent HA product in a coordinated fashion.
Antibiotic biosynthetic gene clusters contain genes required for the process of assembling the natural product and for the generation of specific biosynthetic precursors that are not readily available from primary metabolism (16). In addition, they often contain genes that are responsible for self-resistance, thereby protecting the organism from its own natural product, and regulatory genes that control the expression of biosynthetic genes. Although these gene classifications are useful, they are often incomplete in their descriptions. In fact, a complex interplay between biosynthesis, resistance, and regulation has been observed for many natural product biosynthetic processes, and it is becoming increasingly evident that some of these gene products perform multiple functions.
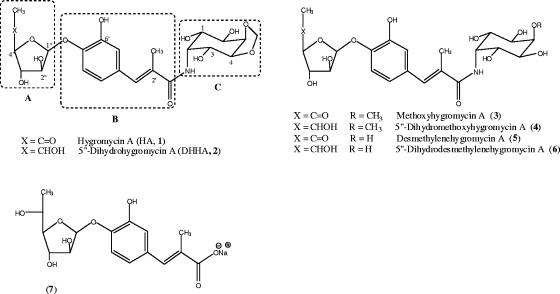
Hygromycin A (HA; compound 1) (Fig. 1) is an aminocyclitol antibiotic that acts by inhibiting the peptidyltransferase reaction of protein synthesis in gram-positive and gram-negative bacteria (7, 15, 25). Biosynthetic studies have shown that HA is assembled from three independently synthesized subunits, 5-dehydro-α-L-fucofuranose (subunit A), (E)-3-(3,4-dihydroxyphenyl)-2-methylacrylic acid (subunit B), and the aminocyclitol, 2l-2-amino-2-deoxy-4,5-O-methylene-neo-inositol (subunit C) (6). The corresponding biosynthetic gene cluster has been cloned and sequenced from Streptomyces hygroscopicus NRRL 2388, and putative functions have been assigned to the 29 open reading frames (22). The proposed role of hyg26 as a short-chain dehydrogenase gene has also been verified at the genetic level (22).
FIG. 1.
Structures of HA and related compounds.
Disruption of hyg26 in S. hygroscopicus abolished the production of HA and methoxyhygromycin A (compound 3) and, instead, yielded their reduced analogs, 5″-dihydrohygromycin A (DHHA; compound 2) and 5″-dihydromethoxyhygromycin A (compound 4), respectively, and small amounts of (E)-3-(3-hydroxy-4-O-α-fucofuranosylphenyl)-2-methylacrylic acid (compound 7). Compound 7 is DHHA without the C subunit (see Fig. 1 for the structures). While these results showed that Hyg26 is required for the dehydrogenation of the C-5″ OH on the fucofuranose moiety, the timing of this reaction in the biosynthetic sequence and, subsequently, the question as to whether DHHA is a shunt metabolite or a precursor of the final product (HA) remain unresolved.
Analysis of the HA biosynthetic gene cluster has also revealed multiple genes likely to be involved in antibiotic resistance. Of these, the function of hyg21 has been biochemically determined (4). hyg21 encodes a phosphotransferase that inactivates HA by phosphorylating the C-2″ OH on the fucofuranose. A Δhyg21 strain has been shown to retain resistance to HA, indicating the presence of other mechanisms of self-resistance. Antibiotic transporters/efflux pumps are additional mechanisms of self-resistance in antibiotic producers and likely operate in S. hygroscopicus also, as all of the HA produced is found outside the cell. The HA biosynthetic gene cluster contains two putative HA transporter genes, hyg19 and hyg28. The hyg19 gene shows similarity to proton gradient-dependent transporter genes. The hyg28 gene encodes a putative type II ATP-binding cassette transporter. The role of these gene products in exporting HA or pathway intermediates and in contributing to self-resistance has not been determined.
In the work described here, we show that a recombinant Hyg26 catalyzes the reversible interconversion of DHHA and HA and that the equilibrium for this reaction lies heavily toward the biologically less active, reduced HA product. The putative proton gradient-dependent transporter encoded by hyg19 is observed to be required for the high-level production of HA and is hypothesized to operate by the selective and efficient exportation of HA. A hyg19 deletion mutant did not show increased sensitivity to HA, but a mutant in which both hyg19 and hyg21 were deleted had markedly increased HA sensitivity. This work has demonstrated that DHHA is likely the final biosynthetic pathway intermediate and that Hyg19 has a role in both self-resistance and biosynthesis, ensuring that the final and most biologically active HA product is generated.
MATERIALS AND METHODS
Chemicals, bacterial strains, and growth conditions.
All antibiotics and chemicals were purchased from Sigma Aldrich unless otherwise stated. PCR primers were obtained from Integrated DNA Technologies. The enzymes used for DNA manipulations were obtained from New England Biolabs. HA was kindly provided by Pfizer Inc. DHHA, 5″-dihydromethoxyhygromycin A (compound 4), and (E)-3-(3-hydroxy-4-O-α-fucofuranosylphenyl)-2-methylacrylic acid (compound 7) were purified from the fermentation broth of mutant strain S. hygroscopicus SCH30 by a semipreparative high-performance liquid chromatography (HPLC) method, as described earlier (22). A Δhyg7 strain of S. hygroscopicus was used as the source for methoxyhygromycin A (compound 3). A Δhyg6 disruption strain was used as the source for desmethylenehygromycin A (compound 5) and 5″-dihydrodesmethylenehygromycin A (compound 6) (submitted for publication). The S. hygroscopicus strains were propagated by using the media and culture conditions described earlier (6, 22). The Escherichia coli strains were cultured by standard protocols (28). E. coli ΔtolC was procured from the E. coli Genetic Stock Center at Yale University.
Nucleotide and protein sequence analysis.
Searches for sequences with homology to the sequences of the hyg19, hyg26, and hyg28 genes were carried out with the BLAST suite of programs at the National Center for Biotechnology Information website (1). Protein sequence alignments were obtained by using the CLUSTALX program at the European Bioinformatics Institute website (14). Protein motif searches were carried out by using the PROSITE database at the Expasy proteomics server (9). Hydropathy analysis for Hyg19 was based on the amino acid hydropathy values of Kyte and Doolittle (13) and carried out at the Transporter Classification Database server (26, 27). The MEMSAT3 program was also used for prediction of the secondary structure and topology of Hyg19 (10).
