Abstract
Structures have been obtained for the complexes that triacetyloleandomycin and mycalamide A form with the large ribosomal subunit of Haloarcula marismortui. Triacetyloleandomycin binds in the nascent peptide tunnel and inhibits the activity of ribosomes by blocking the growth of the nascent peptide chain. Mycalamide A binds to the E site and inhibits protein synthesis by occupying the space normally occupied by the CCA end of E-site-bound tRNAs.
Since 2001, crystal structures have been reported for the complexes that ribosomes form with scores of antiribosomal antibiotics and other inhibitors of protein synthesis (3, 4, 11-14, 20-22, 26, 27, 32-34). Many of them have been obtained using the large ribosomal subunit from Haloarcula marismortui (5, 11-14, 26, 27, 32), which forms crystals that diffract to higher resolution than do the ribosomal crystals obtained from any other organism thus far. Although H. marismortui, which is an archaeal halophile, is not pathogenic, the structures that it forms with protein synthesis inhibitors generally explain why those inhibitors block protein synthesis and are consistent with the relevant genetic and biochemical data, almost all of which derive from experiments done with either eukaryotes or eubacteria. Furthermore, there is ample evidence that protein synthesis inhibitors bind to the ribosomes from H. marismortui at the same sites where and in the same manner that they bind to the ribosomes from pathogenic organisms but (often) with different affinity (32). Thus, inhibitor structures obtained with H. marismortui have provided important insights into the mechanisms of action of many clinically important compounds, as well as the structural basis of both their species specificities and the source of ribosomal drug resistance. Here we present crystal structures of the complexes that the large ribosomal subunit of H. marismortui forms with triacetyloleandomycin and mycalamide A. The triacetyloleandomycin complex structure presented below differs significantly from that reported previously for the same compound bound to the large ribosomal subunit of Deinococcus radiodurans (4). The structure of mycalamide A complex is interesting because it binds to the E site, where normally the CCA end of an E-site tRNA would bind, and there are only two other structures available for inhibitors bound at or near the same site in the ribosome (27).
Triacetyloleandomycin is a semisynthetic derivative of the macrolide oleandomycin (Fig. 1a) (7). Both its structure and its mode of action closely resemble those of other 14-membered macrolides like erythromycin (10), and it is used to treat diseases caused by gram-positive bacteria in some parts of the world. As expected, triacetyloleandomycin binds in the peptide exit tunnel and therefore should inhibit the growth of nascent peptide chains but not the peptidyl transferase reaction as such (17).
FIG. 1.
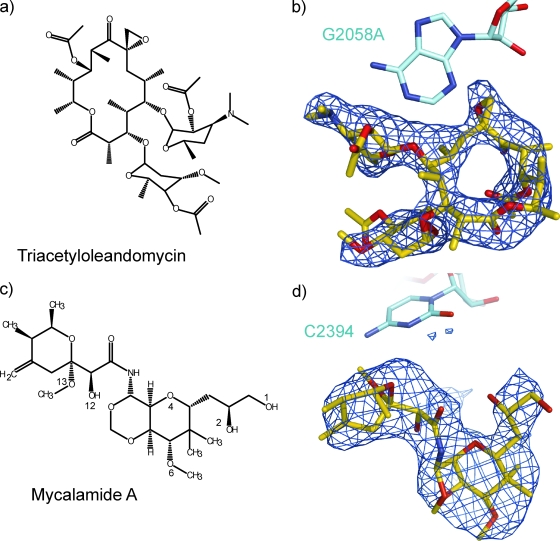
Chemical structures and difference electron density maps. The chemical structures of triacetyloleandomycin and mycalamide A are shown in panels a and c, respectively. The atoms involved in hydrogen bonding interactions with the ribosome are labeled for mycalamide A. The features in the corresponding difference electron density maps that were assigned to triacetyloleandomycin and mycalamide are shown in panels b and d, respectively, with the structures of the drugs superimposed on them. Difference electron density maps are contoured at 3σ.
Mycalamide A was first isolated from a marine sponge of the genus Mycale that is found in the waters of New Zealand, and it is chemically related to pederin, onnamides A to F, and theopederins A to L (Fig. 1c) (29). Mycalamide A disrupts protein synthesis in cell-free protein synthesis systems from rabbit reticulocyte lysates with a 50% inhibitory concentration in the micromolar range. It possesses strong antitumor activity both in vitro and in vivo (8).
