Abstract
Antimicrobial peptides (AMPs) are important components of our first line of defense. Induction of AMPs such as LL-37 of the cathelicidin family might provide a novel approach in treating bacterial infections. In this study we identified 4-phenylbutyrate (PBA) as a novel inducer of AMP expression and investigated affected regulatory pathways. We treated various cell lines with PBA and assessed mRNA expression by real-time reverse transcriptase PCR (RT-PCR). Cathelicidin AMP (CAMP) gene expression was found to be upregulated in all four cell lines tested. Additionally, we found that the beta-defensin 1 gene was upregulated in the lung epithelial cell line VA10 while being downregulated in the monocytic cell line U937. Further we found that PBA induced CAMP gene expression synergistically with 1,25-dihydroxyvitamin D3 at both protein and mRNA levels. The general mechanism of induction of CAMP gene expression by PBA was found to be dependent on protein synthesis. Results from quantitative chromatin immunoprecipitation experiments challenge the common view that histone deacetylase inhibitors directly increase CAMP gene expression. Furthermore, we have demonstrated that inhibition of the mitogen-activated protein kinases MEK1/2 and c-Jun N-terminal kinase attenuate PBA-induced CAMP gene expression. Similarly, α-methylhydrocinnamate (ST7), an analogue of PBA, increases CAMP gene expression. Our findings contribute to understanding of the regulation of AMP expression and suggest that PBA and/or ST7 is a promising drug candidate for treatment of microbial infections by strengthening the epithelial antimicrobial barriers.
The increased prevalence of multidrug-resistant pathogens calls for new approaches in fighting bacterial infections. One approach is to induce the expression of endogenous antimicrobial peptides (AMPs) to strengthen the epithelial antimicrobial barrier. AMPs have broad activity against various pathogens, including viruses, bacteria, fungi, and parasites. In spite of their ubiquity, their effectiveness has been preserved throughout evolution in contrast to fast-evolving resistance to antibiotics. Still, many bacteria have developed countermeasures to escape the activity of certain AMPs. We predict that the success of epithelial protection by AMPs is dependent on the multiplicity of the peptides with different mechanisms of action. This strategy has most likely limited the development of general resistance.
Defensins and cathelicidins are the two major classes of AMPs found in humans. They are abundantly expressed by epithelial and phagocytic cells. Combined with other components of the innate immune system, they form the first line of defense against infections. While we express numerous defensins, LL-37 is the only cathelicidin-derived peptide expressed in humans. LL-37 is an amphipathic α-helical peptide, composed of 37 amino acids (14). In addition to its antimicrobial activity, LL-37 has been shown to bind to lipopolysaccharide (24) and to possess immunomodulatory functions such as chemotactic signaling, induction of dendritic cell differentiation, and modulation of mast cell function (2, 6, 7, 42). Additionally, LL-37 and its mouse homolog have been shown to promote wound healing (18, 30) and angiogenesis (23). The cathelicidin AMP (CAMP) gene encodes the pre-pro-LL-37 protein containing a signal sequence which, upon translocation to the endoplasmic reticulum, is cleaved to the pro-LL-37. Finally, the pro-LL-37 has been shown to be cleaved extracellularly, yielding the mature LL-37 peptide (39). Understanding of the processing mechanisms of pro-LL-37 is still incomplete, and processing of pro-LL-37 appears to happen in different ways depending on cell type and location (14, 39). Fewer studies have investigated the role of the highly conserved cathelin propart. Interestingly, one study shows that it has both protease-inhibitory and direct antimicrobial functions (43).
Most expression studies have focused on the detection of CAMP gene expression in various tissues and the effect of disease states corresponding to LL-37 levels. However, the underlying molecular mechanism of CAMP gene expression has not been resolved, although interest in this topic is steadily increasing. We and others have demonstrated an effect of butyrate and other short chain fatty acid derivatives on CAMP gene expression and proposed that the molecular mechanism may be linked to an increase in histone acetylation and mitogen-activated protein (MAP) kinase signaling (17, 21, 35, 37).
More recently, it was discovered that 1,25(OH)2D3 induces CAMP gene expression through binding of the ligand-vitamin D receptor complex to a vitamin D-responsive element in the CAMP gene proximal promoter (11, 41). The interplay between nuclear receptors and histone deacetylase (HDAC) inhibitors such as butyrate has recently been investigated in several independent studies, all indicating a cooperative effect between butyrate and additional compounds, activating CAMP gene expression through nuclear receptors (12, 34, 36, 38, 40). The use of HDAC inhibitors to enhance nuclear receptor-mediated expression of the CAMP gene may become a novel approach of strengthening innate immunity to treat bacterial infections.
In this study we examined the regulatory mechanism of a novel inducer of AMP expression in several cell lines. We show that clinically achievable doses (31) of 4-phenylbutyrate (PBA) induce the expression of cathelicidin mRNA in four human cell lines. Furthermore, we show that PBA acts synergistically with 1,25-dihydroxyvitamin D3 [1,25(OH)2D3] in inducing CAMP gene expression at the mRNA level. We show that MAP kinase signaling plays an important role in PBA-induced CAMP gene expression and challenge the well-established hypothesis that HDAC inhibitors have a direct effect on CAMP gene regulation. Finally, we show that α-methylhydrocinnamate, a chemical analogue of PBA, has similar effects on CAMP gene expression. Thus, we provide novel insights into the regulation of the CAMP gene and identify two drug candidates for treatment of infections through upregulation of endogenous AMPs.
MATERIALS AND METHODS
Cell culture and reagents.