Cloning, overexpression, and purification of recombinant Hyg26.
The 816-bp hyg26 gene was amplified from cosmid 17E3 (22). Primers were designed to introduce an NdeI restriction site at the initiation codon and a BamHI restriction site 3′ to the termination codon (Table 1). Amplification was performed by using the following temperature program: initial denaturation at 95°C for 5 min, 25 cycles of amplification (45 s of denaturation at 95°C, 45 s of primer annealing at 56°C, 1 min of extension at 72°C), and a final 7-min extension at 72°C. The gene was subsequently cloned into the NdeI and BamHI restriction sites of the pET15b vector (Novagen). The resulting pET15b-hyg26 construct was introduced into the E. coli BL21-CodonPlus (DE3) expression host (Stratagene) by transformation. For protein expression, cells were grown at 37°C in LB medium containing 100 μg/ml ampicillin and 50 μg/ml chloramphenicol. When an A600 of 0.6 was reached, the culture flask was brought to room temperature (∼23°C) and expression was induced by adding 0.5 mM isopropyl thio-β-d-galactoside. After a further 6 h of incubation, cells were harvested by centrifugation and stored at −80°C.
TABLE 1.
Primers used in this study
| Primer purpose and primer | Sequencea |
|---|---|
| Disruption primers for hyg19 gene | |
| Forward | GATCCGACCCGTGCCGGGCGGAAGGGAGCACCAGTGATGATTCCGGGGATCCGTCGACC |
| Reverse | GGTGATGAGCGGGGCGCTGAGCCAGAGGAACATCACGACTGTAGGCTGGAGCTGCTTC |
| Disruption primers for hyg28 gene | |
| Forward | CTGATGACTGCCACTCTCGTCGCCAAGGATCTGGCCGCCATTCCGGGGATCCGTCGACC |
| Reverse | CTCGGTGACCCGGCCGTCCGCCACCTCCAGGCGGCGGGTTGTAGGCTGGAGCTGCTTC |
| Disruption primers for hyg19, hyg20, and hyg21 genes | |
| Forward | GATCCGACCCGTGCCGGGCGGAAGGGAGCACCAGTGATGATTCCGGGGATCCGTCGACC |
| Reverse | ACCGTTGACCGAGAATGGGTAAAGGAGCAGAAAACAATGTGTAGGCTGGAGCTGCTTC |
| Expression primers for hyg26 gene | |
| Forward | CATATGAGTGGACTGATGCGGGAC |
| Reverse | GGATCCTCAGAATTCGCTGACGC |
The 39-nucleotide homologous region flanking the targeted gene is indicated in boldface. The italicized primer region is homologous to pIJ773. The NdeI and BamHI sites in hyg26 expression primers are also indicated in boldface.
The N-terminal His-tagged Hyg26 was purified under native conditions according to Qiagen's Ni-nitrilotriacetic acid protocol. The cells were resuspended in BugBuster master mixture protein extraction reagent (Novagen) and lysed by incubation at room temperature for 30 min. The lysate was cleared by centrifugation; and the supernatant was loaded onto Ni-nitrilotriacetic acid agarose preequilibrated with buffer containing 50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole (pH 8.0). The column was washed with buffer containing 50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole (pH 8.0). The protein was eluted with the same buffer containing a higher concentration of imidazole (250 mM). Fractions containing Hyg26 were pooled, dialyzed overnight against a buffer (50 mM Tris-HCl, 20% glycerol, 5 mM 2-mercaptoethanol at pH 7.5), and stored at −80°C until further use.
Determination of molecular mass of Hyg26.
The molecular mass of purified His-tagged Hyg26 was determined by size-exclusion chromatography with a Phenomenex BioSep-SEC-S4000 size-exclusion column fitted to an Agilent 1100 series HPLC system. The column was equilibrated and eluted with 20 mM sodium phosphate (pH 7.2) at a flow rate of 0.5 ml/min. Carbonic anhydrase (25 kDa), bovine serum albumin (66 kDa), β-amylase (200 kDa), apoferritin (443 kDa), and thyroglobulin (669 kDa) were used as standards.
Hyg26 enzyme assays and kinetic analyses.
Initial enzyme assays were performed at 30°C with 4 μg of purified enzyme in a 100-μl reaction mixture of 50 mM Tris-HCl (pH 7.5) and 2 mM Tris(2-carboxyethyl) phosphine (TCEP). The dehydrogenase activity was studied by incubating Hyg26 with 100 μM DHHA, compound 4, compound 6, or compound 7 and 1,000 μM NAD or NADP. For lactate dehydrogenase-coupled assays, 1,000 μM sodium pyruvate and 5 U of lactate dehydrogenase were added. The reductase activity was detected using 100 μM HA, compound 3, or compound 5 and 1,000 μM NADH or NADPH. After incubation for 2 h at 30°C, the sample was loaded onto a reverse-phase HPLC column (Agilent Eclipse XDB-C18) to detect the oxidized products (with longer retention times) in the dehydrogenase assay or the reduced products (with shorter retention times) in the reductase assay. Samples were separated at a flow rate of 1 ml/min with a methanol-water gradient, as described earlier (4). The optimal temperature and pH for Hyg26 reductase activity were determined with 100 μM HA and 1,000 μM NADH in a 15-min assay by the use of HPLC analysis and the peak area to quantitate the production of DHHA. The temperatures studied were 23°C (room temperature), 30°C, 37°C, and 42°C. For the pH studies, the following buffers were used: 50 mM potassium acetate, pH 4.0 and 5.0; 50 mM potassium phosphate, pH 6.0 and 7.0; and 50 mM Tris-HCl, pH 7.5, 8.0, and 9.0. The metal requirement was probed by conducting the assay in the presence of 5 mM and 10 mM EDTA. Kinetic analyses of reductase activity were carried out at 37°C for 15 min with 800 ng Hyg26 in 100 μl assay buffer comprised of 50 mM phosphate (pH 6) and 2 mM TCEP. The NADH concentration was fixed at 60 mM, while the antibiotic concentration was varied from 2 mM to 60 mM. For kinetic analyses of NADH, the HA concentration was fixed at 20 mM, while the NADH concentration was varied from 0.25 mM to 10 mM. Each assay was performed in duplicate, and the average values of the two measurements were plotted in the GraFit 4 program to determine the kinetic parameters.
