Abstract
In this study, a chromosomally encoded putative drug efflux pump of the SMR family, named AbeS, from a multidrug-resistant strain of Acinetobacter baumannii was characterized to elucidate its role in antimicrobial resistance. Expression of the cloned abeS gene in hypersensitive Escherichia coli host KAM32 resulted in decreased susceptibility to various classes of antimicrobial agents, detergents, and dyes. Deletion of the abeS gene in A. baumannii confirmed its role in conferring resistance to these compounds.
Acinetobacter baumannii is an important nosocomial pathogen frequently reported to be associated with a variety of infections, including respiratory tract infections, urinary tract infections, bacteremia, and skin and skin structure infections (3). Reports of the increased isolation of multidrug-resistant A. baumannii clinical isolates from different regions of the United States are appearing at a startling rate (1, 4, 10, 25, 30, 33).
Antibiotic resistance in A. baumannii has been attributed to either intrinsic or acquired mechanisms (21). The resistance mechanisms in A. baumannii are diverse and include enzymatic modification of antibiotics, target gene mutation, altered outer membrane permeability, and upregulated multidrug efflux pump activity (20).
Efflux systems involve transport proteins that function to reduce the concentration of drugs or toxic substances by transporting them across the inner and outer membranes into the external medium (24). These multidrug efflux systems are classified into five different families: ATP-binding cassette (ABC), major facilitator super family (MFS), resistance/nodulation/cell division (RND), multidrug and toxic-compound extrusion (MATE), and the small multidrug resistance (SMR) family of bacterial integral membrane proteins (22). ABC transporters are ATP-driven efflux pumps; MFS, RND, and SMR are proton driven; and MATE transporters consist of an Na+/H+ drug antiporter system (22, 23). Genome sequence analyses reveal that, on average, efflux pumps constitute at least 10% of the transporters in bacterial species, and they usually are capable of extruding a broad range of structurally unrelated compounds (18).
Multidrug efflux pumps of the SMR type are made of a transport protein located in the inner membrane (19). The polypeptide chains of SMR efflux pumps, found in the inner membranes of gram-negative bacteria, are 110 amino acid residues in length and contain four transmembrane helices (29). Reports show that 52% of currently sequenced genomes of eubacteria and archaea contain at least one SMR homologue (2). Well-studied examples of SMR efflux pumps include EmrE of Pseudomonas aeruginosa, EbrAB of Bacillus subtilis, SsmE of Serratia marcescens, and EmrE of Escherichia coli, which are involved in resistance to a variety of antimicrobial agents and quaternary ammonium compounds (14, 16, 17, 34).
The 3.9-Mb genome of A. baumannii AYE is reported to harbor 46 open reading frames (ORFs) encoding putative efflux pumps of different families (8). The efflux systems functionally characterized so far include AdeABC and AdeIJK (RND type), AbeM (MATE type), and CraA (MFS type) (20, 27). Albeit comparative genomics clearly reveal the existence of several chromosomally borne putative efflux transporters (8, 12), apparently the role of the Acinetobacter SMR efflux pump was never examined. Therefore, the objective of the present study was to investigate the function of one putative SMR efflux pump from a clinical isolate, A. baumannii AC0037.
(Part of this work was presented at the 48th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, 2008 [8a].)
Cloning of a putative SMR efflux pump gene from A. baumannii.
A. baumannii AC0037 used in this study (isolated from the respiratory system of an infected patient in The Ohio State University Medical Center) is a multidrug clinical strain (31) with the following MICs for antibiotics: amikacin, >128 μg/ml; ceftazidime, 32 μg/ml; cefepime, >16 μg/ml; ciprofloxacin, >72 μg/ml; imipenem, 4 μg/ml; and meropenem, 7.5 μg/ml. The MICs were determined using the broth dilution method, and interpretation was done per the criteria approved by the Clinical and Laboratory Standards Institute (CLSI) (7). The drug-hypersusceptible E. coli KAM32 (generously provided by Tomofusa Tsuchiya) that lacks the major multidrug efflux pumps AcrAB and YdhE was used as a host (5).