An immortalized human bronchial epithelial cell line, VA10, was cultured as previously described (15). Briefly, cells were cultured at 37°C under a 5% CO2 humidified air atmosphere in bronchial epithelial cell basal medium supplemented with bronchial epithelial growth medium supplements (BEGM SingleQuots; Lonza). Gentamicin was omitted, and penicillin (25 U/ml) and streptomycin (25 μg/ml) (Gibco) were utilized. HT-29, A498, and U937 cells (ATCC) were all cultured in RPMI 1640 (Gibco), supplemented with 10% fetal bovine serum (Gibco), penicillin (25 U/ml), and streptomycin (25 μg/ml). Cells at 100% confluence were stimulated for different time periods with PBA (Fluka), 1,25(OH)2D3 (Fluka), butyrate (Sigma), α-methylhydrocinnamate (Sigma), or 2,2-dimethylbutyrate (Sigma) or their combinations. All stimulants were dissolved in ethanol. The short chain fatty acid derivates were purchased as solid acids and neutralized with an equimolar amount of sterile filtered NaOH. For inhibition of MAP kinase signaling, cells were incubated with SP600125 (Sigma), SB203580 (Sigma), or U0126 (Cell Signaling Technology); dissolved in dimethyl sulfoxide; and added 1 hour prior to stimulation. Protein synthesis was blocked by adding cycloheximide (CHX; Sigma) dissolved in ethanol 1 hour prior to stimulation.
Real-time reverse transcriptase PCR (RT-PCR).
Total RNA was extracted using the NucleoSpin RNA II kit (Macherey-Nagel) and quantified on a spectrophotometer (Thermo Scientific; NanoDrop). RNA was reverse transcribed using random primers according to the manufacturer's protocol (Fermentas; RevertAid First Strand cDNA synthesis kit), modified by using 100 units of reverse transcriptase per reaction. The cDNA of stimulated cells was quantified relative to controls, using either Power SYBR or TaqMan Universal Master mix (ABI) with respective primers and probes (Table 1) on a real-time PCR thermocycler (ABI). Primers and probes with 6-carboxyfluorescein fluorescent label and black hole quencher 1 were designed using Primer Express (ABI) and PerlPrimer (28) software. All nucleic acid synthesis was performed by Microsynth (Switzerland).
TABLE 1.
Targets and respective primers for real-time PCR
| Target | Primer sequence |
Probea | |
|---|---|---|---|
| Forward | Reverse | ||
| CAMP gene | 5′-TCACCAGAGGATTGTGACTTCAA-3′ | 5′-TGAGGGTCACTGTCCCCATAC-3′ | 5′-FAM-TGCCTCTTCCAGGTGTT-BHQ1-3′ |
| DEFB1 gene | 5′-TGCTGTTTACTCTCTGCTTACTTTTGT-3′ | 5′-CCAAGGCCTGTGAGAAAGTTACC-3′ | 5′-FAM-CTGAGATGGCCTCAGGT-BHQ1-3′ |
| CAMP gene promoter | 5′-TCATACTGAGCTTCACTCTGTTACCC-3′ | 5′-GAATCACTTGAACCCATTGAACCC-3′ | |
FAM, 6-carboxyfluorescein; BHQ1, black hole quencher 1.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis.
Supernatants of cell culture medium were enriched for proteins on Oasis HLB minicolumns and lyophilized. Total cell lysate was prepared by homogenizing the cells in a 1-ml syringe fitted with a 0.9-mm needle. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotting were performed using the NuPAGE blotting kit (Invitrogen), following the manufacturer's instructions. Membrane blocking was carried out with 5% fat-free milk in phosphate-buffered saline with 0.05% Tween 20. The blot was incubated with affinity-purified rabbit anti-LL-37 (Innovagen) overnight at 4°C. After being washed, the membrane was incubated with secondary antibodies (peroxidase-linked goat anti-rabbit immunoglobulin G [IgG]; Sigma). Bands were visualized by chemifluorescence (ECL+; GE Healthcare) detection, utilizing a Typhoon 9400 scanner (GE Healthcare).
Quantitative ChIP.
Quantitative chromatin immunoprecipitation (ChIP) was performed as previously described (29). Briefly, cells were cross-linked in 1.42% formaldehyde for 15 min and collected using a cell scraper and washed. Thereafter, the cells were lysed and sonicated in order to generate DNA fragments of approximately 500 bp, as confirmed by electrophoresis. Chromatin was immunoprecipitated on Sepharose A beads (Zymed) using polyclonal antibodies for acetylated histones H3 and H4 (Upstate; catalog nos. 06-599 and 06-866). After five washes, the precipitate was treated with proteinase K (Invitrogen) and DNA was amplified by PCR using Chelex 100 (Bio-Rad). Specific DNA fragments were quantified using primers designed to amplify a 70-bp sequence about 500 bp upstream from the transcriptional start site of the CAMP gene.
Statistical analysis.
Normally distributed data are presented as means and standard errors of the means (SEM), from at least three independent experiments. Total RNA level per cell was validated to be unaffected by treatment. Outliers were not excluded from statistical analysis. For comparison of differences between groups, a two-sided t test for independent samples was used. P values of less than 0.05 were considered statistically significant. Tests were performed using the R programming language.
RESULTS
PBA induces CAMP gene expression in four human cell lines.
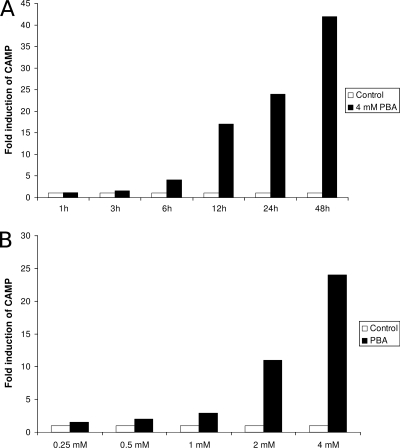
For an initial assessment of time and dose dependency of PBA-induced CAMP gene expression, VA10 cells were treated with PBA and induction of the CAMP gene was assayed. The induction was studied with respect to time over the course of 48 h (Fig. 1A) and PBA concentrations up to 4 mM (Fig. 1B). Total RNA was isolated from the cells, and CAMP mRNA expression levels were analyzed by real-time RT-PCR. Increase of CAMP mRNA expression was dependent on PBA dose and increased with time over the course of 48 h. As stimulation with 4 mM of PBA over 24 h showed a strong response, this time-dose relationship was chosen for most subsequent experiments.