Gene disruption experiments.
The hyg19 and hyg28 genes and a segment of the HA biosynthetic gene cluster comprised of the hyg19, hyg20, and hyg21 genes were individually replaced by the apramycin resistance cassette by the PCR targeted Streptomyces gene replacement method (8). The primers used to amplify the disruption cassette from plasmid pIJ773 are listed in Table 1. The primers used for hyg28 disruption were designed such that the 68-nucleotide overlap between the 3′ end of hyg28 and the 5′ end of the downstream hyg29 gene was unaffected. The hyg19 and the hyg19, hyg20, and hyg21 disruptions were carried out in cosmid 17E3, while the hyg28 gene disruption was done in cosmid 15A10 (22). The recombinant cosmids were introduced into a wild-type S. hygroscopicus strain by conjugation and selected for apramycin resistance. The genotype of the mutant strains was confirmed by PCR amplification of the apramycin cassette from chromosomal DNA and by verifying the sequence of the PCR product. Automated DNA sequencing was done at the Molecular Microbiology and Immunology Research Core Facility at Oregon Health and Science University. The DNA sequence data were analyzed by using Accelrys DS Gene software.
Analysis of antibiotic production in S. hygroscopicus strains.
Fermentation broths of S. hygroscopicus disruption strains were analyzed for antibiotic production by HPLC and liquid chromatography-mass spectrometry methods on a Bruker Daltonics MicroTOF-Q instrument fitted with an Agilent 1100 series HPLC system. Cell extracts were generated by methanol lysis of the mycelia and were examined by HPLC (4). The production yields of HA and related metabolites were estimated from their corresponding peak areas in the chromatograms by using a standard curve of HA as a reference.
Susceptibilities of S. hygroscopicus strains to HA.
The HA susceptibilities of S. hygroscopicus wild-type and mutant strains were determined by the agar plate dilution method, as described previously (4). Briefly, spores were plated on yeast extract malt extract agar plates containing various amounts of HA, incubated at 30°C for 48 h, and scored for growth. The MIC was defined as the lowest concentration of HA that prevented the visible growth of 95% or more of the CFU on the agar plate.
Bioactivity of DHHA and other dihydro analogs.
The ΔtolC E. coli strain was used as the test organism to determine the antibiotic activities of DHHA and compounds 4, 6, and 7. The strain was grown in 200 μl LB medium at 37°C for 2 h in the presence of different concentrations of antibiotic, and the A600 was measured. The MIC was defined as the lowest concentration of the antibiotic at which 95% of bacterial growth was inhibited compared to the amount of growth of a control E. coli culture grown in the absence of any antibiotic.
Coupled transcription-translation assay.
All coupled transcription-translation experiments were performed with an E. coli lysate-based system in the presence and absence of antibiotics, as described previously (5, 30). The reaction mixtures were transferred into 96-well microtiter plates, and the fluorescence of the green fluorescent protein (GFP) was measured with a Typhoon 9400 scanner (Amersham Bioscience) and a Typhoon blue laser module (Amersham Bioscience). The images were then quantified with the ImageQuantTL program (GE Healthcare) and are represented graphically by using the SigmaPlot program (Systat Software, Inc.).
RESULTS
Enzymatic characterization of a short-chain dehydrogenase encoded by hyg26.
The hyg26 gene from the HA biosynthetic gene cluster shows homology to short-chain dehydrogenase/reductase genes. A previous gene disruption study demonstrated that a Δhyg26 mutant accumulates DHHA and that Hyg26 is likely responsible for dehydrogenation of the C-5 OH on the l-fucofuranose moiety (22). It was suggested that this reaction may precede the glycosyltransferase reaction that connects the A subunit of HA to its B subunit. Alternatively, Hyg26 may act at a much later step in the biosynthetic pathway and may be responsible for converting DHHA to HA (Fig. 1). In such a case, DHHA could represent the penultimate pathway intermediate and not a shunt metabolite, and Hyg26 would be responsible for catalyzing the last step of HA biosynthesis. To probe this possibility, we generated and purified recombinant Hyg26 and determined its ability to interconvert HA and DHHA. A pET15b-hyg26 expression vector was constructed and the N-terminal hexahistidine-tagged Hyg26 was purified by Ni2+ affinity chromatography. The purified protein showed the expected size of ∼30 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and an apparent molecular mass of 64,000 Da by size-exclusion chromatography, suggesting that recombinant Hyg26 is a dimeric protein.
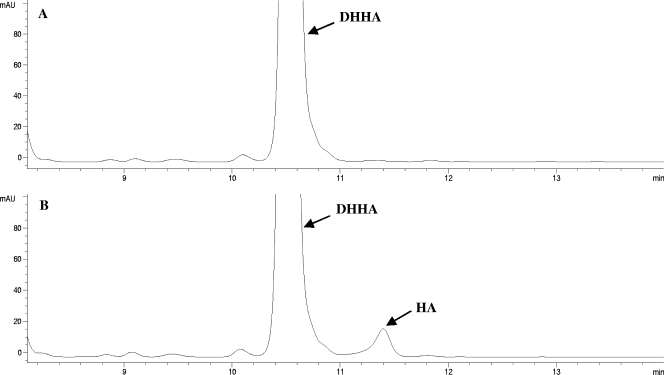
The dehydrogenase activity was assayed by incubation of DHHA, compound 4, or compound 6 in the presence of NAD+ or NADP+ as a cofactor. In assays with NAD+, reverse-phase HPLC analysis of each reaction mixture showed the appearance of a new peak with a retention time longer than that of the substrate (see Fig. 2 for the results of the assay with DHHA). This peak was not observed in a control assay without Hyg26 or in the assay with NADP+. The retention time of the new peak, the results of coinjection experiments with the appropriate standards, and the results of mass spectrometric analyses were all consistent with the formation in the assays of the corresponding C-5″-oxidized analog (HA, compound 3, and compound 5), products of an Hyg26-dependent dehydrogenation. In each case, there was only a low level of product formation (1.2 to 1.5 μM from 100 μM of substrate after 2 h at 30°C). Attempts to increase the conversion level by the use of increased incubation times or by the performance of coupled enzymatic assays that would regenerate NAD+ (by using pyruvate and lactate dehydrogenase) were unsuccessful.