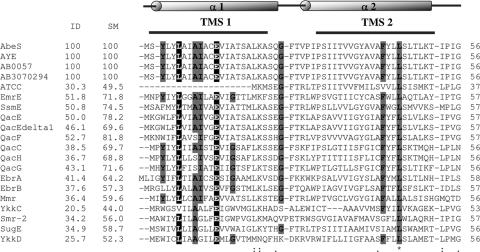
The protein encoded by ORF ABAYE1181 in the AYE genome is annotated as a putative multidrug efflux protein. It exhibits 51.8% identity (E value of 1e−25 and score of 108) to E. coli EmrE (GenBank accession number NC_013364), a well-characterized member of the SMR family (34). Thus, the ABAYE1181 ORF was selected as the putative SMR-type-pump gene to be investigated in this study. The genomic DNA of A. baumannii was extracted using the DNeasy tissue kit (Qiagen, Valencia, CA). The putative efflux gene from AC0037 was amplified by PCR as described previously (28), with minor modifications. The primers used were P1 and P2 (5′-TAGAGAATTCATGTCTTATCTTTATTTAGC-3′ [EcoRI sequence underlined] and 5′-CGCTCTGCAGTTATAGATGGGTGTTTTTAG-3′ [PstI sequence underlined]). The PCR product was purified using the QiaQuick PCR purification kit (Qiagen, Valencia, CA). The amplicon was digested with EcoRI and PstI and subsequently ligated with vector pUC18 (New England Biolabs, MA) to obtain recombinant plasmid pVBS1. At least two independently generated recombinant plasmids were sequenced bidirectionally using the CEQ 8000 capillary electrophoresis system (Beckman Coulter Instruments, Inc., Palo Alto, CA) to rule out mutations introduced during PCR. Analysis revealed the presence of one ORF that was designated abeS (A. baumannii efflux pump of SMR family). Further analysis revealed that abeS was a 330-bp gene encoding a 109-amino-acid protein with a calculated mass of 11.14 kDa. AbeS exhibited 100% identity to the product of ORF ABAYE1181 found in the A. baumannii AYE genome (8). AbeS exhibited different degrees of identity with other SMR transporters from various gram-negative and gram-positive species (Fig. 1).
FIG. 1.
Multiple sequence alignment of AbeS and related SMR efflux proteins. Multiple sequence alignment of AbeS and related homologues was generated using ClustalW (32). Secondary structure elements indicated on top correspond to those observed in the E. coli EmrE protein (Protein Data Bank identification no. 3b5d). Four probable transmembrane segments (TMS) are indicated with horizontal lines above the alignments. Asterisks indicate fully conserved residues, colons indicate strongly similar residues, and dots indicate weakly similar residues. Dashes represent gaps. Essential residues (as shown in EmrE [29]) are highlighted in black. Conserved residues are highlighted in gray. The identities (ID) and similarities (SM) of AbeS with the corresponding SMR transporter are shown in their respective column. Sequences were obtained from the GenBank database by the use of the following (protein) accession numbers: AbeS, A. baumannii AC0037 (FJ843079); A. baumannii AYE (YP_001713109.1); AB0057 (YP_002319996.1); AB307-0294 (YP_002325052.1); ATCC 17978 (YP_001085323.1); EmrE, E. coli (P23895.1); SsmE, S. marcescens (BAF80121.1); QacE, K. aerogenes (P0AGD0.1); QacEdelta1, Klebsiella pneumoniae (ABF48386.1); QacF, Enterobacter aerogenes (Q9X2N9.1); QacC, Staphylococcus aureus (AAM94143.1); QacH, Staphylococcus saprophyticus (O87868.1); QacG, Staphylococcus sp. strain ST94 (O87866.1); EbrA, Bacillus subtilis (O31792.1); EbrB, B. subtilis (O31791.1); Mmr, Mycobacterium tuberculosis (P95094); YkkC, B. subtilis (Q65KV1.1); Smr-2, Pseudomonas aeruginosa (CAH04647.1); SugE, E. coli (AAQ16658.1); and YkkD, B. subtilis (Q65KV0.1).
Antimicrobial susceptibility of E. coli expressing abeS.
To evaluate the role of abeS in antimicrobial resistance, the MICs of various antibiotics, detergents, and dyes (Sigma, St. Louis, MO) were determined in triplicate in Mueller-Hinton broth (Difco, Sparks, MD) by the broth dilution method, according to the guidelines of CLSI (7).