FIG. 1.
Induction of CAMP mRNA expression by PBA. (A) VA10 cells were stimulated with 4 mM PBA or treated with solvent alone and harvested after the indicated period of time. (B) VA10 cells were stimulated with the indicated concentrations of PBA or solvent (Control) for 24 h. These experiments were performed once for initial assessment. CAMP mRNA levels were determined by real-time RT-PCR. Individual samples were normalized to total RNA input. Results were normalized to expression in control samples, where controls were given the arbitrary value of 1.
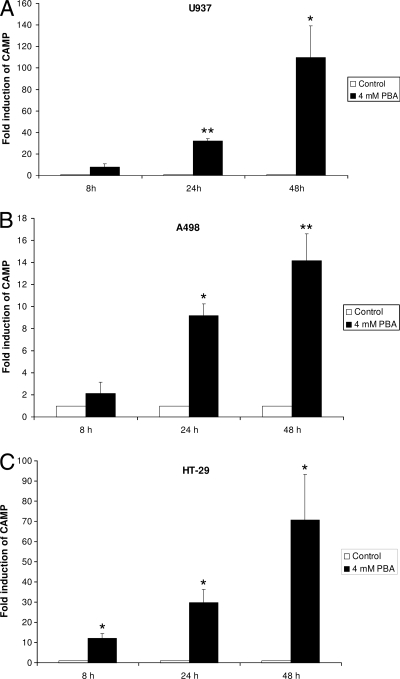
To investigate the effect of PBA on other cell lines, a human colonic adenocarcinoma cell line (HT-29), a human renal carcinoma cell line (A498), and a human leukemic monocyte lymphoma cell line (U937) were stimulated with 4 mM PBA for 8, 24, and 48 h. Total RNA was isolated from the cells, and CAMP mRNA expression levels were analyzed by real-time RT-PCR. While CAMP mRNA expression was significantly increased in all the cell lines tested, the magnitude of induction varied considerably between cell lines (Fig. 2).
FIG. 2.
Induction of CAMP mRNA expression by PBA. U937 (A), A498 (B), and HT-29 (C) cells were stimulated with 4 mM PBA or solvent alone and harvested after the indicated period of time. CAMP mRNA levels were determined by real-time RT-PCR. Individual samples were normalized to total RNA input. Results were plotted as induction over control samples at the respective time points. Error bars indicate SEM from at least three independent experiments. *, P < 0.05; **, P < 0.01.
PBA affects DEFB1 gene expression in human cell lines.
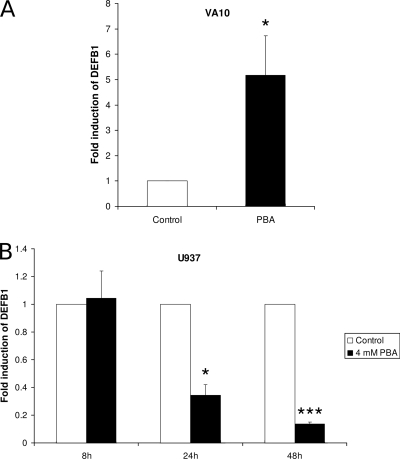
As the expression of the human beta-defensin 1 (DEFB1) gene often correlates with CAMP gene expression (19), we tested the effect of PBA on DEFB1 gene expression. Upon treatment with 4 mM PBA, we detected significant DEFB1 gene upregulation at the mRNA level in VA10 cells by using real-time RT-PCR (Fig. 3A), while the expression of beta-defensin 2, 3, and 4 genes could not be detected at all (data not shown). In contrast to CAMP gene expression, DEFB1 gene expression in U937 cells was significantly downregulated upon PBA stimulation for 24 and 48 h (Fig. 3B). These findings indicate that the impact of PBA on DEFB1 gene expression varies from the effects seen on the CAMP gene and may be cell type specific.
FIG. 3.
Induction of DEFB1 mRNA expression by PBA. (A) VA10 cells were stimulated with 4 mM of PBA or solvent (Control) for 24 h. (B) U937 cells were stimulated with 4 mM of PBA or solvent (Control) for 8, 24, and 48 h. DEFB1 mRNA levels were determined by real-time RT-PCR. Individual samples were normalized to total RNA input. Results were plotted as induction over control samples at the respective time points. Error bars indicate SEM from at least three independent experiments. *, P < 0.05; ***, P < 0.001.
PBA and 1,25(OH)2D3 induce CAMP gene expression synergistically.
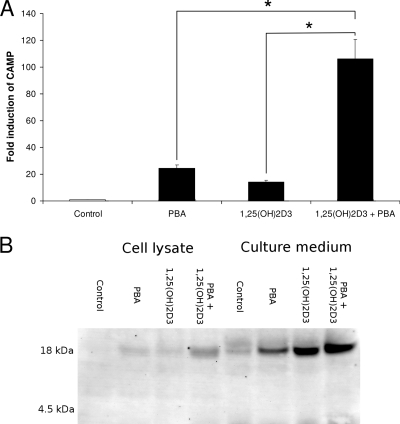
1,25(OH)2D3 is a known direct inducer of CAMP gene expression (41). To assess the cooperative induction of CAMP gene expression by 1,25(OH)2D3 and PBA, VA10 cells were incubated with 20 nM of 1,25(OH)2D3 and 4 mM PBA, either together or with the respective compounds alone. Costimulation resulted in more than a 100-fold increase of CAMP mRNA levels over those in controls. This induction was found to be significantly higher than stimulation with either of the compounds alone, indicating synergistic induction (Fig. 4A). By Western blot analysis of cell lysates and cell culture supernatants, secretion of the pro-LL-37 protein was detected. The measured band intensity indicated additive effects of 1,25(OH)2D3 and PBA on secretion of pro-LL-37 (Fig. 4B). As the pro-LL-37 is not processed in our model system, no increase in antimicrobial activity could be measured in the medium. Processing mechanisms of pro-LL-37 are still poorly understood; we hypothesize that the processing may occur through proteases secreted by leukocytes or other secretory cells.