FIG. 2.
Reverse-phase HPLC analysis of DHHA (compound 2) dehydrogenation by Hyg26. (A) Control reaction of DHHA and NAD+ without Hyg26; (B) reaction of DHHA and NAD+ after incubation with Hyg26 showing the formation of HA. mAU, milli-absorbance units.
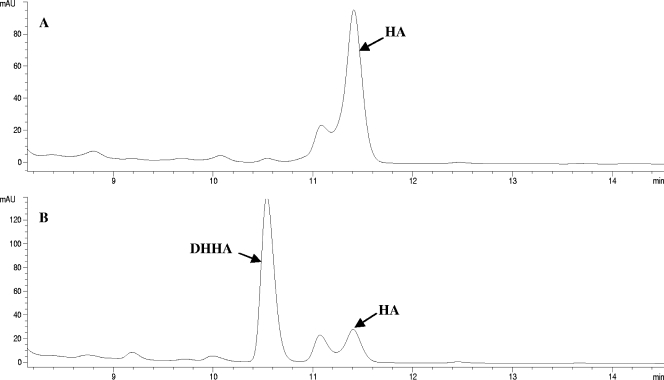
The ability of Hyg26 to catalyze the reverse reaction (reductase activity) was also investigated. Assays were carried out with HA, compound 3, or compound 5 in the presence of NADH or NADPH, followed by HPLC analyses. Compared to the results for a control without Hyg26, each of the NADH assays showed a significant decrease in the amount of substrate and the appearance of a new peak with a retention time shorter than that of the substrate (see Fig. 3 for the results of the assay with HA). This peak was not observed in the reaction with NADPH. Mass spectrometric analyses of the NADH reaction mixtures showed that the compounds under the new peaks had an increase in mass by 2 atomic mass units with respect to the masses of the substrates, consistent with that expected for the C-5″-reduced analogs. Coinjections of the reaction mixtures with authentic materials provided additional support for the characterization of the reaction products. These observations confirm that Hyg26 is an NAD+/NADH-dependent oxidoreductase with broad substrate specificity. These data also demonstrated that under the given assay conditions, the enzyme catalyzed the reverse reaction more efficiently than the forward biosynthetic reaction, suggesting that the equilibrium position favors the DHHA product.
FIG. 3.
Reverse-phase HPLC analysis of hygromycin A (HA) reduction by Hyg26. (A) Control reaction of HA and NADH without Hyg26; (B) reaction of HA and NADH after incubation with Hyg26, showing the formation of DHHA. An additional peak (11.1 min) eluting earlier than HA is the C-4″ epimer of HA and not a substrate for Hyg26. mAU, milli-absorbance units.
The optimal pH for catalysis of the reduction reaction was determined over a pH range of 4.0 to 9.0 and was found to be 6.0. Among the temperatures from 23°C to 42°C that were tested, the optimal temperature was 37°C, although no significant difference was observed between the amounts of product formed at 30°C and 37°C. The metal-chelating reagent EDTA did not reduce the enzyme activity, suggesting that, like other short-chain dehydrogenases, Hyg26 is metal ion independent (24). Analysis of the steady-state kinetic parameters for the reduction of HA, compound 3, and compound 5 showed that Hyg26 has a very low affinity for them, as indicated by the high Km values (in the millimolar range; Table 2). The catalytic efficiency, as indicated by kcat/Km, was found to be comparable for all three substrates. The Km for NADH was determined to be 400 ± 34 μM.
TABLE 2.
Kinetic parameters for Hyg26 reductase activity
| Substrate | kcat (s−1) | Km (mM) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| HA | 5.1 | 5.8 ± 0.8 | 1.0 × 103 |
| Methoxyhygromycin A (compound 3) | 15.5 | 19.8 ± 1.9 | 0.9 × 103 |
| Desmethylenehygromycin A (compound 5) | 31.7 | 63.6 ± 24.4 | 0.9 × 103 |
Predicted roles of hyg19 and hyg28 genes in the HA biosynthetic gene cluster.
The biosynthetic gene cluster of hygromycin A has two genes, hyg19 and hyg28, that encode putative transporter proteins. The predicted translation product of hyg19 has 416 amino acids with a theoretical molecular mass of 43.8 kDa. A search of the National Center for Biotechnology Information protein sequence database by use of the deduced Hyg19 sequence returned several transmembrane proteins belonging to the major facilitator superfamily (MFS). The highest level of sequence similarity (66%) was observed with a presumptive transmembrane protein, Ata9, from the biosynthetic gene cluster of antibiotic A201A from Saccharothrix mutabilis subsp. capreolus. The structure of HA resembles that of A201A, particularly in the A and B subunits, and many genes with similar sequences and possibly similar functions have been identified in the biosynthetic gene clusters of these two antibiotics (22). The N-terminal region of Hyg19 specifically shows a high degree of similarity to the MFS proteins involved in drug efflux, such as the TetV tetracycline resistance determinant from Mycobacterium smegmatis, the MefA macrolide efflux protein from Streptococcus pneumoniae, and the chloramphenicol resistance protein from Streptomyces lividans. Hydropathy analysis and transmembrane topology prediction suggest that Hyg19 has 12 transmembrane helices with cytoplasmic N and C termini and, therefore, is likely a member of the proton gradient-dependent DHA12 family of MFS transporters. Five sequence motifs have been identified in the DHA12 efflux proteins (23). Hyg19 does not show full conservation of all the motifs but shows a partially conserved motif C (gxxxGPxxGGrl) in transmembrane 5 (positions 163 to 181), and a partially conserved motif G (GxxxGPL) in transmembrane 11 (positions 373 to 392) can clearly be distinguished. On the basis of these observations, Hyg19 is hypothesized to be a proton gradient-dependent efflux protein that confers resistance to S. hygroscopicus by exporting HA out of the cell.