The SMR efflux pump-expressing cells exhibited a higher MIC for erythromycin (sixfold) as well as novobiocin (fivefold). Minor increases (twofold) were observed with MICs of aminoglycosides (amikacin), quinolones (ciprofloxacin, norfloxacin), tetracycline, and trimethoprim. No significant differences in MICs were found for chloramphenicol, nalidixic acid, and rifampin (rifampicin) (Table 1). KAM32/pVBS1 also showed increased levels of resistance to detergents, such as deoxycholate (25-fold) and sodium dodecyl sulfate (16-fold), and dyes, such as acridine orange (fivefold), acriflavine (eightfold), and benzalkonium chloride (sixfold). A nearly fourfold increase in resistance was observed for 4′,6-diamidino-2-phenylindole (DAPI), ethidium bromide (EtBr), and rhodamine 123. A marginal decrease in susceptibility toward chlorhexidine, pyronin Y, and tetraphenylphosphonium chloride was also noted (Table 1).
TABLE 1.
MICs of various antimicrobial agents for E. coli (KAM32/pUC18 and KAM32/pVBS1) and A. baumannii (AC0037, AC0037ΔabeS, and AC0037ΔabeSΩabeS) strains used in this study
| Compound | MIC (μg/ml) for E. coli straind: |
Fold changea | MIC (μg/ml) for A. baumannii strain: |
Fold changec | MIC (μg/ml) for A. baumannii AC0037ΔabeSΩabeS | ||
|---|---|---|---|---|---|---|---|
| pUC18/KAM32 | pVBS1/KAM32 | AC0037 | AC0037ΔabeSb | ||||
| Amikacin | 1 (0.15) | 2 (0.15) | 2 | 256 | 256 | 1 | 256 |
| Chloramphenicol | 0.2 | 0.2 | 1 | 256 | 64 | 4 | 128 |
| Ciprofloxacin | 0.002 (0.0005) | 0.004 (0.0005) | 2 | 120 | 40 | 3 | >60 |
| Erythromycin | 4 (<0.05) | 24 (0.25) | 6 | 64 | 8 | 8 | 32 |
| Nalidixic acid | 1 | 1 | 1 | 256 | 128 | 2 | 256 |
| Norfloxacin | 0.06 (0.001) | 0.12 (0.001) | 2 | 512 | 256 | 2 | >128 |
| Novobiocin | 4 (<0.5) | 20 (<0.5) | 5 | 120 | 40 | 3 | 84 |
| Rifampin | 2 | 2 | 1 | 2 | 2 | 1 | 2 |
| Tetracycline | 0.5 | 1 | 2 | 64 | 64 | 1 | 64 |
| Trimethoprim | 0.125 | 0.2 | 2 | 64 | 64 | 1 | 64 |
| Acridine orange | 16 | 80 | 5 | 200 | 50 | 4 | 125 |
| Acriflavine | 2 | 16 | 8 | 128 | 16 | 8 | 64 |
| Benzalkonium chloride | 1 (0.25) | 6 (0.25) | 6 | 48 | 12 | 4 | 24 |
| Chlorhexidine | 2 | 4 | 2 | 32 | 16 | 2 | 32 |
| DAPI | 0.5 | 2 | 4 | 2 | 0.5 | 4 | 2 |
| Deoxycholate | 125 (<0.25) | 3,125 (5) | 25 | >2,048 | 128 | >16 | 512 |
| EtBr | 2 | 8 | 4 | 1,024 | 128 | 8 | >512 |
| Methyl viologen | 64 (<2.5) | 128 (<2.5) | 2 | 800 | 400 | 2 | 800 |
| Pyronin Y | 4 | 8 | 2 | 4 | 2 | 2 | 4 |
| Rhodamine 123 | 8 | 32 | 4 | 128 | 64 | 2 | 128 |
| Sodium dodecyl sulfate | 50 (<5) | 800 (<5) | 16 | >256 | 16 | >16 | 128 |
| Tetraphenylphosphonium chloride | 8 (0.25) | 16 (0.25) | 2 | 1,500 | 300 | 5 | >500 |
Ratio of MIC for pVBS1 to MIC for pUC18.
AC0037ΔabeS did not show any difference in susceptibility toward imipenem, meropenem, and ceftazidime.
Ratio of MIC for AC0037 to MIC for AC0037ΔabeS.