FIG. 4.
Synergistic induction of CAMP mRNA and pro-LL-37 expression by PBA and 1,25(OH)2D3. VA10 cells were stimulated with PBA (4 mM) and/or 1,25(OH)2D3 (20 nM) as well as solvent (Control) for 24 h. (A) CAMP mRNA levels were determined by real-time RT-PCR. Individual samples were normalized to total RNA input. Results were plotted as induction over control samples. Error bars indicate SEM from at least three independent experiments. (B) Western blot analysis showing pro-LL-37 expression in total cell lysates and cell culture supernatants. One representative blot out of three is shown. *, P < 0.05. Synthetic LL-37 and seminal fluid were used as controls for mature peptide and proform, respectively (not shown).
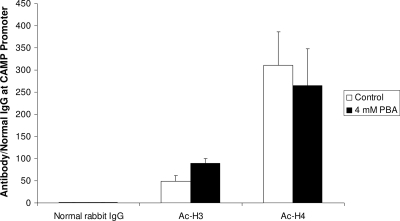
PBA has no effect on histone acetylation on the CAMP gene promoter.
As PBA is known to have an HDAC-inhibitory function, we assessed the acetylation of histones H3 and H4 by quantitative ChIP after treatment of VA10 cells with 4 mM PBA for 24 h. After correction for total chromatin input, enrichment in CAMP gene promoter DNA over precipitation with normal rabbit IgG was calculated using the threshold cycle method (29). No significant change in histone acetylation could be observed on the CAMP gene proximal promoter (Fig. 5). An increase in genome-wide histone acetylation was confirmed by Western blotting (data not shown). These results are in contrast with the well-established hypothesis that HDAC inhibitors increase histone acetylation at the CAMP gene promoter and thereby open the chromatin structure to allow the binding of transcription factors (21, 36).
FIG. 5.
VA10 cells were stimulated with 4 mM of PBA or solvent alone (Control) for 24 h. Acetylation of histones H3 and H4 was analyzed by quantitative ChIP using antibodies against the respective acetylated histones. Results were normalized to normal rabbit IgG and total input and plotted as precipitation over IgG. Normalized data were plotted as means plus SEM from three independent experiments. No significant change in histone acetylation was observed.
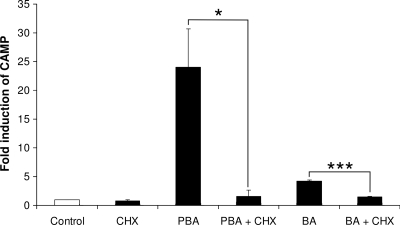
Induction of CAMP gene expression by PBA and butyrate is inhibited by CHX.
In order to assess whether the PBA and butyrate induction pathways of CAMP gene expression are direct, VA10 cells were treated with PBA or butyrate in the presence and absence of CHX, an inhibitor of protein synthesis. After 24 h of incubation, total RNA was isolated and CAMP mRNA levels were measured using real-time RT-PCR. Preincubation of the cells with 20 μg/ml of CHX for 1 hour prior to stimulation effectively blocked both PBA- and butyrate-induced CAMP gene expression (Fig. 6). This suggests that protein synthesis is required for PBA induction of CAMP gene expression. On the other hand, CAMP gene induction by 1,25(OH)2D3 has been shown to be direct and not inhibited by CHX (16, 17). Furthermore, these results support our hypothesis that HDAC inhibitors do not directly affect CAMP gene expression through changes in chromatin structure, since this effect would be expected to be independent of protein synthesis.
FIG. 6.
Inhibition of PBA-induced CAMP gene expression by CHX. VA10 cells were treated with 4 mM PBA or butyrate (BA) in the presence or absence of 20 μg/ml CHX. CAMP mRNA levels were determined by real-time RT-PCR. Individual samples were normalized to total RNA input. Results were plotted as induction over control samples. Error bars indicate SEM from at least three independent experiments. *, P < 0.05; ***, P < 0.001.
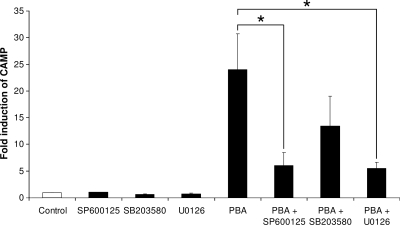
PBA-induced CAMP gene expression is attenuated by MAP kinase inhibitors.
Previous studies (21, 35) have indicated a role of MAP kinase signaling pathways in butyrate-induced CAMP gene expression. We therefore investigated the effect of inhibitors for c-Jun N-terminal kinase (JNK), p38 kinase, and extracellular signal-regulated kinases 1 and 2 (ERK1/2) on PBA-induced CAMP gene expression. One hour prior to stimulation with 4 mM PBA, VA10 cells were preincubated with 20 μM SP600125, SB203580, or U0126 to inhibit the respective kinases. After 24 h of stimulation with 4 mM PBA, total RNA was isolated and analyzed by real-time RT-PCR for CAMP mRNA. Inhibitors for the ERK1/2 and JNK pathways significantly reduced PBA-induced CAMP gene expression (Fig. 7), suggesting that these pathways play an important role in PBA induction of CAMP gene expression. Interestingly these results correlate closely with the above-mentioned studies, where the effect of butyrate on CAMP gene expression was investigated, indicating that the molecular mechanism underlying CAMP gene induction by these compounds may be the same.