The deduced translation product of hyg28, with 570 residues and a theoretical molecular mass of 61.5 kDa, shows homology to several ATP-binding cassette (ABC)-type transporter proteins, including Ard1 from the A201A gene cluster that is known to confer resistance to A201A in S. lividans (2). Two ABC domains containing the Walker A and Walker B motifs characteristic of ATPases are present in Hyg28. The Walker A motif ([AG]-x4-G-K-[ST]) is a glycine-rich loop that binds to the phosphates of ATP or GTP (32). The Walker B motif (hhhhDEPT, where h indicates a hydrophobic residue) coordinates a magnesium ion (32). Therefore, the hyg28 gene is predicted to encode a type II ABC transporter protein, which, in association with an unidentified membrane component, constitutes the second transporter system for HA efflux and subsequent self-resistance.
Effect of hyg19 and hyg28 gene disruptions on HA biosynthesis and efflux.
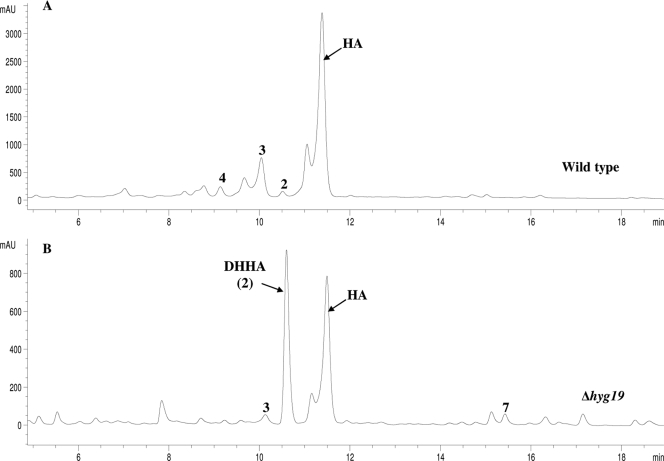
In order to determine whether Hyg19 and Hyg28 are involved in antibiotic biosynthesis and efflux, the corresponding genes were individually replaced with the apramycin resistance cassette in the S. hygroscopicus wild type. The resulting mutant strains, Δhyg19 and Δhyg28, were cultured; the mycelia were removed by centrifugation; and the filtered supernatants were analyzed by HPLC and liquid chromatography-mass spectrometry for antibiotic production. Analysis of the fermentation broth of the Δhyg28 mutant revealed that the antibiotic production profile was unaffected by the disruption of hyg28. The strain produced and excreted nearly the same amount (∼1.5 g/liter) of HA as the wild type (data not shown). This clearly indicates that Hyg28 is not essential for antibiotic biosynthesis and efflux. On the other hand, the disruption of hyg19 caused the level of HA production to drop to ∼509 mg/liter, a threefold decrease compared to the HA yield from the wild type (Fig. 4). The levels of methoxyhygromycin A (compound 3) and 5"-dihydromethoxyhygromycin A (compound 4) (Fig. 1) also appeared to be similarly reduced. Interestingly, the yield of DHHA in Δhyg19 increased 10-fold to ∼380 mg/liter from the ∼37 mg/liter seen for the wild type. A slight increase in the amount of compound 7, detected only at trace levels in the wild type, was also observed in Δhyg19. It was speculated that the reduced amount of HA in the fermentation broth of Δhyg19 could be a consequence of intracellular HA accumulation due to impaired transmembrane transport. To test this possibility, the mycelium of Δhyg19 was lysed by methanol treatment and the lyaste was analyzed by HPLC. The cell lysates of the wild type and Δhyg28 were also examined alongside in the same way. However, neither HA nor any known related compounds, including any phosphorylated forms, were detected in the cell lysates of either the Δhyg19 or the Δhyg28 strain. The data demonstrate that all detectable HA and related products are located outside the cell even in the absence of Hyg19. Nonetheless, the efficient and selective biosynthesis of HA is dependent upon the hyg19 gene product.
FIG. 4.
Reverse-phase HPLC analysis of fermentation broth of the wild-type (A) and Δhyg19 (B) strains. Reduced HA production and accumulation of DHHA were observed in the Δhyg19 strain. mAU, milli-absorbance units.
Contribution of hyg19 and hyg28 gene products to self-resistance of S. hygroscopicus.
The wild-type and mutant strains were assessed for their susceptibilities to HA by growing their spores on agar plates with antibiotics at different concentrations. The MIC of HA for the wild type was found to be 400 μg/ml. The mutants also showed high levels of resistance, with MICs of 400 μg/ml and 300 μg/ml for Δhyg19 and Δhyg28, respectively. These data indicate that the self-resistance of S. hygroscopicus is not entirely conferred by hyg19 or hyg28. Another mutant strain was generated by disrupting hyg19 together with the hyg20 and hyg21 genes present immediately downstream of it (Fig. 5). The hyg20 gene encodes a putative transglucosylase/mutase for the generation of the A subunit, and its disruption has been observed to abolish antibiotic production completely (unpublished data). hyg21 encodes an antibiotic-modifying O-phosphotransferase with a role in self-resistance, and loss of this gene results in reduced HA production (4). The Δ(hyg19-hyg20-hyg21) strain did not produce HA and showed significantly increased susceptibility (MIC, 75 μg/ml). A Δhyg20 strain had a MIC of 300 μg/ml, which was not significantly lower than that of the wild type. The MIC of S. lividans for HA was determined to be 20 μg/ml. These data suggest that the increased susceptibility of Δ(hyg19-hyg20-hyg21) is due to the loss of hyg19 and hyg21, implying a synergistic role of these two genes in self-resistance.
FIG. 5.
Schematic representation of replacement of the hyg19, hyg20, and hyg21 genes with the apramycin resistance cassette. WT, wild type.
Biological activity of C-5″-dihydro analogs.
