Abstract
In this study, application of a dual absorbance/fluorescence assay to a chemical library screen identified several previously unknown inhibitors of mycobacteria. In addition, growth conditions had a significant effect on the activity profile of the library. Some inhibitors such as Se-methylselenocysteine were detected only when screening was performed under nutrient-limited culture conditions as opposed to nutrient-rich culture conditions. We propose that multiple culture condition library screening is required for complete inhibitory profiling and for maximal antimycobacterial compound detection.
Recent data from the World Health Organization show that there are more than 9 million new cases of tuberculosis (TB) each year, half a million of which are caused by drug-resistant Mycobacterium tuberculosis (26, 27, 28). The spread of antibiotic resistance has necessitated the identification of new anti-infective molecules for the treatment of TB (25). TB drug discovery research is often dependent on the robustness of the upstream biological assay used. In terms of whole-cell antimycobacterial assays, screens are commonly performed under optimal growth conditions such as in the presence of excess nutrients. Evidence suggests, however, that M. tuberculosis persists in a nutrient-deprived state in the host lung (4, 12-14, 21). Screening under nutrient-rich conditions may fail to detect compounds that are preferentially active against nutrient-limited M. tuberculosis during infection.
Traditionally, antimycobacterial assays use optical density (OD) as an indicator of growth, which can be distorted by the intrinsic absorbance of some compounds and the propensity of mycobacterial cells to form aggregates. Alternatives to OD measurement include the use of reporter molecules, such as the green fluorescent protein (GFP). Collins et al. (10) demonstrated that when expressed in M. tuberculosis, the levels of GFP paralleled the numbers of CFU during growth. The MICs of a range of antitubercular drugs determined using GFP were consistent with those obtained using Alamar Blue (7) and the BACTEC 460 system (9). In this work, we used both OD and GFP fluorescence to screen the library of pharmacologically active compounds (LOPAC) (LO1280; Sigma-Aldrich, St. Louis, MO) for antimycobacterial compounds.
For expression of GFP, a vector was constructed by PCR amplification of the gfpmut2 gene (11) from pOT11 (19) using primers GFP_RBS_F1 (5′-GGGGGTACCTTTAAGAAGATATACATATGAGTAAAGGAGAA-3′) and GFP_R1 (5′-GGGGGCATGCTTATTATTTGTATAGTTCATCCATGCC-3′). The product was cloned into the KpnI and SphI restriction sites of pTKmx (16), generating plasmid pTKmxGFP. The pAL5000 origin of replication of pTKmxGFP was excised by NheI restriction digestion and replaced with the replicon of the high-copy-number plasmid pHIGH100 (5), amplified using PCR primers OriM_F (5′-GGGGGCTAGCAACGAGGACAGTCGCACGAC-3′) and OriM_R (5′-GGGGGCTAGCATCGAGCCGAGAACGTTATC-3′), generating plasmid pSHIGH. The hsp60 gene promoter from Mycobacterium bovis BCG, amplified using PCR primers Hsp60_F (5′-GGGGGGTACCGGTACCGGTGACCACAACGACGCGCCCGCT-3′) and Hsp60_R (5′-GGGGGGTACCCGCAATTGTCTTGGCCATTGCGAA-3′), was cloned into the KpnI site of pSHIGH, generating plasmid pSHIGH+hsp60. (Underlining indicates position of restriction site for each primer.)
Mycobacterium smegmatis mc2155 harboring plasmid pSHIGH+hsp60 was inoculated into Luria-Bertani broth (LB) containing 50 μg/ml kanamycin and supplemented with 0.1% (vol/vol) Tween 80 and 100 μg/ml d-arabinose to reduce cell aggregation as previously described (2, 20). A selection of first- and second-line antitubercular drugs and tetracycline were used to test the validity of the antimycobacterial assay. The cultures were grown to the mid-logarithmic phase and diluted to an OD at 600 nm of 0.2 (10-mm path length). Two hundred microliters of sterile deionized water was added to each well on the perimeter of the plate to minimize evaporation of the growth medium during the assay. Fifty microliters of LB containing 50 μg/ml kanamycin, 0.1% Tween 80, and 100 μg/ml d-arabinose were added to the remaining wells. Starting at 50 μM, twofold serial dilutions were performed for the experimental compounds and control antibiotics. Fifty microliters of the cell culture, corresponding to approximately 5 × 106 CFU per well, was added to each one of the inner wells, except the medium control wells. The plates were sealed, wrapped in parafilm, and incubated at 37°C for 96 h with 200 rpm shaking. OD and GFP fluorescence measurements were performed at 0- and 96-h incubation using a Wallac Envision multilabel plate reader (Perkin-Elmer). Data were analyzed with SigmaPlot 11 (SYSTAT) using four-parameter logistic standard curve analysis. The MIC and 50% inhibitory concentration (IC50) values were determined with respect to the controls at 96 h. For each of the drugs tested, use of OD and GFP measurements produced identical MICs and good correlation for the IC50s (Table 1 and Fig. 1).