Binding experiments demonstrating that pederins compete with 13-deoxytedanolide (18), which is known to be an E-site inhibitor (27), and the chemical similarity between mycalamide A and pederins suggest that mycalamide A is likely to be an E-site inhibitor. The structure presented here confirms that inference and explains why compounds that inhibit protein synthesis by binding to the E site are likely to discriminate very strongly between eukaryotic and bacterial cells. It remains to be seen if an E-site inhibitor specific for bacteria exists in nature or can be synthesized de novo.
MATERIALS AND METHODS
Inhibitors.
Triacetyloleandomycin was purchased from Sigma. Mycalamide A was provided by Sarath Gunasekera of the Harbor Branch Oceanographic Institution in Fort Pierce, FL (Florida Atlantic University).
Preparation of crystals.
Crystals of the large ribosomal subunit from H. marismortui were prepared as previously described (1, 2, 24). Preformed crystals were soaked in solutions containing inhibitors dissolved in buffer B (12% polyethylene glycol 6000, 20% ethylene glycol, 1.7 M NaCl, 0.5 M NH4Cl, 1 mM CdCl2, 100 mM KCH3COO, 6.5 mM CH3COOH [pH 6.0], and 5 mM β-mercaptoethanol and with 30 mM MgCl2 or 21 mM MgCl2 and 30 mM SrCl2) at a 1 mM concentration for several hours at 4°C, with one exchange of soaking solution.
Crystallography.
Diffraction data were collected at the National Synchrotron Light Source (Brookhaven National Laboratory) on beam line X29 and at the Advanced Photon Source (Argonne National Laboratory) on beam line 24-ID-C, as previously described (1, 2, 23). After data processing with HKL2000 (19), electron density maps were calculated for each complex using (|Fo(hkl)|+ drug − |Fo(hkl)| − drug) differences as amplitudes and phases derived from the structure of the large ribosomal subunit with no drug bound (Protein Data Bank accession no. 3CC2) (5). The starting models for both drugs were created using the Dundee PRODRG2 server (http://davapc1.bioch.dundee.ac.uk/prodrg/). The structures of the ribosome complexes with drugs were refined in CNS (6). Data at resolution shell 3.00 to 2.90 Å, at which I/σ was < 1.2, were included in both data processing and refinement for triacetyloleandomycin. Structures were visualized using the program O (9), and figures were generated using PyMOL (15). Buried surface areas were calculated in CNS (6) using the Lee and Richards algorithm (16).
Protein structure accession numbers.
Coordinates and structure factors have been deposited in the Protein Data Bank with the following accession numbers: 3I56 for triacetyloleandomycin and 3I55 for mycalamide A.
RESULTS
Crystal structures.
The structures of the complexes that mycalamide A and triacetyloleandomycin form with the large ribosomal subunit from H. marismortui were determined by X-ray crystallography using the apo structure for that subunit (2) as the source of initial phase information. Table 1 summarizes the crystallographic statistics that characterize these structures. Their resolutions lie between 3.00 and 3.10 Å.
TABLE 1.
Crystallographic statistics of data collectiona
| Crystal soakb | Resolution (Å) | I/σ | Rmerge (%) | Completeness (%) | Redundancy |
|---|---|---|---|---|---|
| Triacetyloleandomycin | 50.0-3.00 (3.11-3.00) | 6.06 (1.2) | 15.9 (73.9) | 99.8 (99.7) | 6.9 (3.8) |
| Mycalamide A | 50.0-3.10 (3.21-3.10) | 12.11 (1.9) | 17.1 (92.7) | 100.0 (100.0) | 7.5 (6.7) |
All data were collected at wavelengths ranging from 1.07 Å to 1.1 Å. Numbers in Parentheses refer to the value of the statistic in question in the highest-resolution shell of data. The redundancy is the average number of times each reflection was measured.
The concentration of both antibiotics was 1 mM.