DHHA and compounds 4 and 6 are the C-5″-dihydro analogs of HA, methoxyhygromycin A (compound 3), and desmethylenehygromycin A (compound 5), respectively (Fig. 1). These three C-5″-dihydro compounds and compound 7 were studied for their ability both to inhibit the growth of ΔtolC E. coli and to inhibit protein synthesis in an E. coli in vitro-coupled transcription-translation system (5, 30). The MICs of DHHA, compound 2, and compound 4 were determined to be 20 μg/ml, 150 μg/ml, and 150 μg/ml, respectively (Table 3). These values are similar to the MICs for the corresponding 5"-oxidized analogs, albeit for the most active pair, HA appeared to be slightly more active (10 μg/ml) than DHHA. The DHHA analog lacking the C subunit (compound 7) was inactive against E. coli ΔtolC even at 250 μg/ml. The data demonstrate that for in vivo activity, the presence of the methylene bridge on the C subunit is of greater importance than the C-5″ oxidation state of the A subunit. HA was also the most active compound in vitro, having a 50% inhibitory concentration (IC50) and an IC90 of 0.18 μM and 0.25 μM, respectively, whereas compound 7 was inactive (consistent with the MICs), exhibiting no effect on translation at 75 μM (Fig. 6A and B and Table 3). On the basis of the IC50s, compounds 3 and 5 were as effective as DHHA and their activities were similar to the activity of HA. In contrast, the reduced forms of these compounds, compounds 4 and 6, had significantly lower (8 to 15 times) potency (Fig. 6C and Table 3). The data demonstrate that for in vitro activity, the presence of the methylene bridge on the C subunit is of lesser importance than the C-5″ oxidation state of the A subunit.
TABLE 3.
Comparison of MIC and IC50/IC90 data for HA and DHHA analogs
| Compound | MIC (μg/ml) | IC50 (μM) | IC90 (μM) |
|---|---|---|---|
| HA | 10 | 0.18 | 0.25 |
| DHHA | 20 | 0.3 | 1.5 |
| 3 | 150 | 0.5 | 2 |
| 4 | 150 | 7.4 | 50 |
| 5 | 150 | 0.32 | 1 |
| 6 | 150 | 2.7 | 14.1 |
FIG. 6.
Effects of HA and derivatives on in vitro transcription-translation. (A) Detection of template-dependent synthesis of GFP by using fluorescence and native polyacrylamide gel electrophoresis in the presence of template with increasing concentrations (μM) of the antibiotic HA, DHHA, and compound 7 on GFP synthesis; (B) quantitation of effects of HA, DHHA, and compound 7 on GFP synthesis; (C) quantitation of effects of HA and compounds 3, 4, 5, and 6 on GFP synthesis. The experiments whose results are shown in panels B and C were performed in triplicate. GFP fluorescence is given as a percentage, where 100% is defined as the fluorescence detected in the absence of the antibiotic. Rel., relative.
DISCUSSION
hyg26 in the HA biosynthetic gene cluster encodes a protein that is a member of the short-chain dehydrogenase family. The requirement for this gene to obtain HA and not DHHA has previously been demonstrated by a gene disruption study (22). Most short-chain dehydrogenase enzymes are dimers or tetramers and typically do not need a divalent metal ion for activity, unlike the classical metal-dependent medium-chain alcohol dehydrogenases (11, 18). The results of analyses of recombinant Hyg26 in the present study are consistent with it being a dimeric, metal ion-independent protein. The enzyme showed an absolute preference for NAD(H) and did not utilize NADP(H). The coenzyme-binding site of short-chain dehydrogenases preferring NAD(H) has an acidic residue (Asp or Glu) that forms hydrogen bonds with the hydroxyl groups of the adenine ribose, whereas the binding site for NADP(H) has two basic residues (Arg or Lys) that bind to the phosphate (12, 31). The specificity of Hyg26 for NAD(H) is consistent with the presence of an Asp residue at position 38 in the putative coenzyme-binding region within residues 12 to 38 (VTGAARGQGREHAVRMAGEGADVIAID; conserved residues are indicated in boldface). Both the ability of Hyg26 to catalyze the interconversion of DHHA and HA and the production of DHHA by a Δhyg26 mutant are consistent with this being the last step of HA biosynthesis. A previous proposal that Hyg26 may catalyze the conversion of l-fucofuranose to 5-dehydro-α-l-fucofuranose during the formation of just the A subunit appears to be less likely.
Additional support for the hypothesis that DHHA is the penultimate intermediate in the HA pathway comes from the analyses of the Δhyg19 mutant. The equilibrium for the Hyg26-catalyzed process was observed to heavily favor DHHA, even though HA is the primary fermentation product. Loss of the subsequent energetically favorable step, which would shift the equilibrium toward HA, would be predicted to result in the formation of decreased of levels of HA and the formation of increased levels of DHHA, as seen for the Δhyg19 mutant. The possibility that this accumulation of DHHA arises from a polar effect on the adjacent hyg20 is unlikely, as a hyg20 deletion mutant does not produce any detectable levels of HA. There are several examples of this, in which the final steps of antibiotic biosynthesis are linked with efflux. In the case of nystatin biosynthesis by S. noursei, disruption of the nysH and nysG genes for a putative type III ABC transporter resulted in the accumulation of 10-deoxynystatin, a precursor of nystatin lacking the hydroxyl at C-10 (29). Furthermore, expression in the mutant strains of an additional copy of nysL, which presumably encodes a monooxygenase for C-10 hydroxylation, partially restored nystatin production with a concomitant decrease in 10-deoxynystatin. NysL has been expressed and has been shown to catalyze the hydroxylation of 10-deoxynystatin (29). The reaction has not been shown to be reversible (in contrast to the reaction observed for Hyg26), and there does not appear to be detectable inhibition of NysL by the product nystatin. Thus, while the observations indicate that the final biosynthetic step is balanced with efficient efflux, the biochemical basis seems to be less clear than that proposed herein for HA. A linkage between the final proposed step in chromomycin A3 biosynthesis, the conversion of dideacetylated precursor by a membrane-bound acetyltransferase, and exportation by a type I ABC transporter in S. griseus subsp. griseus have also been reported (17).