FIG. 7.
Inhibition of PBA-induced CAMP gene expression by MAP kinase inhibitors. VA10 cells were treated with 4 mM PBA in the presence or absence of 20 μM of the indicated inhibitors. CAMP mRNA levels were determined by real-time RT-PCR. Individual samples were normalized to total RNA input. Results were plotted as induction over control samples. Error bars indicate SEM from at least three independent experiments. *, P < 0.05.
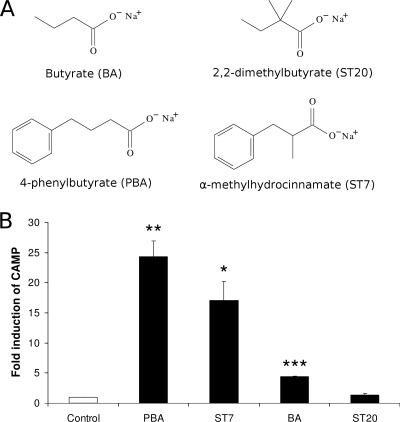
A chemical analogue to PBA induces CAMP gene expression.
Even though PBA is approved for drug use by the United States Food and Drug Administration and has been shown to reach millimolar concentrations in blood after oral administration, it is metabolized within hours (9). Therefore, we tested the abilities of two chemically related compounds that have longer half-lives in vivo to induce CAMP gene expression. VA10 cells were stimulated with 4 mM α-methylhydrocinnamate (ST7), a PBA analogue, or 2,2-dimethylbutyrate (ST20), a butyrate analogue. The chemical structures of the compounds are shown in Fig. 8A. After 24 h of incubation, total RNA was isolated from the cells and CAMP mRNA expression levels were analyzed by real-time RT-PCR. ST7 significantly increased CAMP mRNA expression, whereas ST20 stimulation had no apparent effect on CAMP mRNA expression levels (Fig. 8B).
FIG. 8.
Induction of CAMP mRNA expression by butyrate (BA) and PBA analogues. (A) Structures of utilized chemicals: butyrate (BA), PBA, α-methylhydrocinnamate (ST7), and 2,2-dimethylbutyrate (ST20). (B) Induction of CAMP mRNA expression by indicated chemicals. VA10 cells were stimulated with 4 mM of the respective chemicals or solvent (Control) for 24 h. CAMP mRNA levels were determined by real-time RT-PCR. Individual samples were normalized to total RNA input. Results were plotted as induction over control samples at the respective time points. Error bars indicate SEM from at least three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
In this study we show that PBA, a known HDAC inhibitor, induces CAMP gene expression in four different cell lines (Fig. 1 and 2). The PBA concentrations utilized were close to those achievable in plasma by oral administration (9). Enhanced expression was also detected for the DEFB1 gene. Treatment of lung epithelial cells with PBA resulted in upregulation of the gene, while expression was diminished in a monocytic cell line (Fig. 3), indicating a cell-specific response to PBA.
PBA was originally identified as a drug for treatment of urea cycle disorders (4) and has been investigated as a potential drug candidate for treatment of sickle cell anemia (8), adrenoleukodystrophy (20), cancer (13), and cystic fibrosis (32). PBA is a known HDAC inhibitor, and many of these inhibitors have recently regained attention as drug candidates for cancer and neurodegenerative disorders (1, 22).
The most prominent effect that HDAC inhibitors have on cells is the hyperacetylation of histones, leading to changes in chromatin structure, gene expression, and cellular function. As histone acetylases and HDACs lack specificity for histones, they can modify the function of other molecules (10). Acetylation has been implied to play an important role in modulating transcription factors and other regulatory proteins of innate immunity (5).
Butyrate, another HDAC inhibitor, has been shown to upregulate CAMP gene expression, and this effect was believed to be linked to cell differentiation (16). We, however, demonstrated that butyrate and the synthetic HDAC inhibitor trichostatin A could induce CAMP gene expression independently of cellular differentiation (37). Furthermore, we confirmed that MAP kinase signaling plays an important role in butyrate-induced CAMP gene expression (37). Later, the same pathways were shown to govern butyrate-induced CAMP gene expression in the lung epithelial cell line ECB-1 (21). In the present study, we show that PBA-induced CAMP gene expression relies on MAP kinase signaling in a similar way (Fig. 7).
The effects of butyrate and other HDAC inhibitors on gene expression have been studied using gene expression microarrays (27). The expression profiles show complex reprogramming of gene expression. In line with the general view on HDAC inhibitors, it has been assumed that induction of CAMP gene expression by butyrate occurs through relaxation of chromatin structure, facilitating the binding of transcription factors to the gene and recruitment of RNA polymerase II (21, 36). In this study, we used quantitative ChIP to directly analyze differences in histone acetylation on the proximal CAMP gene promoter upon treatment with PBA. No significant changes in acetylation of histones 3 and 4 were detected, as would be expected if the effects on the gene were direct. To further explore the mechanism of the effect mediated by PBA, we inhibited protein synthesis in VA10 cells with CHX prior to PBA treatment. Interestingly, this treatment suppressed the induction of CAMP gene expression by both butyrate and PBA (Fig. 6). Taken together, these results imply that the induction of CAMP gene expression by PBA is not due to a direct effect on the chromatin structure at the CAMP gene proximal promoter. Rather, it appears that increased histone acetylation facilitates expression of other genes, encoding critical factors for CAMP gene expression. We have not yet succeeded in identifying these genes, but their identification may be of general importance for understanding the regulation of innate immunity.