TABLE 1.
Comparison of the optical density- and fluorescence-based inhibitor assays using known antibiotics in Mycobacterium smegmatisa
| Antibiotic | MIC (μM) by OD and fluorescence assays | IC50 (μM) (mean ± SEM) by the following assay: |
|
|---|---|---|---|
| OD | Fluorescence | ||
| Capreomycin | 12.5 | 2.12 ± 0.69 | 1.78 ± 0.71 |
| Ciprofloxacin | 12.5 | 1.98 ± 0.25 | 1.07 ± 0.24 |
| Ethambutol | 12.5 | 3.66 ± 0.10 | 3.20 ± 0.13 |
| Ethionamide | 100 | 7.66 ± 0.15 | 6.75 ± 0.75 |
| Rifampin | 6.25 | 4.36 ± 0.49 | 3.11 ± 0.51 |
| Streptomycin | 1.56 | 0.50 ± 0.09 | 0.48 ± 0.05 |
| Tetracycline | 6.25 | 0.41 ± 0.20 | 0.33 ± 0.30 |
MIC and IC50s for M. smegmatis were compared using both OD and fluorescence measurements for a suite of known antibiotics including first- and second-line antitubercular drugs. Use of OD and fluorescence measurements produced the same MIC and good correlation in terms of the IC50s for all of the antibiotics tested.
FIG. 1.
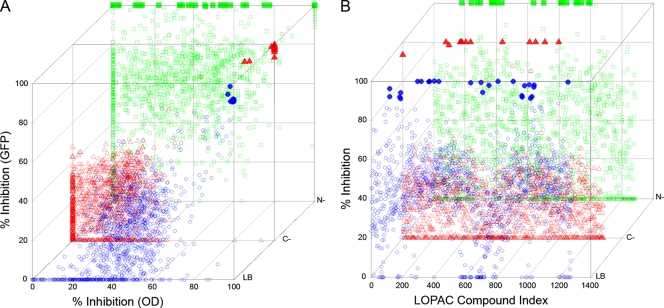
(A) Correlation of the data from the OD and GFP fluorescence-based library screens. The LOPAC inhibitory data from the OD and GFP fluorescence assays were compared to determine the level of correlation in the presence of different growth media. Pearson coefficients of r = 0.64 (nutrient-rich culture conditions), r = 0.5 (carbon-limited culture conditions), and r = 0.33 (nitrogen-limited culture conditions) were obtained indicating medium to large positive correlation between the two assays. (B) Profile of LOPAC inhibitory activity under various growth conditions. Examination of the LOPAC library index determined that a number of compounds were not detected in all of the culture conditions. This resulted in modifications in the inhibitory profile of the LOPAC library as a function of various growth conditions. Mycobacterium smegmatis was grown under nutrient-rich (LB), carbon-limited (C−), and nitrogen-limited (N−) culture conditions with each LOPAC compound present at a single concentration of 20 μM. Inhibitory activity is expressed as percentage inhibition of M. smegmatis growth. Symbols: ○, LB medium; ▵, carbon-limited culture conditions; □, nitrogen-limited culture conditions. The opacity of symbols increases with increasing inhibition. Significant results in both axes are displayed with solid symbols, i.e., LB medium (•), carbon-limited culture conditions (▴), nitrogen-limited culture conditions (▪). Scatter plots were drawn using Scatterplot3d (17).
The LOPAC library was screened for inhibitory activity toward M. smegmatis grown under nutrient-rich conditions. Two microliters of a 1 mM concentration of each compound from the 16 LOPAC stock plates was transferred to the wells of columns 2 to 11 using a Cybi-Well robotic liquid handling station (Cybio) to obtain a final chemical concentration of 20 μM. Two hundred microliters of sterile-distilled water were added to each well in column 1 to minimize evaporation and medium, solvent, and antibiotic controls were established in column 12 of each plate. Solvent controls, consisting of 2% dimethyl sulfoxide, did not produce any significant inhibition of M. smegmatis growth. Antibiotic controls, consisting of rifampin (rifampicin) and ciprofloxacin at 20 μM, yielded complete growth inhibition.