Values in parentheses are MICs in the presence of the efflux pump inhibitor CCCP at 25 μg/ml.
Overall, results demonstrated that AbeS could decrease susceptibility to some antibiotics, disinfectants, dyes, and detergents. The broad substrate specificity displayed by AbeS was very similar to those of other SMR transporters reported previously (6, 14, 16, 17, 34).
To confirm the role of AbeS as an efflux pump, the MICs of substrates with strong specificity were monitored in the presence of 25 μg/ml efflux pump inhibitor carbonyl cyanide 3-chlorophenylhydrazone (CCCP; an uncoupler of oxidative phosphorylation which disrupts the proton gradient on the membrane) (31). Results were consistent with the notion that the abeS gene product conferred resistance to various antimicrobial compounds through an efflux mechanism (Table 1).
Fluorometric efflux assay of E. coli expressing AbeS.
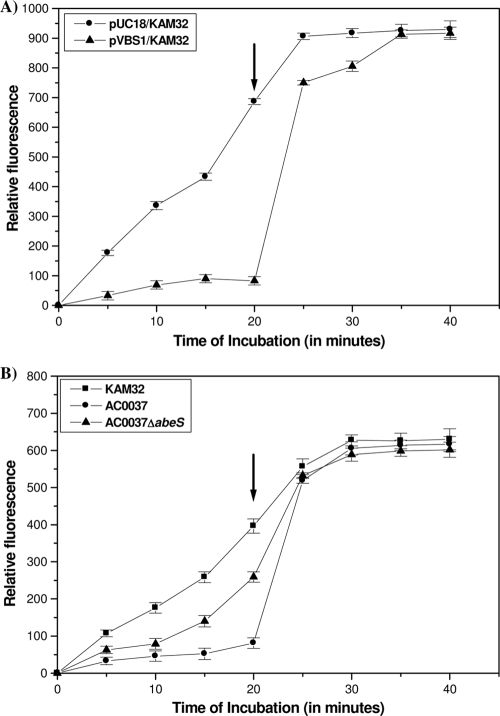
EtBr is highly fluorescent when bound to DNA. Upon being extruded from the cell through an efflux pump, the fluorescence decreases, and this difference can be measured with a spectrofluorometer (13, 17). To confirm the role of AbeS as an efflux pump, we compared the rates of EtBr accumulation in E. coli KAM32 cells carrying either pUC18 or recombinant plasmid pVBS1. We found that the intracellular level of EtBr in E. coli KAM32/pVBS1 (carrying abeS) was significantly lower than that of the control strain E. coli KAM32/pUC18 under energized conditions (in the absence of CCCP) (Fig. 2A). After the addition of CCCP, at 25 μg/ml, there was a rise in the intracellular accumulation level of EtBr in E. coli KAM32/pVBS1, which eventually reached a plateau equal to that of the control strain E. coli KAM32/pUC18 (Fig. 2A). These results showed that AbeS is an energy-dependent active efflux pump. Subsequently, we tested the influence of monovalent cations, such as Na+ and Li+, in the efflux process because the activities of several multidrug efflux pumps are coupled with monovalent cations (9). The addition of either NaCl or LiCl caused no difference in the efflux activity from that of the control strain (data not shown). Thus, results demonstrated that AbeS is an efflux pump and the active extrusion is proton (H+) driven.
FIG. 2.
Fluorometric accumulation assays. The accumulation study was done as described previously (31). (A) Accumulation studies using 10 μg/ml of EtBr with KAM32/pVBS1 and KAM32/pUC18. (B) Accumulation studies using 5 μg/ml of EtBr with A. baumannii AC0037, the A. baumannii abeS null mutant (AC0037ΔabeS), and E. coli KAM32 (lacking AcrAB and YdhE). Instead of sodium phosphate buffer, phosphate-buffered saline (0.136 M NaCl, 0.0026 M KCl, 0.01 M Na2HPO4, 0.00176 M KH2PO4, pH 7.0) was used, and cell lysis was carried out for 6 h. The fluorescence of the supernatant was measured at excitation at 530 nm and emission at 600 nm, using a SpectraMax M2 spectrofluorometer (Molecular Devices Corporation, Sunnyvale, CA). Where indicated (after 20 min of incubation with the substrate), the proton motive force uncoupler CCCP was added to a final concentration of 25 μg/ml. Relative fluorescence intensity on the y axis represents the levels of accumulated EtBr in the bacterial cells. The graph reflects the difference in the fluorescence shown by the bacterial cell in the presence and absence of the inhibitor CCCP. Each data point represents the mean plus the standard deviation of three independent experiments.