Being able to induce the expression of AMPs in a controlled fashion would have considerable medical implications, opening up the possibility to treat or prevent bacterial, viral, and fungal infections by reinforcing our innate immunity. Although we demonstrate the ability of PBA to induce CAMP gene expression in human cells, the clinical usefulness of PBA has been predicted to be limited due to its rapid degradation (9). We therefore tested the CAMP gene expression-inducing effects of ST20 and ST7, synthetic analogues of butyrate and PBA, respectively, that have been shown to have longer half-lives than PBA and butyrate in vivo (31). We found that ST7 stimulation of VA10 cells resulted in a 15- to 20-fold induction of CAMP gene expression. Furthermore, costimulation with ST7 and 1,25(OH)2D3 caused an additive induction of CAMP gene expression, while inhibition of either protein synthesis or MAP kinase signaling attenuated the induction (data not shown). As ST7 has been shown to have a longer half-life at millimolar levels in plasma after a single dose (31), ST7 may be a better candidate for clinical applications than PBA, despite its slightly weaker ability to induce the CAMP gene expression in vitro (Fig. 8).
A further indication that PBA may be useful as an AMP-inducing drug is the observation that PBA induces CAMP gene expression in synergy with 1,25(OH)2D3 (Fig. 4). Recently, the importance of 1,25(OH)2D3-induced LL-37 expression in fighting tuberculosis has been demonstrated (25, 26). Killing of Mycobacterium tuberculosis by macrophages was directly correlated with CAMP gene expression and 1,25(OH)2D3 levels. In this study we demonstrate that PBA enhances CAMP gene expression in synergy with 1,25(OH)2D3 at biologically relevant concentrations. These results indicate that the addition of PBA to the diet of tuberculosis patients may be a way to compensate for low 1,25(OH)2D3 levels and enhance antimicrobial response.
In recent years, PBA has gone through clinical trials for cystic fibrosis patients, as it is believed to function as a molecular chaperone, allowing translocation of the cystic fibrosis transmembrane conductance regulator to the plasma membrane (44). LL-37 has been demonstrated to be an important constituent of the airway surface liquid with high activity against bacteria commonly found in cystic fibrosis patients (3, 33). Our findings therefore have considerable relevance for these clinical trials.
In conclusion, our results contribute to the general understanding of the regulation of AMP expression with specific emphasis on LL-37, suggesting that PBA or ST7 is a promising drug candidate for treatment of microbial infections, by strengthening the innate antimicrobial response.
Acknowledgments
We are grateful for the contribution of Katla Kristjánsdóttir and Kristbjörg Sölvadóttir to this study.
This work was supported by the Icelandic Centre for Research (RANNIS), the University of Iceland research fund, and the Eimskip University Fund.
We disclose that we have a pending patent application for the use of PBA to treat infections.
Footnotes
Published ahead of print on 21 September 2009.
REFERENCES
- 1.Abel, T., and R. S. Zukin. 2008. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr. Opin. Pharmacol. 8:57-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agerberth, B., J. Charo, J. Werr, B. Olsson, F. Idali, L. Lindbom, R. Kiessling, H. Jornvall, H. Wigzell, and G. H. Gudmundsson. 2000. The human antimicrobial and chemotactic peptides LL-37 and alpha-defensins are expressed by specific lymphocyte and monocyte populations. Blood 96:3086-3093. [PubMed] [Google Scholar]
- 3.Bals, R., D. J. Weiner, A. D. Moscioni, R. L. Meegalla, and J. M. Wilson. 1999. Augmentation of innate host defense by expression of a cathelicidin antimicrobial peptide. Infect. Immun. 67:6084-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brusilow, S. W., and J. Finkelstien. 1993. Restoration of nitrogen homeostasis in a man with ornithine transcarbamylase deficiency. Metabolism 42:1336-1339. [DOI] [PubMed] [Google Scholar]
- 5.Calao, M., A. Burny, V. Quivy, A. Dekoninck, and C. Van Lint. 2008. A pervasive role of histone acetyltransferases and deacetylases in an NF-kappaB-signaling code. Trends Biochem. Sci. 33:339-349. [DOI] [PubMed] [Google Scholar]
- 6.Chertov, O., D. F. Michiel, L. Xu, J. M. Wang, K. Tani, W. J. Murphy, D. L. Longo, D. D. Taub, and J. J. Oppenheim. 1996. Identification of defensin-1, defensin-2, and CAP37/azurocidin as T-cell chemoattractant proteins released from interleukin-8-stimulated neutrophils. J. Biol. Chem. 271:2935-2940. [DOI] [PubMed] [Google Scholar]
- 7.Davidson, D. J., A. J. Currie, G. S. D. Reid, D. M. E. Bowdish, K. L. MacDonald, R. C. Ma, R. E. W. Hancock, and D. P. Speert. 2004. The cationic antimicrobial peptide LL-37 modulates dendritic cell differentiation and dendritic cell-induced T cell polarization. J. Immunol. 172:1146-1156. [DOI] [PubMed] [Google Scholar]