The library screens were performed three times, and the resulting data were normalized to control for plate-to-plate variation by linear scaling to match the most extreme plates in each data set and taking their natural log transforms. Control values that were more than 3 standard deviations from the mean were considered outliers and discarded. Z-factors were calculated for each assay control to give an indication of assay reliability, taking into account both dynamic range and assay variability (29). Under standard growth conditions, the Z-factors for capreomycin were Z = 0.87 (OD) and Z = 0.88 (GFP) and the Z-factors for rifampin were Z = 0.68 (OD) and Z = 0.87 (GFP), interpreted as an excellent assay. To enable comparison with previous work (7), the signal-to-noise ratios for capreomycin were 23.21 (OD) and 24.20 (GFP) and the ratios for rifampin were 9.44 (OD) and 22.78 (GFP). Following validation of potential hits using MIC and IC50 determination, 14 compounds that inhibited M. smegmatis growth under nutrient-rich conditions at concentrations of 12.5 μM or below were identified (Table 2). A number of these compounds, e.g., calcimycin, demeclocycline, doxycycline, lomefloxacin, minocycline, ofloxacin, and vancomycin, are known antibacterials, and therefore, activity against mycobacteria was not unexpected. In addition, antimycobacterial activity has previously been recorded for clotrimazole and niclosamide by the Southern Research Institute in Alabama (http://pubchem.ncbi.nlm.nih.gov) and other researchers (6, 22). From our review of the literature, antimycobacterial activity has not been previously reported for the remaining compounds, calmidazolium, diphenyleneiodonium, idarubicin, and methoctramine.
TABLE 2.
Validated inhibitors from the LOPAC chemical library for Mycobacterium smegmatis grown under nutrient-rich and nutrient-limited culture conditionsa
| LOPAC compound | MIC (μM)b,c |
IC50 (μM) (mean ± SEM)b |
||||
|---|---|---|---|---|---|---|
| LB | C− | N− | LB | C− | N− | |
| BAY 11-7085 | 50 | 6.25 | 12.5 | 30.60 ± 0.59 | 2.58 ± 0.48 | 6.22 ± 0.11 |
| Calcimycin | 3.125 | 12.5 | 12.5 | 1.04 ± 0.24 | 5.21 ± 0.16 | 3.43 ± 0.19 |
| Calmidazolium chloride | 12.5 | 50 | 12.5 | 13.20 ± 1.17 | 13.85 ± 0.22 | 7.02 ± 1.51 |
| Carboplatin | 50 | 25 | 50 | 14.07 ± 4.27 | 12.14 ± 0.67 | 7.41 ± 0.25 |
| 4-Chloromercuribenzoic acid | 25 | 6.25 | 12.5 | 7.07 ± 0.75 | 7.65 ± 0.17 | 6.33 ± 0.11 |
| Cisplatin | >50 | 12.5 | 12.5 | >50 | 8.02 ± 3.44 | 10.31 ± 5.45 |
| Clotrimazole | 12.5 | 50 | 50 | 6.54 ± 0.77 | 18.21 ± 3.24 | 12.19 ± 0.07 |
| Demeclocycline | 3.125 | 6.25 | 6.25 | 1.88 ± 0.24 | 5.92 ± 0.28 | 4.40 ± 0.25 |
| Dequalinium analog C-14 | 6.25 | 6.25 | 3.125 | 3.95 ± 0.55 | 2.73 ± 0.40 | 1.74 ± 0.30 |
| Diphenyleneiodonium | 6.25 | 3.125 | 3.125 | 6.77 ± 0.43 | 8.53 ± 1.81 | 1.63 ± 0.30 |
| Doxycycline | 1.56 | 3.125 | 3.125 | 0.76 ± 0.05 | 2.98 ± 0.16 | 1.95 ± 0.57 |
| Idarubicin | 12.5 | 25 | 12.5 | 3.61 ± 2.01 | 6.68 ± 6.53 | 5.13 ± 0.23 |
| Lomefloxacin | 12.5 | 6.25 | 6.25 | 4.33 ± 0.10 | 4.88 ± 0.25 | 1.44 ± 0.15 |
| LY-367265 | 50 | 25 | 50 | 25.11 ± 6.96 | 25.26 ± 1.44 | 22.31 ± 0.34 |
| Methoctramine | 12.5 | 12.5 | 12.5 | 15.90 ± 1.98 | 6.88 ± 1.22 | 7.45 ± 0.40 |
| Minocycline | 12.5 | 12.5 | 12.5 | 6.77 ± 1.12 | 5.02 ± 1.15 | 7.23 ± 0.13 |
| Mitoxantrone | 50 | 50 | 50 | 21.84 ± 5.27 | 9.70 ± 0.84 | 5.02 ± 0.25 |
| Niclosamide | 12.5 | 6.25 | 6.25 | 6.85 ± 0.72 | 3.31 ± 1.21 | 4.73 ± 0.26 |