Construction of an abeS null mutant in A. baumannii.
To evaluate the role of AbeS in A. baumannii AC0037, an abeS deletion mutant was generated. The vector pUC18, not capable of replicating in A. baumannii, was used as a suicide vector (15). To construct the abeS deletion mutant, the 548-bp upstream region (amplicon A) and 542-bp downstream region (amplicon B) of the efflux gene were amplified using specific primers. The primers for amplicon A were P3 (5′-TGATTCTAGATGTGATATGTGCTTACCAGAATGC-3′, with an XbaI linker) and P4 (5′-TCATCTGCAGAGTAAAACCTTGGATGCTTT-3′, with a PstI linker). The primers for amplicon B were P5 (5′-GATACTGCAGTGCATTGGTTTAGCTTTA-3′, with a PstI linker) and P6 (5′-ACAGGGATCCTCTGAGGCGGAAGAACGTGCAC-3′, with a BamHI linker) (enzyme sequences are underlined). PCR products were generated from the genome of AC0037. The digested fragments were ligated with the PstI-kanamycin resistance cassette (K) obtained with the pUC-4K vector (Pharmacia). The generated AKD fragment was inserted into the XbaI- and BamHI-digested plasmid pUC18, resulting in the pabeS-Kan plasmid. It was bidirectionally sequenced to ensure the presence of the kanamycin resistance cassette and to confirm authenticity. The plasmid pabeS-Kan was then introduced into A. baumannii AC0037 for the double homologous recombination event to occur by electroporation as described previously (15).
Selection of transformants was made on 50 μg/ml kanamycin- and 80 μg/ml ticarcillin-containing plates. Inactivation of the abeS gene by insertion of pabeS-Kan was confirmed by PCR amplification and DNA sequencing, and the null mutant was designated AC0037ΔabeS.
The primers used for confirmation were the gene-specific primers P1 and P2, as well as the flanking chromosomal region primers P11 (5′-GCTTATTCAGCGAGTTAAATATG-3′) and P12 (5′-CACATGACAGTACTGGAAAATGT-3′).
Functional characterization of abeS in A. baumannii.
Following the confirmation of the abeS deletion mutant, the role of abeS in A. baumannii was evaluated. Susceptibility testing data showed that deletion of abeS resulted in increased susceptibility to various antimicrobial compounds (Table 1).
The EtBr accumulation experiments demonstrated that the efflux was more efficient in the wild-type AC0037 strain, whereas the efflux was significantly less efficient (increased accumulation of >3-fold for EtBr) in the AC0037ΔabeS deletion mutant. Addition of efflux inhibitor CCCP increased the intracellular accumulation of EtBr in AC0037ΔabeS (Fig. 2B).
The role of the A. baumannii abeS gene was confirmed by gene complementation experiments. Briefly, a DNA fragment containing a functional aadA1 gene was amplified from an AC0019 clinical isolate (GenBank accession number EU977568.1) (26, 31) by PCR using primers P7 (5′-TACAGATATCATGAGGGAAGCGGTGATC-3′ [EcoRV linker underlined]) and P8 (5′-TACGGTCGACTTATTTGCCTACTACCTTGGTGA-3′ [SalI linker underlined]). The cassette was ligated into the E. coli-Acinetobacter shuttle vector pWH1266 (11). The resulting plasmid, pWH-Spc, was modified by cloning a PCR-amplified wild-type abeS gene with the primer pair P1/P2 into the PstI site of the plasmid, yielding a recombinant plasmid, pWH-abeS. Electroporation of the recombinant plasmid pWH-abeS into the A. baumannii AC0037ΔabeS mutant resulted in strain AC0037ΔabeSΩabeS. Selection of the AC0037ΔabeSΩabeS mutant was made on 200 μg/ml streptomycin, 50 μg/ml kanamycin, and 80 μg/ml of ticarcillin-containing plates. Complementing the mutant with the wild-type abeS gene nearly restored reduced susceptibility to antimicrobial agents, detergents, and dyes, similar to that of the parental strain AC0037 (Table 1). These findings confirmed the role of abeS in mediating resistance also in A. baumannii.