- 8.Fibach, E., P. Prasanna, G. P. Rodgers, and D. Samid. 1993. Enhanced fetal hemoglobin production by phenylacetate and 4-phenylbutyrate in erythroid precursors derived from normal donors and patients with sickle cell anemia and beta-thalassemia. Blood 82:2203-2209. [PubMed] [Google Scholar]
- 9.Gilbert, J., S. D. Baker, M. K. Bowling, L. Grochow, W. D. Figg, Y. Zabelina, R. C. Donehower, and M. A. Carducci. 2001. A phase I dose escalation and bioavailability study of oral sodium phenylbutyrate in patients with refractory solid tumor malignancies. Clin. Cancer Res. 7:2292-2300. [PubMed] [Google Scholar]
- 10.Glozak, M. A., N. Sengupta, X. Zhang, and E. Seto. 2005. Acetylation and deacetylation of non-histone proteins. Gene 363:15-23. [DOI] [PubMed] [Google Scholar]
- 11.Gombart, A. F., N. Borregaard, and H. P. Koeffler. 2005. Human cathelicidin antimicrobial peptide (CAMP) gene is a direct target of the vitamin D receptor and is strongly up-regulated in myeloid cells by 1,25-dihydroxyvitamin D3. FASEB J. 19:1067-1077. [DOI] [PubMed] [Google Scholar]
- 12.Gombart, A. F., J. O'Kelly, T. Saito, and H. P. Koeffler. 2007. Regulation of the CAMP gene by 1,25(OH)2D3 in various tissues. J. Steroid Biochem. Mol. Biol. 103:552-557. [DOI] [PubMed] [Google Scholar]
- 13.Gore, S. D., L. J. Weng, S. Zhai, W. D. Figg, R. C. Donehower, G. J. Dover, M. Grever, C. A. Griffin, L. B. Grochow, E. K. Rowinsky, Y. Zabalena, A. L. Hawkins, K. Burks, and C. B. Miller. 2001. Impact of the putative differentiating agent sodium phenylbutyrate on myelodysplastic syndromes and acute myeloid leukemia. Clin. Cancer Res. 7:2330-2339. [PubMed] [Google Scholar]
- 14.Gudmundsson, G. H., B. Agerberth, J. Odeberg, T. Bergman, B. Olsson, and R. Salcedo. 1996. The human gene FALL39 and processing of the cathelin precursor to the antibacterial peptide LL-37 in granulocytes. Eur. J. Biochem. 238:325-332. [DOI] [PubMed] [Google Scholar]
- 15.Halldorsson, S., V. Asgrimsson, I. Axelsson, G. H. Gudmundsson, M. Steinarsdottir, O. Baldursson, and T. Gudjonsson. 2007. Differentiation potential of a basal epithelial cell line established from human bronchial explant. In Vitro Cell. Dev. Biol. Anim. 43:283-289. [DOI] [PubMed] [Google Scholar]
- 16.Hase, K., L. Eckmann, J. D. Leopard, N. Varki, and M. F. Kagnoff. 2002. Cell differentiation is a key determinant of cathelicidin LL-37/human cationic antimicrobial protein 18 expression by human colon epithelium. Infect. Immun. 70:953-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hase, K., M. Murakami, M. Iimura, S. P. Cole, Y. Horibe, T. Ohtake, M. Obonyo, R. L. Gallo, L. Eckmann, and M. F. Kagnoff. 2003. Expression of LL-37 by human gastric epithelial cells as a potential host defense mechanism against Helicobacter pylori. Gastroenterology 125:1613-1625. [DOI] [PubMed] [Google Scholar]
- 18.Heilborn, J. D., M. F. Nilsson, G. Kratz, G. Weber, O. Sørensen, N. Borregaard, and M. Ståhle-Bäckdahl. 2003. The cathelicidin anti-microbial peptide LL-37 is involved in re-epithelialization of human skin wounds and is lacking in chronic ulcer epithelium. J. Investig. Dermatol. 120:379-389. [DOI] [PubMed] [Google Scholar]
- 19.Islam, D., L. Bandholtz, J. Nilsson, H. Wigzell, B. Christensson, B. Agerberth, and G. Gudmundsson. 2001. Downregulation of bactericidal peptides in enteric infections: a novel immune escape mechanism with bacterial DNA as a potential regulator. Nat. Med. 7:180-185. [DOI] [PubMed] [Google Scholar]
- 20.Kemp, S., H. M. Wei, J. F. Lu, L. T. Braiterman, M. C. McGuinness, A. B. Moser, P. A. Watkins, and K. D. Smith. 1998. Gene redundancy and pharmacological gene therapy: implications for X-linked adrenoleukodystrophy. Nat. Med. 4:1261-1268. [DOI] [PubMed] [Google Scholar]
- 21.Kida, Y., T. Shimizu, and K. Kuwano. 2006. Sodium butyrate up-regulates cathelicidin gene expression via activator protein-1 and histone acetylation at the promoter region in a human lung epithelial cell line, EBC-1. Mol. Immunol. 43:1972-1981. [DOI] [PubMed] [Google Scholar]
- 22.Kim, T. Y., Y. J. Bang, and K. D. Robertson. 2006. Histone deacetylase inhibitors for cancer therapy. Epigenetics 1:14-23. [DOI] [PubMed] [Google Scholar]
- 23.Koczulla, R., G. von Degenfeld, C. Kupatt, F. Krotz, S. Zahler, T. Gloe, K. Issbrucker, P. Unterberger, M. Zaiou, C. Lebherz, A. Karl, P. Raake, A. Pfosser, P. Boekstegers, U. Welsch, P. S. Hiemstra, C. Vogelmeier, R. L. Gallo, M. Clauss, and R. Bals. 2003. An angiogenic role for the human peptide antibiotic LL-37/hCAP-18. J. Clin. Investig. 111:1665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larrick, J. W., M. Hirata, R. F. Balint, J. Lee, J. Zhong, and S. C. Wright. 1995. Human CAP18: a novel antimicrobial lipopolysaccharide-binding protein. Infect. Immun. 63:1291-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, P. T., S. Stenger, H. Li, L. Wenzel, B. H. Tan, S. R. Krutzik, M. T. Ochoa, J. Schauber, K. Wu, C. Meinken, D. L. Kamen, M. Wagner, R. Bals, A. Steinmeyer, U. Zügel, R. L. Gallo, D. Eisenberg, M. Hewison, B. W. Hollis, J. S. Adams, B. R. Bloom, and R. L. Modlin. 2006. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science 311:1770-1773. [DOI] [PubMed] [Google Scholar]