| Ofloxacin | 3.125 | 6.25 | 6.25 | 2.03 ± 0.65 | 3.13 ± 1.55 | 1.73 ± 0.37 |
| Pentamidine | 50 | 25 | 50 | 23.53 ± 0.15 | 11.05 ± 0.17 | 9.10 ± 0.26 |
| 1,10-Phenanthroline | 50 | 12.5 | 12.5 | 41.64 ± 0.31 | 4.99 ± 0.28 | 5.76 ± 0.32 |
| Se-methylselenocysteine | >50 | 12.5 | 12.5 | >50 | 2.19 ± 0.18 | 0.60 ± 0.09 |
| Ruthenium red | >50 | 50 | 50 | >50 | 17.71 ± 0.19 | 9.47 ± 0.11 |
| Trifluoperazine | 50 | 50 | 50 | 12.26 ± 0.06 | 12.35 ± 0.18 | 3.15 ± 1.04 |
| U-83836 | 50 | 50 | 50 | >50 | >50 | 34.25 ± 0.38 |
| Vancomycin | 12.5 | 25 | 25 | 3.18 ± 0.43 | 16.69 ± 0.19 | 11.12 ± 0.17 |
| WB64 | 50 | 50 | 25 | 8.15 ± 1.37 | 11.70 ± 5.73 | 6.00 ± 0.12 |
MIC and IC50s for M. smegmatis were determined using OD and/or fluorescence measurements for compounds that were detected as inhibitory in the LOPAC library screens. Validation of the hits was carried out using a starting concentration of 50 μM, and compounds were tested against M. smegmatis grown under nutrient-rich, carbon-limited, and nitrogen-limited culture conditions.
The mycobacteria were grown under nutrient-rich (LB) and carbon-limited (C−) and nitrogen-limited (N−) culture conditions.
Underlined MIC values indicate the growth condition under which an inhibitor was detected in screening.
In the next stage of this work, we tested whether the inhibitory profile of the LOPAC library toward mycobacteria varied significantly under different culture conditions. M. smegmatis was grown in carbon- and nitrogen-limited Hartman-de Bonts (HdeB) medium (18, 21) in place of LB. Many hit compounds detected in rich media were also active under conditions of carbon and nitrogen limitation (Table 2). Clotrimazole was more active under nutrient-rich conditions. However, the screens identified additional inhibitors that were not detected using nutrient-rich culture conditions. These include Bay 11-7085, 4-chloromercuribenzoic, cisplatin, and Se-methylselenocysteine, which exhibit significantly lower MIC values under nutrient limitation (Table 2). These findings indicate that antimycobacterial activity contained within chemical libraries is significantly affected by the growth conditions used for screening. Screening protocols need to be able to detect drugs that are active under nutrient limitation and other conditions that are considered relevant to the host environment.
Of the compounds detected in the library screens against mycobacteria, a number of compounds may have activities relevant to TB drug development. Se-methylselenocysteine (MeSeCys) is used in the dietary chemoprevention of tumors (1). It is metabolized by selenocysteine lyase (β-lyase) producing methylselenol, which causes apoptosis in cancer cells by redox cycling and protein thiol modification (1, 15, 23). Selenocysteine lyases are widely distributed among bacteria (8). Enzymes annotated at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) as selenocysteine lyases are encoded in the genomes of a number of mycobacterial species, including M. smegmatis (e.g., genes MSMEG_1242 and MSMEG_4538) and M. tuberculosis (e.g., genes Rv1464 and Rv3025c). Hence, it is plausible that mycobacteria could convert MeSeCys into toxic selenium products, such as methylselenol. Mycobacteria may potentially use MeSeCys as a source of selenium or other nutrients, which may account for the more pronounced inhibitory activity seen for MeSeCys in nutrient-deprived conditions (Fig. 2).