Structural analysis began for both drugs with the computation of difference electron density maps which were obtained using the differences in the amplitudes between data from crystals into which the drug had been soaked and data obtained from crystals lacking the drug, i.e., [|Fo(hkl)|+ drug − |Fo(hkl)|− drug] (see Materials and Methods). Initial phases were derived from the apo 50S structure. In the difference maps for both drugs, there was only one positive feature large enough to accommodate the relevant drug, and in both cases that feature had an appropriate shape. Figures 1b and 1d show the difference density assigned to each drug with the structure of the drug superimposed. Once the location and orientation of each inhibitor were determined, models were constructed for the corresponding inhibitor-ribosome complex and refined. Table 2 summarizes the statistics of model refinement.
TABLE 2.
Statistics of model refinement
| Crystal soaka | Resolution (Å) | Rcryst (%) | Rfree (%) | Bond (Å) | Angle (o) | Protein Data Bank accession no. |
|---|---|---|---|---|---|---|
| Triacetyloleandomycin | 50.0-2.90 | 20.44 | 24.51 | 0.007 | 1.1 | 3I56 |
| Mycalamide A | 50.0-3.10 | 19.94 | 24.73 | 0.005 | 1.0 | 3I55 |
The concentration of both antibiotics was 1 mM.
(i) Triacetyloleandomycin.
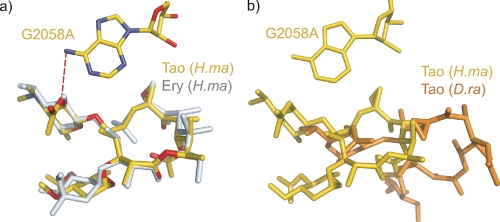
The interaction of triacetyloleandomycin with the ribosome from H. marismortui was investigated using H. marismortui DT917. This strain carries a single rRNA operon with the mutation G2058A (2099), which greatly increases its affinity for macrolide antibiotics (32). (The convention used here for numbering 23S rRNA nucleotides is Escherichia coli number followed by the corresponding H. marismortui number in parentheses.) Triacetyloleandomycin binds to the large ribosomal subunit at the entrance of the nascent peptide tunnel in exactly the same way as erythromycin does. When the two macrolide-ribosome complexes are superimposed using the phosphorus atoms of the rRNA surrounding the binding site, their 14-membered lactone rings align almost perfectly (Fig. 2a). Like erythromycin, triacetyloleandomycin is inserted into the hydrophobic pocket formed by residues A2058 (2099), A2059 (2100), and G2611 (2646), and upon binding, 71% of its surface area is buried.
FIG. 2.
Interaction of triacetyloleandomycin with the H. marismortui ribosome compared with other ribosome-macrolide complexes. (a) Comparison of the position and orientation of triacetyloleandomycin (gold) with those of erythromycin (gray) when both are bound to the large ribosomal subunit from H. marismortui (Protein Data Bank accession no. 1YI2) (31). The heteroatoms of triacetyloleandomycin are color coded, with N shown in blue and O in red. Hydrogen bonds are shown as red dashes. (b) The triacetyloleandomycin complex with the H. marismortui ribosome (gold) compared with the triacetyloleandomycin complex with the D. radiodurans ribosome (orange) (Protein Data Bank accession no. 1OND) (4).
There are some minor differences between the two complexes. In the erythromycin complex, the 2′ OH group of the desosamine forms an H-bond with the N-1 of A2058 (2099). That oxygen is esterified with an acetate group in triacetyloleandomycin, and it is within hydrogen bonding distance from the N-6 of A2058 (2099) (distance, 3.5 Å). Finally, a major reorientation of A2062 (2103) accompanies triacetyloleandomycin binding (not shown). Since the positioning of this base is highly variable, the significance of this observation is uncertain.
(ii) Mycalamide A.
