Abstract
Recent outbreaks of Clostridium difficile infection have been related to the emergence of the NAP1/027 epidemic strain. This strain demonstrates increased virulence and resistance to the C-8-methoxyfluoroquinolones gatifloxacin and moxifloxacin. These antibiotics have been implicated as major C. difficile infection-inducing agents. We investigated by real-time reverse transcription-PCR the impact of subinhibitory concentrations of ampicillin, clindamycin, ofloxacin, and moxifloxacin on the expression of genes encoding three colonization factors, the protease Cwp84, the high-molecular-weight S-layer protein, and the fibronectin-binding protein Fbp68. We have previously shown in six non-NAP1/027 moxifloxacin-susceptible strains that the presence of ampicillin or clindamycin induced an upregulation of these genes, whereas the presence of fluoroquinolones did not. The objective of this study was to analyze the expression of these genes under the same conditions in four NAP1/027 strains, one moxifloxacin susceptible and three moxifloxacin resistant. Two in vitro-selected moxifloxacin-resistant mutants were also analyzed. Moxifloxacin resistance was associated with the Thr82→Ile substitution in GyrA in all but one of the moxifloxacin-resistant strains. The expression of cwp84 and slpA was strongly increased after culture with ampicillin or clindamycin in NAP1/027 strains. Interestingly, after culture with fluoroquinolones, the expression of cwp84 and slpA was only increased in four moxifloxacin-resistant strains, including the NAP1/027 strains and one of the in vitro-selected mutants. The overexpression of cwp84 was correlated with increased production of the protease Cwp84. The historical NAP1/027 moxifloxacin-susceptible strain and its mutant appear to be differently regulated by fluoroquinolones. Overall, fluoroquinolones appear to favor the expression of some colonization factor-encoding genes in resistant C. difficile strains. The fluoroquinolone resistance of the NAP1/027 epidemic strains could be considered an ecological advantage. This could also increase their colonization fitness and promote the infection.
Antibiotic exposure is the most widely recognized risk factor for Clostridium difficile infection (CDI). It is assumed that antibiotics disturb the normal intestinal microbiota and its barrier effect, allowing subsequent C. difficile colonization and CDI. Broad-spectrum antibiotics, such as clindamycin, aminopenicillins, and cephalosporins, possess a high propensity to induce CDI (24, 50). The use of fluoroquinolones has recently been proposed as a significant risk factor for CDI due to emergent strains, such as the NAP1/027 epidemic strain, that are resistant to the C8-methoxyfluoroquinolones gatifloxacin and moxifloxacin (5, 15, 35, 45, 55). This strain displays increased virulence, causing high morbidity and a high mortality rate. This results partly from the hyperproduction of toxins TcdA and TcdB that is caused by a frameshift mutation in the gene encoding negative regulator TcdC (37, 38, 39, 40, 46, 57).
It has been suggested that the increased virulence was probably insufficient to explain the wide spread of this strain (40). One explanation could come from its resistance to the C-8-methoxyfluoroquinolones, which have become the most frequently used antimicrobial class in North America in the last several years, as well as throughout Europe. In France, this epidemic strain emerged a short time after they began to be prescribed. The resistance to fluoroquinolones could then be seen as offering an ecological advantage for this strain. A role of antibiotic resistance in the emergence of epidemic strains has already been suggested for the highly clindamycin-resistant “J strain” in the United States and, more recently, for a fluoroquinolone-resistant, toxin-A-negative, toxin-B-positive clonal strain in Ireland (20, 33). In addition, the increased virulence of strain NAP1/027 and its sudden emergence could also be explained by its considerable ability to adapt to the host gut environment and its greater colonization capacity.
The colonization process, which is the first step of the pathogenesis, involves a variety of cell surface-associated proteins. Among these proteins, the Cwp66 adhesin and the Fbp68 fibronectin-binding protein have been shown to mediate attachment to Vero cells (28, 56). The high-molecular-weight S-layer protein, encoded by the 5′ end of the slpA gene, has been shown to bind to epithelial cell lines (8, 9, 10, 34). The Cwp84 cysteine protease possesses degrading activity on several components of the gut basal lamina and could contribute to the degradation of host tissue integrity and the dissemination of the infection (32). These proteins are able to induce an immune response in patients with CDI, arguing for their putative role in vivo in the colonization process (11, 18, 43, 44).
Antibiotics could also play a direct role in enhancing colonization. In a previous study, we showed that subinhibitory concentrations of ampicillin and clindamycin increased the expression of four colonization factor-encoding genes (cwp66, slpA, fbp68, and especially, cwp84) in six moxifloxacin-susceptible C. difficile strains. In contrast, ofloxacin and moxifloxacin had no effect on the expression of these genes in the fluoroquinolone-susceptible strains (14). In this study, we investigated the impact of the four antibiotics (ampicillin, clindamycin, ofloxacin, and moxifloxacin) on colonization factor expression by six moxifloxacin-susceptible or moxifloxacin-resistant strains, including four NAP1/027 clinical isolates and two in vitro-selected moxifloxacin-resistant mutants.
MATERIALS AND METHODS
C. difficile strains and growth conditions.
Four NAP1/027 C. difficile strains isolated in Europe (CD196 and CD07-259) and the United States (6296 and 6425) and two in vitro-selected moxifloxacin-resistant mutants were used in this study. Among the NAP1/027 isolates, strain CD196 was the historical moxifloxacin-susceptible, nonepidemic French strain (47). The two moxifloxacin-resistant mutants were selected in vitro from strain CD196 and strain ATCC 43603, a nontoxigenic clinical isolate from Belgium (Table 1). Bacteria were grown overnight at 37°C in an anaerobic cabinet (Jacomex), in peptone yeast glucose infusion broth (PYG; Difco Laboratories) containing or not containing subinhibitory concentrations of antibiotic. The antibiotics used in this study were ampicillin (Eurobio), clindamycin (Dalacine; Pfizer), ofloxacin (Sigma-Aldrich), and moxifloxacin (Izilox; Bayer).
TABLE 1.