FIG. 2.
Activity of the compound Se-methylselenocysteine against Mycobacterium smegmatis. M. smegmatis was grown under nutrient-rich (LB), carbon-limited (C−), and nitrogen-limited (N−) culture conditions. (A) Dose-response curve for Se-methylselenocysteine obtained using the optical density-based assay. (B) Dose-response curve for Se-methylselenocysteine obtained using the GFP fluorescence-based assay. (C) Illustration of a possible metabolic reaction through which methyl selenol, a toxic form of selenium, could be generated from Se-methylselenocysteine by selenocysteine lyase in mycobacteria.
We have been unable to find reports of inhibitory activity of MeSeCys or any organic forms of selenium toward mycobacteria or their use in the treatment of TB. Micronutrient supplementation with inorganic selenium can improve health outcomes for patients with TB by improving the nutritional status of the host (3, 24); however, an antibiotic role for selenium-based compounds has not been explored previously. Therefore, further investigations on MeSeCys as a potential antitubercular compound are being conducted.
In conclusion, the use of different growth media exerts a significant effect on the identification of active compounds in a chemical library screen. Incorporation of host-related physicochemical conditions into whole-cell screens could potentially augment the detection of alternative compounds that are active against M. tuberculosis infection.
Acknowledgments
We gratefully acknowledge the support of the Health Research Council of New Zealand (grant 07/379), the Wellington Medical Research Foundation (grant 2006/121), and the University Research Fund of the Victoria University of Wellington (grant 26211/1496). C.H.M. was supported by a postgraduate scholarship from the Victoria University of Wellington.
We are grateful for the valuable advice provided by Paul Teesdale-Spittle and David Bellows.
Footnotes
Published ahead of print on 28 September 2009.
REFERENCES
- 1.Abdulah, R., K. Miyazaki, M. Nakazawa, and H. Koyama. 2005. Chemical forms of selenium for cancer prevention. J. Trace Elem. Med. Biol. 19:141-150. [DOI] [PubMed] [Google Scholar]
- 2.Anton, V., P. Rouge, and M. Daffe. 1996. Identification of the sugars involved in mycobacterial cell aggregation. FEMS Microbiol. Lett. 144:167-170. [DOI] [PubMed] [Google Scholar]
- 3.Benn, C. S., H. Friis, and C. Wejse. 2008. Should micronutrient supplementation be integrated into the case management of tuberculosis? J. Infect. Dis. 197:1487-1489. [DOI] [PubMed] [Google Scholar]
- 4.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 5.Bourn, W. R., Y. Jansen, H. Stutz, R. M. Warren, A. L. Williamson, and P. D. van Helden. 2007. Creation and characterisation of a high-copy-number version of the pAL5000 mycobacterial replicon. Tuberculosis (Edinburgh) 87:481-488. [DOI] [PubMed] [Google Scholar]
- 6.Burguiere, A., P. G. Hitchen, L. G. Dover, A. Dell, and G. S. Besra. 2005. Altered expression profile of mycobacterial surface glycopeptidolipids following treatment with the antifungal azole inhibitors econazole and clotrimazole. Microbiology 151:2087-2095. [DOI] [PubMed] [Google Scholar]
- 7.Changsen, C., S. G. Franzblau, and P. Palittapongarnpim. 2003. Improved green fluorescent protein reporter gene-based microplate screening for antituberculosis compounds by utilizing an acetamidase promoter. Antimicrob. Agents Chemother. 47:3682-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chocat, P., N. Esaki, T. Nakamura, H. Tanaka, and K. Soda. 1983. Microbial distribution of selenocysteine lyase. J. Bacteriol. 156:455-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, H. L., U. E. Schaible, and S. H. Kaufmann. 1998. Early IL-4 induction in bone marrow lymphoid precursor cells by mycobacterial lipoarabinomannan. J. Immunol. 161:5546-5554. [PubMed] [Google Scholar]