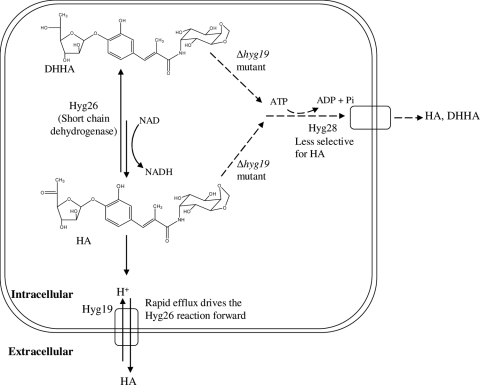
The increased level of production of DHHA by the Δhyg19 mutant is consistent with Hyg19 being the primary efflux pump responsible for the selective and efficient transport of HA. In our working hypothesis, Hyg19 is able to preferentially bind to and export HA over the intermediate DHHA (Fig. 7). These properties would contrast with those observed for Hyg26, which, in the reduction reaction, exhibited poor binding for HA (approximately 6 mM) and showed limited evidence of binding specificity (compounds 3 and 5 were also substrates). The poor HA binding by Hyg26 would permit facile transfer to Hyg19 for export. Hyg19 must also export HA sufficiently fast such that it is not phosphorylated by the resistance element, Hyg21 (this enzyme has an observed kcat of 2.2 min−1 and a Km for HA of 30 μM), and gets trapped inside the cell (4).
FIG. 7.
Working hypothesis for the roles of Hyg19 and Hyg28 in antibiotic efflux. The proton gradient-dependent transporter, Hyg19, is the primary HA transporter in the wild type. Efflux in the Δhyg19 strain is hypothesized to be mediated by the ABC transporter, Hyg28, which is less selective for HA.
The presence of HA and DHHA in the fermentation broth of Δhyg19 implies the existence of an alternative system for antibiotic efflux (Fig. 7). This system would have reduced selectivity (exporting both HA and DHHA) relative to that for the process mediated by Hyg19. The most obvious candidate within the hyg biosynthetic gene cluster is the hyg28-encoded ABC transporter. Hyg28 does not appear to have a significant role in efflux in the wild type, as the Δhyg28 strain continues to produce wild-type levels of HA in the fermentation broth. The Hyg19 process may be significantly faster that that proposed to be catalyzed by Hyg28, or hyg28 expression may increase in the Δhyg19 strain. Attempts to probe these possibilities by the generation of a mutant in which both genes have been deleted have thus far been unsuccessful. There are numerous precedents for the association of multiple efflux systems with antibiotic biosynthetic processes and, in particular, transporters that can excrete pathway intermediates. The 10-deoxynystatin intermediate, produced by deletion of the putative type III ABC transporter in S. noursei, is found in the fermentation broth, suggesting an additional efflux process (29). The premature efflux of landomycin D, an intermediate in landomycin A biosynthesis in S. cyanogenus, has been reported with either the overexpression of the proton gradient-dependent transporter gene lanJ or the disruption of the regulatory gene, lanK, that represses lanJ (19).
In addition to ensuring the production of the final HA product, Hyg19 also contributes to HA resistance in the producing organism. The loss of hyg19 alone did not lead to decreased HA resistance. Similar observations with regard to resistance have been made for the landomycin E producer S. globisporus 1912. Self-resistance was not affected when the proton gradient-dependent transporter gene lndJ and the ABC transporter gene lndW, both of which occur in the landomycin E biosynthetic gene cluster, were independently disrupted. Heterologous expression of these genes, however, conferred resistance to landomycin E, proving their function as resistance genes (20, 21). The intrinsic resistance of an antibiotic producer is often due to the synergistic action of multiple gene products (3). This appears to be the case for HA self-resistance, as the simultaneous deletion of two putative HA resistance determinants, hyg19 and hyg21 (in addition to the hyg20 biosynthetic gene), resulted in an increase in susceptibility greater than that achieved when either of them was individually disrupted (the HA susceptibility of the Δhyg21 mutant has been described separately [4]). In this mutant with triple mutations, both the proposed efficient HA efflux system and an enzyme that can modify and inactivate HA inside the cell are absent. The high wild-type level resistance of the Δhyg20 HA-nonproducing strain [versus the reduced resistance seen for the mutant with triple mutations, Δ(hyg19-hyg20-hyg21)] suggests that HA is not essential for the induction of expression of the resistance genes. It is possible that the hyg28 gene product and a putative methyltransferase encoded by hyg29 may also contribute to resistance in the wild-type strain and to residual self-resistance in the Δ(hyg19-hyg20-hyg21) mutant with triple mutations. A gene inactivation experiment targeting all four putative resistance determinants has been pursued but has been unsuccessful to date.
These analyses provide compelling evidence that Hyg26 catalyzes the final step in HA biosynthesis and that Hyg19 plays a role in ensuring the efficient production of HA (and not the DHHA intermediate) as well as resistance to HA. The MIC and in vitro translation data indicate that HA is the most potent inhibitor (albeit only in tests against E. coli). The presence of the methylene bridge does not significantly affect translation inhibition in vitro but does lead to lower MICs, suggesting the importance of this group for some other aspect of the activity such as cellular uptake. In contrast, changes in the C5″ oxidation state clearly affect the in vitro activity of HA and its analogs, indicating a previously unknown role of this group in interacting with the ribosome target. Thus, the final steps of the HA biosynthetic process lead to a progressively more active compound, culminating in the most active and major product, HA, and have presumably evolved as a result of an ecological advantage to S. hygroscopicus in its natural environment.
Acknowledgments
A part of this work was supported by the Deutsche Forschungsgemeinschaft (grant WI3285/1-1 to D.N.W.).