Bacterial strains used in this study
| Strain | Propertiesc | Resistance to new FQa | Amino acid substitution inb: |
|
|---|---|---|---|---|
| GyrA | GyrB | |||
| ATCC 43603 | TcdA− TcdB−; clinical isolate from a newborn carrier (Belgium) | − | None | None |
| ATCC 43603-M1 | TcdA− TcdB−; in vitro moxifloxacin-resistant mutant of ATCC 43603 | + | Thr82→Ile (ACT→ATT) | Ser366→Ala (TCA→GCA) |
| CD196 (CIP10793) | TcdA+ TcdB+; historical NAP1/027, clinical isolate from CDI (France) | − | None | None |
| CD196-M1 | TcdA+ TcdB+; in vitro moxifloxacin-resistant mutant from CD196 | + | Thr82→Ile (ACT→ATT) | Gln434→Lys (CAA→AAA) |
| CD07-259 | TcdA+ TcdB+; current NAP1/027, clinical isolate from CDI (Nord-Pas-de-Calais, France) | + | Thr82→Ile (ACT→ATT) | None |
| 6296 | TcdA+ TcdB+; current NAP1/027, REA-type BI-11, clinical isolate from CDI (United States) | + | None | None |
| 6425 | TcdA+ TcdB+; current NAP1/027, REA type BI-12, clinical isolate from CDI (United States) | + | Thr82→Ile (ACT→ATT) | None |
New FQ, new fluoroquinolone moxifloxacin; −, susceptible strain (MIC < 8 μg/ml); +, resistant strain (MIC ≥ 8 μg/ml).
Compared to the GyrA and the GyrB sequences in strain 630.
REA, restriction endonuclease analysis.
MICs were determined by the broth dilution method with Bacteroides thetaiotaomicron ATCC 29741 as control and were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) breakpoints.
For RNA and protein extraction experiments, antibiotics were added to the growth medium to a final concentration of 0.5× MIC. All the cultures were incubated until the stationary phase was reached. Optical density measures and viable count determinations were assessed to exclude bias errors due to differential growth.
In vitro selection of moxifloxacin-resistant mutants.
Two moxifloxacin-resistant mutants were selected from the moxifloxacin-susceptible strains CD196 (NAP1/027) and ATCC 43603 (non-NAP1/027). These two parental strains were chosen in order to evaluate the relative roles of the fluoroquinolone resistance and/or the genetic background of these strains in the increased gene expression. Bacteria were grown overnight in antibiotic-free medium and then spread on Columbia cysteine agar containing moxifloxacin at 1× and 2× MIC (2 μg/ml and 4 μg/ml moxifloxacin). After 3 days of incubation at 37°C in an anaerobic cabinet, colonies were subcultured onto Columbia cysteine agar plates containing 2× MIC of moxifloxacin. One colony of each strain was selected, and the MIC was confirmed. The mutants were designated ATCC 43603-M1 and CD196-M1. The stability of the mutants was assessed by repeated passages on PYG medium.
Molecular analysis of the tcdC gene and the quinolone resistance-determining region.
Genomic DNA was extracted using either a GFX genomic DNA purification kit (GE Healthcare) or a QIAamp DNA kit (Qiagen) according to the manufacturer's instructions. Primers OBD81 and OBD82 were used to amplify a 720-bp fragment encompassing the entire tcdC gene as previously described (39). Primers gyrA1 (5′-AATGAGTGTTATAGCTGGAGG-3′) and gyrA1-R1 (5′-TCTTTTAACGACTCATCAAAG-3′) were used to amplify a 390-bp fragment of gyrA (nucleotide positions 71 to 460), and primers gyrB1 (5′-ATGTGATGAACTGGGGTCTT-3′) and gyrB1-R1 (5′-TCAAAATCTTCTCCAATACCA-3′) were used to amplify a 390-bp fragment of gyrB (nucleotide positions 1059 to 1448). Purified amplicons were sequenced by using a BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems) according to the manufacturer's instructions. Analysis was performed either with an ABI PRISM 310 or with a 3100 genetic analyzer (Perkin-Elmer Applied Biosystems).
Gene expression analysis. (i) RNA isolation and cDNA synthesis.
Optical density measures and viable count determinations confirmed that all cultures reached stationary phase and that RNA extraction was performed on equivalent cell numbers. Total RNA was isolated from at least two independent cultures (biological replicate) by using Trizol reagent (Invitrogen) according to a previously described method (14). The RNA concentration and quality were determined using an Agilent Bioanalyzer 2100 (Roche) and RNA 6000 Nano kit reagents & supplies (Roche). For each RNA sample, absence of DNA contamination was controlled by performing a real-time PCR on RNA templates (with the same conditions as for the expression study) using primers specific for the rrs gene (encoding the 16S rRNA subunit). First-strand DNA synthesis was then performed on 5 μg of purified total RNA with random primers by the use of SuperScriptIII reverse transcriptase (Invitrogen) according to the manufacturer's instructions.
(ii) Real-time PCR.
Primers for cwp84, slpA, fbp68, and rrs were designed from the genome sequences of C. difficile 630 and QCD-32g58 (Canadian NAP1/027) (Table 2). Real-time PCR was carried out on a LightCycler instrument using a LightCycler FastStart DNA MasterPLUS SYBR green I kit (Roche). The dilutions of cDNA used in the study were 1:5,000,000 for the rrs gene, 1:50 for cwp84 and fbp68, and 1:5,000 for slpA. Five microliters of cDNA was added to 5 μl of PCR mixture (2 μl master mix, 0.5 μM of each specific primer, and 2 μl RNase-free water). The following PCR profile was used for amplification: initial denaturation for 8 min at 95°C and amplification for 45 cycles of 5 s at 95°C, 5 s at 60°C, and 6 s at 72°C. An additional step from a start at 70 to 95°C (0.1°C/s) was performed to establish a melting curve and was used to verify the specificity of the real-time PCR for each primer pair.
TABLE 2.
Primers sequences used for real-time PCR
| Gene | Forward primer sequence | Reverse primer sequence | Amplicon size (bp) |
|---|---|---|---|
| cwp84 | TGGGCAACTGGTGGAAAATA | TAGTTGCACCTTGTGCCTCA | 151 |
| slpA | AATGATAAAGCATTTGTAGTTGGTG | TATTGGAGTAGCATCTCCATC | 126 |
| fbp68 | AGTTCGTCAAGTTTTACCTGGTC | GGTCCTTCCAATTCCTCTAGGT | 120 |
| rrs | GGGAGACTTGAGTGCAGGAG | GTGCCTCAGCGTCAGTTACA | 120 |
(iii) Analysis of the results.