- 10.Collins, L., and S. G. Franzblau. 1997. Microplate alamar blue assay versus BACTEC 460 system for high-throughout screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob. Agents Chemother. 41:1004-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 12.Gordhan, B. G., D. A. Smith, H. Alderton, R. A. McAdam, G. J. Bancroft, and V. Mizrahi. 2002. Construction and phenotypic characterization of an auxotrophic mutant of Mycobacterium tuberculosis defective in l-arginine biosynthesis. Infect. Immun. 70:3080-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hampshire, T., S. Soneji, J. Bacon, B. W. James, J. Hinds, K. Laing, R. A. Stabler, P. D. Marsh, and P. D. Butcher. 2004. Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: a model for persistent organisms? Tuberculosis (Edinburgh) 84:228-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hondalus, M. K., S. Bardarov, R. Russell, J. Chan, W. R. Jacobs, Jr., and B. R. Bloom. 2000. Attenuation of and protection induced by a leucine auxotroph of Mycobacterium tuberculosis. Infect. Immun. 68:2888-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson, M. I., and G. F. Combs, Jr. 2008. Selenium and anticarcinogenesis: underlying mechanisms. Curr. Opin. Clin. Nutr. Metab. Care 11:718-726. [DOI] [PubMed] [Google Scholar]
- 16.Kenney, T. J., and G. Churchward. 1996. Genetic analysis of the Mycobacterium smegmatis rpsL promoter. J. Bacteriol. 178:3564-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ligges, U., and M. Machler. 2003. Scatterplot3d - an R package for visualizing multivariate data. J. Stat. Software 8:1-20. [Google Scholar]
- 18.O'Toole, R., M. J. Smeulders, M. C. Blokpoel, E. J. Kay, K. Lougheed, and H. D. Williams. 2003. A two-component regulator of universal stress protein expression and adaptation to oxygen starvation in Mycobacterium smegmatis. J. Bacteriol. 185:1543-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Toole, R., J. Von Hofsten, R. Rosqvist, P. E. Olsson, and H. Wolf-Watz. 2004. Visualisation of zebrafish infection by GFP-labelled Vibrio anguillarum. Microb. Pathog. 37:41-46. [DOI] [PubMed] [Google Scholar]
- 20.Parish, T., and N. G. Stoker. 1998. Mycobacteria protocols. Humana Press, Totowa, NJ.
- 21.Smeulders, M. J., J. Keer, R. A. Speight, and H. D. Williams. 1999. Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181:270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun, Z., and Y. Zhang. 1999. Antituberculosis activity of certain antifungal and antihelmintic drugs. Tuber. Lung. Dis. 79:319-320. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki, K. T., K. Kurasaki, and N. Suzuki. 2007. Selenocysteine beta-lyase and methylselenol demethylase in the metabolism of Se-methylated selenocompounds into selenide. Biochim. Biophys. Acta 1770:1053-1061. [DOI] [PubMed] [Google Scholar]
- 24.Villamor, E., F. Mugusi, W. Urassa, R. J. Bosch, E. Saathoff, K. Matsumoto, S. N. Meydani, and W. W. Fawzi. 2008. A trial of the effect of micronutrient supplementation on treatment outcome, T cell counts, morbidity, and mortality in adults with pulmonary tuberculosis. J. Infect. Dis. 197:1499-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.World Health Organization. 2006. The Global Plan to Stop TB 2006-2015. Actions for life: towards a world free of tuberculosis. World Health Organization and Stop TB Partnership. World Health Organization publication no. WHO/HTM/STB/2006.35. World Health Organization, Geneva, Switzerland. [PubMed]
- 26.World Health Organization. 2008. Anti-tuberculosis drug resistance in the world, report no. 4. Fourth global report. The WHO/IUATLD Global Project on Anti-tuberculosis Drug Resistance Surveillance 2002-2007. World Health Organization publication no. WHO/HTM/TB/2008.394. World Health Organization, Geneva, Switzerland.
- 27.World Health Organization. 2008. Global tuberculosis control-surveillance, planning, financing. WHO Report 2008. World Health Organization publication no. WHO/HTM/TB/2008.393. World Health Organization, Geneva, Switzerland.
- 28.World Health Organization. 2009. Global tuberculosis control-epidemiology, strategy, financing. WHO Report 2009. World Health Organization publication no. WHO/HTM/TB/2009.411. World Health Organization, Geneva, Switzerland.
- 29.Zhang, J. H., T. D. Chung, and K. R. Oldenburg. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67-73. [DOI] [PubMed] [Google Scholar]