- 26.Liu, P. T., S. Stenger, D. H. Tang, and R. L. Modlin. 2007. Cutting edge: vitamin D-mediated human antimicrobial activity against Mycobacterium tuberculosis is dependent on the induction of cathelicidin. J. Immunol. 179:2060-2063. [DOI] [PubMed] [Google Scholar]
- 27.Mariadason, J. M., G. A. Corner, and L. H. Augenlicht. 2000. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 60:4561-4572. [PubMed] [Google Scholar]
- 28.Marshall, O. 2007. Graphical design of primers with PerlPrimer. Methods Mol. Biol. 402:403-414. [DOI] [PubMed] [Google Scholar]
- 29.Nelson, J. D., O. Denisenko, and K. Bomsztyk. 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1:179-185. [DOI] [PubMed] [Google Scholar]
- 30.Nizet, V., T. Ohtake, X. Lauth, J. Trowbridge, J. Rudisill, R. A. Dorschner, V. Pestonjamasp, J. Piraino, K. Huttner, and R. L. Gallo. 2001. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414:454-457. [DOI] [PubMed] [Google Scholar]
- 31.Pace, B. S., G. L. White, G. J. Dover, M. S. Boosalis, D. V. Faller, and S. P. Perrine. 2002. Short-chain fatty acid derivatives induce fetal globin expression and erythropoiesis in vivo. Blood 100:4640-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubenstein, R. C., M. E. Egan, and P. L. Zeitlin. 1997. In vitro pharmacologic restoration of CFTR-mediated chloride transport with sodium 4-phenylbutyrate in cystic fibrosis epithelial cells containing delta F508-CFTR. J. Clin. Investig. 100:2457-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saiman, L., S. Tabibi, T. D. Starner, P. S. Gabriel, P. L. Winokur, H. P. Jia, P. B. McCray, and B. F. Tack. 2001. Cathelicidin peptides inhibit multiply antibiotic-resistant pathogens from patients with cystic fibrosis. Antimicrob. Agents Chemother. 45:2838-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schauber, J., and R. L. Gallo. 2008. The vitamin D pathway: a new target for control of the skin's immune response? Exp. Dermatol. 17:633-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schauber, J., K. Iffland, S. Frisch, T. Kudlich, B. Schmausser, M. Eck, T. Menzel, A. Gostner, H. Lührs, and W. Scheppach. 2004. Histone-deacetylase inhibitors induce the cathelicidin LL-37 in gastrointestinal cells. Mol. Immunol. 41:847-854. [DOI] [PubMed] [Google Scholar]
- 36.Schauber, J., Y. Oda, A. S. Büchau, Q.-C. Yun, A. Steinmeyer, U. Zügel, D. D. Bikle, and R. L. Gallo. 2007. Histone acetylation in keratinocytes enables control of the expression of cathelicidin and CD14 by 1,25-dihydroxyvitamin D(3). J. Investig. Dermatol. 128:816-824. [DOI] [PubMed] [Google Scholar]
- 37.Schauber, J., C. Svanholm, S. Termén, K. Iffland, T. Menzel, W. Scheppach, R. Melcher, B. Agerberth, H. Lührs, and G. H. Gudmundsson. 2003. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut 52:735-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwab, M., V. Reynders, Y. Shastri, S. Loitsch, J. Stein, and O. Schröder. 2007. Role of nuclear hormone receptors in butyrate-mediated up-regulation of the antimicrobial peptide cathelicidin in epithelial colorectal cells. Mol. Immunol. 44:2107-2114. [DOI] [PubMed] [Google Scholar]
- 39.Sørensen, O. E., L. Gram, A. H. Johnsen, E. Andersson, S. Bangsbøll, G. S. Tjabringa, P. S. Hiemstra, J. Malm, A. Egesten, and N. Borregaard. 2003. Processing of seminal plasma hCAP-18 to ALL-38 by gastricsin: a novel mechanism of generating antimicrobial peptides in vagina. J. Biol. Chem. 278:28540-28546. [DOI] [PubMed] [Google Scholar]
- 40.Termén, S., M. Tollin, E. Rodriguez, S. H. Sveinsdóttir, B. Jóhannesson, A. Cederlund, J. Sjövall, B. Agerberth, and G. H. Gudmundsson. 2008. PU.1 and bacterial metabolites regulate the human gene CAMP encoding antimicrobial peptide LL-37 in colon epithelial cells. Mol. Immunol. 45:3947-3955. [DOI] [PubMed] [Google Scholar]
- 41.Wang, T.-T., F. P. Nestel, V. Bourdeau, Y. Nagai, Q. Wang, J. Liao, L. Tavera-Mendoza, R. Lin, J. W. Hanrahan, S. Mader, J. H. White, and J. H. Hanrahan. 2004. Cutting edge: 1,25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J. Immunol. 173:2909-2912. [DOI] [PubMed] [Google Scholar]
- 42.Yoshioka, M., N. Fukuishi, Y. Kubo, H. Yamanobe, K. Ohsaki, Y. Kawasoe, M. Murata, A. Ishizumi, Y. Nishii, N. Matsui, and M. Akagi. 2008. Human cathelicidin CAP18/LL-37 changes mast cell function toward innate immunity. Biol. Pharm. Bull. 31:212-216. [DOI] [PubMed] [Google Scholar]
- 43.Zaiou, M., V. Nizet, and R. L. Gallo. 2003. Antimicrobial and protease inhibitory functions of the human cathelicidin (hCAP18/LL-37) prosequence. J. Investig. Dermatol. 120:810-816. [DOI] [PubMed] [Google Scholar]
- 44.Zeitlin, P. L., M. Diener-West, R. C. Rubenstein, M. P. Boyle, C. K. Lee, and L. Brass-Ernst. 2002. Evidence of CFTR function in cystic fibrosis after systemic administration of 4-phenylbutyrate. Mol. Ther. 6:119-126. [DOI] [PubMed] [Google Scholar]