The results were normalized to those for the rrs gene and analyzed using the comparative critical threshold (ΔΔCT) method. For each RNA extract (two biological replicates under each condition), measurements were performed in triplicate for each gene tested. The comparison of the relative expression ratios of the two biological replicates for each strain allowed us to consider that genes were significantly upregulated if their relative expression level was found to be at least 3. Results were expressed as means ± standard deviations.
Analysis of Cwp84 production by immunoblotting.
Glycine extracts containing S-layer proteins were prepared in triplicate from 2 ml of fresh, independent, overnight broth culture as previously described (9). The protein extract concentration was determined by the Bradford method, using Bio-Rad protein assay reagent with bovine serum albumin as a standard. Equivalent amounts of each protein extract (10 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a Hybond-P polyvinylidene difluoride membrane (Amersham, GE Healthcare) for immunoblotting. Immunoblotting with specific polyclonal anti-Cwp84 antibodies (1:4,000 dilution in blocking buffer) was performed as previously described (32). Quantitative differences in Cwp84 production were estimated by densitometric analysis with an imaging system (ImageJ), and results were expressed as means ± standard deviations.
Statistical analysis.
The nonparametric Wilcoxon test was performed to evaluate the significance of differences in the expression levels of colonization factor-encoding genes between (i) in vitro-selected moxifloxacin-resistant mutants and their parental strains and (ii) moxifloxacin-susceptible and moxifloxacin-resistant strains. P values of <0.05, with a two-tailed significance level, were considered significant.
RESULTS
Antimicrobial susceptibilities.
Table 3 shows the MICs of ampicillin, clindamycin, ofloxacin, and moxifloxacin for the five clinical isolates and the two in vitro-selected moxifloxacin-resistant mutants studied. Ampicillin displayed MICs of ≥2 μg/ml for all the strains tested. Among the five clinical isolates, two strains were resistant to clindamycin (MIC = 128 μg/ml). The nontoxigenic strain ATCC 43603 and the historical NAP1/027 strain CD196 were susceptible to moxifloxacin (MIC = 2 μg/ml), and the three current NAP1/027 strains were resistant (MIC ≥ 8 μg/ml). All the strains were resistant to ofloxacin (MIC > 4 μg/ml), but the two moxifloxacin-susceptible strains showed a low level of resistance (MIC = 8 μg/ml).
TABLE 3.
MICs of the antibiotics used in this study
| Strain | MIC (μg/ml)a |
|||
|---|---|---|---|---|
| Ampicillin | Clindamycin | Ofloxacin | Moxifloxacin | |
| ATCC 43603 | 2 | 4 | 8 | 2 |
| ATCC 43603-M1 | ND | ND | 256 | 64 |
| CD196 | 8 | 128 | 8 | 2 |
| CD196-M1 | ND | ND | 128 | 128 |
| CD07-259 | 4 | 4 | 256 | 32 |
| 6296 | 2 | 1 | 32 | 8 |
| 6425 | 4 | 128 | 256 | 32 |
ND, not determined.
Following selection for moxifloxacin resistance, the two mutant strains, ATCC 43603-M1 and CD196-M1, displayed high-level resistance to moxifloxacin (MICs of 64 and 128 μg/ml, respectively) and ofloxacin (MIC ≥ 128 μg/ml).
Detection of gyrA and gyrB mutations.
The quinolone resistance-determining regions of gyrA and gyrB were sequenced from nucleotide codons 40 to 145 and 354 to 482, respectively. No mutation in gyrA or gyrB was found in the two moxifloxacin-susceptible strains, including the historical NAP1/027 strain. In contrast, in two of the three NAP1/027 moxifloxacin-resistant strains (6425 and CD07-259) and in the two in vitro-selected moxifloxacin-resistant mutants, a point mutation (C→T transition) in gyrA was identified, leading to amino acid substitution Thr82→Ile (Table 1). An additional point mutation in gyrB was found in each in vitro-selected moxifloxacin-resistant mutant, resulting in amino acid substitutions Ser366→Ala in mutant ATCC 43603-M1 and Gln434→Lys in mutant CD196-M1 (Table 1). No mutation was found in either gyrA or gyrB in the third NAP1/027 moxifloxacin-resistant clinical isolate (6296), which showed a low level of resistance to moxifloxacin (MIC = 8 μg/ml).
Analysis of partial deletion and frameshift mutation of the tcdC gene.
The tcdC gene was sequenced from the four C. difficile NAP1/027 strains and compared to the published tcdC gene for strain 630. We showed that all NAP1/027 strains studied carry not only the 18-bp deletion in tcdC at positions 330 to 347 but also the 1-bp deletion at position 117 relative to the ATG start codon. In addition, no single nucleotide polymorphism appears when the tcdC genes of the four NAP1/027 strains are compared to each other.
Effects of subinhibitory concentrations of antibiotics on the expression of genes encoding colonization factors in C. difficile NAP1/027 strains.
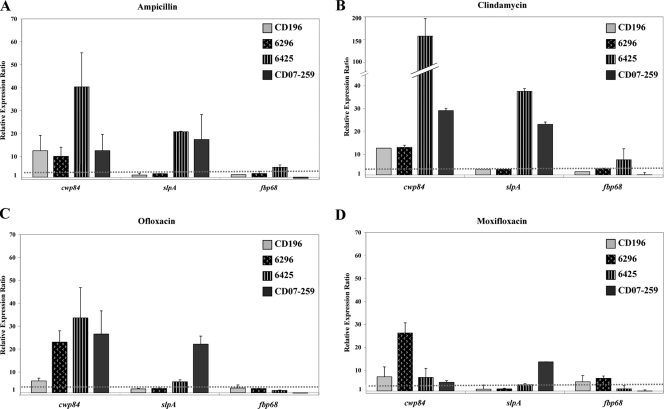
The expression of cwp84, encoding a cysteine protease; slpA, encoding the precursor of the S-layer proteins; and fbp68, encoding a fibronectin-binding protein, was analyzed in the four C. difficile NAP1/027 strains grown in the presence of subinhibitory concentrations of ampicillin, clindamycin, ofloxacin, or moxifloxacin (0.5× MIC). In the presence of the four antibiotics tested, a general upregulation of the cwp84 and slpA genes in the four NAP1/027 strains was observed. However, the level of regulation varied between genes and strains (Fig. 1). The fbp68 gene was marginally influenced by the presence of these antibiotics (Fig. 1).