Footnotes
Published ahead of print on 21 September 2009.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barrasa, M. I., J. A. Tercero, R. A. Lacalle, and A. Jimenez. 1995. The ard1 gene from Streptomyces capreolus encodes a polypeptide of the ABC-transporters superfamily which confers resistance to the aminonucleoside antibiotic A201A. Eur. J. Biochem. 228:562-569. [DOI] [PubMed] [Google Scholar]
- 3.Cundliffe, E. 1989. How antibiotic-producing organisms avoid suicide. Annu. Rev. Microbiol. 43:207-233. [DOI] [PubMed] [Google Scholar]
- 4.Dhote, V., S. Gupta, and K. A. Reynolds. 2008. An O-phosphotransferase catalyzes phosphorylation of hygromycin A in the antibiotic-producing organism Streptomyces hygroscopicus. Antimicrob. Agents Chemother. 52:3580-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinos, G., D. N. Wilson, Y. Teraoka, W. Szaflarski, P. Fucini, D. Kalpaxis, and K. H. Nierhaus. 2004. Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: the universally conserved residues G693 and C795 regulate P-site RNA binding. Mol. Cell 13:113-124. [DOI] [PubMed] [Google Scholar]
- 6.el Habib, S. E., J. N. Scarsdale, and K. A. Reynolds. 2003. Biosynthetic origin of hygromycin A. Antimicrob. Agents Chemother. 47:2065-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerrero, M. D., and J. Modolell. 1980. Hygromycin A, a novel inhibitor of ribosomal peptidyltransferase. Eur. J. Biochem. 107:409-414. [DOI] [PubMed] [Google Scholar]
- 8.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hulo, N., A. Bairoch, V. Bulliard, L. Cerutti, E. De Castro, P. S. Langendijk-Genevaux, M. Pagni, and C. J. Sigrist. 2006. The PROSITE database. Nucleic Acids Res. 34:D227-D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones, D. T. 2007. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 23:538-544. [DOI] [PubMed] [Google Scholar]
- 11.Jornvall, H., B. Persson, M. Krook, S. Atrian, R. Gonzalez-Duarte, J. Jeffery, and D. Ghosh. 1995. Short-chain dehydrogenases/reductases (SDR). Biochemistry 34:6003-6013. [DOI] [PubMed] [Google Scholar]
- 12.Kallberg, Y., U. Oppermann, H. Jornvall, and B. Persson. 2002. Short-chain dehydrogenases/reductases (SDRs). Coenzyme-based functional assignments in completed genomes. Eur. J. Biochem. 269:4409-4417. [DOI] [PubMed] [Google Scholar]
- 13.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 14.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 15.Mann, R. L., R. M. Gale, and R. F. Van Abeele. 1953. Hygromycin. II. Isolation and properties. Antibiot. Chemother. 3:1279-1282. [PubMed] [Google Scholar]
- 16.Martin, M. F., and P. Liras. 1989. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol. 43:173-206. [DOI] [PubMed] [Google Scholar]
- 17.Menendez, N., A. F. Brana, J. A. Salas, and C. Mendez. 2007. Involvement of a chromomycin ABC transporter system in secretion of a deacetylated precursor during chromomycin biosynthesis. Microbiology 153:3061-3070. [DOI] [PubMed] [Google Scholar]
- 18.Oppermann, U., C. Filling, M. Hult, N. Shafqat, X. Wu, M. Lindh, J. Shafqat, E. Nordling, Y. Kallberg, B. Persson, and H. Jornvall. 2003. Short-chain dehydrogenases/reductases (SDR): the 2002 update. Chem. Biol. Interact. 143-144:247-253. [DOI] [PubMed] [Google Scholar]
- 19.Ostash, I., B. Ostash, A. Luzhetskyy, A. Bechthold, S. Walker, and V. Fedorenko. 2008. Coordination of export and glycosylation of landomycins in Streptomyces cyanogenus S136. FEMS Microbiol. Lett. 285:195-202. [DOI] [PubMed] [Google Scholar]
- 20.Ostash, I., B. Ostash, S. Walker, and V. Fedorenko. 2007. Proton-dependent transporter gene lndJ confers resistance to landomycin E in Streptomyces globisporus. Genetika 43:1032-1037. [PubMed] [Google Scholar]
- 21.Ostash, I., Y. Rebets, B. Ostash, A. Kobylyanskyy, M. Myronovskyy, T. Nakamura, S. Walker, and V. Fedorenko. 2008. An ABC transporter encoding gene lndW confers resistance to landomycin E. Arch. Microbiol. 190:105-109. [DOI] [PubMed] [Google Scholar]
- 22.Palaniappan, N., S. Ayers, S. Gupta, S. el Habib, and K. A. Reynolds. 2006. Production of hygromycin A analogs in Streptomyces hygroscopicus NRRL 2388 through identification and manipulation of the biosynthetic gene cluster. Chem. Biol. 13:753-764. [DOI] [PubMed] [Google Scholar]
- 23.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Persson, B., M. Krook, and H. Jornvall. 1991. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur. J. Biochem. 200:537-543. [DOI] [PubMed] [Google Scholar]
- 25.Pittenger, R. C., R. N. Wolfe, P. N. Hoehn, W. A. Daily, and J. M. McGuire. 1953. Hygromycin. I. Preliminary studies in the production and biologic activity on a new antibiotic. Antibiot. Chemother. 3:1268-1278. [PubMed] [Google Scholar]
- 26.Saier, M. H., Jr., C. V. Tran, and R. D. Barabote. 2006. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34:D181-D186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saier, M. H., Jr., M. R. Yen, K. Noto, D. G. Tamang, and C. Elkan. 2009. The Transporter Classification Database: recent advances. Nucleic Acids Res. 37:D274-D278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Sletta, H., S. E. Borgos, P. Bruheim, O. N. Sekurova, H. Grasdalen, R. Aune, T. E. Ellingsen, and S. B. Zotchev. 2005. Nystatin biosynthesis and transport: nysH and nysG genes encoding a putative ABC transporter system in Streptomyces noursei ATCC 11455 are required for efficient conversion of 10-deoxynystatin to nystatin. Antimicrob. Agents Chemother. 49:4576-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szaflarski, W., O. Vesper, Y. Teraoka, B. Plitta, D. N. Wilson, and K. H. Nierhaus. 2008. New features of the ribosome and ribosomal inhibitors: non-enzymatic recycling, misreading and back-translocation. J. Mol. Biol. 380:193-205. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka, N., T. Nonaka, M. Nakanishi, Y. Deyashiki, A. Hara, and Y. Mitsui. 1996. Crystal structure of the ternary complex of mouse lung carbonyl reductase at 1.8 Å resolution: the structural origin of coenzyme specificity in the short-chain dehydrogenase/reductase family. Structure 4:33-45. [DOI] [PubMed] [Google Scholar]
- 32.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]