In conclusion, this study demonstrated for the first time the role of the SMR efflux pump AbeS in mediating resistance to some antibiotics, hydrophobic compounds, detergents, and disinfectants in A. baumannii clinical strain AC0037.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been deposited in the GenBank nucleotide sequence database under the accession number FJ843079.
Acknowledgments
This study was funded by The Ohio State University intramural funding to W.A.G.
We thank the members of the Infectious Diseases Molecular Epidemiology Laboratory (IDMEL) team for technical assistance. We also thank Tomofusa Tsuchiya, Craig Altier, Preeti Pancholi, Kurt Stevenson, and Mario Marcon for providing the E. coli host strain, plasmids, and A. baumannii clinical isolates for this study.
Footnotes
Published ahead of print on 21 September 2009.
REFERENCES
- 1.Adams-Haduch, J. M., D. L. Paterson, H. E. Sidjabat, A. W. Pasculle, B. A. Potoski, C. A. Muto, L. H. Harrison, and Y. Doi. 2008. Genetic basis of multidrug resistance in Acinetobacter baumannii clinical isolates at a tertiary medical center in Pennsylvania. Antimicrob. Agents Chemother. 52:3837-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bay, D. C., K. L. Rommens, and R. J. Turner. 2008. Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim. Biophys. Acta 1778:1814-1838. [DOI] [PubMed] [Google Scholar]
- 3.Bergogne-Bérézin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bratu, S., D. Landman, D. A. Martin, C. Georgescu, and J. Quale. 2008. Correlation of antimicrobial resistance with β-lactamases, the OmpA-like porin, and efflux pumps in clinical isolates of Acinetobacter baumannii endemic to New York City. Antimicrob. Agents Chemother. 52:2999-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J., Y. Morita, M. N. Huda, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2002. VmrA, a member of a novel class of Na+-coupled multidrug efflux pumps from Vibrio parahaemolyticus. J. Bacteriol. 184:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung, Y. J., and M. H. Saier. 2002. Overexpression of the Escherichia coli sugE gene confers resistance to a narrow range of quaternary ammonium compounds. J. Bacteriol. 184:2543-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A7, vol. 26-2. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Fournier, P. E., D. Vallenet, V. Barbe, S. Audic, H. Ogata, L. Poirel, H. Richet, C. Robert, S. Mangenot, C. Abergel, P. Nordmann, J. Weissenbach, D. Raoult, and J. M. Claverie. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Gebreyes, W. A., V. Srinivasan, G. Rajamohan, P. Pancholi, K. Stevenson, and M. Marcon. 2008. Novel secondary active transporters conferring antimicrobial resistance in Acinetobacter baumannii with broad substrate specificity, abstr. C1-1048. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 9.He, G. X., T. Kuroda, T. Mima, Y. Morita, T. Mizushima, and T. Tsuchiya. 2004. An H+-coupled multidrug efflux pump, PmpM, a member of the MATE family of transporters, from Pseudomonas aeruginosa. J. Bacteriol. 186:262-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hujer, K. M., A. M. Hujer, E. A. Hulten, S. Bajaksouzian, J. M. Adams, C. J. Donskey, D. J. Ecker, C. Massire, M. W. Eshoo, R. Sampath, J. M. Thomson, P. N. Rather, D. W. Craft, J. T. Fishbain, A. J. Ewell, M. R. Jacobs, D. L. Paterson, and R. A. Bonomo. 2006. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 50:4114-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunger, M., R. Schmucker, V. Kishan, and W. Hillen. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45-51. [DOI] [PubMed] [Google Scholar]