FIG. 1.
Effects of subinhibitory concentrations of ampicillin (A), clindamycin (B), ofloxacin (C), and moxifloxacin (D) on the expression of C. difficile colonization factor-encoding genes in four NAP1/027 strains: one moxifloxacin susceptible (strain CD196) and three moxifloxacin resistant (strains 6296, 6425, and CD07-259). Bacteria were grown overnight in medium supplemented with 0.5× MIC of antibiotic. The data are the mean relative expression ratios (expression in bacteria grown with antibiotic versus expression in control grown in antibiotic-free medium) ± standard deviations. The dotted line represents the threshold value of upregulation (threefold).
Exposure to subinhibitory concentrations of ampicillin or clindamycin led to the upregulation of slpA and, especially, cwp84. The levels of upregulation were variable and were not correlated with the levels of resistance to ampicillin and clindamycin in the strains tested (Fig. 1A and B).
In contrast, the impact of fluoroquinolones on the regulation of slpA and cwp84 appears to differ in moxifloxacin-susceptible and moxifloxacin-resistant strains. Exposure to ofloxacin led to a strong increase in cwp84 expression (23- to 34-fold) in the three moxifloxacin-resistant strains; exposure to moxifloxacin led to a strong, 26-fold increase in cwp84 expression in one of the three moxifloxacin-resistant strains (6296) and a moderate increase in the two other resistant strains. Exposure to ofloxacin and moxifloxacin led to strong increases in the expression of slpA in strain CD07-259 (22- and 14-fold, respectively) and to weak increases in strain 6425 (6- and 4-fold, respectively). In contrast, in the historical NAP1/027 moxifloxacin-susceptible strain CD196, exposure to ofloxacin and moxifloxacin led to weak increases in the expression of cwp84 (six- and sevenfold, respectively) and did not significantly modify the expression of slpA (Fig. 1C and D).
Effects of subinhibitory concentrations of antibiotics on the expression of genes encoding colonization factors in in vitro-selected moxifloxacin-resistant C. difficile mutants.
In order to determine the differences in regulation observed between C. difficile-susceptible and -resistant strains exposed to fluoroquinolones, we analyzed the expression of cwp84, slpA, and fbp68 in the presence of subinhibitory concentrations of ofloxacin or moxifloxacin (0.5× MIC) in in vitro-selected moxifloxacin-resistant mutants.
As previously observed, the expression of fbp68 was not influenced by the presence of these antibiotics (Table 4).
TABLE 4.
Effects of subinhibitory concentrations of fluoroquinolones on the expression of genes encoding colonization factors of C. difficile in the two in vitro-selected moxifloxacin-resistant mutants and their parental strains
| Gene | Antibiotic stressa | Mean relative expression ratiob ± SD |
|||||
|---|---|---|---|---|---|---|---|
| NAP1/027 strain |
Non-NAP1/027 strain |
||||||
| CD196 | CD196-M1 | P value | ATCC 43603 | ATCC 43603-M1 | P value | ||
| cwp84 | OFX | 6.1 ± 1.3 | 11.7 ± 4.1 | 0.132c | 3.2 ± 1.1 | 67.4 ± 34.0 | 0.009c |
| MXF | 7.1 ± 4.3 | 2.9 ± 0.2 | 0.485 | 3.1 ± 1.8 | 17.3 ± 5.4 | 0.024c | |
| slpA | OFX | 2.7 ± 0.2 | 4.3 ± 2.4 | 0.610 | 3.9 ± 0.1 | 32.8 ± 24.7 | 0.114 |
| MXF | 1.8 ± 1.8 | 1.9 ± 0.8 | 0.575 | 2.9 ± 2.7 | 16.5 ± 11.8 | 0.262 | |
| fbp68 | OFX | 3.1 ± 1.3 | 2.3 ± 1.8 | 0.381 | 4.8 ± 1.7 | 2.1 ± 1.0 | 0.267 |
| MXF | 5.0 ± 2.8 | 1.1 ± 0.9 | 0.048 | 2.1 ± 1.9 | 0.8 ± 0.2 | 0.800 | |
OFX, ofloxacin; MXF, moxifloxacin.
Ratio of expression in bacteria grown with antibiotic versus expression in control grown without antibiotic. P values for the comparison of the upregulation of genes between the moxifloxacin-resistant mutant and its parental strain were derived using the Wilcoxon test.
The difference between expression levels in the moxifloxacin-resistant mutant and its parental strain was significant.
In the NAP1/027 moxifloxacin-resistant mutant strain CD196-M1, exposure to ofloxacin and moxifloxacin led to nonsignificant upregulation in the expression of cwp84 and slpA compared to their expression in its parental strain CD196 (Table 4).
In the nontoxigenic mutant strain ATCC 43603-M1, exposure to fluoroquinolones resulted in the general upregulation of cwp84 and slpA expression (Table 4). The levels of overexpression of cwp84 with ofloxacin and moxifloxacin were significantly higher (P < 0.01 and P < 0.05, respectively) than the level of expression in the moxifloxacin-susceptible parent; the expression of slpA was also upregulated, but not significantly so, compared to its expression in the parental strain (P < 0.3) (Table 4).
These results suggest that the acquisition of antibiotic resistance could modify, at least for those strains, the regulation of colonization factor genes in response to antibiotic stress.
Effects of subinhibitory concentrations of antibiotics on the production of Cwp84 protease.
To correlate levels of gene upregulation and protein production, we analyzed the effects of fluoroquinolones on the production of the Cwp84 protease. Immunoblotting was performed on glycine extracts of the four C. difficile NAP1/027 isolates, the ATCC 43603 parental strain, and the two in vitro-selected moxifloxacin-resistant mutants.
Densitometric analyses of immunoblots revealed that growth in the presence of subinhibitory concentrations of ofloxacin or moxifloxacin has a tendency to increase Cwp84 production in the three NAP1/027 moxifloxacin-resistant strains and the mutant strain ATCC 43603-M1 (Fig. 2). These results correlate with the increase of cwp84 transcriptional expression previously observed.