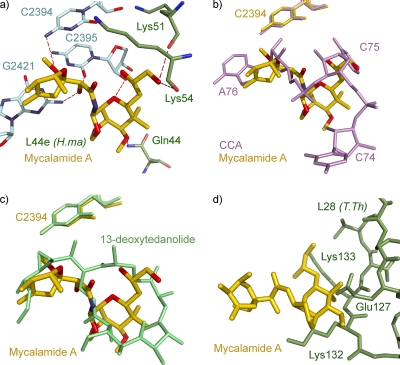
Mycalamide A binds in the E site of the large ribosomal subunit, where it makes extensive interactions both with the 23S rRNA and with the ribosomal protein L44e (Fig. 3a). Of its surface, 64% becomes solvent inaccessible upon binding to the ribosome, and it forms several hydrogen bonds with the ribosome. The O-13 of mycalamide A is 3.37 Å away from the N-4 of C2394 (2431). Its O-12 is within hydrogen bonding distance of the N-2 of G2421 (2459) and the O-2 of C2395 (2432), two bases that form a Watson-Crick base pair. In addition, the O-4 of mycalamide A is 3.21 Å away from the 2′ hydroxyl of the sugar of C2395 (2432), and the O-1 of mycalamide is hydrogen bonded to the backbone carbonyl oxygens of Lys51 and Lys54 of L44e (2.88 Å and 3.44 Å, respectively). Finally, the O-6 of mycalamide A may hydrogen bond with the NH2 of Gln44 of L44 (3.57 Å).
FIG. 3.
Interaction of mycalamide A with the ribosome. (a) The complex that mycalamide A forms with the H. marismortui ribosome is displayed. Mycalamide A is shown with C in gold, N in blue, and O in red. 23S rRNA residues involved in hydrogen bonding are shown with C in light cyan, N in blue, and O in red. L44e residues involved in drug interactions are shown with C in green, N in blue, and O in red. Hydrogen bonds are shown as red dashes. E. coli base numbering is used for 23S rRNA residues, and H. marismortui numbering is used for L44e protein. (b) Superposition of structures of mycalamide A bound to H. marismortui and the deacylated tRNA mimic CCA oligonucleotide bound to the same ribosome (Protein Data Bank accession no. 1QVG) (25). Mycalamide A is shown with C in gold, N in blue, and O in red. CCA residues are shown in pink. (c) Superposition of the mycalamide A structure with the structure of the complex that 13-deoxytedanolide forms with the H. marismortui ribosome (Protein Data Bank accession no. 2OTJ) (27). Mycalamide A is shown with C in gold, N in blue, and O in red, and 13-deoxytedanolide is shown in green. (d) Superposition of the mycalamide A structure (gold) with the structure of the 70S ribosome from T. thermophilus (Protein Data Bank accession no. 2J03) (28). T. thermophilus protein L28 is shown in green, and T. thermophilus numbers are used for labeling of the protein L28.
DISCUSSION
As typical of the macrolides, triacetyloleandomycin binds in the peptide exit tunnel (17). Since the mode of binding of triacetyloleandomycin to the H. marismortui ribosome is very similar to that of erythromycin and its structural relatives, oleandomycin and erythromycin, both inhibit the progression of nascent peptide chains down the exit tunnel, triacetyloleandomycin presumably inhibits translation in the same way (30-33).
The position and orientation of triacetyloleandomycin in the ribosome complex reported here are quite different from those reported for the drug in the complex that it forms with the large ribosomal subunit from D. radiodurans (Fig. 2b) (4). In addition, the rearrangement of L22 reported to occur in D. radiodurans when triacetyloleandomycin binds is not evident in the H. marismortui complex. Since the strain of H. marismortui used for the study reported here carries the mutation G2058A (2099), the difference in the modes of triacetyloleandomycin binding to the ribosomes of the two species cannot be ascribed to a sequence difference at that position, which is known to be critical for macrolide binding.
Interestingly, other macrolides like erythromycin and telithromycin, which are close relatives of triacetyloleandomycin structurally, have also been reported to bind to the D. radiodurans ribosome in a manner different from what is seen in H. marismortui (4, 22, 32). However, a more recent publication, which includes a description of the redetermination of the structures of both erythromycin and telithromycin bound to the D. radiodurans ribosome, states that the discrepancies between the structures of drug-ribosome complexes reported for H. marismortui and the structures of those reported for D. radiodurans are in fact much smaller than they originally appeared (33). Indeed, the revised coordinates of the D. radiodurans erythromycin-ribosome complex are superimposed almost exactly on those of the H. marismortui erythromycin-ribosome complex, whereas the original erythromycin model was nearly orthogonal. Thus, it would be interesting to know if the structure first reported for triacetyloleandomycin bound to the D. radiodurans ribosome can be confirmed.