- 12.Iacono, M., L. Villa, D. Fortini, R. Bordoni, F. Imperi, R. J. Bonnal, T. Sicheritz-Ponten, G. De Bellis, P. Visca, A. Cassone, and A. Carattoli. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 52:2616-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonas, B. M., B. E. Murray, and G. M. Weinstock. 2001. Characterization of emeA, a norA homolog and multidrug resistance efflux pump, in Enterococcus faecalis. Antimicrob. Agents Chemother. 45:3574-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, X.-Z., K. Poole, and H. Nikaido. 2003. Contributions of MexAB-OprM and an EmrE homologue to intrinsic resistance of Pseudomonas aeruginosa to aminoglycosides and dyes. Antimicrob. Agents Chemother. 47:27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchand, I., L. Damier-Piolle, P. Courvalin, and T. Lambert. 2004. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 48:3298-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masaoka, Y., Y. Ueno, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2000. A two-component multidrug efflux pump, EbrAB, in Bacillus subtilis. J. Bacteriol. 182:2307-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minato, Y., F. Shahcheraghi, W. Ogawa, T. Kuroda, and T. Tsuchiya. 2008. Functional gene cloning and characterization of the SsmE multidrug efflux pump from Serratia marcescens. Biol. Pharm. Bull. 31:516-519. [DOI] [PubMed] [Google Scholar]
- 18.Paulsen, I. T. 2003. Multidrug efflux pumps and resistance: regulation and evolution. Curr. Opin. Microbiol. 6:446-451. [DOI] [PubMed] [Google Scholar]
- 19.Paulsen, I. T., R. A. Skurray, R. Tam, M. H. Saier, Jr., R. J. Turner, J. H. Weiner, E. B. Goldberg, and L. L. Grinius. 1996. The SMR family: a novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol. Microbiol. 19:1167-1175. [DOI] [PubMed] [Google Scholar]
- 20.Peleg, A. Y., H. Seifert, and D. L. Paterson. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez, F., A. M. Hujer, K. M. Hujer, B. K. Decker, P. N. Rather, and R. A. Bonomo. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3471-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piddock, L. J. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piddock, L. J. 2006. Multidrug-resistance efflux pumps-not just for resistance. Nat. Rev. Microbiol. 4:629-636. [DOI] [PubMed] [Google Scholar]
- 24.Putman, M., H. W. Van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi, C., M. Malczynski, M. Parker, and M. H. Scheetz. 2008. Characterization of genetic diversity of carbapenem-resistant Acinetobacter baumannii clinical strains collected from 2004 to 2007. J. Clin. Microbioal. 46:1106-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajamohan, G., V. B. Srinivasan, and W. A. Gebreyes. 2009. Biocide tolerant multidrug resistant Acinetobacter baumannii clinical strains are associated Iwith higher biofilm formation. J. Hosp. Infect. 73:287-289. [DOI] [PubMed] [Google Scholar]
- 27.Roca, I., S. Marti, P. Espinal, P. Martínez, I. Gibert, and J. Vila. 6 July 2009, posting date. CraA: an MFS efflux pump associated with chloramphenicol resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. doi: 10.1128/AAC.00584-09. [DOI] [PMC free article] [PubMed]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Schuldiner, S. 2007. When biochemistry meets structural biology: the cautionary tale of EmrE. Trends Biochem. Sci. 32:252-258. [DOI] [PubMed] [Google Scholar]
- 30.Shelburne, S. A., III, K. V. Singh, A. C. White, Jr., L. Byrne, A. Carmer, C. Austin, E. Graviss, C. Stager, B. E. Murray, and R. L. Atmar. 2008. Sequential outbreaks of infections by distinct Acinetobacter baumannii strains in a public teaching hospital in Houston, Texas. J. Clin. Microbiol. 46:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srinivasan, V. B., G. Rajamohan, P. Preeti, K. Stevenson, D. Tadesse, P. Patchanee, M. Marcon, and W. A. Gebreyes. 2009. Genetic relatedness and molecular characterization of resistance determinants in multidrug resistant Acinetobacter baumannii isolated in central Ohio, USA. Ann. Clin. Microbiol. Antimicrob. 8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson, J. D., T. J. Gibson, and D. G. Higgins. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics 2:2.3. [DOI] [PubMed] [Google Scholar]
- 33.Valentine, S. C., D. Contreras, S. Tan, L. J. Real, S. Chu, and H. H. Xu. 2008. Phenotypic and molecular characterization of Acinetobacter baumannii clinical isolates from nosocomial outbreaks in Los Angeles County, California. J. Clin. Microbiol. 46:2499-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yerushalmi, H., M. Lebendiker, and S. Schuldiner. 1995. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J. Biol. Chem. 270:6856-6863. [DOI] [PubMed] [Google Scholar]