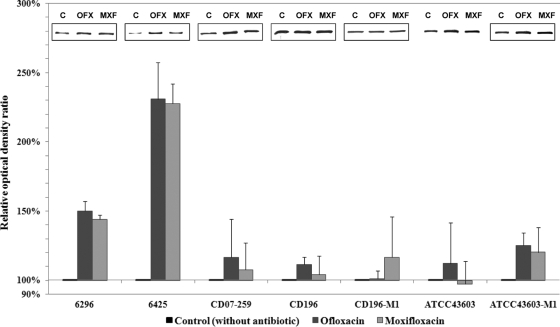
FIG. 2.
Effect of subinhibitory concentrations of ofloxacin and moxifloxacin on the production of protease Cwp84 in three moxifloxacin-resistant NAP1/027 strains (6296, 6425, and CD07-259), a moxifloxacin-susceptible NAP1/027 strain (CD196), a non-NAP1/027 strain (ATCC 43603), and the two in vitro-selected moxifloxacin-resistant mutants (CD196-M1 and ATCC 43603-M1). Immunoblots and densitometry analyses of glycine extracts were performed. Anti-Cwp84 antibodies reacted with a unique band of 85-kDa protein. Histograms show mean relative optical density ratios (optical density of extract from bacteria grown with antibiotic versus optical density of extract from control grown in antibiotic-free medium) ± standard deviations; optical densities were determined by densitometry scanning. The inserts show examples of Cwp84 immunoblots for each strain grown in the absence (C, control without antibiotic) or presence of 0.5× MIC of ofloxacin (OFX) or moxifloxacin (MXF).
The production of the protease was increased strongly in strain 6425 (approximately 130% in the presence of ofloxacin or moxifloxacin) and moderately in strains 6296, CD07-259, and ATCC 43603-M1. The production of the protease was also moderately increased in the historical NAP1/027 moxifloxacin-susceptible strain CD196, as well as in the mutant strain CD196-M1 (Fig. 2).
On the other hand, in the mutant strain ATCC 43603-M1, growth in the presence of ofloxacin or moxifloxacin, respectively, led to 25% and 20% increases in Cwp84 production, while the level of production was not significantly modified in its parental strain, ATCC 43603 (Fig. 2).
DISCUSSION
In this study, we investigated the influence of antibiotics on colonization factor expression in six moxifloxacin-susceptible or moxifloxacin-resistant C. difficile strains, including four NAP1/027 strains and two in vitro-selected moxifloxacin-resistant mutants.
No mutation was found in gyrA or gyrB in the two moxifloxacin-susceptible strains tested (Table 1). The high-level fluoroquinolone resistance of NAP1/027 strains CD07-259 and 6425 and of the in vitro-selected moxifloxacin-resistant mutants was associated with the usual amino acid substitution in GyrA (Thr82→Ile) (1, 2, 16, 19, 31, 51). An additional amino acid substitution in GyrB was found in each in vitro-selected mutant (20). In contrast, no mutation was found in gyrA or in gyrB in the NAP1/027 epidemic strain 6296. Since C. difficile lacks genes for topoisomerase IV, the moderate moxifloxacin resistance of this strain may then be explained by the reduction of the intracellular accumulation of the antibiotic, mediated by a drug efflux mechanism (1, 16). Nevertheless, several attempts to identify an efflux mechanism in C. difficile have failed (16, 20, 36).
There is increasing evidence that antibiotics at concentrations below the MIC possess biological activities that could influence the expression of virulence factors in many pathogenic gram-positive bacteria (13). In Staphylococcus aureus, the expression of the gene encoding the alpha-toxin Hla is induced to different degrees by exposure to subinhibitory concentrations of beta-lactams and fluoroquinolones and repressed by clindamycin (41). In contrast, clindamycin has been shown to stimulate the production of some exoproteins from staphylococci and Streptococcus pyogenes (29, 41, 49, 54).
Concerning C. difficile, the impact of subinhibitory concentrations of antibiotics on the production of TcdA and TcdB was subjected to numerous investigations, sometimes leading to disparate results (3, 21, 25, 30, 42, 48). However, some authors have suggested that the presence of antibiotics could facilitate toxin production (4, 23).
In contrast to these numerous studies of toxin expression, few studies have been performed on the impact of antibiotics on colonization factor expression. It has been previously shown that C. difficile grown in the presence of ampicillin produced greater levels of GroEL, a heat shock protein with adhesive properties (27). More recently, a study of the entire transcriptome of C. difficile strain 630 showed that antibiotic stress increased the transcription of genes encoding surface-associated proteins, including putative colonization factors (22).
We previously showed that exposure to ampicillin and clindamycin increased the expression of colonization factor-encoding genes by non-NAP1/027 moxifloxacin-susceptible C. difficile strains. This increase was correlated with increased adherence to Caco2-TC7 cells. In contrast, ofloxacin and moxifloxacin did not have such an effect (14). In this study, we showed that fluoroquinolones could have a different effect on C. difficile moxifloxacin-resistant strains. No significant difference in response to ampicillin and clindamycin stress was observed between moxifloxacin-resistant and moxifloxacin-susceptible strains. In contrast, the influence of ofloxacin on cwp84 expression in moxifloxacin-resistant strains was strongly and significantly greater than in moxifloxacin-susceptible strains, including the historical NAP1/027 strain CD196 (32.5-fold mean increase in moxifloxacin-resistant strains versus 3.4-fold mean increase in moxifloxacin-susceptible strains, P < 0.01). In addition, the influence of ofloxacin on slpA expression was also greater in moxifloxacin-resistant strains (13.6-fold mean increase versus 3-fold mean increase in moxifloxacin-susceptible strains), but this difference was not significant (P = 0.052). The influence of moxifloxacin on the expression of cwp84 and slpA in moxifloxacin-resistant strains also appeared to be greater, but the changes in expression levels were not significantly different from those in moxifloxacin-susceptible strains.