The structure reported here for mycalamide A bound to the ribosome offers a simple explanation for why it inhibits protein synthesis. When bound to the ribosome, it completely fills the space that is normally occupied by C75 and A76 of E-site-bound tRNAs (Fig. 3b). In this regard it is similar to 13-deoxytedanolide, another inhibitor of eukaryotic protein synthesis that binds to the E site (Fig. 3c) (27). Thus, this structure is in perfect accord with the competitive binding experiments reported by Nishimura and colleagues demonstrating that pederin, theopederin, and onnamide A, compounds that are structurally related to mycalamide A, all compete with 13-deoxytedanolide for binding in Saccharomyces cerevisiae ribosomes (18).
The structure reported here also explains the species specificity of mycalamide A. As shown in Fig. 3a, the drug interacts extensively with protein L44e, which is found at the E site of archaea and eukaryotes. In eubacteria, L28, a protein that is structurally unrelated to L44e, is found in about the same location. In order to investigate what might happen if mycalamide A were to bind to a eubacterial ribosome, the structure described above was superimposed onto 70S ribosome structure from Thermus thermophilus (Protein Data Bank accession no. 2J01) (28). This superposition reveals that mycalamide A is unlikely to be able to bind to eubacterial ribosomes. Residues Glu127, Lys132, and Lys133 of the eubacterial protein L28 extend too far into the drug's binding site (Fig. 3d).
Acknowledgments
We thank Larysa Vasylenko for technical assistance, Jimin Wang for useful discussions during structure refinement, the staff at the Center for Structural Biology Core Laboratory for computational help, and the staff of beam line X29 (National Synchrotron Light Source, Brookhaven National Laboratory) and that of beam line 24-ID-C (Advanced Light Source, Argonne National Laboratory) for their support during data collection.
This work was supported by a grant from the NIH (GM-022778).
Footnotes
Published ahead of print on 8 September 2009.
REFERENCES
- 1.Ban, N., B. Freeborn, P. Nissen, P. Penczek, R. A. Grassucci, R. Sweet, J. Frank, P. B. Moore, and T. A. Steitz. 1998. A 9 angstrom resolution X-ray crystallographic map of the large ribosomal subunit. Cell 93:1105-1115. [DOI] [PubMed] [Google Scholar]
- 2.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 angstrom resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 3.Berisio, R., J. Harms, F. Schluenzen, R. Zarivach, H. A. S. Hansen, P. Fucini, and A. Yonath. 2003. Structural insight into the antibiotic action of telithromycin against resistant mutants. J. Bacteriol. 185:4276-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berisio, R., F. Schluenzen, J. Harms, A. Bashan, T. Auerbach, D. Baram, and A. Yonath. 2003. Structural insight into the role of the ribosomal tunnel in cellular regulation. Nat. Struct. Biol. 10:366-370. [DOI] [PubMed] [Google Scholar]
- 5.Blaha, G., G. Gurel, S. J. Schroeder, P. B. Moore, and T. A. Steitz. 2008. Mutations outside the anisomycin-binding site can make ribosomes drug-resistant. J. Mol. Biol. 379:505-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 7.Bryskier, A. J., J.-P. Butzler, H. C. Neu, and P. M. Tulkens. 1993. Macrolides. Chemistry, pharmacology and clinical uses. Arnette Blackwell, Paris, France.
- 8.Burres, N. S., and J. J. Clement. 1989. Antitumor activity and mechanism of action of the novel marine natural products mycalamide-A and -B and onnamide. Cancer Res. 49:2935-2940. [PubMed] [Google Scholar]
- 9.DeLano, W. L. 2002. The Pymol molecular graphics system. De Lano Scientific, San Carlos, CA.
- 10.Evans, R. M. 1965. The chemistry of the antibiotics used in medicine. Pergamon Press, Oxford, United Kingdom.