In order to evaluate the respective roles of the acquisition of fluoroquinolone resistance and/or the genetic background of the strains in these differences in colonization factor gene regulation, we selected moxifloxacin-resistant mutants in vitro from one non-NAP1/027 strain (ATCC 43603) and the historical NAP1/027 moxifloxacin-susceptible strain (CD196). In the mutant strain ATCC 43603-M1, fluoroquinolones appear to upregulate colonization factor genes cwp84 and slpA.
The regulation of genes appears to vary according to the gene. In our study, cwp84 was the most upregulated gene in the presence of fluoroquinolones. The increased production of the protease Cwp84 observed in the glycine extracts did not correlate perfectly with the gene's overexpression. However, Cwp84 is not only associated with the cell wall but is also secreted (unpublished data). This could explain some of the discrepancies between gene upregulation and protein production.
In addition, an interstrain variability was observed despite the limited number of strains studied. This variability could concern the genetic environment of target genes and, also, regulatory genes. Although these NAP1/027 strains display the same tcdC organization and are clonally related, they appear to be differently regulated regarding their response to fluoroquinolone stress. This microheterogeneity and microevolution between hypervirulent NAP1/027 strains has already been suggested (52). We have shown here, for the NAP1/027 nonepidemic strain CD196, that the acquisition of fluoroquinolone resistance in the in vitro-selected mutant was insufficient to increase the expression of colonization factor genes in response to fluoroquinolone stress. The regulation of expression in this historical NAP1/027 strain appears to differ from that in the current NAP1/027 epidemic strains. This diversity of gene regulation between epidemic and nonepidemic strains has to be confirmed by the analysis of a larger panel of strains.
The molecular mechanisms of the regulatory effects of fluoroquinolones on colonization factor gene expression are not yet defined. Fluoroquinolones interact with DNA gyrase, leading to alterations in DNA supercoiling, which is known to affect gene transcription (17, 31). In addition, fluoroquinolones can induce the SOS response, which is recognized as a critical component of the response to environmental stress (12). Several studies have previously suggested that the stimulation or depression of bacterial gene expression by subinhibitory concentrations of many antibiotics, whatever their chemical structures and inhibitory actions, occurs at the transcriptional level (6, 26, 29).
In quinolone-resistant S. aureus, it was shown that the expression of the two fibronectin-binding proteins was increased by subinhibitory concentrations of ciprofloxacin. This was correlated with an increased bacterial adhesion, and the authors have suggested that this may have contributed to the emergence of highly fluoroquinolone-resistant strains (6, 7).
In conclusion, our results suggest that antibiotics could lead to CDI not only by disruption of the barrier microbiota and its colonization resistance but also by inducing a bacterial stress response leading to enhanced expression of colonization factors, at least in the epidemic fluoroquinolone-resistant NAP1/027 strains studied. Antibiotics, then, may help C. difficile to colonize recently vacated niches. After oral administration, fluoroquinolones achieve high concentrations in human stools but the major part is bound to fecal material, leading to reduced amounts of active drug (53). These residual concentrations of fluoroquinolones may favor the adaptation of some C. difficile fluoroquinolone-resistant strains to the host environment. The genetic specificity and the colonization fitness of the C. difficile NAP1/027 epidemic strains combined with their antibiotic resistance may have contributed to their selection over other strains and their worldwide spread.
Acknowledgments
We thank Marc Monot from the Institut Pasteur (Paris, France) for statistical analysis assistance. We also thank Michel R. Popoff from the Institut Pasteur (Paris, France) and Dale N. Gerding from the Hines Veterans Affairs Hospital (Chicago, IL) for providing C. difficile CD196 and NAP1/027 clinical isolates 6296 and 6425, respectively.
Footnotes
Published ahead of print on 5 October 2009.
REFERENCES
- 1.Ackermann, G., Y. J. Tang, R. Kueper, P. Heisig, A. C. Rodloff, J. Silva, Jr., and S. H. Cohen. 2001. Resistance to moxifloxacin in toxigenic Clostridium difficile isolates is associated with mutations in gyrA. Antimicrob. Agents Chemother. 45:2348-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackermann, G., Y. J. Tang-Feldman, R. Schaumann, J. P. Henderson, A. C. Rodloff, J. Silva, and S. H. Cohen. 2003. Antecedent use of fluoroquinolones is associated with resistance to moxifloxacin in Clostridium difficile. Clin. Microbiol. Infect. 9:526-530. [DOI] [PubMed] [Google Scholar]
- 3.Adams, D. A., M. M. Riggs, and C. J. Donskey. 2007. Effect of fluoroquinolone treatment on growth of and toxin production by epidemic and nonepidemic Clostridium difficile strains in the cecal contents of mice. Antimicrob. Agents Chemother. 51:2674-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines, S. D., J. Freeman, and M. H. Wilcox. 2005. Effects of piperacillin/tazobactam on Clostridium difficile growth and toxin production in a human gut model. J. Antimicrob. Chemother. 55:974-982. [DOI] [PubMed] [Google Scholar]
- 5.Biller, P., B. Shank, L. Lind, M. Brennan, L. Tkatch, G. Killgore, A. Thompson, and L. C. McDonald. 2007. Moxifloxacin therapy as a risk factor for Clostridium difficile-associated disease during an outbreak: attempts to control a new epidemic strain. Infect. Control Hosp. Epidemiol. 28:198-201. [DOI] [PubMed] [Google Scholar]