- 11.Gürel, G., G. Blaha, P. B. Moore, and T. A. Steitz. 2009. U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J. Mol. Biol. 389:146-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen, J. L., J. A. Ippolito, N. Ban, P. Nissen, P. B. Moore, and T. A. Steitz. 2002. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10:117-128. [DOI] [PubMed] [Google Scholar]
- 13.Hansen, J. L., P. B. Moore, and T. A. Steitz. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 330:1061-1075. [DOI] [PubMed] [Google Scholar]
- 14.Ippolito, J. A., Z. F. Kanyo, D. P. Wang, F. J. Franceschi, P. B. Moore, T. A. Steitz, and E. M. Duffy. 2008. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J. Med. Chem. 51:3353-3356. [DOI] [PubMed] [Google Scholar]
- 15.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 16.Lee, B., and F. M. Richards. 1971. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55:379-400. [DOI] [PubMed] [Google Scholar]
- 17.Mankin, A. S. 2008. Macrolide myths. Curr. Opin. Microbiol. 11:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimura, S., S. Matsunaga, M. Yoshida, H. Hirota, S. Yokoyama, and N. Fusetani. 2005. 13-Deoxytedanolide, a marine sponge-derived antitumor macrolide, binds to the 60S large ribosomal subunit. Bioorg. Med. Chem. 13:449-454. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 20.Schlunzen, F., J. M. Harms, F. Franceschi, H. A. S. Hansen, H. Bartels, R. Zarivach, and A. Yonath. 2003. Structural basis for the antibiotic activity of ketolides and azalides. Structure 11:329-338. [DOI] [PubMed] [Google Scholar]
- 21.Schlunzen, F., E. Pyetan, P. Fucini, A. Yonath, and J. M. Harms. 2004. Inhibition of peptide bond formation by pleuromutilins: the structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin. Mol. Microbiol. 54:1287-1294. [DOI] [PubMed] [Google Scholar]
- 22.Schlunzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 23.Schmeing, T. M., K. S. Huang, D. E. Kitchen, S. A. Strobel, and T. A. Steitz. 2005. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol. Cell 20:437-448. [DOI] [PubMed] [Google Scholar]
- 24.Schmeing, T. M., K. S. Huang, S. A. Strobel, and T. A. Steitz. 2005. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 438:520-524. [DOI] [PubMed] [Google Scholar]
- 25.Schmeing, T. M., P. B. Moore, and T. A. Steitz. 2003. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA 9:1345-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schroeder, S. J., G. Blaha, and P. B. Moore. 2007. Negamycin binds to the wall of the nascent chain exit tunnel of the 50S ribosomal subunit. Antimicrob. Agents Chemother. 51:4462-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroeder, S. J., G. Blaha, J. Tirado-Rives, T. A. Steitz, and P. B. Moore. 2007. The structures of antibiotics bound to the E site region of the 50 S ribosomal subunit of Haloarcula marismortui: 13-deoxytedanolide and girodazole. J. Mol. Biol. 367:1471-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selmer, M., C. M. Dunham, F. V. Murphy, A. Weixlbaumer, S. Petry, J. R. Weir, A. C. Kelley, and V. Ramakrishnan. 2006. Structure of the Thermus thermophilus 70S ribosome complexed with mRNA and tRNA. Science 313:1935-1942. [DOI] [PubMed] [Google Scholar]
- 29.Sohn, J. H., N. Waizumi, H. M. Zhong, and V. H. Rawal. 2005. Total synthesis of mycalamide A. J. Am. Chem. Soc. 127:7290-7291. [DOI] [PubMed] [Google Scholar]
- 30.Tenson, T., M. Lovmar, and M. Ehrenberg. 2003. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 330:1005-1014. [DOI] [PubMed] [Google Scholar]
- 31.Thompson, J., C. A. Pratt, and A. E. Dahlberg. 2004. Effects of a number of classes of 50S inhibitors on stop codon readthrough during protein synthesis. Antimicrob. Agents Chemother. 48:4889-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tu, D., G. Blaha, P. B. Moore, and T. A. Steitz. 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121:257-270. [DOI] [PubMed] [Google Scholar]
- 33.Wilson, D. N., J. M. Harms, K. H. Nierhaus, F. Schlunzen, and P. Fucini. 2005. Species-specific antibiotic-ribosome interactions: implications for drug development. Biol. Chem. 386:1239-1252. [DOI] [PubMed] [Google Scholar]
- 34.Wilson, D. N., F. Schluenzen, J. M. Harms, A. L. Starosta, S. R. Connell, and P. Fucini. 2008. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci. USA 105:13339-13344. [DOI] [PMC free article] [PubMed] [Google Scholar]