- 6.Bisognano, C., P. Vaudaux, P. Rohner, D. P. Lew, and D. C. Hooper. 2000. Induction of fibronectin-binding proteins and increased adhesion of quinolone-resistant Staphylococcus aureus by subinhibitory levels of ciprofloxacin. Antimicrob. Agents Chemother. 44:1428-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bisognano, C., P. E. Vaudaux, D. P. Lew, E. Y. Ng, and D. C. Hooper. 1997. Increased expression of fibronectin-binding proteins by fluoroquinolone-resistant Staphylococcus aureus exposed to subinhibitory levels of ciprofloxacin. Antimicrob. Agents Chemother. 41:906-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calabi, E., F. Calabi, A. D. Phillips, and N. F. Fairweather. 2002. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect. Immun. 70:5770-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calabi, E., S. Ward, B. Wren, T. Paxton, M. Panico, H. Morris, A. Dell, G. Dougan, and N. Fairweather. 2001. Molecular characterization of the surface layer proteins from Clostridium difficile. Mol. Microbiol. 40:1187-1199. [DOI] [PubMed] [Google Scholar]
- 10.Cerquetti, M., A. Molinari, A. Sebastianelli, M. Diociaiuti, R. Petruzzelli, C. Capo, and P. Mastrantonio. 2000. Characterization of surface layer proteins from different Clostridium difficile clinical isolates. Microb. Pathog. 28:363-372. [DOI] [PubMed] [Google Scholar]
- 11.Cerquetti, M., A. Pantosti, P. Stefanelli, and P. Mastrantonio. 1992. Purification and characterization of an immunodominant 36 kDa antigen present on the cell surface of Clostridium difficile. Microb. Pathog. 13:271-279. [DOI] [PubMed] [Google Scholar]
- 12.Cirz, R. T., M. B. Jones, N. A. Gingles, T. D. Minogue, B. Jarrahi, S. N. Peterson, and F. E. Romesberg. 2007. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J. Bacteriol. 189:531-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies, J., G. B. Spiegelman, and G. Yim. 2006. The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol. 9:445-453. [DOI] [PubMed] [Google Scholar]
- 14.Deneve, C., C. Delomenie, M. C. Barc, A. Collignon, and C. Janoir. 2008. Antibiotics involved in Clostridium difficile-associated disease increase colonization factor gene expression. J. Med. Microbiol. 57:732-738. [DOI] [PubMed] [Google Scholar]
- 15.Deshpande, A., C. Pant, A. Jain, T. G. Fraser, and D. D. Rolston. 2008. Do fluoroquinolones predispose patients to Clostridium difficile associated disease? A review of the evidence. Curr. Med. Res. Opin. 24:329-333. [DOI] [PubMed] [Google Scholar]
- 16.Dridi, L., J. Tankovic, B. Burghoffer, F. Barbut, and J. C. Petit. 2002. gyrA and gyrB mutations are implicated in cross-resistance to ciprofloxacin and moxifloxacin in Clostridium difficile. Antimicrob. Agents Chemother. 46:3418-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drudy, D., E. Calabi, L. Kyne, S. Sougioultzis, E. Kelly, N. Fairweather, and C. P. Kelly. 2004. Human antibody response to surface layer proteins in Clostridium difficile infection. FEMS Immunol. Med. Microbiol. 41:237-242. [DOI] [PubMed] [Google Scholar]
- 19.Drudy, D., L. Kyne, R. O'Mahony, and S. Fanning. 2007. gyrA mutations in fluoroquinolone-resistant Clostridium difficile PCR-027. Emerg. Infect. Dis. 13:504-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drudy, D., T. Quinn, R. O'Mahony, L. Kyne, P. O'Gaora, and S. Fanning. 2006. High-level resistance to moxifloxacin and gatifloxacin associated with a novel mutation in gyrB in toxin-A-negative, toxin-B-positive Clostridium difficile. J. Antimicrob. Chemother. 58:1264-1267. [DOI] [PubMed] [Google Scholar]
- 21.Drummond, L. J., D. G. Smith, and I. R. Poxton. 2003. Effects of sub-MIC concentrations of antibiotics on growth of and toxin production by Clostridium difficile. J. Med. Microbiol. 52:1033-1038. [DOI] [PubMed] [Google Scholar]
- 22.Emerson, J. E., R. A. Stabler, B. W. Wren, and N. F. Fairweather. 2008. Microarray analysis of the transcriptional responses of Clostridium difficile to environmental and antibiotic stress. J. Med. Microbiol. 57:757-764. [DOI] [PubMed] [Google Scholar]
- 23.Freeman, J., F. J. O'Neill, and M. H. Wilcox. 2003. Effects of cefotaxime and desacetylcefotaxime upon Clostridium difficile proliferation and toxin production in a triple-stage chemostat model of the human gut. J. Antimicrob. Chemother. 52:96-102. [DOI] [PubMed] [Google Scholar]
- 24.Freeman, J., and M. H. Wilcox. 1999. Antibiotics and Clostridium difficile. Microbes Infect. 1:377-384. [DOI] [PubMed] [Google Scholar]
- 25.Gerber, M., C. Walch, B. Loffler, K. Tischendorf, U. Reischl, and G. Ackermann. 2008. Effect of sub-MIC concentrations of metronidazole, vancomycin, clindamycin and linezolid on toxin gene transcription and production in Clostridium difficile. J. Med. Microbiol. 57:776-783. [DOI] [PubMed] [Google Scholar]
- 26.Goh, E. B., G. Yim, W. Tsui, J. McClure, M. G. Surette, and J. Davies. 2002. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. USA 99:17025-17030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hennequin, C., A. Collignon, and T. Karjalainen. 2001. Analysis of expression of GroEL (Hsp60) of Clostridium difficile in response to stress. Microb. Pathog. 31:255-260. [DOI] [PubMed] [Google Scholar]
- 28.Hennequin, C., C. Janoir, M. C. Barc, A. Collignon, and T. Karjalainen. 2003. Identification and characterization of a fibronectin-binding protein from Clostridium difficile. Microbiology 149:2779-2787. [DOI] [PubMed] [Google Scholar]
- 29.Herbert, S., P. Barry, and R. P. Novick. 2001. Subinhibitory clindamycin differentially inhibits transcription of exoprotein genes in Staphylococcus aureus. Infect. Immun. 69:2996-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honda, T., I. Hernadez, T. Katoh, and T. Miwatani. 1983. Stimulation of enterotoxin production of Clostridium difficile by antibiotics. Lancet 1:655. [DOI] [PubMed] [Google Scholar]
- 31.Hooper, D. C. 1999. Mechanisms of fluoroquinolone resistance. Drug Resist. Updat. 2:38-55. [DOI] [PubMed] [Google Scholar]
- 32.Janoir, C., S. Pechine, C. Grosdidier, and A. Collignon. 2007. Cwp84, a surface-associated protein of Clostridium difficile, is a cysteine protease with degrading activity on extracellular matrix proteins. J. Bacteriol. 189:7174-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, S., M. H. Samore, K. A. Farrow, G. E. Killgore, F. C. Tenover, D. Lyras, J. I. Rood, P. DeGirolami, A. L. Baltch, M. E. Rafferty, S. M. Pear, and D. N. Gerding. 1999. Epidemics of diarrhea caused by a clindamycin-resistant strain of Clostridium difficile in four hospitals. N. Engl. J. Med. 341:1645-1651. [DOI] [PubMed] [Google Scholar]
- 34.Karjalainen, T., A. J. Waligora-Dupriet, M. Cerquetti, P. Spigaglia, A. Maggioni, P. Mauri, and P. Mastrantonio. 2001. Molecular and genomic analysis of genes encoding surface-anchored proteins from Clostridium difficile. Infect. Immun. 69:3442-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuijper, E. J., B. Coignard, and P. Tull. 2006. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin. Microbiol. Infect. 12(Suppl. 6):2-18. [DOI] [PubMed] [Google Scholar]
- 36.Lebel, S., S. Bouttier, and T. Lambert. 2004. The cme gene of Clostridium difficile confers multidrug resistance in Enterococcus faecalis. FEMS Microbiol. Lett. 238:93-100. [DOI] [PubMed] [Google Scholar]
- 37.Loo, V. G., L. Poirier, M. A. Miller, M. Oughton, M. D. Libman, S. Michaud, A. M. Bourgault, T. Nguyen, C. Frenette, M. Kelly, A. Vibien, P. Brassard, S. Fenn, K. Dewar, T. J. Hudson, R. Horn, P. Rene, Y. Monczak, and A. Dascal. 2005. A predominantly clonal institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442-2449. [DOI] [PubMed] [Google Scholar]
- 38.MacCannell, D. R., T. J. Louie, D. B. Gregson, M. Laverdiere, A. C. Labbe, F. Laing, and S. Henwick. 2006. Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from eastern and western Canada. J. Clin. Microbiol. 44:2147-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matamouros, S., P. England, and B. Dupuy. 2007. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol. Microbiol. 64:1274-1288. [DOI] [PubMed] [Google Scholar]
- 40.McDonald, L. C., G. E. Killgore, A. Thompson, R. C. Owens, Jr., S. V. Kazakova, S. P. Sambol, S. Johnson, and D. N. Gerding. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433-2441. [DOI] [PubMed] [Google Scholar]
- 41.Ohlsen, K., W. Ziebuhr, K. P. Koller, W. Hell, T. A. Wichelhaus, and J. Hacker. 1998. Effects of subinhibitory concentrations of antibiotics on alpha-toxin (hla) gene expression of methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 42:2817-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onderdonk, A. B., B. R. Lowe, and J. G. Bartlett. 1979. Effect of environmental stress on Clostridium difficile toxin levels during continuous cultivation. Appl. Environ. Microbiol. 38:637-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pechine, S., A. Gleizes, C. Janoir, R. Gorges-Kergot, M. C. Barc, M. Delmee, and A. Collignon. 2005. Immunological properties of surface proteins of Clostridium difficile. J. Med. Microbiol. 54:193-196. [DOI] [PubMed] [Google Scholar]
- 44.Pechine, S., C. Janoir, and A. Collignon. 2005. Variability of Clostridium difficile surface proteins and specific serum antibody response in patients with Clostridium difficile-associated disease. J. Clin. Microbiol. 43:5018-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pepin, J., N. Saheb, M. A. Coulombe, M. E. Alary, M. P. Corriveau, S. Authier, M. Leblanc, G. Rivard, M. Bettez, V. Primeau, M. Nguyen, C. E. Jacob, and L. Lanthier. 2005. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin. Infect. Dis. 41:1254-1260. [DOI] [PubMed] [Google Scholar]
- 46.Pepin, J., L. Valiquette, and B. Cossette. 2005. Mortality attributable to nosocomial Clostridium difficile-associated disease during an epidemic caused by a hypervirulent strain in Quebec. CMAJ 173:1037-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Popoff, M. R., E. J. Rubin, D. M. Gill, and P. Boquet. 1988. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect. Immun. 56:2299-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pultz, N. J., and C. J. Donskey. 2005. Effect of antibiotic treatment on growth of and toxin production by Clostridium difficile in the cecal contents of mice. Antimicrob. Agents Chemother. 49:3529-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sawai, J., T. Hasegawa, T. Kamimura, A. Okamoto, D. Ohmori, N. Nosaka, K. Yamada, K. Torii, and M. Ohta. 2007. Growth phase-dependent effect of clindamycin on production of exoproteins by Streptococcus pyogenes. Antimicrob. Agents Chemother. 51:461-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spencer, R. C. 1998. The role of antimicrobial agents in the aetiology of Clostridium difficile-associated disease. J. Antimicrob. Chemother. 41(Suppl. C):21-27. [DOI] [PubMed] [Google Scholar]
- 51.Spigaglia, P., F. Barbanti, P. Mastrantonio, J. S. Brazier, F. Barbut, M. Delmee, E. Kuijper, and I. R. Poxton. 2008. Fluoroquinolone resistance in Clostridium difficile isolates from a prospective study of C. difficile infections in Europe. J. Med. Microbiol. 57:784-789. [DOI] [PubMed] [Google Scholar]
- 52.Stabler, R. A., D. N. Gerding, J. G. Songer, D. Drudy, J. S. Brazier, H. T. Trinh, A. A. Witney, J. Hinds, and B. W. Wren. 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 188:7297-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sullivan, A., C. Edlund, and C. E. Nord. 2001. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 1:101-114. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka, M., T. Hasegawa, A. Okamoto, K. Torii, and M. Ohta. 2005. Effect of antibiotics on group A Streptococcus exoprotein production analyzed by two-dimensional gel electrophoresis. Antimicrob. Agents Chemother. 49:88-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walbrown, M. A., S. L. Aspinall, N. K. Bayliss, R. A. Stone, F. Cunningham, C. L. Squier, and C. B. Good. 2008. Evaluation of Clostridium difficile-associated diarrhea with a drug formulary change in preferred fluoroquinolones. J. Manag. Care Pharm. 14:34-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waligora, A. J., C. Hennequin, P. Mullany, P. Bourlioux, A. Collignon, and T. Karjalainen. 2001. Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect. Immun. 69:2144-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warny, M., J. Pepin, A. Fang, G. Killgore, A. Thompson, J. Brazier, E. Frost, and L. C. McDonald. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079-1084. [DOI] [PubMed] [Google Scholar]